Abstract
The involvement of macrophages (Mφs) as host, accessory, and effector cells in the development of infectious diseases, together with their central role in iron homeostasis, place these immune cells as key players in the interface between iron and infection. Having previously shown that the functional expression of NRAMP-1 results in increased protein phosphorylation mediated in part by an iron-dependent inhibition of Mφ protein-tyrosine phosphatase (PTP) activity, we sought to study the mechanism(s) underlying this specific event. Herein we have identified the mononuclear dicitrate iron complex [Fe(cit)2H4-x](1+x)− as the species responsible for the specific inhibition of Mφ PTP activity. By using biochemical and computational approaches, we show that [Fe(cit)2]5− targets the catalytic pocket of the PTP SHP-1, competitively inhibiting its interaction with an incoming phosphosubstrate. In vitro and in vivo inhibition of PTP activity by iron-citrate results in protein hyperphosphorylation and enhanced MAPK signaling in response to LPS stimulation. We propose that iron-citrate-mediated PTP inhibition represents a novel and biologically relevant regulatory mechanism of signal transduction.
Keywords: Iron, Macrophage, MAP Kinases (MAPKs), Phosphatase, Signal Transduction
Introduction
The balance of protein phosphorylation, maintained by the concerted action of protein kinases and protein phosphatases, is fundamental in determining the outcome of multiple cellular functions ranging from cell proliferation to cell death (1). In recent years the importance of protein tyrosine phosphatases (PTPs)3 as coordinators of signaling pathways and the immune response has become evident (2). Progress has been achieved in understanding the mechanisms of PTP regulation, including receptor PTP dimerization (3), oxidation (4), and PTP phosphorylation. However, with more than 100 family members, a broad structural diversity, varying subcellular localizations and substrate specificities (5), the complexity of PTP activity regulation is far from being completely unraveled.
Natural resistance-associated macrophage protein-1, NRAMP-1, is a divalent metal transporter localized to the late endosome/lysosomal compartment of Mϕs (6), and present in gelatinase positive tertiary granules of neutrophils (7). Upon phagocytosis, NRAMP-1 is recruited to the phagolysosomal membrane where it mediates the export of Mn2+, Fe2+, Co2+, and potentially other metals including Zn2+ from the vesicle into the cytoplasmic compartment (8, 9). Functional NRAMP-1 expression has been linked to innate resistance to Leishmania donovani and other unrelated intracellular pathogens (10). In addition, it has been associated with the up-regulation of pro-inflammatory Mϕ functions such as MHC II expression (11), IL-1β (12), TNF-α (13), and NO production (14). We have recently reported that functional expression of NRAMP-1 results in lower Mϕ PTP activity and increased protein phosphorylation, in part explaining the pleiotropic effects of NRAMP-1 (15). Our findings suggest a model linking metal transport and the regulation of Mϕ functions: NRAMP-1 mediated iron transport directly or indirectly (by catalyzing the formation of reactive oxygen species, ROS) inhibits PTP activity, promoting protein phosphorylation and positive signal transduction. This results in the up-regulation of effector Mϕ functions such as NO production, which contributes to the control of intracellular pathogen infections (15).
Here we have extended our observations and studied the mechanisms underlying the iron-dependent Mϕ PTP regulation. The involvement of Mϕs as host, accessory and effector cells in the development of infectious diseases, together with their central role in iron homeostasis, place these immune cells as key players in the interface between iron and infection (16). Herein show that Mϕs regulate MAPK signaling in part by modulating PTP activity in an iron-dependent manner. We propose that this novel mechanism may represent a biologically relevant PTP regulatory event, which could be exploited as a therapeutic approach.
EXPERIMENTAL PROCEDURES
Materials
4-Nitrophenylphosphate disodium salt hexahydrate (pNPP), LPS (Escherichia coli, serotype 0111:B4), catalase, superoxide dismutase, poly(Glu,Tyr), iron (III) chloride (FeCl3), and iron (II) sulfate (FeSO4)were purchased from Sigma-Aldrich. [γ-32P]dATP (3000 Ci/mmol) was obtained from GE Healthcare. Guanidine hydrochloride and citric acid were purchased from Laboratoire MAT (Beauport, QC, Canada).
Cell Culture
B10R murine Mϕ cell line was kept in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 100 μg/ml streptomycin, 100 units/ml penicillin, and 2 mm l-glutamine at 37 °C and 5% CO2.
Phosphatase Assays
As previously described (17), B10R Mϕs were collected, lysed in PTP lysis buffer (50 mm Tris pH 7.0, 0.1 mm EDTA, 0.1 mm EGTA, 0.1% 2-mercaptoethanol (ME), 1% Igepal, 25 μg/ml aprotinin, and 25 μg/ml leupeptin) and kept on ice 45 min. Lysates were cleared by centrifugation, and protein content determined by Bradford's method. 10 μg of protein extract were incubated in phosphatase reaction buffer (50 mm Hepes pH 7.5, 0.1% β-ME, 10 mm pNPP) for 30 min. Absorbance was taken at 405 nm.
Preparation of Iron-Citrate Solutions
Three different stock solutions of iron-citrate (Fe-cit) were prepared with variations in the iron-to-citrate (Fe:cit) molar ratio: 1:1, 1:4, and 1:10. A stock solution of 0.5 m FeCl3 was prepared and added dropwise to citric acid (H4cit) solutions to obtain a final concentration of 100 mm Fe3+ in all preparations. 100 mm, 400 mm, and 1 m citric acid solutions were used to generate 1:1, 1:4, and 1:10 Fe:cit preparations respectively. Solutions were slowly stirred for 1 h at room temperature and protected from light to avoid photoreduction of Fe3+. Fe-cit preparations were kept at room temperature and used for a maximum of one month after preparation.
Electrospray Ionization Mass Spectrometry (ESI-MS)
The ESI-MS experiments were performed on a LCQ-ion trap (Finnigan-Thermoquest, San Jose, CA) equipped with an electrospray source. Electrospray full scan spectra, in the range of m/z 50–1200, were obtained by infusion through fused silica tubing at 2–10 μl min−1. The LCQ calibration (m/z 50–2000) was achieved according to the standard calibration procedure from the manufacturer (mixture of caffeine, MRFA and Ultramark 1621). The temperature of the heated capillary of the LCQ was set to 100 °C, the ion spray voltage was in the range of 1–6 kV with an injection time of 5–200 ms. Solutions were analyzed in the negative mode. Experimental peak values throughout this study were identified by the m/z ratio of the most abundant peak in the parent group. Calculated m/z values tabulated are those based on the most abundant isotopes. Peak intensities are cited as percentages of the base major peak intensity. When citric acid was in excess in the solution (1:4 and 1:10), the peaks ([H3cit]− m/z 191; ([H4cit][H3cit]− m/z 383) become the major species, so spectra were represented in the range of 200 (or 400) to 900 for clarity. As protonation occurs during the ionization process in the spectrometer, a single species can be detected with different degrees of protonation. Mass spectrometry allows to establish the FeIII:citrate stoichiometry of the complexes. The nuclearity of complexes is in accordance with the result of isotopic pattern calculation.
For ESI-MS analyses, 1:1, 1:4, and 1:10 Fe:cit solutions (1 mm Fe3+) were prepared 20 h prior to be analyzed, and kept in the dark. The pH of the 1:1 and 1:4 solutions was 3.5, and 2.4 for Fe:cit 1:10. Results do not vary with time, showing that the mixture has reached equilibrium at the time when the experiments were performed, consistent with data obtained from solutions prepared with the iron(III) salt (Fe(ClO4)3·9H2O) used in the previous analysis (18).
Molecular Docking
The three-dimensional structures of the catalytic domain of the PTP SHP-1 (PDB code: 1GWZ) (19) and its complex with the phosphotyrosyl decapeptide PY469 derived from SHPS-1 (PDB code: 1FPR) (20) were retrieved from the Protein Data Bank. Molecular docking simulations of the Fe-cit complexes with SHP-1 were carried out using the program FlexX (21) embedded in the SYBYL7.3 (Tripos, Inc). The water molecules were removed from the proteins and the essential hydrogen atoms were added prior to docking. Protein conformations were kept rigid during all experiments. H2O, NH4, and Him molecules were removed from the Fe-cit complexes (C7, (NH4)5[Fe(Cit)2]·2H2O; C2, (Him)2[Fe2(Cit)2(H2O)2]· 2H2O) (18). Ligand charges were calculated using Gasteiger-Huckel method and their structures were minimized using Tripos Force Field. For each individual ligand the first thirty docking solutions were ranked and the results of top ranked conformation were analyzed.
In Vitro and in Vivo Iron Treatment
10 μg of total B10R Mϕ cell lysates were incubated for 10 min at RT with FeCl3, Fe:cit 1:1, Fe:cit 1:4, Fe:cit 1:10, or FeSO4 to a final iron concentration of 500 μm, and PTP activity assay performed as described above. Alternatively, B10R Mϕs were incubated for different time periods with Fe:cit 1:1, Fe:cit 1:10, FeCl3 or FeSO4 in serum-free DMEM. Cells were then lysed in PTP lysis buffer and PTP activity evaluated by pNPP hydrolysis.
In-gel PTP Assay
In-gel PTP assay was performed as previously described (15, 22). Briefly, poly(Glu,Tyr) substrate was tyrosine-phosphorylated by overnight (O/N) incubation with GST-FER protein kinase (10 μg) and 150 μCi [γ-32P]dATP. The substrate was then incorporated in a 10 SDS-polyacrylamide gel mixture at a concentration of 2 × 105 cpm/ml. Mϕ protein extracts, prepared as described above, were denatured for SDS-PAGE and loaded onto the gel. After electrophoresis, the gel was incubated O/N in Buffer A (50 mm Tris-HCl pH 8.0, 20% isopropanol), washed twice with Buffer B (50 mm Tris-HCl pH 8.0, 0.3% β-ME), and followed by full protein denaturation in Buffer B containing 6 m guanidine hydrochloride and 1 mm EDTA. Gels were washed twice in Buffer C (50 mm Tris-HCl pH 8.0, 1 mm EDTA, 0.3% β-ME, and 0.04% Tween 20) and incubated for final renaturation O/N in Buffer C with or without 500 μm Fe:cit 1:10. Gels were dried and exposed to x-ray film. Active PTPs were detected as clear bands on the film.
Western Blotting and GST Pulldown
Western blots were performed as previously described (23). Primary antibodies used were α-phosphotyrosine clone 4G10 (Millipore), α-phospho-ERK1/2 (Thr-202/Tyr-204), α-ERK1/2, α-phospho-SAPK/JNK (Thr-183/Tyr-185), and α-SAPK/JNK (Cell Signaling). Band densitometry was performed using the alpha-DigiDoc software. GST-tagged PTP1B, 1–321 amino acids, PTP-PEST, 1–453 amino acids, and full-length TCPTP and SHP-1 constructs were expressed in E. coli BL21. Fusion proteins were isolated by 1 h of incubation of bacterial lysates and glutathione-Sepharose beads (GE Healthcare) at 4 °C. Isolated fusion proteins were incubated with Fe-cit for 10 min at room temperature and PTP activity evaluated by pNPP hydrolysis.
Statistical Analysis
Data were analyzed by one-way ANOVA and t test. Statistically significant difference between groups was considered when p < 0.05 or p < 0.01. All data are presented as the means ± S.E.
RESULTS
Iron-dependent PTP Inhibition Is Specific to Iron-Citrate Chelates
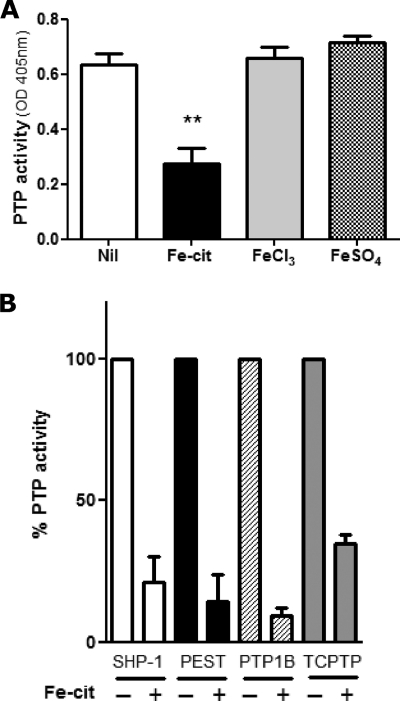
Having previously observed that Fe-cit strongly inhibits PTP activity in vitro (15), we investigated whether different iron donors excreted a similar effect. Only iron in the form of Fe-cit, but not FeSO4 or FeCl3, was able to inhibit total Mϕ PTP activity (Fig. 1A). Moreover, Fe-cit inhibited by more than 50% the specific enzymatic activities of the classic PTPs SHP-1, TCPTP, PTP1B, and PTP-PEST (Fig. 1B).
FIGURE 1.
Iron-dependent PTP inhibition is specific to Fe-cit. Mϕ PTP activity was determined following incubation of (A) 10 μg of B10R total protein lysates (B) or GST-purified recombinant PTPs for 10 min at RT with Fe-cit, FeSO4 or FeCl3 to a final concentration of 500 μm. PTP activity was evaluated by pNPP hydrolysis and absorbance reading at 405 nm. Statistically significant differences (**) were considered when p < 0.01.
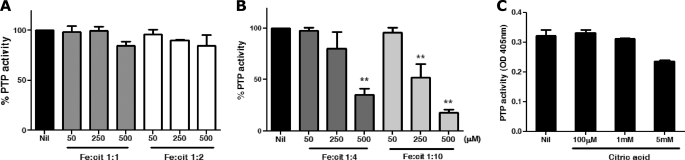
The complexity of Fe-cit solutions is evidenced by the formation of numerous Fe-cit species found in equilibrium in any given solution. These may vary in the number of coordinated iron nuclei, citrate molecules, and protonation states (24). These complexes can range from simple mononuclear complexes such as [Fe(cit)2]5− (25) or [Fe(Hcit)(cit)]4− (18), binuclear complexes such as [Fe2(cit)2(H2O)2]2− or [Fe2(Hcit)3]3− (18, 26), or trinuclear species characterized in solid state as the [Fe9O(cit)8(H2O)3]7− nonanuclear complex (18, 27). The formation of specific Fe-cit complexes is dependent on the pH, temperature, and particularly on the molar iron-to-citrate ratio (Fe:cit), among others (18). In this line of thought, we evaluated the impact of Fe:cit ratios on the PTP inhibitory capacity of different Fe-cit solutions. As shown in Fig. 2A, the Fe:cit ratio is critical for the capacity of Fe-cit solutions to inhibit Mϕ PTP activity. Whereas Fe-cit solutions prepared with 1:1 and 1:2 ratios did not inhibit PTP activity (50–500 μm), strong enzymatic inhibition was observed with 1:4 and 1:10 Fe:cit preparations (Fig. 2A). The Fe-cit-mediated inhibition of PTPs was specific to Fe-cit complexes as neither citric acid (Fig. 2B) nor FeCl3 (Fig. 1A) alone could inhibit PTP activity.
FIGURE 2.
Effect of iron-to-citrate ratio on PTP inhibition. Three Fe-cit solutions were prepared modifying the iron-to-citrate ratios to obtain 1:1, 1:4, and 1:10 Fe:cit solutions. A, dose response curve of B10R Mϕ protein extracts incubated with 50–500 μm 1:1, 1:2, B, 1:4 and 1:10 Fe:cit or C, 0.1–5 mm citric acid was performed. PTP activity was evaluated by pNPP hydrolysis and absorbance reading at 405 nm. Statistically significant differences (**) were considered when p < 0.01.
Identification of a Mononuclear Dicitrate Iron Complex in PTP Inhibitory Preparations
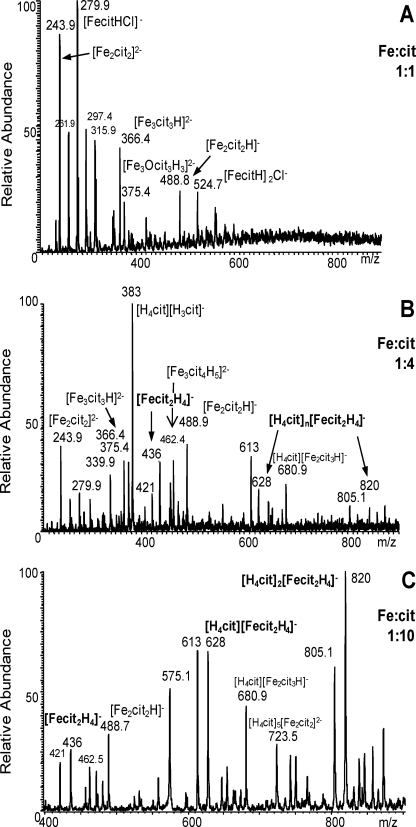
The composition of each individual preparation was investigated in an attempt to unravel putative Fe-cit complexes responsible for the inhibitory effect. Electrospray ionization mass spectrometry (ESI-MS) spectra of 1:1, 1:4, and 1:10 Fe:cit solutions (Fig. 3) revealed the multicomplex nature of these preparations (Table 1).
FIGURE 3.
Specific inhibition of PTPs by mononuclear iron dicitrate. Electrospray mass spectra of (A) 1:1, (B) 1:4, and (C) 1:10 Fe:cit solutions.
TABLE 1.
Fe-cit species detected by ESI-MS
Species with major peak intensity are in bold characters. In italics are represented mononuclear iron dicitrate species. When the concentration of citric acid increases in the solution, free citric acid is associated with Fe-cit species, or other ions present in solution: ([H3cit]− m/z 191) ([H4cit][H3cit]− m/z 283) ([H4cit]2[H3cit]− m/z 575.5) ((K[H2cit])− m/z 229) ((K[H3cit]2)− m/z 421) ((K[H4cit][H3cit]2)− m/z 613) ((K[H4cit]2[H3cit]2)− m/z 805.1).
| Fe:cit ratio | m/z calculated | m/z measured | Species* |
|---|---|---|---|
| 1:1 | 279.9 | 279.9 | [FecitHCl]− |
| 315.9 | 315.9 | [FecitH2Cl2]2− | |
| 524.8 | 524.7 | ([FecitH]2Cl)− | |
| 243.9 | 243.9 | [Fe2(cit)2]2− | |
| 261.9 | 261.9 | [Fe2(cit)2H Cl]2− | |
| 488.9 | 488.8 | [Fe2(cit)2H]− | |
| 297.4 | 297.4 | [Fe3O3(cit)2H3]2− | |
| 366.4 | 366.4 | [Fe3(cit)3H]2− | |
| 375.4 | 375.3 | [Fe3O(cit)3H3]2− | |
| 1:4 | 279.9 | 279.9 | [Fe(cit)H Cl]− |
| 436.0 | 435.9 | [Fe(cit)>2H4]− | |
| 628.0 | 627.7 | [H4cit] [Fe(cit)2H4]− | |
| 820.0 | 819.3 | [H4cit]2 [Fe(cit)2H4]− | |
| 243.9 | 243.9 | [Fe2(cit)2]2− | |
| 339.9 | 339.9 | [H4cit] [Fe2(cit)2]2− | |
| 488.9 | 488.8 | [Fe2(cit)2H]− | |
| 462.4 | 462.3 | [Fe3(cit)4H5]2− | |
| 366.4 | 366.4 | [Fe3(cit)3H]2− | |
| 375.4 | 375.3 | [Fe3O(cit)3H3]2− | |
| 680.9 | 680.8 | [H4cit] [Fe2(cit)2H]− | |
| 1:10 | 436.0 | 435.9 | [Fe(cit)2H4]− |
| 628.0 | 627.7 | [H4cit] [Fe(cit)2H4]− | |
| 820.0 | 819.3 | [H4cit]2 [Fe(cit)2H4]− | |
| 488.9 | 488.8 | [Fe2(cit)2H]− | |
| 680.9 | 680.8 | [H4cit] [Fe2(cit)2H]− | |
| 724.0 | 723.5 | [H4cit]5[Fe2(cit)2]2− |
The composition of 1:1 Fe:cit solution consisted mainly of binuclear and trinuclear iron complexes. The peaks at m/z 243.9 and 488.9 were assigned to [Fe2(cit)2]2− and [Fe2(cit)2H]− anions respectively, corresponding to the [Fe2(cit)2(H2O)2]2− binuclear complex characterized in solid state. The peaks at m/z 366.5 and 375.5 were assigned to [Fe3(cit)3H]2−, [Fe3O(cit)3H3]2− trinuclear species, which are the precursor building blocks observed in the [Fe9O(cit)8(H2O)3]7− nonanuclear complex. Some mononuclear species (([Fe(Hcit)Cl]− m/z 279.9); ([Fe(H2cit)Cl2]2− m/z 315.9); ([Fe(Hcit)]2Cl)− m/z 524.8)) associated with chloride coming from the iron salt were observed. Chloride anion is a strong ligand and coordinates the iron at acidic pH. In these species, the iron is in tetrahedral geometry as suggested by the presence of the [FeCl4]− species (m/z 198) in the 1:1 Fe:cit solution, citrate ligand being bidentate or tridentate. The intensity of the [Fe(Hcit)Cl]− peak is lower in the 1:4 Fe:cit solution, and is not observed in 1:10 Fe:cit.
The 1:4 Fe:cit solution had a predominant abundance of bi and trinuclear complexes. Interestingly, peaks at m/z 436, 628, and 820 were detected, representing the mononuclear iron dicitrate species [Fe(H2cit)2]−, [H4cit][Fe(H2cit)2]−, and [H4cit]2[Fe(H2cit)2]−, respectively, these last two associated with free citric acid. This mononuclear dicitrate complex has been characterized in its solid state as [Fe(cit)2]5− and [Fe(Hcit)(cit)]4− crystallized from solution at pH >7 and pH 6, respectively (18). At more acidic pH, in solution, this species should be [Fe(Hcit)2]3−, [Fe(H2cit)(Hcit)]2−, or [Fe(H2cit)2]−, and for the remaining of the text we refer to the mononuclear iron dicitrate species with the general formula [Fe(cit)2H4-x](1+x)−.
The 1:10 Fe:cit preparation, which in vitro exerts the strongest PTP inhibitory activity, contained mononuclear dicitrate complexes in greater relative abundance compared with binuclear or trinuclear species (Fig. 3). Most interestingly is the absence of mononuclear dicitrate species in the 1:1 Fe:cit preparation, which in vitro has no PTP inhibitory activity, strongly suggests that mononuclear dicitrate complexes could represent the active PTP inhibitory species.
Mechanism of PTP Inhibition
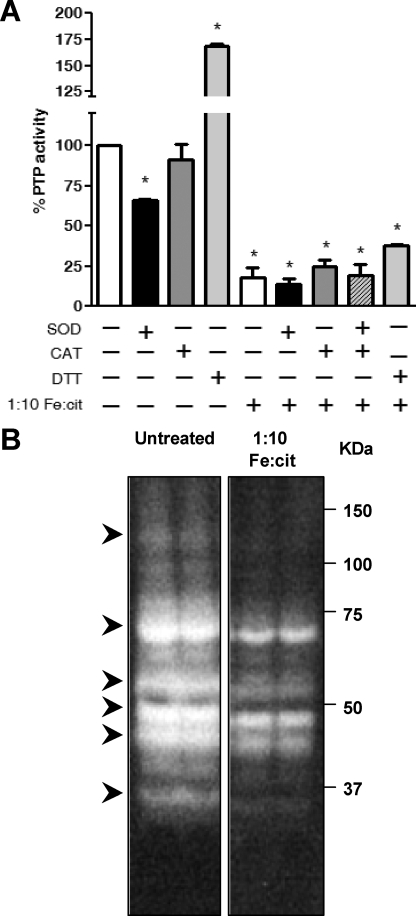
The importance of iron as a biologically active transition metal stems from its ability to engage in one-electron oxidation-reduction reactions. However, this same property is responsible for its toxic effects, where free reactive ions can catalyze the formation of ROS via Habber Weiss and Fenton chemistry (1) O2˙̄ + Fe3+ ⇆ Fe2+ + O2; (2) 2 O2˙̄ + 2H+ → H2O2 + O2; (3) Fe2+ + H2O2 → Fe3+ + OH· + OH− (28). ROS play an important role in the regulation of PTP activity. Oxidation of the conserved catalytic cysteine residue in the PTP domain impairs enzymatic activity (4). We hypothesized that the Fe-cit PTP inhibition could result from PTP oxidation mediated by ROS. Incubation of Mϕ protein lysates with 1:10 Fe:cit solution in the presence or absence of superoxide dismutase (SOD), catalase (CAT), SOD+CAT or DTT, did not rescue PTP activity (Fig. 4A), suggesting that generation of ROS is likely not the mechanism of Fe-cit PTP inhibition. Increased PTP activity observed upon treatment with 5 mm DTT could result from total reduction of the PTPs catalytic Cys after protein lysis.
FIGURE 4.
Fe-cit-dependent PTP inhibition is independent of Fe-catalyzed ROS. A, B10R Mϕ protein lysates were incubated for 10 min at room temperature with 500 μm 1:10 Fe:cit, in the presence or absence of 5 mm dithiothreitol, 50 units/ml superoxide dismutase (SOD), and/or 50 units/ml catalase (CAT). PTP activity was evaluated by pNPP hydrolysis and absorbance reading at 405 nm. Statistically significant differences (*) were considered when p < 0.05. B, 50 μg of total B10R protein lysate were subjected to in-gel PTP assay. During the final renaturation step, gels were incubated with 500 μm 1:10 Fe:cit (right panel) or left untreated (left panel). Samples are loaded in duplicates. Bands of dephosphorylation represent active PTPs. Arrows represent PTP targeted by Fe-cit.
In-gel PTP activity assays have provided a good tool for studying the regulation of cellular PTPs. In this assay, PTPs are denatured and subsequently renatured within the gel, recovering PTP enzymatic activity (15, 22, 29). Addition of 1:10 Fe:cit during the renaturation steps in an in-gel PTP assay of total Mϕ PTPs abolished complete recovery of enzymatic activity (Fig. 4B). This suggests that Fe-cit could physically interact with the PTP active site (Fig. 4B).
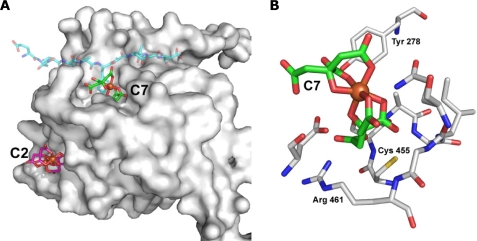
Computational molecular docking of Fe-cit complexes on the macrophage PTP SHP-1 (PDB code: 1GWZ) showed that the most energetically favorable interaction with the mononuclear dicitrate complex C7 ([Fe(cit)2]5−) occurred within the catalytic domain (Fig. 5). The mononuclear dicitrate complex occupied this pocket, forming favorable interactions with residues lining the wall, such as Tyr278, Ser456, and Arg461, the later involved in both substrate binding and stabilization of the phosphoenzyme intermediate (30) Molecular docking of [Fe(cit)2]5− and the crystal structure of SHP-1 in complex with a tyrosine phosphopeptide (PDB code: 1FPR.pdb) suggests that mononuclear dicitrate complexes block access to the PTP active site, potentially acting as competitive PTP inhibitors (supplemental Fig. S1).
FIGURE 5.
Computational docking of SHP-1 and Fe-cit complexes. Docking solutions of SHP-1 (PDB code: 1GWZ) and the mononuclear iron dicitrate complex C7 [Fe(cit)2]5− and with the dinuclear dicitrate iron complex C2 [Fe2(cit)2(H2O)2]2−. A, shown is a surface representation of SHP-1 and the docked C7 (green base color) and C2 (purple base color) molecules in stick representation. For reference the phosphotyrosine-containing substrate analog (PDB code: 1FPR) is also shown (transparent blue base color) illustrating that C7 and the phosphotyrosine occupy the same binding site. B, detailed view of the SHP-1 active site, highlighting the catalytically important residues Cys-455 and Arg-461, and other residues lining the wall of the pocket, as well as the inhibitory Fe-cit molecule C7.
In line with our biochemical data, modeling experiments of SHP-1 and the binuclear Fe-cit complex C2 ([Fe2(cit)2(H2O)2]2−) showed no convincing interaction with the PTP catalytic domain. A nonspecific, presumably low affinity, binding site could be modeled, but its location was far away from the mononuclear binding pocket (∼20 Å). This correlates with the inability of the binuclear Fe-cit complex to inhibit PTP activity. Collectively, these results corroborate our biochemical observations, and identify the mononuclear dicitrate complex [Fe2(cit)2]5− (or in other protonated state [Fe(cit)2H4-x](1+x)−) as the active PTP inhibitory species.
Fe-cit Transiently Inhibits Intracellular Mϕ PTP Activity Resulting in Protein Hyperphosphorylation
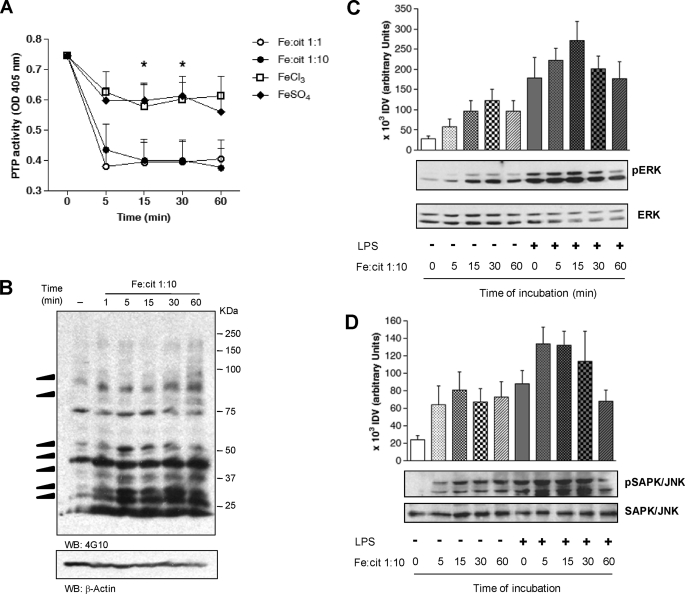
Incubation B10R murine Mϕs with 1:1 and 1:10 Fe:cit solutions showed ∼50% inhibition of PTP activity as early as 5 min following Fe:cit delivery (Fig. 6A). Surprisingly, intracellular PTP inhibition also occurred with 1:1 Fe:cit preparation (which shows no inhibitory capacity in vitro, Fig. 2A), possibly attributable to intracellular metabolism of multinuclear Fe:cit complexes to more stable mononuclear species. Substantiating the specificity of the regulatory effect of Fe:cit complexes, Mϕ incubation with FeCl3 or FeSO4 only marginally inhibited Mϕ PTP activity (Fig. 6A).
FIGURE 6.
Fe-cit inhibits PTP activity in Mϕs and modulates protein phosphorylation. A, B10R Mϕs were incubated with 500 μm 1:10 and 1:1 Fe:cit, FeCl3, or FeSo4 in serum-free DMEM for 5–60 min. PTP activity was evaluated by pNPP hydrolysis and absorbance reading at 405 nm. Data represent the mean ± S.E. of five independent experiments performed in duplicate. Statistically significant differences (**) were considered when p < 0.01. B, 4G10 anti-phosphotyrosine Ab was used to evaluate the phosphotyrosine content of 1:10 Fe:cit-treated Mϕs. Loading control was performed using anti-β-actin Ab. C, Mϕs were pretreated with 500 μm 1:10 Fe:cit in serum-free medium for 5–60 min or left untreated. Next, cells were stimulated with 100 ng/ml LPS and SAPK/JNK and ERK1/2 phosphorylation was evaluated by Western blot. Shown are the loading controls using anti-JNK and anti-ERK1/2 antibodies. Data from densitometric analyses of three independent experiments are presented as the mean ± S.E. of integrated densitometric values (IDV).
Western blot of total phosphotyrosine showed hyperphosphorylation of Mϕ proteins as early as 5 min of incubation with 1:10 Fe:cit (Fig. 6B). Specific phosphorylation of ERK 1/2 and SAPK/JNK MAP kinases was detected, respectively, after 15 min and 5 min of 1:10 Fe:cit treatment (Fig. 6, C and D). When Mϕs were incubated for different time periods with 1:10 Fe:cit prior to LPS stimulation, potentiation of ERK1/2 and SAPK/JNK hyperphosphorylation was observed in response to LPS-Fe:cit co-treatment, similarly observed at early treatment time points (5–15 min). Of interest, MAPK phosphorylation decreased after 1 h of LPS-Fe:cit co-stimulation, but not during Fe:cit treatment alone, consistent with the regulatory role of inflammation on iron homeostasis. Taken together, these data agree with an iron-dependent PTP inhibition favoring protein kinase activity and protein phosphorylation, ultimately leading to positive signal transduction.
DISCUSSION
Knowledge on the mechanisms underlying intracellular PTP activity regulation is pivotal for understanding the course of signal transduction in response to extracellular and intracellular stimuli, and provides insights into how and when can this group of enzymes be considered as potential targets for therapeutic interventions. The PTP family encompasses a group of enzymes diverse in structural features, cellular distribution, subcellular localization, and substrate specificities, however, maintaining a conserved catalytic domain (5). Similarly, the mechanisms of activity regulation can be diverse, specific and broad. To date, three main events controlling PTP activity have been described: protein phosphorylation, dimerization and oxidation. Whereas the first two have been shown to play a role in the regulation of specific PTPs or PTP subfamilies (3, 31–33), ROS-mediated PTP oxidation is thought to represent a broad mechanism of PTP regulation, as it involves oxidation of the conserved catalytic cysteine within the active site ([I/V]HCXXGXXR[S/T]) of all PTPs (4).
Metals such as zinc and vanadium have been known to play an important role in the regulation of PTP activity (34, 35); however, only modulation by Zn2+ has been proposed to play a biologically relevant role (36, 37). Herein we reveal a novel mechanism of PTP regulation which involves the inhibition of PTP activity by mononuclear iron dicitrate (with the general formula [Fe(cit)2H4-x](1+x)−). We have shown both, in vitro and intracellularly, that 1:10 Fe:cit solution inhibits PTP activity promoting general protein tyrosine hyperphosphorylation. Of particular interest was the finding that following LPS stimulation, the phosphorylation levels of both ERK 1/2 and SAPK/JNK were significantly increased in 1:10 Fe:cit-treated cells (Fig. 6C-D), indicating that Fe-cit-mediated PTP inhibition enhances pro-inflammatory Mϕ signaling.
As shown in Table 1 and Fig. 3, Fe:cit consists of a variety of iron citrate complexes found in equilibrium within any given solution. Statistically significant PTP inhibition was observed with high concentrations of Fe:cit solutions (>500 μm and >250 μm for the Fe:cit 1:4 and 1:10, respectively), reflecting the low and moderate relative abundance of mononuclear dicitrate complexes (the PTP inhibitory species) in the 1:4 and 1:10 solutions, respectively. Targeting the generation of isolated mononuclear dicitrate iron complexes should circumvent treatment with high total iron concentrations.
The intracellular labile iron pool (LIP) is a transient and redox-active pool of LMW·Fe complexes which allow iron bioavailability at neutral pH and are readily available for extracellular export, association with iron-containing proteins and regulation of iron-dependent functions (38). Although the exact composition of the LIP is still a matter of controversy, iron can complex intracellularly with LMW chelates including ascorbate, citrate (39, 40), small polypeptides, and lipid moieties (38). To date, the identity of the bioactive Fe-cit complex(es) remains unknown. However, based on intracellular citrate concentrations ranging from 0.1–0.4 mm (39) and labile iron concentrations of 0.4–16 μm (41), the intracellular Fe:cit ratios could range from 1:6–1:1000. As previously shown, high Fe:cit ratios (≥ 1:10) allow for increased relative abundance of mononuclear Fe-cit complexes (18). This, together with the co-crystallization of the E. coli outer membrane receptor FecA complexed with the binuclear complex [Fe2(cit)2]2− (42, 43), and the importance of mononuclear Fe-cit species as bioavailable iron sources, suggests that mononuclear and binuclear Fe-cit complexes could represent the biologically relevant species.
Using ESI-MS and computational modeling (Figs. 3 and 5), we have identified the mononuclear iron dicitrate complex [Fe(cit)2H4-x](1+x)− as the active species in the inhibition of Mϕ PTP activity, providing further evidence of its physiological relevance. We show that the mechanism of PTP inhibition by mononuclear iron dicitrate is not dependent on ROS-mediated oxidation, but rather, follows the direct interaction of the iron complex with the PTP catalytic domain, impairing the entrance of an incoming phosphoprotein substrate (Fig. 5 and supplemental Fig. S1).
In contrast to what is observed in vitro, the 1:1 Fe:cit preparation (which mainly contains bi and trinuclear complexes) inhibits PTP activity in B10R Mϕs (Fig. 6A). However, consistent with the in vitro data, FeCl3 and FeSO4 did not significantly inhibit Mϕ PTP activity. This suggests that internalized Fe-cit complexes, but not iron salts (either as iron oxohydroxo oligomers precipitates or as complexes with LMW chelates), may be metabolized to mononuclear Fe-cit species, either spontaneously, enzymatically, or as part of the process of iron mobilization. Considering that internalization of Fe-cit complexes may expose these species to a different environment that may promote dissociation of multinuclear complexes to mononuclear Fe-cit (pH changes from an acidic solution to the neutral intracellular pH and variation in the intracellular molar ratio of citrate), intracellular metabolism is a plausible explanation for the apparent discrepancy between in vivo and in vitro data. Given the current unavailability of precise methodological approaches to determine the composition of intracellular Fe-cit species, intracellular metabolism of Fe-cit complexes toward mononuclear iron dicitrate remains thus far hypothetical.
Several lines of evidence point to an important interplay between the immune system and iron metabolism; the regulation of different aspects of iron metabolism by inflammatory cytokines (44), the altered response to infectious diseases in the context of iron deficiency and iron overload (45), and the central role of immune cells in the control of iron homeostasis (46). However the molecular events underlying the iron-mediated regulation of immune functions are just beginning to be unraveled. We have recently reported that functional expression of NRAMP-1, a divalent metal transporter (including Fe2+) whose expression has been associated with innate resistance to unrelated intracellular pathogens, results in lower Mϕ PTP activity and increased protein phosphorylation. Our results supported an iron-dependent inhibition of PTP activity and up-regulation of macrophage functions, favoring intracellular pathogen clearance (15). Here we have extended our findings and present a molecular mechanism of mononuclear dicitrate mediated PTP inhibition, which links NRAMP-1 mediated iron transport from endolysosomal compartments into the cytoplasm, to the regulation of pro-inflammatory signaling pathways that may be responsible for the pleitropic effects of NRAMP-1 expression.
Recently, Wang et al. have shown that the attenuated Salmonella and LPS-mediated inflammatory response in hfe−/− mice (which display reduced intramacrophage iron accumulation due to low circulating hepcidin and increased phagocyte ferroportin expression) resulted from lower production of IL-6 and TNF-α by macrophage, an effect that could be mimicked by iron chelation in WT Mϕs. The authors showed that reduced pro-inflammatory cytokine levels were a result of altered translational regulation (47). These observations agree with an upstream iron-mediated inhibition of PTPs; reduced intracellular iron results in increased PTP activity, down-regulating signaling pathways and downstream pro-inflammatory cytokine production. In light of the interplay between immune regulation and iron homeostasis and our current data, it seem interesting to revisit the role of iron homeostasis modulation (as chelation therapies or supplemental iron treatment) in the treatment of diseases such as visceral leishmaniasis, where strong pathogen-induced down-regulation of cell functions leads to disease establishment and progression (48). It is pivotal, however, to keep into consideration the toxic side effects of iron overload, the differential outcomes of other infectious diseases in the context of host iron status, and the different degrees of host iron metabolism (44, 45).
Collectively, our data revealed a novel mechanism of PTP activity regulation by [Fe(cit)2H4-x](1+x)− mononuclear iron dicitrate. Of utmost interest, is the biological relevance of this finding, given the nature of iron-citrate as a ubiquitous bioavailable iron source. Similarly to ROS, Fe-cit-dependent regulation of cellular PTP activity may represent an important mechanism for the control of signal transduction both, at resting state and in the scenario of an inflammatory response, which could be potentially exploited for the manipulation of signaling pathways by iron chelation or iron delivery therapies.
Supplementary Material
Acknowledgment
We thank Dr. Keith Burridge (University of North Carolina, Chapel Hill) for kindly providing us with the GST-FER construct.
This work was supported by a Canadian Institute of Health Research (CIHR) operating grant (to M. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PTP
- protein tyrosine phosphatase
- MAPK
- mitogen-activated protein kinase
- Mϕ
- macrophage
- ROS
- reactive oxygen species
- GST
- glutathione S-transferase
- ME
- mercaptoethanol
- ANOVA
- analysis of variance
- ESI-MS
- electrospray ionization mass spectrometry.
REFERENCES
- 1.Johnson L. N., Lewis R. J. (2001) Chem. Rev. 101, 2209–2242 [DOI] [PubMed] [Google Scholar]
- 2.Mustelin T., Vang T., Bottini N. (2005) Nat. Rev. Immunol. 5, 43–57 [DOI] [PubMed] [Google Scholar]
- 3.Bilwes A. M., den Hertog J., Hunter T., Noel J. P. (1996) Nature 382, 555–559 [DOI] [PubMed] [Google Scholar]
- 4.Tonks N. K. (2005) Cell 121, 667–670 [DOI] [PubMed] [Google Scholar]
- 5.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 6.Gruenheid S., Pinner E., Desjardins M., Gros P. (1997) J. Exp. Med. 185, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canonne-Hergaux F., Calafat J., Richer E., Cellier M., Grinstein S., Borregaard N., Gros P. (2002) Blood 100, 268–275 [DOI] [PubMed] [Google Scholar]
- 8.Forbes J. R., Gros P. (2003) Blood 102, 1884–1892 [DOI] [PubMed] [Google Scholar]
- 9.Goswami T., Bhattacharjee A., Babal P., Searle S., Moore E., Li M., Blackwell J. M. (2001) Biochem. J. 354, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal S., Tremblay M. L., Govoni G., Gauthier S., Sebastiani G., Malo D., Skamene E., Olivier M., Jothy S., Gros P. (1995) J. Exp. Med. 182, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojciechowski W., DeSanctis J., Skamene E., Radzioch D. (1999) J. Immunol. 163, 2688–2696 [PubMed] [Google Scholar]
- 12.Blackwell J. M., Roach T. I., Atkinson S. E., Ajioka J. W., Barton C. H., Shaw M. A. (1991) Immunol. Lett. 30, 241–248 [DOI] [PubMed] [Google Scholar]
- 13.Roach T. I., Kiderlen A. F., Blackwell J. M. (1991) Infect. Immun. 59, 3935–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritsche G., Dlaska M., Barton H., Theurl I., Garimorth K., Weiss G. (2003) J. Immunol. 171, 1994–1998 [DOI] [PubMed] [Google Scholar]
- 15.Gomez M. A., Li S., Tremblay M. L., Olivier M. (2007) J. Biol. Chem. 282, 36190–36198 [DOI] [PubMed] [Google Scholar]
- 16.Theurl I., Fritsche G., Ludwiczek S., Garimorth K., Bellmann-Weiler R., Weiss G. (2005) Biometals 18, 359–367 [DOI] [PubMed] [Google Scholar]
- 17.Blanchette J., Racette N., Faure R., Siminovitch K. A., Olivier M. (1999) Eur. J. Immunol. 29, 3737–3744 [DOI] [PubMed] [Google Scholar]
- 18.Gautier-Luneau I., Merle C., Phanon D., Lebrun C., Biaso F., Serratrice G., Pierre J. L. (2005) Chem. Eur. J. 11, 2207–2219 [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Liang X., Niu T., Meng W., Zhao Z., Zhou G. W. (1998) J. Biol. Chem. 273, 28199–28207 [DOI] [PubMed] [Google Scholar]
- 20.Yang J., Cheng Z., Niu T., Liang X., Zhao Z. J., Zhou G. W. (2000) J. Biol. Chem. 275, 4066–4071 [DOI] [PubMed] [Google Scholar]
- 21.Rarey M., Kramer B., Lengauer T., Klebe G. (1996) J. Mol. Biol. 261, 470–489 [DOI] [PubMed] [Google Scholar]
- 22.Burridge K., Nelson A. (1995) Anal. Biochem. 232, 56–64 [DOI] [PubMed] [Google Scholar]
- 23.Jaramillo M., Godbout M., Olivier M. (2005) J. Immunol. 174, 475–484 [DOI] [PubMed] [Google Scholar]
- 24.Pierre J. L., Gautier-Luneau I. (2000) Biometals 13, 91–96 [DOI] [PubMed] [Google Scholar]
- 25.Matzapetakis M., Raptopoulou C. P., Tsohos A., Papaefthymiou V., Moon N., Salifoglou A. (1998) J. Am. Chem. Soc. 120, 13266–13267 [Google Scholar]
- 26.Shweky I., Bino A., Goldberg D. P., Lippard S. J. (1994) Inorganic Chem. 33, 5161–5162 [Google Scholar]
- 27.Bino A., Shweky I., Cohen S., Bauminger E. R., Lippard S. J. (1998) Inorganic Chem. 37, 5168–5172 [Google Scholar]
- 28.Halliwell B., Gutteridge J. M. (1990) Methods Enzymol. 186, 1–85 [DOI] [PubMed] [Google Scholar]
- 29.Markova B., Gulati P., Herrlich P. A., Böhmer F. D. (2005) Methods 35, 22–27 [DOI] [PubMed] [Google Scholar]
- 30.Andersen J. N., Mortensen O. H., Peters G. H., Drake P. G., Iversen L. F., Olsen O. H., Jansen P. G., Andersen H. S., Tonks N. K., Møller N. P. (2001) Mol. Cell. Biol. 21, 7117–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dadke S., Kusari A., Kusari J. (2001) Mol. Cell. Biochem. 221, 147–154 [DOI] [PubMed] [Google Scholar]
- 32.Lorenz U., Ravichandran K. S., Pei D., Walsh C. T., Burakoff S. J., Neel B. G. (1994) Mol. Cell. Biol. 14, 1824–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strack V., Krützfeldt J., Kellerer M., Ullrich A., Lammers R., Häring H. U. (2002) Biochemistry 41, 603–608 [DOI] [PubMed] [Google Scholar]
- 34.Brautigan D. L., Bornstein P., Gallis B. (1981) J. Biol. Chem. 256, 6519–6522 [PubMed] [Google Scholar]
- 35.Swarup G., Cohen S., Garbers D. L. (1982) Biochem. Biophys. Res. Commun. 107, 1104–1109 [DOI] [PubMed] [Google Scholar]
- 36.Haase H., Maret W. (2003) Exp. Cell Res. 291, 289–298 [DOI] [PubMed] [Google Scholar]
- 37.Haase H., Maret W. (2005) Biometals 18, 333–338 [DOI] [PubMed] [Google Scholar]
- 38.Kakhlon O., Cabantchik Z. I. (2002) Free Rad. Biol. Med. 33, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 39.Gautier-Luneau I., Bertet P., Jeunet A., Serratrice G., Pierre J. L. (2007) Biometals 20, 793–796 [DOI] [PubMed] [Google Scholar]
- 40.Bakkeren D. L., de Jeu-Jaspars C. M., van der Heul C., van Eijk H. G. (1985) Int. J. Biochem. 17, 925–930 [DOI] [PubMed] [Google Scholar]
- 41.Petrat F., de Groot H., Sustmann R., Rauen U. (2002) Biol. Chem. 383, 489–502 [DOI] [PubMed] [Google Scholar]
- 42.Ferguson A. D., Chakraborty R., Smith B. S., Esser L., van der Helm D., Deisenhofer J. (2002) Science 295, 1715–1719 [DOI] [PubMed] [Google Scholar]
- 43.Yue W. W., Grizot S., Buchanan S. K. (2003) J. Mol. Biol. 332, 353–368 [DOI] [PubMed] [Google Scholar]
- 44.Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004) Cell 117, 285–297 [DOI] [PubMed] [Google Scholar]
- 45.Schaible U. E., Kaufmann S. H. (2004) Nat. Rev. 2, 946–953 [DOI] [PubMed] [Google Scholar]
- 46.Knutson M. D., Oukka M., Koss L. M., Aydemir F., Wessling-Resnick M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 1324–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Johnson E. E., Shi H. N., Walker W. A., Wessling-Resnick M., Cherayil B. J. (2008) J. Immunol. 181, 2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olivier M., Gregory D. J., Forget G. (2005) Clin. Microbiol. Rev. 18, 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.