Abstract
SNAP25 plays an essential role in neuronal exocytosis pathways. SNAP25a and SNAP25b are alternatively spliced isoforms differing by only nine amino acids, three of which occur within the palmitoylated cysteine-rich domain. SNAP23 is 60% identical to SNAP25 and has a distinct cysteine-rich domain to both SNAP25a and SNAP25b. Despite the conspicuous differences within the palmitoylated domains of these secretory proteins, there is no information on their comparative interactions with palmitoyl transferases. We report that membrane association of all SNAP25/23 proteins is enhanced by Golgi-localized DHHC3, DHHC7, and DHHC17. In contrast, DHHC15 promoted a statistically significant increase in membrane association of only SNAP25b. To investigate the underlying cause of this differential specificity, we examined a SNAP23 point mutant (C79F) designed to mimic the cysteine-rich domain of SNAP25b. DHHC15 promoted a marked increase in membrane binding and palmitoylation of this SNAP23 mutant, demonstrating that the distinct cysteine-rich domains of SNAP25/23 contribute to differential interactions with DHHC15. The lack of activity of DHHC15 toward wild-type SNAP23 was not overcome by replacing its DHHC domain with that from DHHC3, suggesting that substrate specificity is not determined by the DHHC domain alone. Interestingly, DHHC2, which is closely related to DHHC15, associates with the plasma membrane in PC12 cells and can palmitoylate all SNAP25 isoforms. DHHC2 is, thus, a candidate enzyme to regulate SNAP25/23 palmitoylation dynamics at the plasma membrane. Finally, we demonstrate that overexpression of specific Golgi-localized DHHC proteins active against SNAP25/23 proteins perturbs the normal secretion of human growth hormone from PC12 cells.
Keywords: Membrane Fusion, Membrane Trafficking, Protein Acylation, Protein Motifs, Protein Palmitoylation
Introduction
SNAP25a2 and SNAP25b are SNARE proteins that are highly expressed in the brain, where they perform essential functions in presynaptic neurotransmitter release. SNAP25a/b are present at the presynaptic plasma membrane and form a complex with syntaxin 1, an additional SNARE present at the plasma membrane, and the synaptic vesicle SNARE protein VAMP2 (1). The formation of this trans-SNARE complex between the plasma membrane and the vesicle membrane is an essential step for subsequent membrane fusion (exocytosis) and secretion of neurotransmitters into the synaptic cleft. In addition to neurons, SNAP25a/b display a restricted tissue distribution and have limited functions outside the nervous system, most notably in regulated exocytosis pathways in pancreatic beta cells and adrenal medullary chromaffin cells (2, 3). SNAP25a and SNAP25b are derived from a single gene by differential splicing of exon 5, and the proteins differ by only 9 of 206 amino acids (4). Despite their high sequence conservation, the proteins are not functionally identical; SNAP25b supports more exocytosis than SNAP25a when overexpressed in embryonic adrenal medullary chromaffin cells (5), and replacement of SNAP25b with an extra copy of SNAP25a in mice leads to developmental defects, seizures, and impairment of learning (6). Thus, the nine non-conserved amino acids in SNAP25a and SNAP25b must somehow have a direct impact on the functionality of the proteins; to date, the mechanism whereby these amino acid substitutions affect the function of the SNAP25 isoforms is not clear.
Another member of the SNAP25 protein family is SNAP23; this protein is derived from a separate gene to SNAP25a/b and is ∼60% identical to the SNAP25 isoforms at the amino acid level (7, 8). Unlike SNAP25a/b, SNAP23 has a ubiquitous tissue distribution and functions in regulated exocytosis pathways in cell types such as mast cells (9) and adipocytes (10) and possibly in constitutive exocytosis pathways throughout the body. In addition, a recent study reported that SNAP23 was targeted to dendritic spines in hippocampal neurons, where it regulated trafficking of NMDA receptor subunits (11).
Whereas syntaxin and VAMP2 are anchored to membranes by transmembrane sequences, SNAP25 and SNAP23 are synthesized as soluble proteins and become membrane-associated via palmitoylation of their respective cysteine-rich domains (12, 13). Palmitoylation is a post-translational modification of proteins that involves the attachment of the C16 saturated fatty acid palmitate most often to cysteine residues via a thioester linkage (S-palmitoylation) (14–17). The majority of cellular palmitoylation events are enzyme-mediated, and recent work identified a family of 23 DHHC proteins that function as palmitoyl transferases (18). The defining feature of DHHC proteins is a 51-amino acid domain containing a DHHC motif (aspartate-histidine-histidine-cysteine) within a cysteine-rich (CR) domain. This DHHC-CR domain is thought to contain the catalytic site of DHHC proteins. SNAP25b is palmitoylated by DHHC3, DHHC7, and DHHC17 (18–20), although a complete screen of all of these 23 DHHC proteins for activity against SNAP25b has not been reported. There is little information available on the enzymes that modify SNAP25a and SNAP23.
Despite their overall similarity, SNAP25a, SNAP25b, and SNAP23 display conspicuous differences in their cysteine-rich domains; SNAP25a and SNAP25b each have four cysteines, although the configuration of these residues is different for the two isoforms, whereas SNAP23 has five cysteine residues (Fig. 1) (21). These differences are important functionally, as SNAP25b engineered to have five cysteines supports less exocytosis than wild-type SNAP25b in PC12 cells, and two different SNAP23 mutants, each with four cysteines, are both more active in exocytosis than wild-type SNAP23 (22, 23). Furthermore, SNAP25 mutants with fewer cysteines than wild-type SNAP25 are more active in exocytosis in pancreatic beta cells (24).
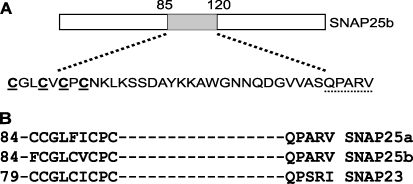
FIGURE 1.
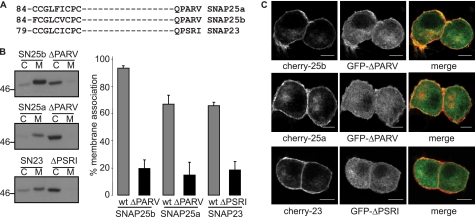
Minimal membrane targeting domain of SNAP25b. A, SNAP25b contains a minimal membrane targeting sequence that is located between amino acids 85 and 120. Palmitoylated cysteines in this domain are shown in bold and underlined. The QPARV motif, which is underlined with a dashed line is important for DHHC interactions of SNAP25b. B, comparison is shown of the cysteine-rich domains of SNAP25a, SNAP25b, and SNAP23. The QPARV motif in SNAP25b is conserved in SNAP25a, whereas mouse SNAP23 contains the sequence QPSRI. Amino acid numbers are shown.
These differences in functionality of SNAP25/23 mutated in the cysteine-rich domains suggest that the extent of palmitoylation or the palmitoylation recognition sites of the proteins regulate their function. Indeed, our previous work found that exocytotic activity of the cysteine mutants inversely correlated with association with cholesterol-rich membranes (22).
The aim of this study was to perform a systematic analysis of the DHHC proteins that promote membrane association of each of the SNAP25/23 proteins. This analysis will provide an important insight into the comparative interactions between DHHCs and SNAP25 proteins. Furthermore, we have extended this analysis by probing the palmitoylation of SNAP25/23 by DHHC proteins that are associated with the plasma membrane; such DHHC proteins may be important in dynamic remodeling of SNAP25 palmitoylation. Finally, we have examined how overexpression of selected DHHC proteins impacts on secretory processes in PC12 cells.
MATERIALS AND METHODS
Plasmids
DHHC proteins cloned into pEFBOS-HA were kindly provided by Masaki Fukata (National Institute of Physiological Sciences, Japan) (18). Rat SNAP25a and SNAP25b and mouse SNAP23 (lacking their initiating ATG) were cloned into pEGFP-C2. Site-directed mutants were generated using PCR. The DHHC15/3 chimeric construct was produced by inserting the DHHC domain from murine DHHC3 (amino acids 126–176) between amino acids 128 and 180 of murine DHHC15. The validity of all clones was confirmed by sequencing.
Antibodies, Kits, and Chemicals
Monoclonal GFP antibody (JL8) was obtained from Clontech. Rat anti-HA for immunoblotting and the growth hormone enzyme-linked immunosorbent assay kits were from Roche Applied Science. A SPEK fractionation kit was from Merck. Anti-HA Alexa Fluor 488-conjugated monoclonal antibody and Lipofectamine 2000 were from Invitrogen. Triton X-100 and all other chemicals were from Sigma (Poole, UK).
Cell Culture and Transfection
HEK293T cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum in a humidified atmosphere containing 5% CO2. PC12 cells were grown in RPMI1640 with 10% horse serum and 5% fetal calf serum in a humidified atmosphere containing 7.5% CO2. HEK293T and PC12 cells were transfected using Lipofectamine 2000; HEK293T cells were examined 24 h post-transfection, and PC12 cells were used 48 h post-transfection.
Analysis of DHHC Effects on SNAP25/23 Membrane Association
HEK293T cells were co-transfected with 0.8 μg of SNAP25/23 and 1.6 μg of each of the 23 DHHC proteins. Empty pEFBOS-HA vector was used as a control. Transfections were performed in 24-well plates in a volume of 500 μl. Twenty-four hours after the transfections the cells were fractionated into cytosol and membrane fractions using an SPEK fractionation kit (Merck) as previously described (20, 25, 26). Equal volumes of the recovered fractions were resolved by SDS-PAGE and transferred to nitrocellulose for immunoblotting analysis. Band densities were calculated using Image J software and the percentage membrane association of SNAP25/23 calculated in the presence of each of the 23 DHHC proteins. This experiment was repeated three times, and statistical analysis was performed using a one-way ANOVA.
[3H] Palmitate Labeling
HEK293T cells growing on 24-well plates were co-transfected with GFP-tagged SNAP25/23 (0.8 μg) and HA-DHHC (1.6 μg) constructs. Twenty-four hours post-transfection the cells were incubated in [3H]palmitic acid (0.5 mCi/ml) in Dulbecco's modified Eagle's medium with 1% de-fatted bovine serum albumin for 4 h at 37 °C. The cells were washed and lysed in 150 mm NaCl, 1% Triton X-100, 1% n-octylglucoside, 50 mm Tris, pH 8.0. The lysate was centrifuged at 14,000 × g for 10 min, and the recovered supernatant was mixed with SDS sample buffer. The samples were resolved by SDS-PAGE and transferred to duplicate nitrocellulose membranes, which were either probed with anti-GFP antibody or exposed to film with the aid of a Kodak Biomax Transcreen LE intensifier screen for detection of [3H]palmitate incorporation.
Immunofluorescence and Confocal Imaging
For comparison of the intracellular localizations of SNAP25/23 wild-type and mutant proteins, PC12 cells were co-transfected with 0.2 μg each of mcherry-tagged wild-type plasmid and GFP-tagged mutant plasmid. For analysis of GFP-tagged DHHC2 proteins, 1 μg of plasmid DNA was used for transfection. Cells were fixed in 4% formaldehyde 48 h post-transfection.
In other experiments PC12 cells were transfected with 1 μg of HA-DHHC proteins for 48 h. Cells were fixed in 4% formaldehyde and permeabilized for 6 min in 0.2% Triton X-100 (in PBS with 0.3% bovine serum albumin). HA-DHHC was detected using an Alexa Fluor 488-conjugated mouse anti-HA antibody (1:100). For analysis of SNAP25 localization in DHHC-transfected cells, anti-SNAP25 (SMI81, 1:100) was added for 1 h at room temperature. The cells were then washed and incubated for 1 h in anti-mouse secondary antibody conjugated to Alexa647 (1:400). After this, cells were washed extensively and incubated for 1 h in mouse anti-HA-488 (1:100). Cells were then washed and mounted in Mowiol. Imaging was performed on a Zeiss LSM510 confocal laser scanning microscope. Image data acquired at Nyquist sampling rates were deconvolved using Huygens software (Scientific Volume Imaging).
PCR Analysis
To examine DHHC2 mRNA expression, total RNA was purified from PC12 cells and from rat brain using an RNeasy kit (Qiagen). Reverse transcription was performed using In Prom-II reverse transcriptase (Promega, Madison, WI). PCR amplifications were set up using either 20 ng of DHHC plasmid DNA or 5 μl from a 20-μl reverse transcription reaction. Primers used were designed based on the sequence of rat DHHC2. Primers (10 pmol) and GoTaq PCR master mix (Promega) were added to the DNA templates. PCR consisted of 30 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 90 s.
Growth Hormone Release Assays
PC12 cells growing on 24-well plates were transfected with 0.5 μg of human growth hormone (hGH) plasmid together with 1 μg of pEFBOS-HA (control), HA-DHHC3, HA-DHHC7, or HA-DHHC17. Forty-eight hours post-transfection the cells were washed 3 times in Krebs buffer and then incubated for 15 min in 300 μl of Krebs buffer (basal) or Krebs buffer containing 300 μm ATP. The buffer was then removed and centrifuged to remove any detached cells. The remaining cells were solubilized in 0.5% Triton X-100. The amount of secreted and cell-associated human growth hormone was calculated using an enzyme-linked immunosorbent assay kit (Roche Applied Science), and growth hormone secretion was expressed as a percentage of total cell content. The results obtained from 3 independent experiments of this type (with n = 3 for each experiment) were analyzed for statistical significance using one-way ANOVA.
RESULTS
Identification of DHHC Proteins That Enhance Membrane Binding of SNAP25/23
The minimal membrane targeting sequence of SNAP25b was mapped to amino acids 85–120 (27) (Fig. 1A). This region is well conserved in all SNAP25/23 proteins, although there are conspicuous differences both in the cysteine-rich domains and in downstream residues that are important for palmitoylation (116QPARV120 in SNAP25b; Refs. 20 and 27) (Fig. 1B).
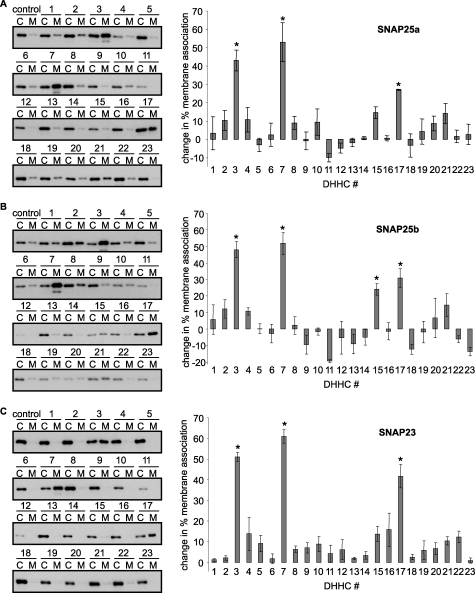
Our previous study reported that membrane binding of SNAP25b was inefficient in HEK293 cells and could be rescued by co-expression of DHHC3, DHHC7, and DHHC17 (20). However, as yet no complete screen has been reported to identify all DHHC proteins active against SNAP25b in this assay, and there has been no comparative analysis of the SNAP25 protein family. Given the differences in the respective cysteine-rich regions of SNAP25a, SNAP25b, and SNAP23 (Fig. 1B), it is important to define the subset of DHHC proteins active against SNAP25/23 and to highlight any differences in DHHC specificity. Thus, we co-transfected GFP-tagged SNAP25a, SNAP25b, or SNAP23 together with HA-tagged versions of each of 23 murine DHHC proteins into HEK293T cells. The cells were then fractionated into cytosol and membrane fractions, and the percent association of SNAP25/23 with membranes was calculated. By examining membrane binding as a functional readout, this assay should detect DHHC proteins that are involved in the initial palmitoylation reactions that attach soluble SNAP25/23 to membranes. In the absence of DHHC co-expression, membrane association of all three SNAP25/23 proteins was inefficient (13.5 ± 4.8% for SNAP25a, 22.2 ± 4.7% for SNAP25b, and 2.6 ± 0.7% for SNAP23). The effect of DHHC co-expression on membrane association of SNAP25/23 is shown in Fig. 2. The experiment was repeated three times for each SNAP25/23 protein, and the average change in membrane binding promoted by each of the DHHCs is presented. Statistical analyses revealed that significant increases in membrane binding of all three SNAP25/23 proteins were induced by DHHC3/7/17. In addition, DHHC15 significantly increased membrane binding of SNAP25b but not SNAP25a or SNAP23. It is noticeable that all DHHC proteins that promote membrane binding of SNAP25/23 are localized to the Golgi (25). This implies that palmitoylation of these proteins after their synthesis occurs at the Golgi.
FIGURE 2.
Regulation of SNAP25/23 membrane binding by DHHC proteins. HEK293T cells were co-transfected with EGFP-tagged versions of SNAP25a, SNAP25b, or SNAP23 together with HA-tagged constructs encoding each of the 23 murine DHHC constructs (or empty HA vector as a control). Twenty-four hours post-transfection, the cells were fractionated into cytosol (C) and membrane (M) fractions, which were resolved by SDS-PAGE and transferred to nitrocellulose for immunoblotting analysis with anti-GFP. The left panel of the figure shows representative immunoblots (top, SNAP25a; middle, SNAP25b; bottom, SNAP23). The numbering corresponds to the specific DHHC protein that was co-transfected with SNAP25/23. The right panel shows averaged data and S.E. (n = 3) for the increase in % membrane association of the SNAP25/23 proteins in the presence of each of the DHHCs. For clarity, the basal level of membrane association in the absence of DHHC co-expression was subtracted for each condition. The data were analyzed using a one-way ANOVA, which revealed significant increases in membrane association with DHHC3/7/17 for each SNAP25/23 protein and for DHHC15 specifically with SNAP25b when compared with transfections in the absence of DHHCs (*, p < 0.05).
Factors Regulating the Specificity of DHHC15 SNAP25/23 Interactions
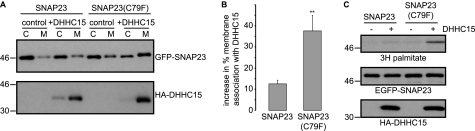
Having determined that DHHC15 is more active against SNAP25b than SNAP25a or SNAP23, we analyzed the intrinsic features of SNAP25/23 that underlie this difference. For this, we focused on the cysteine-rich domain of SNAP23, examining the effect of a single point mutation (C79F), which made the number and configuration of cysteines in the cysteine-rich domain of this SNAP23 mutant identical to that found in SNAP25b (Fig. 1B). Interestingly, membrane binding of this mutant in the presence of DHHC15 was substantially increased compared with wild-type SNAP23 (Fig. 3, A and B). Similarly, [3H]palmitate incorporation into SNAP23(C79F) in the presence of DHHC15 was increased compared with wild-type SNAP23 (Fig. 3C). These observations clearly show that the exact amino acid sequence of the cysteine-rich domains of SNAP25b and SNAP23 determine interaction specificity with DHHC15.
FIGURE 3.
A point mutation in SNAP23 enhances interaction with DHHC15. HEK293T cells were co-transfected with EGFP-tagged SNAP23 or a SNAP23(C79F) mutant together with HA-tagged DHHC15 or empty vector (control). Twenty-four hours post-transfection the cells were fractionated into cytosol (C) and membrane (M) fractions, which were resolved by SDS-PAGE and transferred to nitrocellulose for immunoblotting analysis with anti-GFP and anti-HA. A, representative immunoblots are shown. B, averaged data and S.E. (n = 4) for the increase in % membrane association of the SNAP23 proteins in the presence of DHHC15 are shown. ** indicates a statistically significant increase in membrane binding (p < 0.01, Student's t test) compared with wild-type SNAP23. C, HEK293T cells were transfected with EGFP-SNAP23 wild-type or C79F mutant together with pEFBOS-HA (control) or HA-DHHC15. Cells were labeled with [3H]palmitic acid, lysed, and resolved by SDS-PAGE. The top panel shows [3H]palmitate incorporation, the middle panel shows expression levels of EGFP-SNAP23, and the bottom panel shows HA-DHHC15 expression. Molecular weight markers are shown on the left of all figure panels.
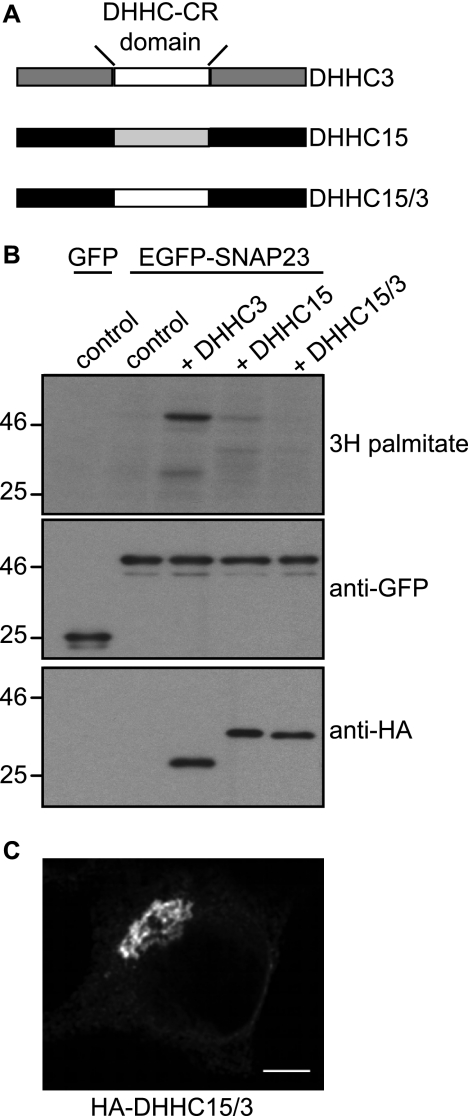
Having identified features within SNAP25/23 that contribute to the specificity of interaction with DHHC15, we next examined if specificity at the level of the DHHC protein is dependent on the DHHC-CR domain. Thus, we generated a DHHC15 construct containing the DHHC-CR domain of DHHC3 (Fig. 4A) and examined whether this chimeric protein was capable of palmitoylating SNAP23. Fig. 4B reveals that this DHHC15/3 chimera did not palmitoylate SNAP23 despite the protein displaying a typical Golgi-like distribution (Fig. 4C) similar to DHHC3 and DHHC15 (25). Thus, the differential specificity of DHHC3 and DHHC15 against SNAP23 appears not to be regulated by the DHHC domains alone and likely involves interplay between the DHHC domains and other regions of the proteins.
FIGURE 4.
A DHHC15 protein containing the DHHC-CR domain from DHHC3 is inactive against SNAP23. A, schematic highlighting the make-up of the DHHC15/3 chimeric protein is shown. B, HEK293T cells were transfected with EGFP-SNAP23 (or GFP) together with pEFBOS-HA (control), HA-DHHC3, HA-DHHC15, or HA-DHHC15/3. Cells were labeled with [3H]palmitic acid acid, lysed, and resolved by SDS-PAGE. The top panel shows [3H]palmitate incorporation, the middle panel shows expression levels of EGFP-SNAP23, and the bottom panel shows DHHC protein expression. Molecular weight markers are shown on the left. C, HA-DHHC15/3 was expressed in HEK293T cells, labeled with Alexa Fluor 488-conjugated anti-HA, and visualized using confocal microscopy. The scale bar represents 5 μm.
Palmitoylation of All SNAP25/23 Proteins Is Dependent on a Conserved Motif Downstream of the Cysteine-rich Domain
In addition to the cysteine rich domains of SNAP25/23, a short stretch of downstream amino acids (116QPARV120) is also essential for palmitoylation and membrane binding of SNAP25b (20, 27). We previously reported that this region of SNAP25b is important for recognition by DHHC17 but not DHHC3 (20). This sequence is identical in SNAP25a but only partially conserved in SNAP23 (mouse sequence is QPSRI). Thus, to further investigate any intrinsic features of the SNAP25/23 proteins that may differentially affect palmitoylation, we compared the effect of mutating this region in SNAP25a, SNAP25b, and SNAP23. Mutation of this region in SNAP25b inhibits membrane binding and plasma membrane targeting in PC12 cells (20, 27), and we, thus, used these assays as readouts to assess the importance of this region in all three SNAP25/23 proteins. Thus, the corresponding regions of these proteins were mutated (in SNAP25a and SNAP25b the PARV sequence was changed to ALAA, and in SNAP23 the QSRI sequence was mutated to AAAA). Fig. 5, A and B, demonstrates that mutation of this region in all SNAP25/23 proteins results in a significant reduction in membrane binding. Furthermore, GFP-tagged forms of these mutant proteins were co-transfected into PC12 cells with mcherry-tagged forms of the wild-type proteins, allowing a direct comparison of membrane targeting to be made in the same cell. Fig. 5C demonstrates that the mutant proteins all showed a more dispersed localization compared with wild-type proteins, showing that this region of all three SNAP25/23 proteins is essential for efficient membrane targeting.
FIGURE 5.
A conserved motif downstream of the cysteine-rich domain is important for membrane targeting of all SNAP25/23 proteins. A, comparison is shown of the cysteine-rich domains and downstream QPARV motif in SNAP25/23. B, analysis of the importance of the PARV/PSRI sequences in SNAP25/23 for membrane binding is shown. Wild-type (wt) and mutant EGFP-tagged SNAP25/23 proteins were transfected into PC12 cells before cell fractionation into cytosol (C) and membrane (M) fractions. The recovered fractions were probed with a GFP antibody. The left panel shows representative immunoblots, whereas the right panel is averaged data (n = 4). The reduction in membrane binding of mutant proteins compared with the respective wild-type control was statistically significant for all SNAP25/23 proteins (p < 0.00002 for SNAP25b, p < 0.02 for SNAP25a, and p < 0.0005 for SNAP23, Student's t test). C, PC12 cells were co-transfected with mcherry-tagged versions of wild-type SNAP25a, SNAP25b, or SNAP23 together with EGFP-tagged proteins in which the PARV and PSRI sequences were mutated to either ALAA or AAAA, respectively. Scale bars represent 5 μm.
Palmitoylation of SNAP25/23 by Plasma Membrane-associated DHHC Proteins
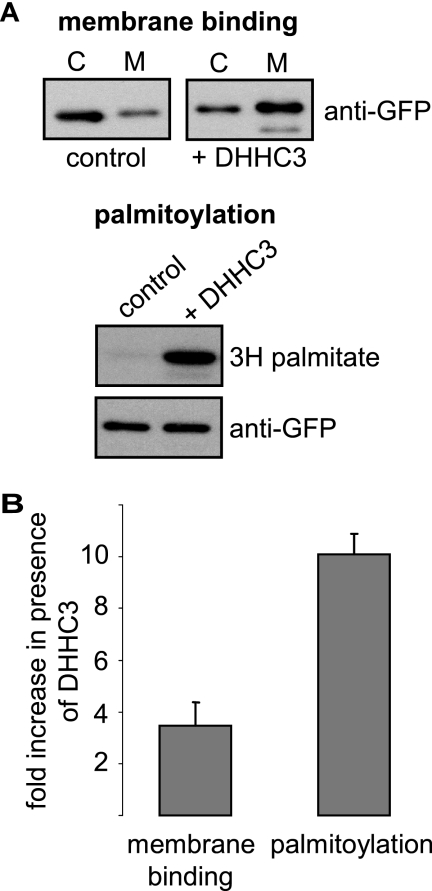
An important question is whether membrane association of SNAP25 proteins at the Golgi always correlates with full palmitoylation of the cysteine-rich domain or whether the level of palmitoylation can be lower or indeed heterogeneous. To examine this point, we compared the levels of membrane binding and palmitoylation of SNAP25b in cells co-transfected with either DHHC3 or empty vector. The mean -fold increase in membrane binding of SNAP25b in the presence of DHHC3 was 3.47-fold. By contrast, DHHC3 promoted a 10.08-fold increase in [3H]palmitate incorporation into SNAP25b (Fig. 6). Thus, the ratio of palmitoylation to membrane association was almost 3-fold greater in cells transfected with DHHC3 than in control cells. This result is consistent with the idea that the membrane-associated pool of SNAP25b in HEK cells without DHHC co-expression is only partially palmitoylated. Thus, stable membrane attachment of SNAP25b does not always correlate with complete palmitoylation of the protein, and it is, therefore, possible that SNAP25 undergoes additional palmitoylation steps after exit from the Golgi.
FIGURE 6.
Comparison of SNAP25b membrane binding and palmitoylation in the presence and absence of DHHC3 overexpression. A, HEK293T cells were transfected with EGFP-SNAP25b together with HA-DHHC3 or empty HA vector (control). Twenty-four hours post-transfection the cells were fractionated into cytosol (C) and membrane (M) fractions (top panel). Alternatively, the cells were incubated with [3H]palmitic acid for 4 h. Samples were then resolved by SDS-PAGE and transferred to nitrocellulose for immunoblotting with anti-GFP or detection of the 3H signal with the aid of an intensifier screen (bottom panel). B, -Fold increase in membrane binding and palmitoylation in the presence of DHHC3 was calculated (n = 3).
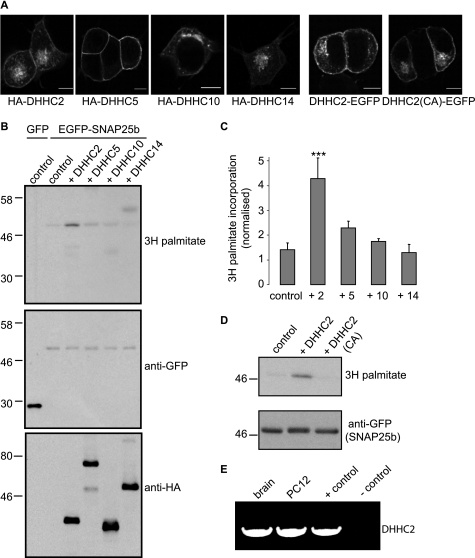
An intriguing question then is whether there are DHHC proteins that might influence SNAP25 palmitoylation dynamics at the plasma membrane. These plasma membrane DHHC proteins would be predicted not to increase stable membrane binding of SNAP25/23 (which occurs at the Golgi) in the assay shown in Fig. 2. We expressed DHHCs 1–23 in PC12 cells and identified a subset that showed varying degrees of plasma membrane association (Fig. 7A). DHHC5 was highly enriched at the plasma membrane. DHHC2 was also readily detected at the plasma membrane but was additionally present in an intracellular compartment. DHHC10 and DHHC14 were mainly intracellular, but plasma membrane staining could be visualized in some cells (Fig. 7A). To examine whether any of these DHHC proteins were active against SNAP25, we co-expressed the proteins in HEK293T cells and analyzed the extent of [3H]palmitate incorporation into SNAP25b. Fig. 7, panels B and C, reveals that DHHC2 promoted a marked and significant increase in the level of [3H]palmitate incorporation into SNAP25b. Note that the palmitoylation of SNAP25b by DHHC2 required an intact DHHC domain and was abolished when the DHHC domain was mutated to DHHA (Fig. 7D). Thus, DHHC2, which is present at the plasma membrane in PC12 cells, is active against SNAP25b. DHHC2 has been suggested to be widely expressed, and as we did not have access to a DHHC2 antibody, we confirmed the expression of DHHC2 mRNA in PC12 cells and rat brain by using RT-PCR analysis (Fig. 7E).
FIGURE 7.
SNAP25b is palmitoylated by DHHC2. A, a subset of DHHC proteins are localized at the plasma membrane in PC12 cells. Cells were transfected with HA-tagged or GFP-tagged DHHCs and fixed 48 h later. Localization of HA-tagged proteins was visualized by staining with an HA antibody conjugated to Alexa Fluor 488. The DHHC2(CA)-GFP protein has a cysteine-to-alanine mutation in the DHHC domain (C156A). The scale bar represents 5 μm. B, EGFP-SNAP25b was co-transfected into HEK293T cells together with each of the indicated DHHC proteins (empty HA vector was used for control). Twenty-four hours after transfection, the cells were incubated in [3H]palmitic acid for 4 h, washed, and lysed. The resulting lysates were resolved by SDS-PAGE and transferred to nitrocellulose, and the 3H signal was detected using an intensifier screen. Duplicate nitrocellulose membranes were probed with anti-GFP and anti-HA antibodies. Other bands labeled with 3H are DHHC proteins, which undergo autopalmitoylation. C, shown are mean values and S.E. for the 3H signal intensity of EGFP-SNAP25b as a fraction of the signal detected with the GFP antibody (n = 4). *** denotes a p value of <0.001 for the level of palmitoylation of EGFP-SNAP25b when co-expressed with DHHC2 compared with control, analyzed by one-way ANOVA. D, cells were transfected with EGFP-SNAP25b together with pEFBOS-HA (control), HA-DHHC2, or HA-DHHC2 (C156A). Cells were labeled with [3H]palmitic acid acid, lysed, and resolved by SDS-PAGE. The top panel shows [3H]palmitate incorporation, and the bottom panel shows expression levels of EGFP-SNAP25b. E, RT-PCR analysis of DHHC2 mRNA expression in rat brain and PC12 cells. Controls used were HA-DHHC plasmid (+ control) and no template (− control). Position of molecular weight markers are shown on the left side of all immunoblots.
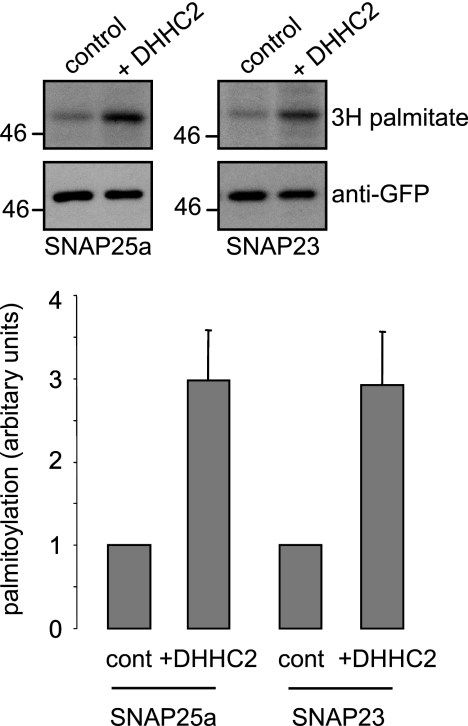
DHHC2 belongs to the same phylogenetic DHHC subfamily as DHHC15 (18). As we noted differences in the ability of DHHC15 to promote membrane association of the SNAP25/23 proteins (Fig. 2), we next tested whether DHHC2 was effective at enhancing palmitoylation of SNAP25a and SNAP23. Thus, HEK293T cells were co-transfected with EGFP-tagged SNAP25a or SNAP23 together with HA-DHHC2 or empty HA vector. [3H]Palmitate incorporation into SNAP25a and SNAP23 was quantified, revealing that DHHC2 increased palmitoylation of these isoforms to a similar extent as SNAP25b (Fig. 8). Thus, DHHC2 may play an important role in regulating palmitoylation of all three SNAP25/23 proteins.
FIGURE 8.
Palmitoylation of SNAP25a and SNAP23 by DHHC2. EGFP-SNAP25a and EGFP-SNAP23 were co-transfected into HEK293T cells together with HA-DHHC2 or empty vector (control). Twenty-four hours after transfection, the cells were incubated in [3H]palmitic acid for 4 h, washed, and lysed. The resulting lysates were resolved by SDS-PAGE and transferred to nitrocellulose. The 3H signal was detected using an intensifier screen, and duplicate nitrocellulose membranes were probed with anti-GFP. The top panel shows representative blots, with the molecular weight standards indicated on the left side. The bottom panel shows averaged data ± S.E. (n = 6) for palmitoylation in the presence and absence of DHHC2. The palmitoylation level in the absence of DHHC co-expression was set as 1.
Effects of DHHC Overexpression on Exocytosis in PC12 Cells
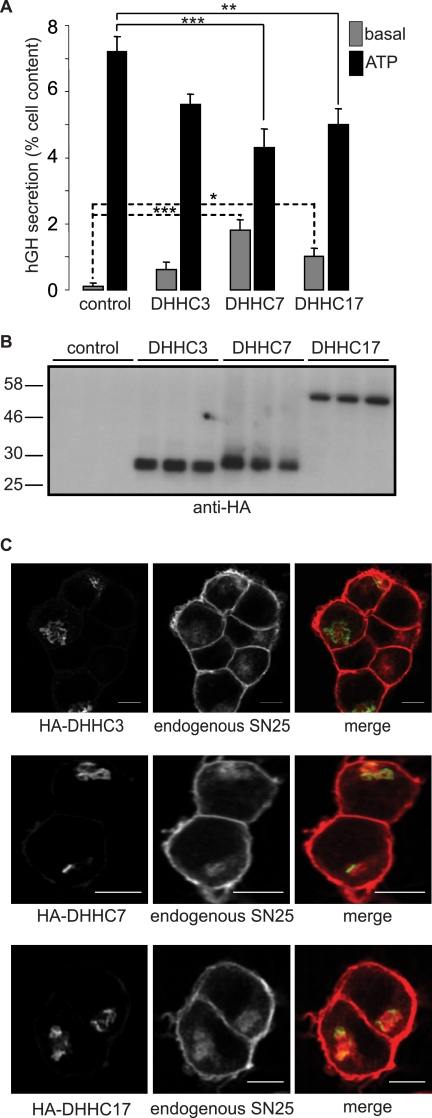
Finally, we sought to determine whether modulation of DHHC expression levels in PC12 cells had any functional effect on cellular secretion, a pathway that is regulated by SNAP25 proteins. For this analysis we focused on the subset of Golgi DHHC proteins that are active against all SNAP25/23 isoforms. Thus, we co-transfected PC12 cells with DHHC3, DHHC7, or DHHC17 together with a plasmid encoding hGH. HGH is packaged into secretory granules in PC12 cells and is released from cells after activation of regulated exocytosis, a pathway that is stimulated by increased cytoplasmic Ca2+ levels (23). This pathway is completely dependent upon SNAP25 and is potently blocked by botulinum neurotoxin E (BoNTE), a toxin that specifically cleaves SNAP25 (23). Interestingly, overexpression of DHHC3, DHHC7, or DHHC17 each caused a marked reduction in regulated exocytosis evoked by 300 μm ATP (Fig. 9A). Furthermore, the effects of DHHC overexpression were not limited to regulated secretion, as basal release of hGH was elevated by expression of all three DHHC proteins, a result that was statistically significant for DHHC7 and DHHC17. Thus, overexpression of Golgi DHHC proteins perturbs the normal secretion of hGH.
FIGURE 9.
Effects of DHHC overexpression on exocytosis in PC12 cells. A, PC12 cells were co-transfected with a plasmid encoding hGH together with HA-DHHC3, DHHC7, DHHC17, or empty HA plasmid (control). Forty-eight hours post-transfection, the cells were washed and incubated in Krebs buffer in the absence (basal) and presence of 300 μm ATP. The secreted and cell-associated hGH was assayed using an enzyme-linked immunosorbent assay kit, and secretion determined as a percentage of the total cell content. Results from three independent experiments (n = 3 for each experiment) were averaged, and the means ± S.E. are presented. One-way ANOVA tests revealed that DHHC7 and DHHC17 significantly reduced ATP-stimulated secretion compared with control transfected cells. The basal secretion level for DHHC7- and DHHC17-transfected cells was significantly increased compared with control transfected cells. *, indicates a p value of < 0.05; **, denotes a p value of < 0.01; ***, is p < 0.001. B, PC12 cells transfected with the HA-DHHC constructs were lysed, separated by SDS-PAGE, and transferred to nitrocellulose for immunoblotting analysis with anti-HA. Molecular weight standards are shown on the left. C, PC12 cells transfected with HA-DHHC3, HA-DHHC7, or HA-DHHC17 were fixed and permeabilized. The cells were incubated with anti-SNAP25 (SMI81) and anti-mouse antibody conjugated to Alexa Fluor 647 to stain endogenous SNAP25 and anti-HA conjugated to Alexa Fluor 488 to stain the HA-tagged DHHC constructs. Scale bars represent 5 μm.
It has been previously reported that overexpression of Golgi-localized DHHC proteins can trap substrate proteins including SNAP25 at the Golgi (for example, see Refs. 18 and 19), although most of these analyses have examined cells that overexpressed tagged substrates rather than looking at endogenous proteins. Thus, one possibility to explain the results of the hGH assay could be that substrates such as SNAP25 are trapped at the Golgi when DHHC3/7/17 are overexpressed, reducing their level at the plasma membrane and, thus, inhibiting exocytosis. To test this possibility, we examined the distribution of SNAP25 in PC12 cells that express HA-tagged DHHC proteins. In untransfected cells, endogenous SNAP25 was localized at the plasma membrane and also in an intracellular compartment (see untransfected cells in Fig. 9C, top panel, and also mcherry-SNAP25b distribution shown in Fig. 5E), which was previously shown to represent recycling endosomes/trans Golgi network (28). As shown in Fig. 9C, we did not detect trapping of endogenous SNAP25 in the Golgi of PC12 cells that express any of these DHHC proteins; the intracellular pool of SNAP25 is clearly distinct from the Golgi DHHC proteins (merged images) This result confirms that the effect of DHHC overexpression on hGH secretion is not a consequence of a modified intracellular localization of endogenous SNAP25.
DISCUSSION
SNAP25 and its ubiquitous homologue SNAP23 play essential roles as SNARE proteins in membrane fusion events that occur at the plasma membrane. Most SNARE proteins contain a single SNARE motif and a transmembrane anchor. Thus, SNAP25 and SNAP23 are unusual in having two SNARE domains that are joined by a palmitoylated linker domain. The primary role of the palmitoylated cysteine-rich domain is undoubtedly to stably attach SNAP25/23 to intracellular membranes and, thus, facilitate its trafficking to the plasma membrane. However, the cysteine-rich domains of SNAP25a, SNAP25b, and SNAP23 are notably distinct, suggesting that they might impart differential regulation to the three protein isoforms. Indeed, we have previously shown that SNAP23 associates to a greater extent with cholesterol-rich membranes in vitro than does SNAP25 (22); this was directly related to differences in the cysteine-rich domains (22). Given the potential for differential regulation of SNAP25/23 by their respective cysteine-rich domains, we considered it important to undertake a comprehensive analysis of DHHC activity against these proteins. In the process we hoped to highlight any differences in DHHC interaction that might contribute to differential regulation of the proteins. Indeed, although we identified a core subset of DHHC proteins that were able to drive membrane association of all SNAP25/23 proteins, we also found that DHHC15 was more active against the SNAP25b isoform in this assay.
There is little known about how DHHC specificity is encoded by palmitoylated substrates. Our own work identified proline-117 in SNAP25b as important for interaction with DHHC17, whereas this region did not appear to be required for interaction with DHHC3 (20). Work from Nadolski and Linder (29) also showed that DHHC specificity can be influenced by residues outside the palmitoylated domains of substrate proteins. In this study we report that the amino acid sequence of the cysteine-rich domains of SNAP25/23 influences DHHC interaction. In this case, replacement of one of the five cysteines in SNAP23 with a phenylalanine (as present in SNAP25b) led to a marked increase in membrane binding and palmitoylation of SNAP23 induced by DHHC15 co-expression. Thus, DHHC interaction specificity at the level of the substrate protein is influenced by residues both within and outside the palmitoylated domains.
We further extended this analysis by investigating whether DHHC specificity toward SNAP25/23 in determined by the DHHC-CR domains. Replacement of the DHHC-CR domain of DHHC15 with that from DHHC3 produced a chimeric protein that was expressed and targeted correctly but that was unable to palmitoylate SNAP23. This result suggests that essential factors regulating DHHC interaction with SNAP25/23 proteins are located outside the DHHC-CR domains and is consistent with previous work showing that the N-terminal ankyrin-repeat domain of DHHC17 contributes toward specificity of interaction with huntingtin (30).
What is the underlying reason for DHHC15 selectivity toward SNAP25b? We previously reported that the hydrophobicity of the cysteine-rich domain of SNAP25b is important to allow transient membrane interactions before palmitoylation by membrane-associated DHHC proteins (20). Thus, there are at least two factors that are required for SNAP25 palmitoylation in cells; (i) an underlying membrane affinity to permit proximity with membrane-bound DHHC proteins and (ii) recognition and palmitoylation by specific DHHC proteins, which promotes stable membrane attachment. It is most likely then that the cysteine-to-phenylalanine mutation in SNAP23 either increases the direct affinity of DHHC15 for SNAP23 or, alternatively, that it enhances the membrane affinity of SNAP23, allowing increased membrane residency of the unpalmitoylated protein and enhancing the likelihood of productive association with DHHC15. Whatever the reason for the isoform selectivity of DHHC15, the preferential palmitoylation of SNAP25b by DHHC15 may be functionally relevant, perhaps contributing to differences in palmitoylation or sorting of the different isoforms.
The recent report that SNAP23 is enriched in dendritic spines, whereas SNAP25 is axonally distributed raises interesting questions about the mechanisms that underlie the distinct polarized sorting of these proteins. As SNAP25 and SNAP23 are only ∼60% identical, it is possible that the different localizations are a consequence of interactions with distinct targeting factors. However, another possibility is that the distinct cysteine-rich domains and differences in palmitoylation may contribute to the differential sorting of these proteins in neurons. Indeed, interactions of SNAP25/23 with different Golgi DHHC proteins might facilitate their association with specific Golgi bud sites that control protein sorting. As a first step to understanding how this differential targeting is achieved, it will be interesting to test whether the distinct sorting patterns of SNAP25 and SNAP23 is recapitulated by the isolated membrane targeting domains of these proteins.
It is interesting to speculate on the HEK293 cell DHHC proteins that mediate the basal level of SNAP25/23 membrane association in the absence of DHHC co-expression. We suggest that DHHC proteins active against SNAP25/23 are expressed at low levels in human embryonic kidney cells sufficient to catalyze palmitoylation of a small amount of overexpressed SNAP25/23.
There are conflicting reports in the literature concerning the half-life of SNAP25 palmitoylation (12, 31, 32), and there is very little information available on the degree of SNAP25 palmitoylation or its heterogeneity (12). Comparing the extent of membrane association and palmitoylation of SNAP25b in HEK293T cells with and without co-transfection with DHHC3 suggested that the palmitoylation of membrane-associated SNAP25b was significantly increased in the presence of DHHC3. This result suggests that SNAP25b associated with membranes in HEK293T cells in the absence of DHHC co-expression may only be partially palmitoylated. Although this analysis was performed in a heterologous cell line, it nevertheless suggests that stable membrane association of SNAP25 does not require palmitoylation of every cysteine residue. This notion is in agreement with our previous work showing that membrane binding is preserved when single cysteine residues in the cysteine-rich domain are mutated (20). As stable membrane binding does not require palmitoylation of every cysteine, significant changes in SNAP25/23 palmitoylation status may occur in cells without the proteins detaching from membranes. DHHC2 emerged from this study as a candidate palmitoylating enzyme that might regulate the palmitoylation status of SNAP25/23 at the plasma membrane.
There have been very few studies that have examined the effects of modulating DHHC expression levels on the functionality of a defined cellular pathway. Interestingly, overexpression of DHHC3/7/17 all perturbed the secretion of hGH from PC12 cells. This was seen as an increase in basal secretion and a decrease in regulated release. We clearly showed that these effects were not caused by trapping of endogenous SNAP25 on Golgi membranes. It is interesting that DHHC proteins localized to the Golgi can impact on exocytotic events at the plasma membrane. There are a number of possibilities that could explain these results including the following. (i) Overexpression of these DHHC proteins increases the extent of SNAP25 palmitoylation during its biosynthetic delivery to the plasma membrane. As increased palmitoylation of SNAP25 has been suggested to inhibit its function (23); this could explain the reduced level of regulated exocytosis. (ii) Changes in the palmitoylation of other exocytotic proteins could modulate the secretion of hGH. (iii) Overexpression of these DHHC proteins might modulate the sorting of hGH at the Golgi, for example, by increasing the packaging of hGH into constitutive rather than regulated secretory vesicles. (iv) DHHC overexpression could somehow lead to an increased secretion of regulated secretory vesicles under resting conditions. Indeed it is noticeable that the reduced level of regulated hGH secretion corresponds in all cases with an increased level of basal hGH release. Whatever the reason for these effects, it is intriguing that changes in the expression of Golgi DHHC proteins impacts on secretory processes. By identifying SNAP25/23-DHHC interactions and determining how specificity is regulated, this study represents an important step in defining how palmitoylation of SNAP25 is controlled and in understanding DHHC-substrate specificity in general.
Acknowledgments
Image analysis was performed at the IMPACT imaging facility at the University of Edinburgh. We are very grateful to Masaki Fukata for the DHHC constructs.
This work was supported by a Medical Research Council Senior Research fellowship (to L. H. C.).
- SNAP25
- synaptosomal-associated protein of 25 kDa
- SNAP23
- SNAP25-related protein of 23 kDa
- DHHC
- aspartic acid, histidine, histidine, cysteine
- CR
- cysteine-rich
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- HA
- hemagglutinin
- hGH
- human growth hormone
- ANOVA
- analysis of variance.
REFERENCES
- 1.Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 2.Sadoul K., Lang J., Montecucco C., Weller U., Regazzi R., Catsicas S., Wollheim C. B., Halban P. A. (1995) J. Cell Biol. 128, 1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence G. W., Weller U., Dolly J. O. (1994) Eur. J. Biochem. 222, 325–333 [DOI] [PubMed] [Google Scholar]
- 4.Bark I. C., Wilson M. C. (1994) Gene 139, 291–292 [DOI] [PubMed] [Google Scholar]
- 5.Sørensen J. B., Nagy G., Varoqueaux F., Nehring R. B., Brose N., Wilson M. C., Neher E. (2003) Cell 114, 75–86 [DOI] [PubMed] [Google Scholar]
- 6.Johansson J. U., Ericsson J., Janson J., Beraki S., Stanić D., Mandic S. A., Wikström M. A., Hökfelt T., Ogren S. O., Rozell B., Berggren P. O., Bark C. (2008) PLOS Genetics 4, e1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravichandran V., Chawla A., Roche P. A. (1996) J. Biol. Chem. 271, 13300–13303 [DOI] [PubMed] [Google Scholar]
- 8.Wang G., Witkin J. W., Hao G., Bankaitis V. A., Scherer P. E., Baldini G. (1997) J. Cell Sci. 110, 505–513 [DOI] [PubMed] [Google Scholar]
- 9.Guo Z., Turner C., Castle D. (1998) Cell 94, 537–548 [DOI] [PubMed] [Google Scholar]
- 10.Rea S., Martin L. B., McIntosh S., Macaulay S. L., Ramsdale T., Baldini G., James D. E. (1998) J. Biol. Chem. 273, 18784–18792 [DOI] [PubMed] [Google Scholar]
- 11.Suh Y. H., Terashima A., Petralia R. S., Wenthold R. J., Isaac J. T., Roche K. W., Roche P. A. (2010) Nat. Neurosci. 13, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veit M., Söllner T. H., Rothman J. E. (1996) FEBS Lett. 385, 119–123 [DOI] [PubMed] [Google Scholar]
- 13.Vogel K., Roche P. A. (1999) Biochem. Biophys. Res. Comm. 258, 407–410 [DOI] [PubMed] [Google Scholar]
- 14.Resh M. D. (2006) Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 15.Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 16.Greaves J., Chamberlain L. H. (2007) J. Cell Biol. 176, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves J., Prescott G. R., Gorleku O. A., Chamberlain L. H. (2009) Mol. Membr. Biol. 26, 67–79 [DOI] [PubMed] [Google Scholar]
- 18.Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 19.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R. R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C. A., Hayden M. R., El-Husseini A. (2004) Neuron 44, 977–986 [DOI] [PubMed] [Google Scholar]
- 20.Greaves J., Prescott G. R., Fukata Y., Fukata M., Salaun C., Chamberlain L. H. (2009) Mol. Biol. Cell 20, 1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott G. R., Gorleku O. A., Greaves J., Chamberlain L. H. (2009) J. Neurochem. 110, 1135–1149 [DOI] [PubMed] [Google Scholar]
- 22.Salaün C., Gould G. W., Chamberlain L. H. (2005) J. Biol. Chem. 280, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salaün C., Gould G. W., Chamberlain L. H. (2005) J. Biol. Chem. 280, 19449–19453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonelle-Gispert C., Molinete M., Halban P. A., Sadoul K. (2000) J. Cell Sci. 113, 3197–3205 [DOI] [PubMed] [Google Scholar]
- 25.Greaves J., Salaun C., Fukata Y., Fukata M., Chamberlain L. H. (2008) J. Biol. Chem. 283, 25014–25026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves J., Chamberlain L. H. (2006) Mol. Biol. Cell 17, 4748–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalo S., Greentree W. K., Linder M. E. (1999) J. Biol. Chem. 274, 21313–21318 [DOI] [PubMed] [Google Scholar]
- 28.Aikawa Y., Xia X., Martin T. F. J. (2006) Mol. Biol. Cell 17, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadolski M. J., Linder M. E. (2009) J. Biol. Chem. 284, 17720–17730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. (2009) FASEB J. 23, 2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane S. R., Liu Y. (1997) J. Neurochem. 69, 1864–1869 [DOI] [PubMed] [Google Scholar]
- 32.Kang R., Swayze R., Lise M. F., Gerrow K., Mullard A., Honer W. G., El-Husseini A. (2004) J. Biol. Chem. 279, 50524–50536 [DOI] [PubMed] [Google Scholar]