Abstract
Chronic inflammation is now accepted to have a critical role in the onset of several diseases as well as in vascular pathology, where macrophage transformation into foam cells contributes in atherosclerotic plaque formation. Endothelial cells (EC) have a critical function in recruitment of immune cells, and proinflammatory cytokines drive the specific expression of several adhesion proteins. During inflammatory responses several cells produce hyaluronan matrices that promote monocyte/macrophage adhesion through interactions with the hyaluronan receptor CD44 present on inflammatory cell surfaces. In this study, we used human umbilical chord vein endothelial cells (HUVECs) as a model to study the mechanism that regulates hyaluronan synthesis after treatment with proinflammatory cytokines. We found that interleukin 1β and tumor necrosis factors α and β, but not transforming growth factors α and β, strongly induced HA synthesis by NF-κB pathway. This signaling pathway mediated hyaluronan synthase 2 (HAS2) mRNA expression without altering other glycosaminoglycan metabolism. Moreover, we verified that U937 monocyte adhesion on stimulated HUVECs depends strongly on hyaluronan, and transfection with short interference RNA of HAS2 abrogates hyaluronan synthesis revealing the critical role of HAS2 in this process.
Keywords: Extracellular Matrix, Glycosaminoglycan, Inflammation, Interleukin, Proteoglycan Synthesis
Introduction
Hyaluronan (HA)3 is a linear glycosaminoglycan consisting of a disaccharide (glcUA-β1,3-glcNAc-β1,4) repeated several thousand times without any other chemical modifications (i.e. sulfation and epimerization) that are typical of the other glycosaminoglycans (1). HA is a multifunctional molecule in the extracellular matrix. In addition to its viscoelastic properties that modulate tissue hydration, HA can interact with cell surface receptors, including CD44, receptor for HA-mediated motility (RHAMM), Lyve-1 (lymphatic vessel endothelial receptor 1), HARE (HA receptor for endocytosis), intercellular adhesion molecule-1 (ICAM-1), and Toll-like receptor 4 (TLR4), and HA can initiate several signal transduction pathways (1). Chain lengths can depend on the activity of different isoforms of HA synthases (HAS1, -2, and -3) (2), or from the activity of degrading enzymes (i.e. hyaluronidases) (1). Short HA fragments produced after injuries or inflammation can interact with TLR4 and stimulate synthesis of macrophage chemokines and cytokines (3).
In vascular pathologies, HA accumulation can regulate the behavior of smooth muscle cells and contribute to vessel wall thickening by inducing cell migration and proliferation (4). Moreover, in the media and neointima, HA exerts a proatherosclerotic effect by promoting adhesion of immune cells and by recruiting monocytes/macrophages (5) that, through cholesterol rich lipoproteins endocytosis, contribute to progression of atherosclerotic plaque. The molecular mechanism involved in the interaction of immune cells with HA depends on CD44. Interestingly, the organization of HA in the extracellular matrix has a critical role in this process, and cells subjected to various stresses (endoplasmic reticulum stress, viral stimulus, and hyperglycemia) synthesize HA cable-like structures that interact with CD44 on monocyte surfaces and mediate adhesion (6). The organization of the HA cable structure largely is unknown and can contain additional molecules, including versican and the covalently linked heavy chains transferred from the chondroitin sulfate chain in inter-α-trypsin inhibitor to HA catalyzed by tumor necrosis factor α (TNF-α)-induced protein 6 (7). The recruited monocytes/macrophages are required to remove this abnormal matrix, and CD44 also is required. Mice null in CD44 do not survive a bleomycin-induced inflammation in the lung due to the continued accumulation of HA extracellular matrix and despite continued influx of monocytes/macrophages (8). Interestingly, dividing cells in hyperglycemia initiate HA synthesis in intracellular compartments (endoplasmic reticulum/Golgi/transport vesicles), which induces autophagy and cyclin D3 responses that initiate formation of HA cable structures after completion of cell division (9). This suggests that HA synthases located in these intracellular compartments can be activated and utilize the UDP-sugar substrates in the cytosol during HAS transport to and/or from the plasma membrane where it is normally active (6, 10, 11). Such HA cables have been described both in vitro and in vivo and appear to be associated to smooth muscle cells in gut mucosa of Crohn disease patients, in the lung of asthmatic patients, and in glomeruli of kidneys in the streptozotocin diabetic rat model (6). However, immune cells that circulate in the bloodstream have to cross the endothelium to reach the inflamed sites. Therefore, the role of endothelial cells is critical to “capture” circulating inflammatory cells only at specific sites where the immune response is required through interactions with cell membrane structures.
Proinflammatory mediators, including TNF-α and interleukin 1β (IL-1β), significantly alter expression of adhesion molecules on EC surfaces that determine the tethering, rolling, activation, arrest, and extravasation (diapedesis) of leukocytes from the bloodstream into the inflamed tissue. This complex set of successive events depends on many proteins (i.e. selectins and integrins) expressed on both ECs and the immune cells (12, 13). In addition, HA is known to have a critical role in this process (14). Interaction of leukocyte CD44 with HA on the EC surface determines their adhesion and activation (15). Interestingly, ECs normally synthesize very little HA, whereas its production dramatically increases after stimulation by proinflammatory cytokines (16). However, evidence for HA cable formation in ECs is lacking, and our data support a model in which EC synthesis of HA forms a structure bound to CD44 on the EC surface that can be recognized by CD44 on immune cells (17). In this study, we investigated the mechanism by which ECs increase HA synthesis at the molecular level and identify the pathway involved in this process.
EXPERIMENTAL PROCEDURES
Cell Culture
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and were grown for 2–6 passages in EGM2 culture medium (Lonza) supplemented with 10% fetal bovine serum. The cultures were maintained in an atmosphere of humidified 95% air, 5% CO2 at 37 °C. Six h before treatments, subconfluent HUVECs were cultured in EGM2 with 1% fetal bovine serum. The medium was then changed to EGM2 plus 10% fetal bovine serum with 5 μg/ml of IL-1β (Peprotech), or 0.1 μg/ml of TNF-α (Euroclone), or 0.3 μg/ml of TNF-β (Euroclone), or 1 μg/ml of tumor growth factor α (TGF-α Euroclone), or 0.01 μg/ml of tumor growth factor β (TGF-β, Euroclone) and incubated for 24 h. In some experiments, 2 μm (final concentration) of pyrolidine dithiocarbamate (PDTC, Sigma) was added to the cells. By using these cytokine concentrations, we did not measure any mortality effects detected by trypan blue staining (results not shown).
Glycosaminoglycan Quantification
HA and chondroitin sulfate released into the culture medium were quantified by polyacrylamide gel electrophoresis of fluorophore-labeled saccharides (PAGE-FS) and by HPLC analyses as described previously (18, 19). Briefly, proteins in the culture medium were digested with proteinase K, and the glycosaminoglycans were purified by ethanol precipitation. The specific unsaturated disaccharides of HA and chondroitin sulfate (ΔHA and ΔCS, respectively) were obtained by specific glycosidase digestions and derivatization with 2-aminoacridone. 2-Aminoacridone-labeled ΔHA and ΔCS disaccharides were separated and quantified by PAGE-FS or HPLC and normalized to the cell number. Agarose gel electrophoresis, as described by Vigetti et al. (5), was used to assess the molecular weight distributions of synthesized HA after IL-1β treatments.
HA Visualization
To visualize HA, HUVECs were grown on glass coverslips and either untreated or treated with IL-1β. They were processed using a biotinylated HA-binding protein and a green fluorescent reporter as described previously (20).
Quantitative RT-PCR
Total RNA samples were extracted from untreated or cytokine-treated HUVECs with TRIzol (Invitrogen), and contaminating DNA was removed by DNase (Ambion). cDNAs were generated by using the High Capacity cDNA synthesis kit (Applied Biosystems) and were amplified on an Abi Prism 7000 instrument (Applied Biosystems) using the Taqman Universal PCR Master Mix (Applied Biosystems) following the manufacturer's instructions. The following human TaqMan gene expression assays were used: HAS1 (Hs00155410_m1), HAS2 (Hs00193435_m1), HAS3 (Hs00193436_m1), CD44 (HS00174139_m1), and β-actin (Hs99999903_m1). The relative quantification of gene expression levels was determined by comparing ΔCt (5).
HAS Activity
To quantify the activity of HAS on cell membranes, we used the nonradioactive assay of HA synthesis as described previously (11).
Human HAS2 Silencing
Short interfering RNA (siRNA) was used to reduce the expression of human HAS2 in HUVECs by using HAS2 siRNA (ID117326, 5′-GCUGCUUAUAUUGUUGGCUtt-3′) and negative control siRNA kit (scramble, catalog no. 4611), both from Ambion. The transfections were done using a Nucleofector apparatus (Lonza) and the human HUVEC Nucleofector kit as described previously (21). After 48 h of incubation, the silencing efficiency was quantified by quantitative RT-PCR measuring the HAS2 mRNA transcript.
Monocytes Adhesion Assay
Adhesion of U937 monocytes to HUVEC cultures was assayed as described previously (22). 1 × 106 U937 monocytes were labeled with fluorescent green CytoTracker (Invitrogen) vital staining and plated on cytokine-treated or untreated HUVEC cultures followed by incubation at 4 °C for 30 min. After phosphate-buffered saline washing at 4 °C, the numbers of adherent cells were assessed under a fluorescent microscope (Olympus). As controls for binding to HA and CD44, HUVECs were pretreated with 2 units/μl of Streptomyces hyaluronidase (Seikagaku), or were incubated with monocytes in the presence of an antibody against CD44 (BRIC235, 5 μg/ml) that inhibits CD44/HA interaction, or were incubated with unrelated antibodies. Furthermore, monocytes were placed on HUVECs in the presence of 25 μg/ml high molecular mass HA (Healon, 4 × 106 Da) to compete with HA produced by treated cells.
Statistical Analyses
Statistical analysis of the data were done using analysis of variance, followed by post hoc tests (Bonferroni) using Origin 7.5 software (OriginLab). Probability values of p < 0.01 were considered statistically significant. Experiments were repeated three times each time in duplicate, and data are expressed as means ± S.E.
RESULTS AND DISCUSSION
Proinflammatory Cytokines Induce HA Synthesis
One of the main roles of ECs in inflammation is to regulate recruitment of immune cells from the bloodstream into the inflamed site. This mechanism involves not only expression of adhesive molecules on EC surfaces but also a coordinated expression of the corresponding ligands on the circulating cells. Diffusible alerting molecules (i.e. cytokines) bind to EC receptors and drive the expression of specific adhesion proteins, selectins, and integrins. Cytokines such as IL-1β and TNF-α are among the common proinflammatory-alerting molecules that are synthesized by immune cells such as macrophages in response to infection in an early phase of the process. The pioneering works of Siegelmen's research group clearly showed that HA is also involved in lymphocyte recruitment in vivo as well as in vitro (16, 23–26). Circulating leukocytes are activated by HA through CD44 interaction, whereas proinflammatory cytokines stimulate ECs to produce HA together with other well known adhesion molecules such as ICAM-1 and VCAM-1.
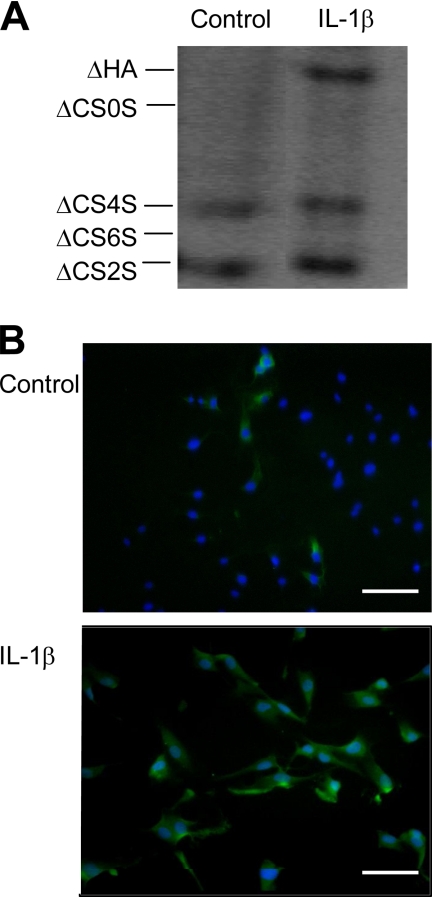
In our study, we focused on the mechanism of HA induction after IL-1β and TNF-α treatment. We used HUVECs because they are a convenient and broadly accepted artery EC model. After preliminary dose-response experiments, we found that a 24-h treatment of HUVEC cultures with 5 μg/ml of IL-1β induced a clear accumulation of HA secreted into the culture medium, which was quantitatively measured by PAGE-FS (Fig. 1A), and HA in the cell layer was qualitatively visualized by immunofluorescence (Fig. 1B). The band corresponding to HA disaccharides dramatically increased after IL-1β addition, whereas the bands corresponding to chondroitin sulfate disaccharides remained unchanged, indicating that IL-1β induces a specific alteration in the HA content without modifying chondroitin sulfate synthesis. The organization of HA in treated cells revealed a diffuse staining around HUVEC plasma membranes without showing the cable-like structures that are known to be adhesive toward monocytes in other cell models (6, 14). Moreover, analysis of the size of HA induced by IL-1β by agarose gel electrophoresis revealed the synthesis of a high molecular weight polymer of >2 × 106 Da (results not shown).
FIGURE 1.
IL-1β induces HA biosynthesis in HUVECs. A, PAGE-FS analysis of the HUVEC culture medium in untreated (CNT) and 24-h treated cells with 5 μg/ml of IL-1β. ΔHA and ΔCS0S indicate HA and chondroitin disaccharides, respectively, while ΔCS4S, ΔCS6S, and ΔCS2S indicate chondroitin 4, 6, and 2 sulfate disaccharides, respectively. B, visualization of HA (green) and nuclei (blue) on untreated (Control) or 24-h treated cells with 5 μg/ml of IL-1β. The microphotographs show representative results of different experiments. Scale bars, 70 μm.
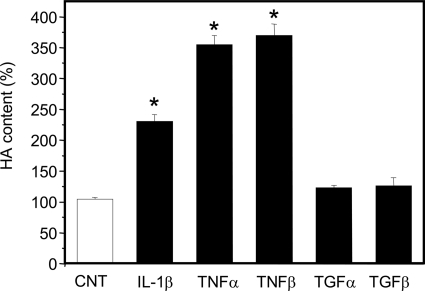
Furthermore, we tested the responses of HUVECs to other proinflammatory cytokines (i.e. TNF-α and TNF-β) or to anti-inflammatory cytokines (i.e. TGF-α and TGF-β) (27). After preliminary dose-response experiments (data not shown), the proinflammatory TNF-α and TNF-β at plateau concentrations induced the highest HA secretion, ∼3.5-fold over control, whereas IL-1β increased HA ∼2-fold over control. In contrast, anti-inflammatory TGF-α and TGF-β treatments did not increase HA secretion over control (Fig. 2). These results suggest that macrovascular artery ECs, such as HUVECs, together with previously reported microvascular ECs (16), can respond to microenvironmental changes by activating HA synthesis after treatment with proinflammatory cytokines. Previous studies and our unpublished data have shown that HUVECs induce ICAM-1 and VCAM-1 expression after treatment with proinflammatory cytokines, whereas Mohamadzadeh et al. (16) reported that HUVECs did not increase HA synthesis after stimulation with 10 ng/ml of TNF-α or IL-1β for 4 h. These apparent differences in results can be explained considering the different cytokine concentrations and incubation times, as well as the different sensitivity of the HA quantification methods that we used in our experiments. Although the cytokine concentrations used in this study appear to be higher than physiological level, it was discussed elsewhere that as cytokines act locally, their concentration in vivo may be difficult to define and higher than expected (28).
FIGURE 2.
Effects of several cytokines on HA biosynthesis in HUVECs. Relative quantification by HPLC of ΔHA secreted into the culture medium in untreated (CNT, white bar) and 24-h treated (black bars) cells with 5 μg/ml of IL-1β, 0.1 μg/ml of TNF-α, 0.3 μg/ml of TNF-β, 1 μg/ml of TGF-α, and 0.01 μg/ml of TGF-β. Results are expressed as mean ± S.E. *, p < 0.01 control versus treated samples.
Proinflammatory Cytokine-induced HA Depends on HAS2 and NF-κB
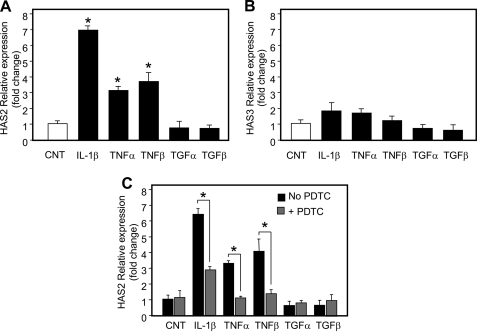
As the synthesis of HA depends on the activity of one or more of the three HA synthases, HAS1, -2, and -3, we measured their mRNA expression after cytokine treatments by using quantitative RT-PCR (Fig. 3, A and B). As HAS protein only is present at low levels at the plasma membrane (29), quantitative assessment at the protein level remains a technical challenge and could not be performed in a reliable manner. HAS1 mRNA was not detected in HUVECs, whereas HAS2 and HAS3 mRNAs were present at similar levels. Interestingly, only HAS2 mRNA significantly increased, ∼7-fold after IL-1β treatment and ∼3-fold after TNF-α and TNF-β treatments. In contrast, HAS2 mRNA did not change after treatment with the anti-inflammatory cytokines. It should be noted that the induction of HAS2 mRNA and HA levels after the cytokine treatments did not match exactly. This could be due to an earlier expression of HAS2 that we did not detect after 24 h of incubation, whereas HA accumulates during the whole incubation time. HAS2 enzymes synthesize HA of high molecular weight in agreement with the size that we observed after IL-1β stimulation. These results indicate that increased HA in response to the proinflammatory cytokines is regulated by expression of HAS2. In addition, we measured the HA synthetic activity in purified plasma membrane vesicles (11) isolated from control, IL-1β-, TNF-α-, and TNF-β-treated HUVEC cultures and found values of 22.4 ± 3, 91.1 ± 7, 78.9 ± 8, and 83.7 ± 5 pmol ΔHA/μg of protein/min, respectively. The cytokine treated cultures showed statistically significant increases ∼4-fold over control of HAS activity in purified plasma membranes, consistent with the increases in HA in the culture medium. Interestingly, the proinflammatory cytokine treatments induced similar HAS activities, whereas IL-1β induced a much greater HAS2 mRNA response than did TNF-α and TNF-β. Therefore, this activity increase was not related completely to HAS2 gene expression as shown in Fig. 3A, because cytokine treatments caused a different degree of HAS2 messenger accumulation. These data could suggest the existence of complex post-translational processing of HAS mRNA as has been shown for the stability of other TNF-α-induced transcripts (30). Moreover, other additional factors, such as hyaluronidase activity and HA turnover could contribute to this issue. Previous data from TNF-α-treated lymph node ECs reported that increased HA synthesis did not involve a significant change in HAS gene expression (16). However, only HAS1 mRNA was investigated at that time as the other isoenzymes were just discovered, and their sequences were not available in public databases.
FIGURE 3.
Gene expression alteration after treating HUVECs with cytokines. Relative quantification of transcripts coding for HAS2 (A) or HAS3 (B) in untreated (CNT, white bar) and after 24 h treated (black bars) cells with 5 μg/ml of IL-1β, 0.1 μg/ml of TNF-α, 0.3 μg/ml of TNF-β, 1 μg/ml of TGF-α, and 0.01 μg/ml of TGF-β. The lowest HAS2 or HAS3 mRNA level in three different untreated samples was set at 1, and the S.E. is shown on each bar. *, p < 0.01 control versus treated samples. C, relative quantification of HAS2 mRNA was done after cytokines exposure in the presence (gray bars) or absence (black bars) of PDTC (final concentration, 2 μm). The lowest HAS2 mRNA level in three different untreated samples was set at 1, and the S.E. is shown on each bar. *, p < 0.01 PDTC-untreated versus PDTC-treated samples.
A large body of evidence (31) has demonstrated that many cellular responses to proinflammatory signals are mediated by NF-κB. This transcription factor normally is present in the cytoplasm in a complex with the protein IκBα that maintains NF-κB in an inactive state. IκBα phosphorylation, mediated by IκB kinase, induces a rapid degradation of IκBα via the ubiquitin-proteosome pathway, which leads to translocation of NF-κB into the nucleus and its transcriptional functions (31). IκB kinase activation can be induced by several stimuli including TNF receptors. Therefore, we investigated whether NF-κB is involved in the increased HAS2 transcript expression after proinflammatory cytokines treatment by using PDTC, a widely used chemical inhibitor of NF-κB (32). As shown in Fig. 3C, the simultaneous addition of IL-1β, or TNF-α, or TNF-β with PDTC caused a reduction of HAS2 transcription. Interestingly, in TNF-α- and TNF-β-treated cells, NF-κB inhibition inhibited HAS2 transcription to the level of control untreated cells. However, in IL-1β-treated cells, NF-κB inhibition did not lower HAS2 transcription to the level of control, which suggests a more complex mechanism of HAS2 transcription activation that could involve other factors in addition to NF-κB. In fact, in addition to NF-κB sites, the HAS2 promoter region contains putative binding sequences for other transcription factors (33). A previous study on human synoviocytes found that PDTC inhibited HAS1 transcription after IL-1β stimulation, confirming the critical role of NF-κB in HA metabolism (34). Interestingly, HA itself is able to modulate cytokine production via NF-κB, confirming its critical role in inflammation (35).
HAS2 and CD44 Mediate HUVEC Monocyte Adhesive Property
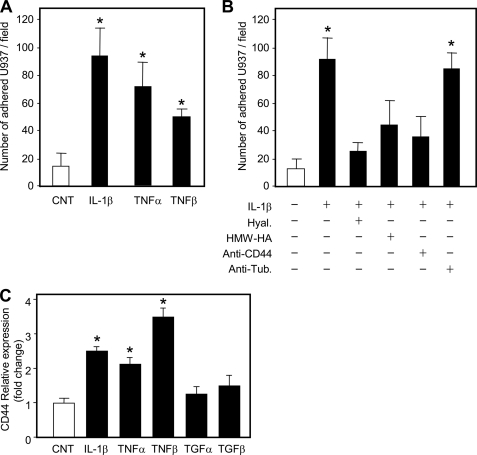
Because some of the effect of HA is to modulate adhesion of immune cells on microvascular ECs (16), we investigated whether monocyte adhesion to HUVECs after treatment with proinflammatory cytokines could be modulated by HA. Fig. 4A shows that IL-1β, TNF-α, and TNF-β increased the number of adherent monocytes ∼5-, ∼4-, and ∼3-fold, respectively. Furthermore, hyaluronidase treatment of HUVECs after IL-1β treatment decreased U937 monocyte binding to near control level (Fig. 4B). Moreover, the simultaneous addition of the monocytes with a pure HA preparation of high molecular weight (Healon) also inhibited binding. These data indicate the critical role of HA in monocytes adhesion in that removal of HA by hyaluronidase treatment as well as the competition with exogenous HA both reduced monocyte adhesion. This point is critical because lowering immune cell adhesion to vascular endothelium would reduce the number of inflammatory cells that reach an inflamed site, therefore reducing the inflammation. Several studies have highlighted the anti-inflammatory and protective effect of HA administered in an experimental model of restenosis (36) as well as in osteoarthritic joints injected with HA (37).
FIGURE 4.
Proinflammatory cytokine effects on HUVEC adhesiveness. A, the adhesion assay was done on untreated (CNT, white bars) and after 24 h-treated (black bars) cells with 5 μg/ml of IL-1β, 0.1 μg/ml of TNF-α, and 0.3 μg/ml of TNF-β. Cytotracker-labeled monocytes (U937) were plated on treated and untreated HUVEC cultures, washed with phosphate-buffered saline, and counted under a fluorescent microscope. Results are expressed as the number of adherent U937 per field and represented as mean ± S.E. of six independent fields. *, p < 0.01; untreated versus the treated sample. B, the adhesion assay was done on untreated (CNT, white bar) and after 24 h-treated (black bars) cells with 5 μg/ml of IL-1β. Before adding monocytes, IL-1β-treated cells were incubated with 2 units/ml of hyaluronidase (Hyal.) for 20 min at 37 °C, or the monocytes were added with high molecular mass HA (Healon, 4 × 106 Da), or with 5 μg/ml CD44-blocking antibody (anti-CD44), or with 5 μg/ml anti-tubulin antibody (anti-Tub.). Results are expressed as the number of adherent U937 per field and represented as mean ± S.E. *, p < 0.01; untreated versus the treated sample. C, relative quantification of CD44 mRNA in untreated (CNT, white bar) and after 24 h-treated (black bars) cells with 5 μg/ml of IL-1β, 0.1 μg/ml of TNF-α, 0.3 μg/ml of TNF-β, 1 μg/ml of TGF-α, and 0.01 μg/ml of TGF-β. The lowest CD44 mRNA level in three different untreated samples was set at 1, and the S.E. is shown on each bar. *, p < 0.01 control versus treated samples.
The molecular mechanism through which immune cells interact with HA is mainly mediated by CD44 (38). To confirm CD44 involvement, we simultaneously added monocytes together with a monoclonal antibody against CD44 known to block HA/CD44 binding and found a clear reduction of monocytes adherent to IL-1β-treated HUVECs, whereas an unrelated control antibody did not modify adhesion (Fig. 4B). We also measured CD44 mRNA expression by quantitative RT-PCR. Interestingly, proinflammatory cytokine treatments induced a significant increase of CD44 mRNA in HUVEC cultures, whereas anti-inflammatory cytokines maintained control CD44 levels (Fig. 4C). This finding could support the hypothesis that CD44 on the plasma membrane of HUVECs binds HA and presents HA to CD44 on circulating immune cells that can attach and form a “sandwich” structure (17). Notably, in this model, HA could remain in a soluble form without forming cable structures, as we found from the immunofluorescent experiments (Fig. 1B). It is of note that a previous work using an immortalized HUVEC cell line (i.e. Ea.hy.926) found an inhibition of neutrophil-EC adhesion by HA independent of CD44 and probably involving ICAM-1 (39), suggesting that in vivo multiple HA-interacting proteins may modulate interactions of immune cells with endothelium.
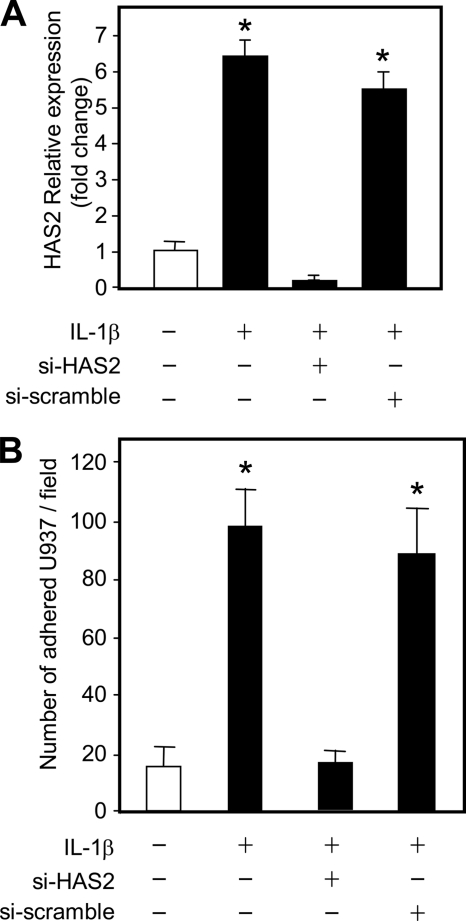
The main HAS isoform responsible for increased HA after treatment of HUVECs with proinflammatory cytokines was HAS2. Therefore, we assessed its involvement in monocyte binding by using an siRNA approach. HUVECs were transfected with commercial siRNA against HAS2 as well as with a scrambled control siRNA by using the Nucleofector apparatus to obtain a high transfection efficiency with a low mortality rate. After 24 h of incubation, nucleofected HUVECs were treated with or without IL-1β and incubated for 24 h. HUVEC cultures were then used for monocytes adhesion experiments or for RNA extraction and cDNA preparation. The effectiveness of HAS2 mRNA silencing was assessed with quantitative RT-PCR and, as shown in Fig. 5A, after IL-1β treatment, HAS2 siRNA-nucleofected cells greatly reduced HAS2 expression. Control siRNA with a random sequence did not perturb the HAS2 mRNA increase due to IL-1β treatment. In parallel cultures, siRNA of HAS2 completely inhibited U937 binding, whereas control siRNA did not alter the adhesive property of HUVECs after IL-1β treatment (Fig. 5B).
FIGURE 5.
HAS2 effects on IL-1β-induced monocyte adhesion. A, relative quantification of the transcript coding for HAS2 in HUVECs silenced for HAS2 and treated with IL-1β (black bars). Cells were left untreated (white bars) or transfected with 50 μm of scramble siRNA (si-scramble) or with 50 μm of HAS2 siRNA (si-HAS2). After 24 h of incubation, 5 μg/ml of IL-1β was added, and, after 24 h, RNA was extracted for quantitative RT-PCR experiments. The lowest HAS2 mRNA level in three different untreated samples was set at 1, and the S.E. is shown on each bar. Results are expressed as mean ± S.E. *, p < 0.01 untreated versus treated samples. B, the adhesion assay was done on HUVECs silenced for HAS2 and treated with IL-1β. 24 h after siRNA transfections and subsequently after 24-h treatment with 5 μg/ml IL-1β, fluorescent monocytes were added, washed, and counted. Results are expressed as the number of adherent U937 per field and represented as mean ± S.E. of six independent fields. *, p < 0.01; untreated versus the treated samples.
Our results clearly demonstrate that HA synthesis in HUVECs is regulated strictly at the transcriptional level by proinflammatory cytokines. In vivo, this issue is of great importance, as HA can participate together with other adhesion molecules to recruitment of immune cells at inflamed sites. Although the generally accepted model foresees the formation of HA cables (6), we demonstrate that monocytes can bind to HA via CD44 without such filamentous structures, consistent with the recent model of Ruffell and Johnson (17). They proposed that HA can be maintained above the endothelium by CD44 on ECs, and leukocytes can bind to HA though their own CD44. Our findings highlight the critical role of HAS2 that could be involved to maintain the nascent HA chain tethered on EC surfaces. In vivo, this model can be modulated by other players that include other cell types, other molecules, and other microenvironmental conditions such as free radical presence, enzyme activity, and shear stress. In fact, recently it has been shown that platelets can produce hyaluronidase 2 that can fragment HA and generate bioactive oligosaccharides that can trigger many cellular responses involving all the vasculature (40).
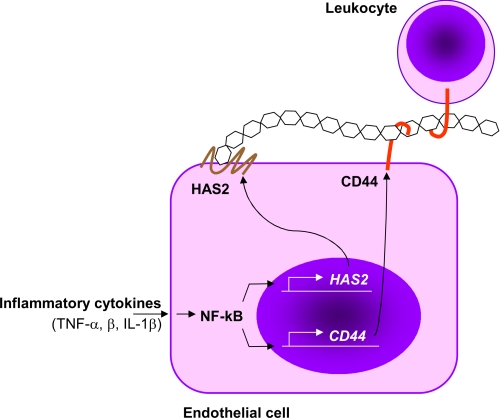
In conclusion, in this work, we provide evidence at molecular and functional levels that HA synthesis can modulate immune cell adhesion on HUVECs that, in turn, could regulate inflammatory responses (Fig. 6). In fact, proinflammatory cytokines induce HA synthesis through the NF-κB mediated expression of HAS2 that modulates monocyte adhesion by CD44, which represents the first step to start an inflammatory process. The central role of NF-κB in inflammation is highlighted by the induction also of other adhesion molecules (such as CD44, ICAM-1, E-selectin, VCAM-1, and MHC class I) (41), which, in vivo, may orchestrate all of the inflammatory responses that include cell adhesion, rolling, extravasation, migration, and specific cellular activity. In light of this model, the clinical relevance of HA administration to ameliorate inflammation (42) can be due to the competition with CD44 on ECs and leukocytes, thereby inhibiting the recruitment process.
FIGURE 6.
Schematic representation of the line of events by which HA modulates the leukocyte-EC interaction. Proinflammatory cytokines, through their receptors, activate NF-κB pathway that has a pivotal role in the inflammatory response by activating several genes including HAS2 (this work), CD44, and other adhesive molecules (ICAM-1, E-selectin, VCAM-1, and MHC class I genes) (41). High molecular weight HA synthesized by HAS2 interacts with CD44 present both on ECs and leukocytes in the “sandwich model” proposed by Ruffell and Johnson (17), which drives immune cells to adhere to ECs that eventually contribute to inflammation.
Acknowledgments
We are grateful to Eleonora Solari (University of Insubria) and Giuseppe Campo (University of Messina) for technical support, Carol De La Motte (The Cleveland Clinic) for U937 cells, and Frances Spring (National Blood Service) for BRIC235 monoclonal antibodies. We acknowledge the “Centro Grandi Attrezzature per la Ricerca Biomedica” Università degli Studi dell'Insubria, for instruments availability.
This work was supported by Fondazione Comunitaria del Varesotto-ONLUS, PRIN 2007 (to G. D. L.) and a young researcher award from the Centro Insubre di Biotecnologie per la Salute Umana (to D. V.).
- HA
- hyaluronan
- HAS
- HA synthase(s)
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- HPLC
- high pressure liquid chromatography
- TNF
- tumor necrosis factor
- TGF
- tumor growth factor
- HUVEC
- human umbilical vein endothelial cells
- ICAM-1
- intercellular adhesion molecule-1
- PAGE-FS
- polyacrylamide gel electrophoresis of fluorophore-labeled saccharides
- VCAM-1
- vascular cell adhesion molecule-1
- EC
- endothelial cells
- MHC
- major histocompatibility complex.
REFERENCES
- 1.Jiang D., Liang J., Noble P. W. (2007) Annu. Rev. Cell Dev. Biol. 23, 435–461 [DOI] [PubMed] [Google Scholar]
- 2.Itano N., Sawai T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., Miyauchi S., Spicer A. P., McDonald J. A., Kimata K. (1999) J. Biol. Chem. 274, 25085–25092 [DOI] [PubMed] [Google Scholar]
- 3.Termeer C. C., Hennies J., Voith U., Ahrens T., Weiss J. M., Prehm P., Simon J. C. (2000) J. Immunol. 165, 1863–1870 [DOI] [PubMed] [Google Scholar]
- 4.Vigetti D., Viola M., Karousou E., Genasetti A., Rizzi M., Clerici M., Bartolini B., Moretto P., De Luca G., Passi A. (2008) ScientificWorldJournal 8, 1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vigetti D., Rizzi M., Viola M., Karousou E., Genasetti A., Clerici M., Bartolini B., Hascall V. C., De Luca G., Passi A. (2009) Glycobiology 19, 537–546 [DOI] [PubMed] [Google Scholar]
- 6.Hascall V. C., Majors A. K., De La Motte C. A., Evanko S. P., Wang A., Drazba J. A., Strong S. A., Wight T. N. (2004) Biochim. Biophys. Acta 1673, 3–12 [DOI] [PubMed] [Google Scholar]
- 7.Selbi W., de la Motte C. A., Hascall V. C., Day A. J., Bowen T., Phillips A. O. (2006) Kidney Int. 70, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 8.Jiang D., Liang J., Fan J., Yu S., Chen S., Luo Y., Prestwich G. D., Mascarenhas M. M., Garg H. G., Quinn D. A., Homer R. J., Goldstein D. R., Bucala R., Lee P. J., Medzhitov R., Noble P. W. (2005) Nat. Med. 11, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 9.Ren J., Hascall V. C., Wang A. (2009) J. Biol. Chem. 284, 16621–16632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majors A. K., Austin R. C., de la Motte C. A., Pyeritz R. E., Hascall V. C., Kessler S. P., Sen G., Strong S. A. (2003) J. Biol. Chem. 278, 47223–47231 [DOI] [PubMed] [Google Scholar]
- 11.Vigetti D., Genasetti A., Karousou E., Viola M., Clerici M., Bartolini B., Moretto P., De Luca G., Hascall V. C., Passi A. (2009) J. Biol. Chem. 284, 30684–30694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantovani A., Bussolino F., Dejana E. (1992) FASEB. J. 6, 2591–2599 [DOI] [PubMed] [Google Scholar]
- 13.Pober J. S., Cotran R. S. (1990) Physiol. Rev. 70, 427–451 [DOI] [PubMed] [Google Scholar]
- 14.Day A. J., de la Motte C. A. (2005) Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 15.Johnson P., Ruffell B. (2009) Inflamm Allergy Drug Targets 8, 208–220 [DOI] [PubMed] [Google Scholar]
- 16.Mohamadzadeh M., DeGrendele H., Arizpe H., Estess P., Siegelman M. (1998) J. Clin. Invest. 101, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffell B., Johnson P. (2009) in Science of Hyaluronan Today (Hascall V. C., Yanagishita M., Toole B. P. eds.) [Google Scholar]
- 18.Karousou E. G., Viola M., Genasetti A., Vigetti D., Luca G. D., Karamanos N. K., Passi A. (2005) Biomed. Chromatogr. 19, 761–765 [DOI] [PubMed] [Google Scholar]
- 19.Viola M., Karousou E. G., Vigetti D., Genasetti A., Pallotti F., Guidetti G. F., Tira E., De Luca G., Passi A. (2006) J. Pharm. Biomed Anal. 41, 36–42 [DOI] [PubMed] [Google Scholar]
- 20.Vigetti D., Viola M., Karousou E., Rizzi M., Moretto P., Genasetti A., Clerici M., Hascall V. C., De Luca G., Passi A. (2008) J. Biol. Chem. 283, 4448–4458 [DOI] [PubMed] [Google Scholar]
- 21.Vigetti D., Moretto P., Viola M., Genasetti A., Rizzi M., Karousou E., Pallotti F., De Luca G., Passi A. (2006) Faseb. J. 20, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 22.de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., Strong S. A. (2003) Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeGrendele H. C., Estess P., Picker L. J., Siegelman M. H. (1996) J. Exp. Med. 183, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeGrendele H. C., Estess P., Siegelman M. H. (1997) Science 278, 672–675 [DOI] [PubMed] [Google Scholar]
- 25.Estess P., Nandi A., Mohamadzadeh M., Siegelman M. H. (1999) J. Exp. Med. 190, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandi A., Estess P., Siegelman M. H. (2000) J. Biol. Chem. 275, 14939–14948 [DOI] [PubMed] [Google Scholar]
- 27.Letterio J. J., Roberts A. B. (1998) Annu. Rev. Immunol. 16, 137–161 [DOI] [PubMed] [Google Scholar]
- 28.Kaplan D. (1996) Immunol. Today 17, 303–304 [DOI] [PubMed] [Google Scholar]
- 29.Rilla K., Siiskonen H., Spicer A. P., Hyttinen J. M., Tammi M. I., Tammi R. H. (2005) J. Biol. Chem. 280, 31890–31897 [DOI] [PubMed] [Google Scholar]
- 30.Anderson P. (2009) Nat. Immunol. 10, 233–234 [DOI] [PubMed] [Google Scholar]
- 31.Pasparakis M. (2009) Nat. Rev. Immunol. 9, 778–788 [DOI] [PubMed] [Google Scholar]
- 32.Schreck R., Meier B., Männel D. N., Dröge W., Baeuerle P. A. (1992) J. Exp. Med. 175, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monslow J., Williams J. D., Fraser D. J., Michael D. R., Foka P., Kift-Morgan A. P., Luo D. D., Fielding C. A., Craig K. J., Topley N., Jones S. A., Ramji D. P., Bowen T. (2006) J. Biol. Chem. 281, 18043–18050 [DOI] [PubMed] [Google Scholar]
- 34.Kao J. J. (2006) Exp. Gerontol. 41, 641–647 [DOI] [PubMed] [Google Scholar]
- 35.Campo G. M., Avenoso A., Campo S., D'Ascola A., Nastasi G., Calatroni A. (2010) Biochimie 92, 204–215 [DOI] [PubMed] [Google Scholar]
- 36.Savani R. C., Turley E. A. (1995) Int. J. Tissue React. 17, 141–151 [PubMed] [Google Scholar]
- 37.Miltner O., Schneider U., Siebert C. H., Niedhart C., Niethard F. U. (2002) Osteoarthritis Cartilage 10, 680–686 [DOI] [PubMed] [Google Scholar]
- 38.Khan A. I., Kerfoot S. M., Heit B., Liu L., Andonegui G., Ruffell B., Johnson P., Kubes P. (2004) J. Immunol. 173, 7594–7601 [DOI] [PubMed] [Google Scholar]
- 39.Alam C. A., Seed M. P., Freemantle C., Brown J., Perretti M., Carrier M., Divwedi A., West D. C., Gustafson S., Colville-Nash P. R., Willoughby D. A. (2005) Inflammopharmacology 12, 535–550 [DOI] [PubMed] [Google Scholar]
- 40.de la Motte C., Nigro J., Vasanji A., Rho H., Kessler S., Bandyopadhyay S., Danese S., Fiocchi C., Stern R. (2009) Am. J. Pathol. 174, 2254–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackay F., Loetscher H., Stueber D., Gehr G., Lesslauer W. (1993) J. Exp. Med. 177, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ialenti A., Di Rosa M. (1994) Agents Actions 43, 44–47 [DOI] [PubMed] [Google Scholar]