Abstract
The ethanol-inducible cytochrome P450 2E1 (CYP2E1) is also induced under different pathological and physiological conditions. Studies including ours have shown that CYP2E1 is bimodally targeted to both the endoplasmic reticulum (microsomes) (mc CYP2E1) and mitochondria (mt CYP2E1). In this study we investigated the role of mtCYP2E1 in ethanol-mediated oxidative stress in stable cell lines expressing predominantly mt CYP2E1 or mc CYP2E1. The ER+ mutation (A2L, A9L), which increases the affinity of the nascent protein for binding to the signal recognition particle, preferentially targets CYP2E1 to the endoplasmic reticulum. The Mt+ (L17G) and Mt++ (I8R, L11R, L17R) mutant proteins, showing progressively lower affinity for signal recognition particle binding, were targeted to mitochondria at correspondingly higher levels. The rate of GSH depletion, used as a measure of oxidative stress, was higher in cells expressing Mt++ and Mt+ proteins as compared with cells expressing ER+ protein. In addition, the cellular level of F2-isoprostanes, a direct indicator of oxidative stress, was increased markedly in Mt++ cells after ethanol treatment. Notably, expression of Mt++ CYP2E1 protein in yeast cells caused more severe mitochondrial DNA damage and respiratory deficiency than the wild type or ER+ proteins as tested by the inability of cells to grow on glycerol or ethanol. Additionally, liver mitochondria from ethanol-fed rats containing high mt CYP2E1 showed higher levels of F2-isoprostane production. These results strongly suggest that mt CYP2E1 induces oxidative stress and augments alcohol-mediated cell/tissue injury.
Keywords: Cytochrome P450, Drug Metabolism, Glutathione, Mitochondria, Mitochondrial Transport, Oxidative Stress, Protein Translocation, Reactive Oxygen Species (ROS)
Introduction
The cytochrome P450 2E1 (CYP2E1)5 is involved in the metabolism of alcohol, aldehydes, and an array of small molecular weight carcinogens. CYP2E1 is induced under diverse pathophysiological conditions including diabetes, obesity, fasting, cancer, alcohol liver disease, and non-alcoholic hepatic steatosis (1, 2). The structure and function of CYP2E1 has been extensively studied because of its potential role in chemical toxicity and carcinogenesis and highly conserved nature of the gene in mammals. Experiments with knock-out animal models have revealed that CYP2E1 plays an important role in benzene-, acrylonitrile-, azoxymethane-, and acetaminophen-induced toxicity as well (3–5). A major interest in CYP2E1 stems from its ability to oxidize ethanol into reactive products, acetaldehyde and 1-hydroxyethyl radical, and also its ability to activate small molecular weight chemicals including CCl4, acetaminophen, benzene, halothane, halogenated alkanes, etc. into electrophilic reactive products (6). Additionally, a large number of studies implicate a role for CYP2E1 in reactive oxygen species (ROS) production and oxidative stress (7–11).
A large fraction of hepatocellular CYP2E1 is located in the endoplasmic reticulum (ER), although significant levels are detected in other cell compartments including lysosomes (12), plasma membrane (13, 14), Golgi apparatus (15), peroxisomes (16), and mitochondria (17–20). Studies have shown increased mitochondrial CYP2E1 content in streptozotocin-treated and alcohol-treated rats and mice (21, 22). Despite the suggested role of CYP2E1 in ROS production, oxidative stress, and lipid peroxidation, it is not clear if the major contribution comes from the microsomal or mitochondrial compartments or both.
In this study we have assessed the role of mitochondria targeted CYP2E1 in inducing oxidative stress. We used a mutational approach to modify the N-terminal chimeric signal of CYP2E1 to mostly mitochondria- or mostly microsome-targeted proteins (mtCYP2E1 and mcCYP2E1, respectively) and expressed them stably in COS cells. Our results show that mtCYP2E1 induces ROS production, oxidative stress, and augments ethanol induced cellular injury. MtCYP2E1 in yeast cells induced respiratory incompetence and ρo phenotypes indicating extensive mitochondrial DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Transfection
COS-7 cells and cells transduced with retroviral vectors (stable expression cells) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (v/v) in presence of added gentamycin (50 μg/ml).
Transient transfection of COS-7 cells with WT and mutant ER+, Mt+, Mt++ rat CYP2E1 cDNAs (6 μg/100 mm2 plate) were carried out using the lipophilic reagent FuGENE 6 (Roche Applied Sciences). The cells were harvested 48 h post-transfection and used for isolating subcellular fractionations.
Generation of Stable Cell Lines
WT and mutated 2E1 cDNAs cloned in a retroviral vector (pBABE-puro) were transduced into 293T cells along with gag-pol and VSV-G plasmids to produce viral particles (23). COS-7 cells were transduced with the viral particles, and the single colonies were selected by screening in presence of puromycin (2 μg/ml) as the selection marker.
Antibodies
Polyclonal antibody to rat CYP2E1 (anti-goat) was purchased from Oxford Biomedical Research, Oxford, MI. Antibody to human cytochrome c oxidase subunit 1 (CcO 1) was from Mitosciences, Eugene, OR. Antibody to human calreticulin (CRT) was from Affinity Bioreagents, Golden, CO. Antibody to yeast dolicholphosphate mannose synthase was from Invitrogen. Antibodies to TIM23, TOM20, the 70-kDa subunit of complex II, β-actin, cytochrome P450 reductase (NPR), and the 70-kDa subunit of succinate dehydrogenase were from Santa Cruz Biotechnology, Santa Cruz, CA. Anti-mouse FLAG antibody was purchased from Sigma.
Ethanol Feeding Experiments
Sprague-Dawley rats (about 150 gm) were fed with ethanol for 2, 4, 6, and 8 weeks, and pair-fed controls received an isocaloric diet. The standard procedure for alcohol feeding was based on the Lieber and De Carli protocol (24). Animals were fed ad libitum a nutritionally balanced liquid diet containing 36% ethanol, 18% protein, 35% fat, and 11% carbohydrate as % of total calories (supplied by BioServe Corp., San Diego CA). Control pair-fed animals received the same diet except that ethanol was isocalorically replaced by carbohydrates. Feeding was carried out in the Animal Resource Facility of Thomas Jefferson University Medical College, Philadelphia, PA under their approved animal care protocol.
Subcellular Fractionation and Immunoblot Analysis
Mitochondria from stable cells and transiently transfected cells were isolated in the presence of protease inhibitors (1 mm phenylmethanesulfonyl fluoride and 50 μg/ml each of leupeptin, pepstatin, aprotinin, and antipain) as described before (25, 26). Mitochondria from ethanol-fed and pair-fed control rat livers were isolated essentially as described before (27), and the mitochondrial isolates were sedimented through 1 m sucrose to minimize contamination (28). Mitochondria were subjected to digitonin fractionation as described before (20, 27). Proteins were dissociated in Laemmli sample buffer (29) at 95 °C for 5 min, resolved by electrophoresis on 10% SDS-polyacrylamide gels (29), and subjected to immunoblot analysis (26). Blots were developed using Super Signal West Femto maximum sensitivity substrate from Pierce.
Limited Trypsin Treatment of Cell Organelles
Freshly isolated mitochondria or microsome (150 μg protein) were subjected to trypsin digestion (30 μg/mg) at 25 °C for 30 min as described before (25, 26).
Assay of N,N-Dimethylnitrosamine N-Demethylation Activity
N-Demethylation of N,N-dimethylnitrosamine was assayed according to Eliasson et al. (13, 30) in the presence of added 0.2 nmol of adrenodoxin (Adx), 0.02 nmol of NADPH-Adx reductase (Adr) and 300 μg mitochondrial protein/ml as enzyme source. In assays with the microsomal fractions, the microsome-associated NADPH cytochrome P450 reductase (NPR) served as electron donor for the reaction. Details were essentially similar to erythromycin N-demethylase assay published before (31).
GSH Measurements
The intracellular and mitochondrial GSH levels were measured as described by Tietze (32) with the fluorometric substrate 5,5′-dithiobis-(2-nitrobenzoic acid) using an NWSS kit from Northwest Lifesciences, Vancouver, WA, following the manufacturer's protocol. Assays were carried out with and without added glutathione reductase and NADPH. Values without added glutathione reductase and NADPH were used to determine the reduced GSH levels, and those with glutathione reductase and NADPH yielded total GSH (reduced plus oxidized) pool.
Measurement of ROS Production Using Dichlorofluorescein Diacetate (DCFH-DA)
For measurement in whole cells, 15 × 103 cells were seeded per well of a black 96-well plate in phenol-free medium and grown overnight. Cells were pretreated with various antioxidants or inhibitors in phenol-free medium followed by incubation with 10 μm DCFH-DA (Molecular Probes, Eugene, OR) for 15 min. The fluorescence of the oxidative product (DCF) was measured using Microwin Chameleon microplate reader with excitation at 485 nm and an emission wavelength of 535 nm. ROS generation in mitochondria was measured by the DCFH fluorescence method, modified from LeBel et al. (33) as described earlier (34).
Assay of F2-Isoprostanes
Cellular levels of F2-isoprostanes were determined using a gas chromatography-mass spectrometry-based method as described previously (35).
Assay for ER Membrane Association
Membrane association assays were carried out as described previously by Hegde et al. (36). Proteins were translated in the presence of 5 units of unwashed dog pancreatic microsome/50 μl of translation mix supplemented with protein kinase A as described previously (37). The translation was stopped by the addition of 1 mm cycloheximide. Half of the reaction mixture was sedimented through 0.5 m sucrose at 120,000 × g for 5 min. The membrane pellets were analyzed by SDS gel electrophoresis (36, 37). The second one-half of reaction mix was analyzed directly as the input protein counts.
Immunofluorescence Microscopy
Immunofluorescence microscopy was carried out with 0.1% Triton X-100 permeabilized cells as described before (25, 38) using primary CYP2E1 (anti-goat), CcO 1 (anti-mouse), and calreticulin (anti-rabbit) antibodies at 1:100 dilutions each. The cells were then stained with 1:100 dilution of Alexa 488-conjugated anti-goat antibody and Alexa 594-conjugated anti-mouse IgG (Molecular Probes). Slides were viewed through a Leica TCS SP5 Confocal Microscope, and Pearson coefficient for colocalization was calculated using Volocity software 4.0.
cDNA Cloning and CYP2E1 Expression in Yeast Cells
The cDNAs (WT, ER+, and Mt++) were cloned in the 2-μm vector pTEF-URA3 or the centromeric pTEF-URA3 (39). The Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 strain BY4741 was obtained from Research Genetics, Huntsville, AL. BY4741 was used to express rat CYP2E1 cDNAs (Mock, WT, ER+, Mt++) driven by the elongation factor promoter either using a 2-μm or centromeric URA3 or Leu2 plasmids. Plasmids were transformed into BY4741 by using the standard LioAc method. The cells were grown in minimal medium containing 2% dextrose (synthetic dextrose, w/v) and other details were as described before (20). Yeast cell mitochondria and microsomes were isolated as described previously (20).
For plate assays, cells grown to ∼2.0 absorbance units at 600 nm were washed and serially diluted in 10-fold steps, and 10 μl of each dilution was spotted on SD−URA and SL−URA (2% lactate, w/v) plates. Plates were photographed after incubation at 30 °C for 4 days.
Statistical Analysis
The means ± S.D. were calculated from three to five experimental values. Statistical significance (p values) between control and experimental or paired experiments was calculated using Student's t test. A p value of <0.05 is considered significant.
RESULTS
Altering the Targeting Efficiency of the N-terminal Chimeric Signal of CYP2E1
We have used a mutational approach to test the hypothesis that SRP binding affinity of nascent chains inversely correlates with the level of mt-targeting (40, 41). The N-terminal signal domain mutations were designed using the WOLFPSORT program with the objective of altering the hydrophobicity of the signal region of the rat CYP2E1.
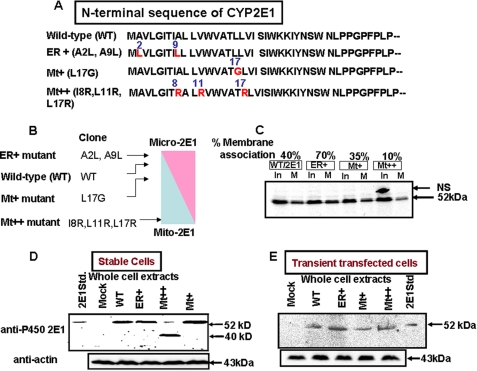
The N-terminal 40-amino acid sequence of the rat CYP2E1 and mutations (ER+, Mt+, and Mt++) targeted to this region are shown in Fig. 1A. ER+ mutation (A2L/A9L) markedly increased the ER targeting potential as predicted by the WOLFPSORT program (Fig. 1B). The Mt+ (L17G) and Mt++ (I8R/L11R/L17R) mutations, on the other hand, showed progressively higher potential for mitochondria targeting (Fig. 1B). The SRP binding affinities of the WT and mutant proteins were tested by the membrane flotation method (36), which measures the SRP-dependent ER membrane association of nascent proteins. WT protein associated with added ER to the extent of ∼40%, whereas ER+ protein, with increased hydrophobicity, associated at a higher level (70%) of input protein (Fig. 1C). The Mt+ and Mt++ proteins, with reduced hydrophobicity, associated with the ER membrane at lower levels (10–30%) (Fig. 1C). These results confirmed the widely varying SRP binding affinities of mutant proteins we generated. Fig. 1D shows the levels of expression of WT and mutant CYP2E1 in stably expressing COS cells. Immunoblot analysis of whole cell extracts show that cells transduced with viral vectors carrying WT, ER+, and Mt+ cDNA constructs express CYP2E1 protein, which comigrated with microsomal CYP2E1 from alcohol-induced rat livers. Cells transduced with Mt++ cDNA construct, however, showed a minor species of nearly intact protein and a prominent ∼40-kDa species. COS cells transiently transfected with all cDNA constructs (including Mt++), on the other hand, expressed nearly intact mtCYP2E1 protein, which comigrated with the mcCYP2E1 protein from alcohol-treated rat liver. These results suggest that all of the cDNA constructs are expressed at similar levels in both stable and transiently transfected cells. The results also suggest that the Mt++ mutant protein is susceptible to an unknown proteolytic activity induced in stably transduced COS cells.
FIGURE 1.
A mutational approach for altering the bimodal targeting efficiency of CYP2E1. A, the WOLFPSORT program was utilized to alter the SRP binding and mitochondria-targeting efficiencies of the N-terminal signal regions of CYP2E1. B, shown is the predicted targeting efficiencies of WT and mutant CYP2E1 proteins. C, ER membrane binding efficiencies of WT, ER+, Mt+, and Mt++ proteins is shown. Proteins were co-translated with added ER membranes as described under “Experimental Procedures.” Membranes from half of the reaction mixture were sedimented through sucrose, and the pellets were dissolved in 2× Laemmli sample buffer (29) and loaded on the gel (M). The other half of the reaction mix was loaded directly as input (In). The gels were fluorographed and also imaged through a GE Healthcare STORM system for quantification of radioactivity in bands. NS means non-specific band. The numbers at the top of the blot show the percentages of 35S protein bound to ER in relation to the input. D, shown is an immunoblot of whole cell extracts (50 μg protein each) of COS cell lines stably expressing the WT and mutant CYP2E1 proteins and also a cell line expressing the empty (mock) retroviral vector. The level of β-actin was used as a loading control. Stable cell lines were generated by transducing COS-7 cells with CYP2E1 cDNA-containing retroviral plasmid (pBABE-puro) using FuGENE transfection (Roche Applied Sciences) reagent. E, immunoblot of COS cell extracts transiently transfected with WT and mutant CYP2E1 constructs. COS-7 cells were transfected with cDNAs cloned in PCMV4 expression vector as described under “Experimental Procedures.” β-Actin levels were used as loading controls. In Parts C, D, and E, proteins were resolved by electrophoresis through 12% SDS-polyacrylamide gels. In C, 25 μg of protein each from flotation gradients was used; in D and E, 50 μg of protein of each whole cell extract were used.
Levels of mtCYP2E1 and mcCYP2E1 in Stable Cell Lines
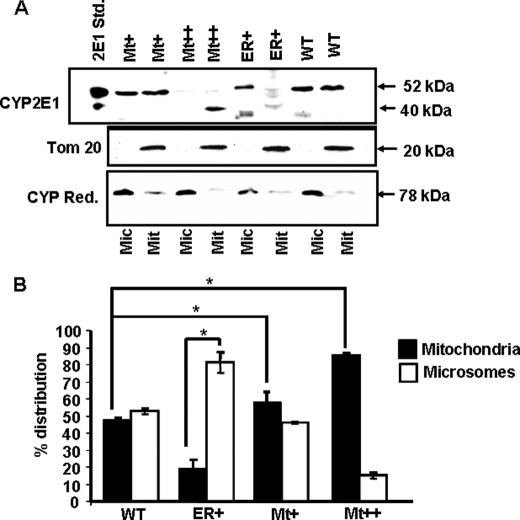
The purity of mitochondrial preparation was routinely tested by immunoblot analysis of mitochondria-specific Tom20 protein and microsomal-specific NPR. As shown in Fig. 2A, mitochondrial preparations contained Tom20, whereas the microsomal fraction contained negligible Tom20 protein. As expected, the microsomal fraction contained a high level of NPR, whereas the mitochondrial preparations contained markedly reduced (<0.1 of the microsomal fraction) 78-kDa NPR. Fig. 2, A and B, also show that in cells expressing WT CYP2E1, the distribution was nearly even (about 50% in each membrane fraction). In cells expressing ER+ mutant protein, the mitochondrial level was ∼20%, whereas the microsomal level of CYP2E1 was >80%. In Mt+ cells, the mitochondrial level was significantly higher (∼58%), whereas in Mt++ cells it was markedly higher (∼90%). Additionally, similar to that shown in Fig. 1, the mitochondria-associated Mt++ CYP2E1 was smaller (∼40 kDa), as opposed to ∼52-kDa protein in other cases. The mcCYP2E1 from ER+ and WT cells also contained a much faster migrating component that we believe is a proteolytic degradation product generated during isolation and storage. The stable cell lines will be hereafter referred to with the respective mutant forms of CYP2E1 they express.
FIGURE 2.
Subcellular distribution of CYP2E1 protein in stable cells. A, shown is an immunoblot analysis of microsomal and mitochondrial proteins from Mock and ER+-, WT-, Mt+-, and Mt++-expressing stable cell lines. Proteins (50 μg each) were resolved by SDS-PAGE on a 12% gel and subjected to immunoblot analysis with anti-CYP2E1 antibody (CYP Ref., 1:1500 dilution). The blots were also probed with an antibody to the mitochondria-specific marker TOM 20 and microsome-specific marker NPR. B, the gel was imaged through a Bio-Rad Florus imager, and the band densities were quantified using the Volume analysis software. Means ± S.D. were calculated based on three separate experiments. Asterisks represent significant difference (p < 0.05) in CYP2E1 contents between WT, Mt+, and Mt++ cells.
Evidence for Intramitochondrial Localization of CYP2E1
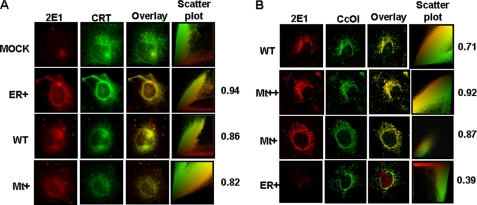
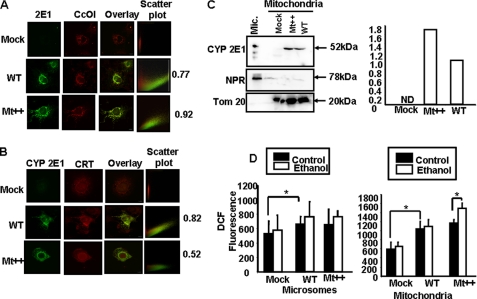
The mitochondrial localization of the CYP2E1 was investigated by colocalization with the ER-specific protein CRT and mitochondria-specific protein CcO I using immunofluorescence microscopy. As expected, cells transduced with a mock vector (Fig. 3A, top panels) show very little CYP2E1-specific staining and insignificant colocalization with CRT antibody (Pearson coefficient of 0.24). However, ER+, Mt+, and WT cells showed significant staining with CYP2E1 antibody, which co-localized with the ER-specific calreticulin with Pearson correlation coefficients of 0.94, 0.82, and 0.86, respectively, suggesting higher level of ER localization of CYP2E1 in ER+ cells. In WT cells, CYP2E1 was also colocalized significantly with mitochondria-specific marker, CcO I, with a Pearson correlation coefficient of 0.71 (Fig. 3B, top panels). The mitochondrial colocalization was markedly higher in Mt++ cells and Mt+ cells with Pearson correlation coefficients of 0.92 and 0.87, respectively (Fig. 3B, second and third panels from the top). In ER+ cells, however, the mitochondrial colocalization was not significant, with a Pearson correlation coefficient score of 0.39. These immunocytochemical results fully support the results of immunoblot analysis in Fig. 2.
FIGURE 3.
Immunocytochemical analysis of stable cell lines. Immunofluorescence microscopy was carried out in 0.1% Triton X-100-permeabilized stable cells incubated with primary CYP2E1 (anti-goat) antibody. The slides were co-stained either with CcO I (anti-mouse) antibody as the mitochondria-specific marker or CRT (anti-rabbit) antibody as the microsomal marker. The cells were subsequently incubated with Alexa 488-conjugated anti-rabbit antibody and Alexa 594-conjugated anti-mouse goat IgG for colocalization of fluorescence signals. Slides were examined by confocal microscopy through Leica TCS SP5 microscope.
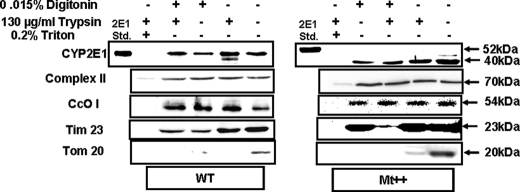
Intramitochondrial localization of CYP2E1 was ascertained using digitonin and trypsin treatment of isolated mitochondria. Fig. 4A shows an immunoblot of mitochondrial fraction from cells expressing WT CYP2E1 (left panel) and Mt++ CYP2E1 (right panel). Results show that mtCYP2E1 was relatively resistant to a combination of digitonin plus trypsin. This is consistent with a protein residing inside the innermembrane compartment such as the 70-kDa subunit of complex II and CcO I subunit. In support of this possibility, treatment with Triton X-100 rendered all these three proteins sensitive to trypsin treatment. The specificity of cell fractionation is further evident from the results that the addition of trypsin to digitonin-treated mitochondria reduced the level of TIM23 but not that of the 70-kDa subunit of complex II or CcO I. A significant portion of Tim23 protein is exposed to the intermembrane space, whereas the other two proteins do not have such exposed domains. These results suggest that the innermembrane integrity is preserved under these treatment conditions. The outer membrane protein, TOM20, which contains a cytosolic extended domain, was substantially reduced by digitonin treatment and degraded by trypsin treatment of whole mitochondria. It is also seen that both nearly intact WT CYP2E1 and the truncated Mt++ CYP2E1 behaved similarly with respect to resistance to digitonin and trypsin treatment. These results confirm that mitochondrial CYP2E1 is localized inside the innermembrane-matrix compartment.
FIGURE 4.
Intramitochondrial localization of CYP2E1 in stable expression cells. Mitochondria from WT (left hand panel) and Mt++ cells (right panel) were treated with digitonin and or trypsin as described under “Experimental Procedures.” In the indicated experiments, mitochondria were treated with 0.2% Triton X-100 (v/v) before treatment with digitonin or trypsin. Proteins (50 μg each) were resolved on 12% SDS-PAGE and subjected to immunoblot analysis with CYP2E1 antibody. The blots were co-developed with antibodies to the 70-kDa subunit of complex II and Tim23. Blots from identically run companion blots were used for developing with antibodies to CcO I subunit and TOM20.
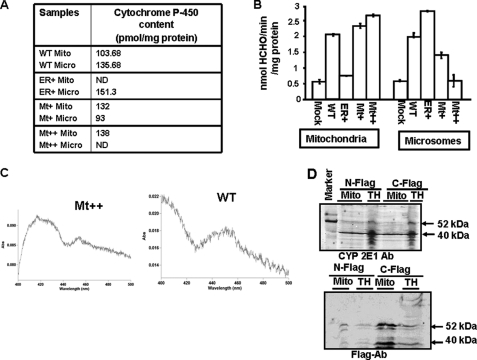
Catalytic Activities of Intact and Truncated mtCYP2E1
The heme contents and N,N-dimethylnitrosamine N-demethylation activity of mitoplasts and microsomes from cells expressing the mock vector and also the four CYP cDNAs are presented in Fig. 5, A and B. Mitochondria from WT, Mt+, and Mt++ cells showed heme contents in the range of 103–138 pmol/mg of protein. Interestingly, mitochondria from Mt++ cells showed a high heme content of 138 pmol/mg protein but undetectable levels in the microsomal fraction. Similarly, microsomes from ER+ cells showed a high level of 151 pmol/mg of protein but undetectable levels in the mitochondrial fraction. The mitochondrial and microsomal heme contents are consistent with the levels of CYP targeting observed in the stable cells (Fig. 2).
FIGURE 5.
Metabolic activities of mitochondria and microsomes from WT and mutant CYP2E1-expressing stable cells. A, the heme contents of mitochondrial and microsomal isolates were determined by using the dithionite reduced and CO-bound difference spectra by the method of Omura and Sato (56). ND, not detected. B, microsomes and mitoplasts from stable cells were assayed for N-demethylation activities using dimethylnitrosamine as the substrate. Assays with mitochondria were carried out with added mitochondria-specific electron transport proteins, Adx and Adr (0.2 nmol of Adx and 0.02 nmol of Adr, respectively). Microsomal activities were driven by microsomal NPR as electron donor system. HCHO is formaldehyde. Values represent the mean ± S.D. of three separate assays. C, representative CO difference spectra for the WT and MT++ cell mitochondria are shown. D, immunoblot analysis of total cell extract (TH) or mitochondrial proteins (Mito) from Mt++ cells transfected with either C-terminal FLAG-tagged (C-FLAG) or N-terminal FLAG-tagged (N-Flag) Mt++ CYP2E1 cDNA is shown. The blot in the top panel was developed with CYP2E1 antibody, and the blot in the bottom panel was developed with FLAG antibody (1:1000 dilutions in each case). Ab, antibody.
The N,N-dimethylnitrosamine N-demethylation activities of mitochondrial and microsomal fractions ranged from 2.1 to 2.6 nmol/mg of protein. Consistent with the heme contents, mitochondria from WT, Mt+, and Mt++ cells showed high activity, whereas those from ER+ cells showed a very low activity. Conversely, microsomes from the WT and ER+ cells showed high activity, whereas those from Mt+ and Mt++ cells showed progressively lower activities. Although not shown, the specific activities of the mitochondrial and microsomal samples were nearly equal and ranged from 16 to 19.6 nmol/nmol CYP/min for the mitochondria and 15 to 18.5 nmol/nmol CYP/min for microsomal samples. Fig. 5C shows the CO absorbance spectra of mitochondrial proteins from WT and Mt++ cells, which were used for calculating the heme contents. These results show that the truncated ∼40-kDa CYP2E1 protein from the Mt++ cell mitochondria is fully assembled with the heme and is as active as the full-length CYP2E1.
The nature of the N-demethylation activity was further verified in control experiments shown in Table 1. The enzyme activity with Mt++ mitoplasts was dependent upon added NADPH and was inhibited by general CYP inhibitor SKF 525-A (Proadifen hydrochloride) and a catalytic site-inhibitory antibody to CYP2E1.
TABLE 1.
DMNA-demethylase activity of Mt++ mitoplast (Mito) supported by Adx and Adr electron donor systems
| Electron transfer system | Inhibitor | % Activity |
|---|---|---|
| No mitoplast | None | 3.9 |
| Mito + NADPH | None | 13.1 |
| Mito + Adx/Adr | None | 3.4 |
| Mito + Adx/Adr + NADPH | None | 100 |
| Mito + Adx/Adr + NADPH | SKF525A | 18.9 |
| Mito + Adx/Adr NADPH | Anti-P450 2E1 antibody | 8.5 |
The possible site of processing of CYP2E1 in Mt++ cells was investigated by transient expression of N-terminal and C-terminal FLAG-tagged Mt++ CYP2E1 in Mt++ cells. The rationale was that by following the FLAG tag with the ∼40-kDa protein, it should be possible to determine whether the processing occurs close to the N terminus or the C terminus. Immunoblots in Fig. 5D show that in mitochondria from cells transfected with both N- and C-terminal- tagged Mt++ CYP2E1 cDNAs, a prominent ∼40-kDa protein is stained with CYP2E1 antibody (top panel). The FLAG antibody, on the other hand, stained the ∼40-kDa species only in cells transfected with the C-terminal FLAG-tagged protein but not the N-terminal-tagged protein. The FLAG antibody also stained a larger 52-kDa protein in the total homogenate and also in mitochondria from cells transfected with both constructs, which likely represent unprocessed proteins. These results suggest that Mt++ CYP2E1 is most likely processed at a site close to the N terminus in Mt++ cells. This possibility is also supported by the functional integrity of the ∼40-kDa CYP2E1 in these cells.
The Role of mt and mc CYP2E1 in Ethanol-mediated Oxidative Stress
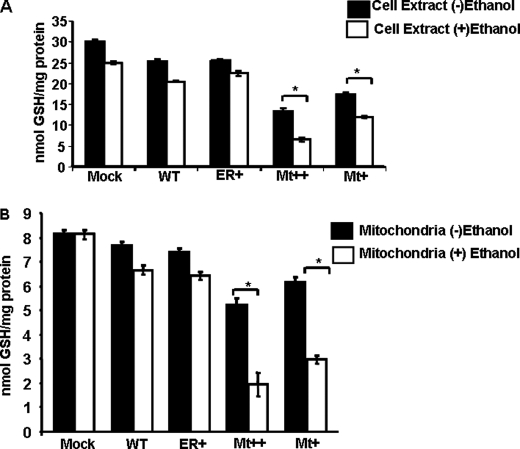
We assessed mitochondrial and extramitochondrial GSH levels and ROS production in subcellular fractions and whole cells. All cell lines except Mt++ did not show significant cell death either in the presence or absence of added ethanol (100 mm for 48 h) (results not shown,). The Mt++ cells, however, showed about 15–17% cell death, which was increased to about 30% by ethanol treatment. Only living monolayer cells were used for analysis.
The reduced GSH pool in whole cell extract was marginally reduced by about 15–20% in WT and ER+ cells, whereas there was a marked (40–50%) reduction in Mt+ and Mt++ cells (Fig. 6A). The GSH levels were further reduced by 10–20% in mock-transfected, WT, and ER+ cells by treatment with ethanol. In Mt+ and Mt++ cells, the ethanol-mediated depletion of GSH was even more pronounced (∼35–55%) compared with untreated cells. As expected, the mitochondrial GSH pool was substantially smaller than the whole cell pool (Fig. 6B). Ethanol did not have any detectable effect on the mitochondrial GSH pool in mock-transfected cells. As with the total cell pool, the mitochondrial GSH pools were marginally lower in WT and ER+ cells but markedly reduced in the Mt+ and Mt++ cells. These results show that Mt+ and Mt++ CYP2E1 exert marked inhibition of both mitochondrial and cellular GSH pools. It is noteworthy that ethanol had a marked inhibitory effect on the mitochondrial GSH pool in Mt+ and Mt++ cells. Although not shown, a decline in reduced GSH pool was accompanied by a corresponding increase in the GSSG pool in different cells and under different treatment conditions.
FIGURE 6.
Cellular and mitochondrial GSH levels in stable cells expressing WT and mutant CYP2E1. A, shown are GSH levels in stable expression cell lines. GSH levels were assayed using fluorometric substrate 5,5′-dithiobis-(2-nitrobenzoic acid). Briefly, cells (2 × 106) were treated with or without 100 mm ethanol. After 48 h of treatment, the total cellular extracts (200 μg of protein), 200 μl of phosphate buffer provided in the kit, and 25 μl of the 5,5′-dithiobis-(2-nitrobenzoic acid) solution were first incubated at 37 °C for 5 min, and the absorbance (412 nm) was measured using a Cary E1 spectrophotometer for 3 min. GSH levels were calculated using a standard curve, and values are presented in nmol/mg protein/min. B, mitochondrial isolates from control and ethanol-treated cells were assayed for GSH levels as described above. Asterisks represent significant difference in GSH (p < 0.05) levels between ethanol-treated and untreated controls. Values represent the mean ± S.D. from six assays of the same cell fractions.
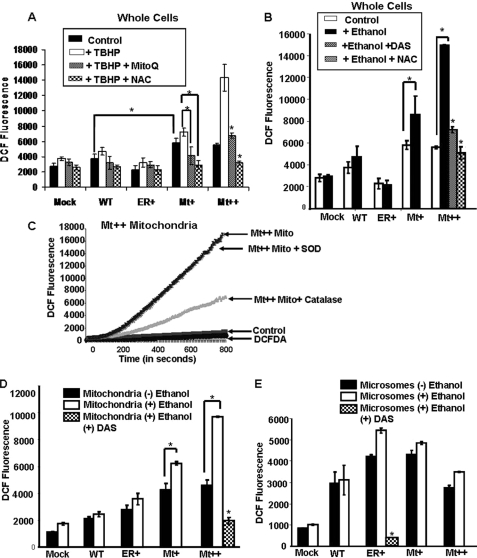
The level of H2O2 produced in whole cells (Fig. 7, A and B) was measured using the DCFH-DA method. Cells were treated with 100 mm ethanol for 48 h with or without the added ROS inducer tert-butyl hydroperoxide (TBHP) and with or without the added CYP2E1 inhibitor diallylsulfide (DAS) and antioxidants, N-acetylcysteine (NAC) or MitoQ. The Mt+ and Mt++ cells showed nearly 2-fold higher ROS as compared with mock-transfected cells (Fig. 7A). The WT CYP2E1-expressing cells showed a marginal increase over the mock-transfected cells, whereas ER+ cells showed marginal reduction. TBHP (Fig. 7A) and alcohol (Fig. 7B) induced ROS production in Mt+ cells, whereas the increase was more than 2-fold in Mt++ cells. The effects on other cell types (Mock, WT, and ER+) were only marginal. Interestingly, the TBHP-induced ROS increase in both Mt+ and Mt++ cells was markedly inhibited by NAC. The mitochondria-specific antioxidant MitoQ inhibited ROS production in these two cell types by >50%. The effects of these inhibitors on other cells, i.e. mock, WT, and ER+ cells, was marginal. The ethanol induced ROS production in both Mt+ and Mt++ cells but not in ER+ cells. The ethanol-induced ROS production was also attenuated in Mt++ cells by NAC and CYP2E1 selective inhibitor, DAS (Fig. 7B).
FIGURE 7.
ROS measurements by DCFH-DA method in whole cells and subcellular fractions from CYP2E1 expressing cells. A, shown are ROS levels in whole cells grown with or without the added oxidant TBHP (400 μm) and/or antioxidants MitoQ (2.5 μm) and NAC (25 mm). B, shown are ROS levels in whole cells grown with or without added ethanol (100 mm) with or without the added CYP2E1 inhibitor DAS (10 μm) or NAC (25 mm). Details are as described under “Experimental Procedures.” C represents a control experiment using Mt++ mitochondria ruptured by freezing and thawing. Catalase (10 units/ml) and superoxide dismutase (SOD, 30 units/ml) were added. The assays were run as described under “Experimental Procedures” using 10 μl of brain cytosolic preparation per 100 μl of reaction volumes. D and E, ROS measurements in isolated mitochondria and microsomes, respectively, from the indicated cells are shown. DAS was added in indicated tubes at 10 μm levels. In all cases, the fluorescence was recorded at an excitation at 488 nm and emissions at 525 nm for 15 min. Values represent the mean ± S.D. values from four separate assays in A, B, D, and E. Asterisks represent significant increase in ROS production (p < 0.05) or reduction by added antioxidants or CYP2E1 inhibitor, DAS.
The level of mitochondrial ROS production in different cell lines was further verified by assaying the DCFH-DA oxidation in an in vitro system described previously (34). In this system, rat brain cytosolic extract is used as the source of deacetylase for converting DCFH-DA into DCFH (33, 34). The fluorometric patterns in Fig. 7C are from a control experiment to ensure the specificity of fluorescence signal. The complete system with added mitochondria from Mt++ cells yielded a linear increase up to 800 s. The addition of superoxide dismutase to the reaction mix had no effect, whereas catalase inhibited the signal, suggesting that the fluorescence signal is mostly due to H2O2. Furthermore, a control reaction with added DCFH-DA alone (or with the brain cytosolic extract without added mitochondria) yielded no significant fluorescence signal, suggesting the specificity of the reaction.
Both the mitochondrial and microsomal membranes from CYP2E1-expressing cells produced 2–3-fold higher ROS compared with mock-transfected controls (Fig. 7, D and E). It is seen that the microsomes from alcohol-treated cells produced marginal (mock, WT, and ER+ cells) to moderately higher (Mt++ and Mt+) signal reflective of levels of ROS production. The mitochondrial isolates from Mt+ and Mt++ cells yielded 60–120% higher ROS production in the presence of added ethanol (Fig. 7D). In both the microsomes and mitochondria, DAS markedly reduced ethanol-induced ROS production (Fig. 7, D and E).
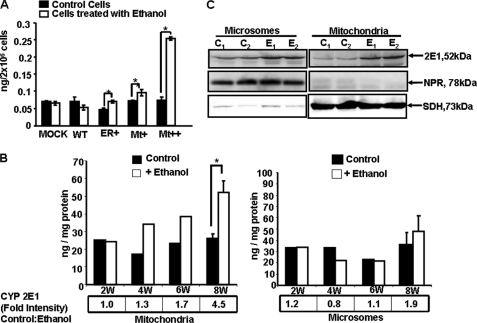
Lipid peroxidation is an immediate consequence of oxidative stress (42–44). Recent multicenter studies propose that the levels of F2-isoprostanes are probably the most reliable measure of oxidative stress (41, 54, 55). We have, therefore, assayed the levels of F2-isoprostanes in alcohol-treated and untreated cells. Results (Fig. 8A) show that in both mock-transfected and WT CYP2E1-expressing cell lines, no significant increase in F2-isoprostanes was observed even after ethanol treatment. In ER+ cells, the basal level of F2-isoprostanes is lower than WT cells. In both ER+ and Mt+ cells, there was a significant increase after ethanol treatment. In Mt++ cells, however, the increase in F2-isoprostane levels was 3.5-fold after ethanol treatment. Together these results show that mitochondria-targeted CYP2E1 plays a prominent role in alcohol-mediated oxidative stress.
FIGURE 8.
Ethanol-induced F2-isoprostanes in CYP2E1-expressing cells and liver fractions from ethanol-fed rats. A, F2-isoprostanes were assayed by using a gas chromatograph-mass spectroscopy method cited under “Experimental Procedures.” In each case 2 × 106 total cells were used for the assay. Asterisks represent significant increase in F2-isoprostanes in ER+, Mt+, and Mt++ cells after ethanol treatment (p < 0.05). Values represent the mean ± S.D. of three assays. B, F2-isopreostanes were measured in mitochondria and microsomes isolated from the livers of rats fed with alcohol for 2–8 weeks (W) and pair-fed controls. In each case 100 μg of protein was used. The means ± S.D. in the 8-week-fed rats were based on assays carried out in three rats each in control and fed groups. Asterisks represent significant difference (p < 0.05) from pair-fed controls. The values presented in boxes below the graph indicate the ratios of CYP2E1 contents between pair-fed controls and alcohol-fed rat livers. The CYP2E1 antibody interactive bands from immunoblots were quantified by imaging through the Bio-Rad Florus 5 gel documentation system. C, shown is an immunoblot analysis of mitochondrial and microsomal proteins (25 μg each) from livers of rats fed with alcohol for 8 weeks and pair-fed controls. Samples from two rats were analyzed in each case. Duplicate blots were developed with NPR antibody and succinate dehydrogenase (SDH) antibody for assessing cross-contamination and loading levels.
The in vivo relevance of these results was investigated using mitochondria and microsomes isolated from rats fed with alcohol from 2 to 8 weeks and also from pair-fed control rats. It is seen that mitochondria from ethanol-fed rats produced progressively increasing levels of F2-isoprostanes compared with pair controls (Fig. 8A). The levels of mtCYP2E1 in relation to the pair-fed controls also increased during this treatment regimen (Fig. 8B). Notably, in 8 weeks of alcohol-fed liver mitochondria, there was a 4.5-fold increase that was also accompanied by a more than 2-fold increase in F2-isoprostanes formation. In the case of the microsomal fraction, there was no significant increase in alcohol-fed rats until 8 weeks of feeding compared with pair-fed controls. Only in microsomes from 8-week-fed rats there was a significant increase in F2-isoprostanes. Immunoblot analysis in Fig. 8C shows a typical pattern of mt- and mc-CYP2E1 induction in 8 week-fed rat livers. The results from these immunoblots were used to determine the fold increase presented in Fig. 8, A and B. These results on mtCYP2E1 induction correlate well with increased oxidative stress in ethanol-treated rats.
Role of Intact mtCYP2E1 in ROS Production
The results that N-terminal-truncated mtCYP2E1 in Mt++ cells induced oxidative stress raised the question if mitochondria targeting of this CYP or N-terminal truncation was responsible for increased oxidative stress. To address this question we overexpressed WT and Mt++ CYP2E1 proteins in COS cells by transient transfection and compared both the levels of mitochondria targeting and levels of ROS production with the two cDNA constructs. In keeping with the results on stable cell lines (Fig. 3), immunocytochemical analysis of transfected cells (Fig. 9, A and B) showed that WT CYP2E1 colocalized with both the ER-specific marker, CRT, and mitochondria-specific marker, CcO I. The Mt++ CYP2E1 protein colocalized poorly with CRT (Pearson coefficient of 0.52) but more extensively with the CcO I, suggesting a higher level of mitochondrial localization (Fig. 9A). The immunoblot of mitochondrial extract (Fig. 9C) shows that both mt- and mc-CYP2E1 comigrated with rat liver microsomal CYP2E1, suggesting that intact CYP2E1 is targeted to mitochondria in both cases. Additionally, the level of mtCYP2E1 with the Mt++ mutant cDNA was significantly higher than with WT protein. Fig. 9D shows that the level of ROS production in microsomes from WT and Mt++ cells was significantly higher than in mock-transfected cells. Alcohol treatment only modestly increased the levels in both WT and Mt++ cDNA-transfected cells. Mitochondria from cells transfected with WT and Mt++ cDNAs showed about a 70–100% increase in ROS production. In mock-transfected and WT cDNA-transfected cells, mitochondrial ROS production was only marginally increased by ethanol treatment. However, a marked increase of ROS production in response to ethanol treatment was observed in mitochondria from Mt++ cDNA-transfected cells. A generally lower level of ROS production in these cells in comparison to stable cells is probably due to lower transfection rates. These results show that intact CYP2E1 targeted to mitochondria is effective in inducing ROS production and alcohol-mediated oxidative stress.
FIGURE 9.
Mitochondrial targeting of intact CYP2E1 in transiently transfected COS cells. A and B, shown is immunocytochemical analysis of transfected cells using mitochondria-specific CcO I and CYP2E1 antibodies (A) and microsome-specific CRT and CYP2E1 antibodies (B). Immunocytochemical analysis and colocalization of stained membrane structures were carried out as described under “Experimental Procedures” and in Fig. 3. C, shown is immunoblot analysis of mitochondrial proteins (50 μg each) from cells transfected with mock vector, WT, and Mt++ cDNAs. Microsome from ethanol-treated rat liver was used as a positive control for CYP2E1 and also NPR. The blots were co-developed with antibodies to mitochondrial marker, TOM20, and microsomal marker NPR. The graph next to the blot represents CYP2E1 contents. ND, not detected. D, shown is the level of ROS production in the microsome and mitochondria isolated from transfected cells. The ROS production was measured fluorometrically by DCFH-DA oxidation method as described in Fig. 7 and under “Experimental Procedures.” The means ± S.D. values were calculated from six assays carried out with cell fractions from two separate transfections. Asterisks represent significant difference (p < 0.05) between the paired values.
Role of mt CYP2E1 in Inducing Respiratory Damage in Yeast Cells
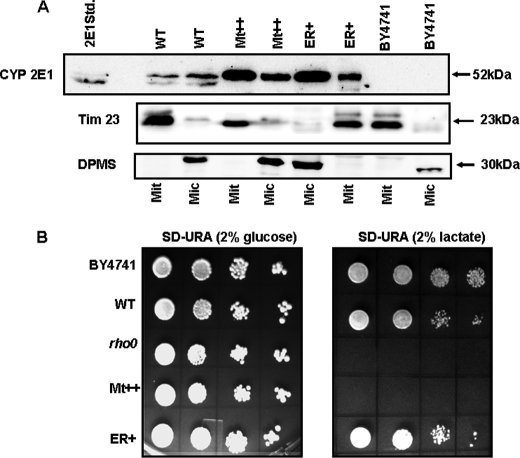
With a view to ascertain the role of mitochondria-targeted CYP2E1 in inducing oxidative damage, we used a heterologous yeast cell system and generated stable cell lines expressing WT, ER+, and Mt++ CYP2E1. Immunoblot analysis of mitochondria and microsome fractions from various cell lines is shown in Fig. 10A. It is seen that both mitochondria and microsomes from WT CYP2E1 cells showed the presence of CYP2E1, although the microsomal fraction contained nearly 2-fold higher levels. The Mt++ cells, on the other hand, contained nearly 3-fold higher levels of mtCYP2E1 than mcCYP2E1. In contrast, the ER+ cells contained more than 3-fold higher levels of mcCYP2E1 compared with mtCYP2E1. These results indicate that the altered targeting property of mutant CYP2E1 signal is also preserved in unicellular eukaryotes. It should be noted that Mt++ CYP2E1 is targeted to mitochondria as a full-length protein, similar to the WT and ER+ proteins. All of the mitochondrial samples showed nearly equal levels of Tim23, and all the microsomal samples showed nearly equal levels of the microsomal marker dolicholphosphate mannose synthase, suggesting nearly equal loading. The blots for Tim23 and dolicholphosphate mannose synthase also indicated the relative purity of the two subcellular membrane fractions.
FIGURE 10.
Mitochondrial CYP2E1-induced respiratory deficiency in yeast cells. A, shown are mitochondrial and microsomal CYP2E1 contents in yeast cells stably expressing WT and mutant CYP2E1 cDNA constructs. The mitochondrial and microsomal proteins (50 μg each) were resolved by SDS-PAGE on a 12% gel and subjected to immunoblot blot analysis with anti-CYP2E1 antibody. Two identically run (parallel) blots were probed with antibody to mitochondria-specific marker Tim 23 and microsome-specific marker dolicholphosphate mannose synthase (DPMS). B, yeast cells expressing ER+, WT, and Mt++ CYP2E1 were grown in yeast extract/peptone/dextrose medium supplemented with appropriate amino acids. Cells corresponding to 2.0 absorbance units at 600 nm were pelleted and resuspended in 1 ml of sterile water. The culture was serially diluted 10 times, and 10 μl of each dilution was spotted onto SD−URA plates containing 2% glucose (w/v) (left panel) and 2% lactate (w/v) (right panel). Plates were incubated at 30 °C for 4 days and photographed.
All of the cell types including the ρo cells (positive control) grew efficiently on the SD−URA medium containing glucose (Fig. 10B). Remarkably, Mt++ cells failed to grow on SD−URA medium, containing lactate, suggesting respiratory incompetence. The later characteristic is similar to the ρo cells with known respiratory deficiency, which also failed to grow on SD−URA media containing lactate. The WT and ER+ cells also showed a marginal growth inhibition on the lactate-containing medium. Mitochondria from Mt++ cells had petite phenotype similar to the ρo cells (results not shown). These cells were significantly smaller than the colonies produced by WT and ER+ cells on the same plate. These results provided a direct and unequivocal evidence for the role of mitochondria targeted CYP2E1 in inducing mitochondrial DNA damage and respiratory deficiency.
DISCUSSION
Previously we observed that nascent CYP1A1, 2B1, and 2E1 proteins exhibit different affinities for SRP binding and showed different levels of mitochondrial targeting (19, 25, 28, 37, 45, 46). In the present study we tested the hypothesis on the regulation of bimodal targeting by altering the SRP binding affinity of the signal region (40, 41) to more hydrophobic (A2L/A9L in ER+) or more hydrophilic (L17G or I8R/L11R/L17R in Mt+ and Mt++, respectively). We show here that the ER+ mutant CYP2E1, with increased hydrophobicity of the N-terminal signal and increased ER association in vitro, is targeted at a higher level to the ER than the WT protein. On the other hand, the Mt+ and Mt++ proteins, with reduced hydrophobicity and reduced affinity for SRP binding (Figs. 1C and 3B), were targeted to mitochondria at higher levels in intact cells.
Several studies over the past decade suggested that CYP2E1 plays a role in oxidative stress in addition to carcinogenesis and chemical toxicity (1–11, 47–49). Also, studies have implicated a role for CYP2E1 in inducing ROS production under in vitro conditions (50–53). Studies from Lieber et al. (51) initially suggested a role for CYP2E1 in inducing oxidative stress and alcohol-mediated liver injury. In vitro studies with stable expression cell lines or isolated microsomes show a role for CYP2E1 in inducing ROS production and oxidative stress (52–54). These latter results have been essentially supported in a CYP2E1 overexpression mouse model generated by Morgan et al. (55). Studies from two groups using this mouse model show that alcohol treatment induced the serum levels of cytokeratin 8 and cytokeratin 18, the biomarkers for alcoholic liver disease (48, 49). Using stable cell lines expressing different levels of mitochondria- and microsome-targeted CYP2E1, we show that mtCYP2E1 plays a role in ethanol-mediated mitochondrial dysfunction, mtDNA damage and ROS production as well as lipid peroxide production. These results also correlate with high levels of mtCYP2E1 with high levels of F2-isoprostanes in alcohol-treated rat livers.
In general, the mitochondrial CYP contents of rodent and human tissues represent only 10–20% of those found in the microsomal fraction. However, in ethanol-treated rats and mice, the liver mitochondrial CYP2E1 contents ranged from about 50% to nearly equal to the microsomal contents (22, 52), suggesting that the mitochondrial CYP2E1 could contribute significantly to drug metabolism, drug-induced toxicity, and oxidative stress.
Detection of truncated protein in Mt++ cells was unexpected and surprising in view of many cell-based studies and analysis of mouse and rat tissues from alcohol-treated or streptozotocin-treated rodents showing full-length CYP2E1 being targeted to mitochondria. We have carried out a series of experiments to show that the truncated form seen in the Mt++ cell lines is very specific to this mutant and specific to stably transduced COS cells. 1) Other CYP2E1 cDNA constructs, such as the WT, ER+, and Mt+ mutant proteins, are targeted to mitochondria as intact proteins. 2) Transient transfection of CYP2E1 cDNAs (including the Mt++) in COS cells resulted in the targeting of intact CYP2E1 to mitochondria, which also induced ROS production in response to ethanol treatment. 3) Although not shown, in vitro import studies in isolated rat liver mitochondria showed that the protease-protected form is a full-length protein. 4) Stable expression of CYP2E1 cDNAs in yeast cells resulted in the mitochondrial targeting of intact CYP2E1 proteins in all cases including the Mt++ mutant form, which induced respiratory stress. These results not only demonstrate that intact CYP2E1 is the natural form of protein targeted to mitochondria but also that mtCYP2E1 causes a higher level of oxidative damage to cells.
Based on the detection of the FLAG containing ∼40-kDa protein in Mt++ cells transfected with the C-terminal FLAG-tagged cDNA, we believe that the processing of the Mt++ mutant protein occurs at a site close to the N terminus. This is consistent with the results showing that the truncated mtCYP2E1 in Mt++ cells is fully folded and assembled with heme because of its characteristic CO-reduced difference spectra. Furthermore, the truncated Mt++ CYP2E1 is catalytically active for N,N-dimethylnitrosamine N-demethylation activity in an Adx + Adr-supported system. It is possible that an aberrant protease activated in Mt++ cells is responsible for this processing. Results clearly show that the processing of CYP2E1 in this cell line occurs at a region that did not affect heme binding, Adx binding, and the catalytic function of the protein.
Results presented here also show that Mt++ cells expressing predominantly mtCYP2E1 are prone to oxidant (TBHP)- and ethanol-mediated ROS production, F2-isoprostane production, depletion of cellular GSH levels, and mitochondrial respiratory defect. We propose that altered structural features of the mt CYP2E1 in the mitochondrial matrix environment, different from the cytosol-exposed microsomal organization, may be responsible for the difference in function and induction of oxidative stress. This possibility is in keeping with our previous CD spectroscopy studies showing a more unfolded conformation of mitochondrial CYP2B1 and 2E1 and markedly reduced α helical contents compared with the microsomal CYPs (17, 37). A role for the mitochondrial CYP2E1 in mediating ethanol-induced oxidative stress is shown by the effects of the CYP inhibitor DAS. Also, the role of mitochondrially produced ROS in mediating oxidative stress was confirmed by the effects of mitochondria specific antioxidant Mito-Q (Fig. 7, A and B).
We show here that intact CYP2E1 is targeted to yeast cell mitochondria, and mutations designed for preferential targeting to the ER (ER+ mutant) and mitochondria (Mt+ and Mt++) show targeting efficiency similar to that observed in stably transduced COS cells. Interestingly, predominantly mitochondria-targeted Mt++ CYP2E1 induced respiratory deficiency as seen by the inability of cells to grow on lactate, a non-fermentable carbon source. The predominantly ER-targeted ER+ CYP2E1, on the other hand, induced only marginal respiratory deficiency in yeast cells. Although not shown, the respiratory deficiency in Mt++ cells is likely associated with petite phenotype indicative of extensive mitochondrial DNA damage (Fig. 10, A and B). These results provide evidence for the role of mt CYP2E1 in ROS production.
In summary alcohol toxicity has been linked to a number of factors including increased CYP2E1 expression, NADPH oxidase activity, and ferrous iron levels (53). Our results show that mitochondria-targeted CYP2E1 can play a direct role in augmenting alcohol-mediated oxidative stress, mtDNA damage, and mitochondrial dysfunction in COS cells. Correlative evidence also suggests that mt CYP2E1 may also play a role in inducing oxidative stress in the livers of ethanol-fed rats.
Acknowledgments
We thank members of the Avadhani laboratory for comments and suggestions on this work.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM034883 and R01 AA017749 (to N. G. A.) and R37 CA090426 (to F. P. G.).
- CYP
- cytochrome P450
- ROS
- reactive oxygen species
- NPR
- NADPH cytochrome P450 reductase
- Adx
- bovine adrenal ferredoxin
- Adr
- bovine adrenal ferredoxin reductase
- DCFH-DA
- 2′,7′-dichlorofluorescein (DCF) diacetate
- CcO
- cytochrome c oxidase
- DAS
- diallylsulfide
- MitoQ
- coenzyme Q cross-linked to diphenylphosphonium ion
- CRT
- calreticulin
- TBHP
- tert-butyl hydroperoxide
- NAC
- N-acetylcysteine
- SRP
- signal recognition particle
- mc
- microsomes
- mt
- mitochondria.
REFERENCES
- 1.Bondoc F. Y., Bao Z., Hu W. Y., Gonzalez F. J., Wang Y., Yang C. S., Hong J. Y. (1999) Biochem. Pharmacol. 58, 461–463 [DOI] [PubMed] [Google Scholar]
- 2.Ronis M. J., Lindros K. O., Ingelman-Sundberg M. (1996) in Cytochromes P450, Pharmacological and Toxicological Aspects (Ioannides C. ed.) CRC Press. Inc., Boca Raton, FL [Google Scholar]
- 3.Sohn O. S., Fiala E. S., Requeijo S. P., Weisburger J. H., Gonzalez F. J. (2001) Cancer Res. 61, 8435–8440 [PubMed] [Google Scholar]
- 4.Sumner S. C., Fennell T. R., Moore T. A., Chanas B., Gonzalez F., Ghanayem B. I. (1999) Chem. Res. Toxicol. 12, 1110–1116 [DOI] [PubMed] [Google Scholar]
- 5.Zaher H., Buters J. T., Ward J. M., Bruno M. K., Lucas A. M., Stern S. T., Cohen S. D., Gonzalez F. J. (1998) Toxicol. Appl. Pharmacol. 152, 193–199 [DOI] [PubMed] [Google Scholar]
- 6.Guengerich F. P., Kim D. H., Iwasaki M. (1991) Chem. Res. Toxicol. 4, 168–179 [DOI] [PubMed] [Google Scholar]
- 7.Caro A. A., Cederbaum A. I. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 27–42 [DOI] [PubMed] [Google Scholar]
- 8.Albano E., Clot P., Morimoto M., Tomasi A., Ingelman-Sundberg M., French S. W. (1996) Hepatology 23, 155–163 [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez F. J. (2007) Drug Metab. Dispos. 35, 1–8 [DOI] [PubMed] [Google Scholar]
- 10.Lieber C. S. (1997) Physiol. Rev. 77, 517–544 [DOI] [PubMed] [Google Scholar]
- 11.Nanji A. A., Zhao S., Sadrzadeh S. M., Dannenberg A. J., Tahan S. R., Waxman D. J. (1994) Alcohol Clin. Exp. Res. 18, 1280–1285 [DOI] [PubMed] [Google Scholar]
- 12.Ronis M. J., Johansson I., Hultenby K., Lagercrantz J., Glaumann H., Ingelman-Sundberg M. (1991) Eur. J. Biochem. 198, 383–389 [DOI] [PubMed] [Google Scholar]
- 13.Eliasson E., Kenna J. G. (1996) Mol. Pharmacol. 50, 573–582 [PubMed] [Google Scholar]
- 14.Neve E. P., Ingelman-Sundberg M. (2000) J. Biol. Chem. 275, 17130–17135 [DOI] [PubMed] [Google Scholar]
- 15.Neve E. P., Eliasson E., Pronzato M. A., Albano E., Marinari U., Ingelman-Sundberg M. (1996) Arch. Biochem. Biophys. 333, 459–465 [DOI] [PubMed] [Google Scholar]
- 16.Pahan K., Smith B. T., Singh A. K., Singh I. (1997) Free Radic. Biol. Med. 23, 963–971 [DOI] [PubMed] [Google Scholar]
- 17.Robin M. A., Anandatheerthavarada H. K., Fang J. K., Cudic M., Otvos L., Avadhani N. G. (2001) J. Biol. Chem. 276, 24680–24689 [DOI] [PubMed] [Google Scholar]
- 18.Neve E. P., Ingelman-Sundberg M. (1999) FEBS Lett. 460, 309–314 [DOI] [PubMed] [Google Scholar]
- 19.Robin M. A., Anandatheerthavarada H. K., Biswas G., Sepuri N. B., Gordon D. M., Pain D., Avadhani N. G. (2002) J. Biol. Chem. 277, 40583–40593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sepuri N. B., Yadav S., Anandatheerthavarada H. K., Avadhani N. G. (2007) FEBS J. 274, 4615–4630 [DOI] [PubMed] [Google Scholar]
- 21.Raza H., Prabu S. K., Robin M. A., Avadhani N. G. (2004) Diabetes 53, 185–194 [DOI] [PubMed] [Google Scholar]
- 22.Robin M. A., Sauvage I., Grandperret T., Descatoire V., Pessayre D., Fromenty B. (2005) FEBS Lett. 579, 6895–6902 [DOI] [PubMed] [Google Scholar]
- 23.Morgenstern J. P., Land H. (1990) Nucleic Acids Res. 18, 3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieber C. S., De Carli L. M. (1973) Res. Commun. Chem. Pathol. Pharmacol. 6, 983–991 [PubMed] [Google Scholar]
- 25.Addya S., Anandatheerthavarada H. K., Biswas G., Bhagwat S. V., Mullick J., Avadhani N. G. (1997) J. Cell Biol. 139, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anandatheerthavarada H. K., Vijayasarathy C., Bhagwat S. V., Biswas G., Mullick J., Avadhani N. G. (1999) J. Biol. Chem. 274, 6617–6625 [DOI] [PubMed] [Google Scholar]
- 27.Niranjan B. G., Wilson N. M., Jefcoate C. R., Avadhani N. G. (1984) J. Biol. Chem. 259, 12495–12501 [PubMed] [Google Scholar]
- 28.Anandatheerthavarada H. K., Addya S., Dwivedi R. S., Biswas G., Mullick J., Avadhani N. G. (1997) Arch. Biochem. Biophys. 339, 136–150 [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Eliasson E., Mkrtchian S., Ingelman-Sundberg M. (1992) J. Biol. Chem. 267, 15765–15769 [PubMed] [Google Scholar]
- 31.Boopathi E., Anandatheerthavarada H. K., Bhagwat S. V., Biswas G., Fang J. K., Avadhani N. G. (2000) J. Biol. Chem. 275, 34415–34423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tietze F. (1969) Anal. Biochem. 27, 502–522 [DOI] [PubMed] [Google Scholar]
- 33.LeBel C. P., Ischiropoulos H., Bondy S. C. (1992) Chem. Res. Toxicol. 5, 227–231 [DOI] [PubMed] [Google Scholar]
- 34.Prabu S. K., Anandatheerthavarada H. K., Raza H., Srinivasan S., Spear J. F., Avadhani N. G. (2006) J. Biol. Chem. 281, 2061–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrow J. D., Roberts L. J., 2nd (1999) Methods Enzymol. 300, 3–12 [DOI] [PubMed] [Google Scholar]
- 36.Hegde R. S., Voigt S., Lingappa V. R. (1998) Mol. Cell 2, 85–91 [DOI] [PubMed] [Google Scholar]
- 37.Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., Avadhani N. G. (1999) EMBO J. 18, 5494–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biswas G., Adebanjo O. A., Freedman B. D., Anandatheerthavarada H. K., Vijayasarathy C., Zaidi M., Kotlikoff M., Avadhani N. G. (1999) EMBO J. 18, 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mumberg D., Müller R., Funk M. (1994) Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasari V. R., Anandatheerthavarada H. K., Robin M. A., Boopathi E., Biswas G., Fang J. K., Nebert D. W., Avadhani N. G. (2006) J. Biol. Chem. 281, 30834–30847 [DOI] [PubMed] [Google Scholar]
- 41.Dong H., Dalton T. P., Miller M. L., Chen Y., Uno S., Shi Z., Shertzer H. G., Bansal S., Avadhani N. G., Nebert D. W. (2009) Mol. Pharmacol. 75, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadiiska M. B., Gladen B. C., Baird D. D., Graham L. B., Parker C. E., Ames B. N., Basu S., Fitzgerald G. A., Lawson J. A., Marnett L. J., Morrow J. D., Murray D. M., Plastaras J., Roberts L. J., 2nd, Rokach J., Shigenaga M. K., Sun J., Walter P. B., Tomer K. B., Barrett J. C., Mason R. P. (2005) Free Radic. Biol. Med. 38, 711–718 [DOI] [PubMed] [Google Scholar]
- 43.Dostalek M., Brooks J. D., Hardy K. D., Milne G. L., Moore M. M., Sharma S., Morrow J. D., Guengerich F. P. (2007) Mol. Pharmacol. 72, 1419–1424 [DOI] [PubMed] [Google Scholar]
- 44.Dostalek M., Hardy K. D., Milne G. L., Morrow J. D., Chen C., Gonzalez F. J., Gu J., Ding X., Johnson D. A., Johnson J. A., Martin M. V., Guengerich F. P. (2008) J. Biol. Chem. 283, 17147–17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhagwat S. V., Mullick J., Raza H., Avadhani N. G. (1999) Toxicol. Appl. Pharmacol. 156, 231–240 [DOI] [PubMed] [Google Scholar]
- 46.Boopathi E., Srinivasan S., Fang J. K., Avadhani N. G. (2008) Mol. Cell 32, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradford B. U., Kono H., Isayama F., Kosyk O., Wheeler M. D., Akiyama T. E., Bleye L., Krausz K. W., Gonzalez F. J., Koop D. R., Rusyn I. (2005) Hepatology 41, 336–344 [DOI] [PubMed] [Google Scholar]
- 48.Butura A., Nilsson K., Morgan K., Morgan T. R., French S. W., Johansson I., Schuppe-Koistinen I., Ingelman-Sundberg M. (2009) J. Hepatol. 50, 572–583 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Millonig G., Nair J., Patsenker E., Stickel F., Mueller S., Bartsch H., Seitz H. K. (2009) Hepatology 50, 453–461 [DOI] [PubMed] [Google Scholar]
- 50.Albano E. (2006) Proc. Nutr. Soc. 65, 278–290 [DOI] [PubMed] [Google Scholar]
- 51.Lieber C. S., Rubin E., DeCarli L. M. (1970) Biochem. Biophys. Res. Commun. 40, 858–865 [DOI] [PubMed] [Google Scholar]
- 52.Bai J., Cederbaum A. I. (2006) J. Biol. Chem. 281, 5128–5136 [DOI] [PubMed] [Google Scholar]
- 53.Kono H., Rusyn I., Yin M., Gäbele E., Yamashina S., Dikalova A., Kadiiska M. B., Connor H. D., Mason R. P., Segal B. H., Bradford B. U., Holland S. M., Thurman R. G. (2000) J. Clin. Invest. 106, 867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekström G., Ingelman-Sundberg M. (1989) Biochem. Pharmacol. 38, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 55.Morgan K., French S. W., Morgan T. R. (2002) Hepatology 36, 122–134 [DOI] [PubMed] [Google Scholar]
- 56.Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]