Abstract
The proton pumping activity of the eukaryotic vacuolar ATPase (V-ATPase) is regulated by a unique mechanism that involves reversible enzyme dissociation. In yeast, under conditions of nutrient depletion, the soluble catalytic V1 sector disengages from the membrane integral Vo, and at the same time, both functional units are silenced. Notably, during enzyme dissociation, a single V1 subunit, C, is released into the cytosol. The affinities of the other V1 and Vo subunits for subunit C are therefore of particular interest. The C subunit crystal structure shows that the subunit is elongated and dumbbell-shaped with two globular domains (Chead and Cfoot) separated by a flexible helical neck region (Drory, O., Frolow, F., and Nelson, N. (2004) EMBO Rep. 5, 1148–1152). We have recently shown that subunit C is bound in the V1-Vo interface where the subunit is in contact with two of the three peripheral stators (subunit EG heterodimers): one via Chead and one via Cfoot (Zhang, Z., Zheng, Y., Mazon, H., Milgrom, E., Kitagawa, N., Kish-Trier, E., Heck, A. J., Kane, P. M., and Wilkens, S. (2008) J. Biol. Chem. 283, 35983–35995). In vitro, however, subunit C binds only one EG heterodimer (Féthière, J., Venzke, D., Madden, D. R., and Böttcher, B. (2005) Biochemistry 44, 15906–15914), implying that EG has different affinities for the two domains of the C subunit. To determine which subunit C domain binds EG with high affinity, we have generated Chead and Cfoot and characterized their interaction with subunit EG heterodimer. Our findings indicate that the high affinity site for EGC interaction is Chead. In addition, we provide evidence that the EGChead interaction greatly stabilizes EG heterodimer.
Keywords: ATPases, Circular Dichroism (CD), Protein Stability, Protein Structure, Vacuolar ATPase
Introduction
The vacuolar ATPase (V-ATPase; V1Vo-ATPase)2 is a ubiquitous multisubunit enzyme that couples the free energy of ATP hydrolysis to active proton transport. In all eukaryotic cells, V-ATPase function sets up an electrochemical potential and acidifies intracellular compartments (1–5). These functions make the V-ATPase a crucial enzyme involved in intracellular traffic, vesicular transport, endo/exocytosis, and pH homeostasis. In the specialized cells of higher eukaryotes, plasma membrane-associated V-ATPases serve to acidify the extracellular space. The malfunction of the V-ATPase in these cells has been linked to a range of diseases including osteoporosis, diabetes, renal tubular acidosis, and cancer (6–9). As shown in EM images and reconstructions, the V-ATPase is bilobular in overall structure, typical of the rotary ATPases (10–13). These two lobes are composed of distinct functional domains, a soluble catalytic sector, V1, and a membrane integral proton pore, Vo. The V1 sector contains subunits A3B3(C)DE3FG3H, and the Vo is made of ac3–4c′c″de. Most V-ATPase subunits have functional and structural counterparts in the A- and F-type motors; however, only the nucleotide-binding and proteolipid subunits share significant sequence identity. Much like in the F-ATP synthase, the AB subunits come together in an alternating arrangement to form a catalytic hexamer with nucleotide-binding sites located at the AB interfaces (14, 15). ATP hydrolysis-driven rotation of a central rotor domain, composed of subunits DFd and a ring of the proteolipid subunits, c3–4c′c″, is coupled to proton translocation along the interface of the proteolipid ring and the C-terminal domain of the a subunit. During catalysis, three peripheral stalks (each formed by a subunit EG heterodimer) in conjunction with subunits C and H and the N-terminal domain of subunit a (aNT), resist the torque of rotation, thus keeping the A3B3 hexamer static and allowing for rotation to be productive.
Unlike F-ATPase, the activity of eukaryotic V-ATPase is regulated by a unique mechanism of reversible dissociation, a process first described for the enzymes from yeast and insect (16, 17) but more recently also found in cells of higher animals (18–20). In yeast under conditions of nutrient deprivation, the V1 sector dissociates from the Vo sector, and both are functionally silenced (21, 22). Although dissociated, a single V1 subunit, C, is released into the cytosol and is reincorporated into the enzyme upon nutrient readdition (16). Because the C subunit has been proposed to be a molecular switch for controlled enzyme dissociation, the interactions between the C subunit and its intraenzyme binding partners are of particular interest.
The crystal structure of the Saccharomyces cerevisiae C subunit revealed an elongated bone-shaped molecule with two globular domains separated by an α helical coiled coil region (23). One of the globular domains, termed the “foot” domain (Cfoot), encompasses the N and C termini of the protein (residues 1–151 and 287–392), whereas the other globular domain, known as the “head” (Chead), includes residues 166–263. The α helical neck domain includes amino acids 152–165 and 264–286. In vivo cross-linking showed that both Cfoot and Chead are in proximity to subunits E and/or G (24). Cross-links were formed between Cfoot and both the E and a subunits, whereas Chead cross-linked to both E and G. This interaction between EG and C has been shown to be mediated by the N termini of the EG heterodimer (25, 26). Subsequently, immunoelectron microscopy was employed to localize the C subunit within the holoenzyme (27). In accord with the earlier cross-linking study, the C subunit was found to reside at the V1-Vo interface, parallel to the plane of the membrane and in an orientation consistent with one EG binding to each of its ends, an arrangement also seen in recent three-dimensional EM reconstructions of the yeast and insect V-ATPase (11–13).
In contrast to the above in vivo experiments, reconstitution and co-expression studies involving EG and the C subunit have shown that subunit C binds only one subunit EG heterodimer in vitro (12, 28). Combined, these data suggest that the two different EG heterodimers contacting Cfoot and Chead have different affinities for binding the C subunit globular domains. Because subunit C is released from the enzyme during dissociation, the differential affinities for EG may play an important role in the mechanism of regulation by reversible dissociation.
Here we have generated isolated Cfoot and Chead domains and studied their interaction with subunit EG heterodimer. The data show that the high affinity interaction between subunit C and EG is mediated by Chead and that the interaction of Chead or full-length C subunit with EG greatly stabilizes the heterodimer.
EXPERIMENTAL PROCEDURES
Materials
All of the reagents and buffer components were of analytical grade. Restriction endonucleases were from New England Biolabs. PreScission protease and the QuikChange kit were from GE Healthcare and Stratagene, respectively.
Plasmid Construction
The Vma5 (subunit C) gene from S. cerevisiae in a pMal-c2e plasmid (New England Biolabs) with a PreScission protease cleavage site in place of the native enterokinase site (Vma5pMalPPase) was a kind gift of Dr. Patricia Kane and Sheena Claire Li (SUNY Upstate Medical University). The head domain (Chead) was constructed using the above as a template for site-directed mutagenesis employing the Stratagene QuikChange kit. The primers used were: M5Δ1–157 forward, GACAAGGTACCGGAATTCGGATCCATGAACTTGGCTGC; M5Δ1–157 reverse, CTTTCTCTCAGCAGCAGCCAAGTTCATGGATCCGAATTC; M5A278stop forward, GAGCATGACTCTGCTTAGAGTTTAGAACAATCTTTGCGCG; and M5A278stop reverse, GATTGTTCTAAACTCTAAGCAGAGTCATGCTCTTTTTTCAACTC. The resulting protein product encompasses amino acids 158–277. The C subunit foot domain (Cfoot) was constructed using Vma5pMalPPase as a template for fusion PCR wherein the regions of the gene corresponding to the C subunit N and C termini were fused together via a four-amino acid flexible linker (GAAA). The restriction enzyme cut sites 5′ KpnI and 3′ HindIII were introduced in the primer sequence for ligation into pMalPPase. The primers used were: M51–151 forward, GCTCA GGTAC C G GCTACTG CGTTATATAC TGCAAAC; M51–151 reverse, TGC GGC GGC TCC ATT TGCATAAGTA GCTCTGACGT C; M5287–392 forward, GGA GCC GCC GCA CA GTTGGTAAGA TTGGCCAAGA C; and M5287–392 reverse, CGAGTC AAGCT T TTATAAATT GATTATATAC ATCACAAATG. The underlined sequences correspond to the linker sequence.
Construction of the plasmid for expression of subunits E and G will be described elsewhere.3 Briefly, the open reading frames for Vma10p (G) and Vma4p (E) separated by a Shine-Dalgarno sequence in a pBAD vector (kindly provided by Dr. Patricia Kane, SUNY Upstate Medical University) were PCR-amplified to introduce 5′ KpnI and 3′ PstI restriction sites. The amplified bicistronic gene construct was ligated into T-vector (Promega) from where it was excised and ligated into the modified pMal-c2e plasmid. The resultant construct yields subunit G with an N-terminal MBP tag and untagged subunit E. The sequences of all of the constructs were verified by DNA sequencing at the SUNY Upstate Medical University Core DNA sequencing facility using the MalE primer (New England Biolabs).
Protein Expression and Purification
All of the proteins in this study were expressed in Escherichia coli strain Rosetta2 grown to mid-log phase in RB medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). Protein expression was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside (1 mm in the case of EG) for 6 h at 30 °C. All of the proteins were purified using amylose affinity chromatography followed by cleavage with PreScission protease. Briefly, the cells were harvested by centrifugation, resuspended in 200 mm NaCl, 20 mm Tris, and 1 mm EDTA, pH 7.4, and lysed by sonication. Lysate was cleared by centrifugation at 12,000 × g and passed over an amylose affinity column. Bound protein was eluted in 25 ml of 200 mm NaCl, 20 mm Tris, 1 mm EDTA, and 10 mm maltose, and the MBP tag was cleaved with PreScission protease for 2 h in presence of 5 mm dithiothreitol according to the manufacturer's instructions. The pH of the protease cleavage product was adjusted to the isoelectric point (pI) of the C subunit (or its domains) by dialysis and passed over an anion exchange (DEAE) column for removal of MBP. The isolated protein was then dialyzed to readjust the pH away from the pI, concentrated, and subjected to size exclusion chromatography for further purification. The EG-MBP cleavage product was purified using cation (carboxy methyl) followed by anion (DEAE) exchange chromatography with subsequent gel filtration. This purification strategy removed both MBP and any excess G subunit from the complex.
Complex formation between subunit C (domains) and EG was accomplished by reconstitution of purified proteins followed by size exclusion chromatography. Protein concentration was determined by measuring absorbance at 280 nm ± guanidinium HCl on a Varian Cary spectrophotometer. Scans were taken from 400 to 200 nm with base-line correction. Prediction of pI and estimation of protein charge versus pH was done using the Scripps protein calculator. Structure analysis including distance measurement was done using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (29) and Wincoot (30) software.
Circular Dichroism Spectroscopy
Far UV CD spectra were recorded on an Aviv 202 Spectrometer in 25 mm sodium phosphate, 0.5 mm EDTA, pH 7.0 (pH 7.5 for Chead). Wavelength scans were taken from 250 to 195 nm at 25 °C for the C subunit and Cfoot and 20 °C for Chead. The same wavelength range was used for EG, but data were collected at 10 °C. The thermal stabilities of EG, C, Cfoot, Chead, EGC, and EGChead were measured at 222 nm. A 0.1-cm-path length cuvette was used for C, EG, and EGChead, whereas a 0.2-cm-path length cuvette was used for Cfoot, Chead, and CEG measurements. The spectra shown were plotted as mean molar ellipticity (θ) as a function of wavelength or temperature. Apparent unfolding temperatures were estimated by fitting the melting curves with a two-state transition equation (31).
Isothermal Titration Calorimetry
The thermodynamics of the EG interaction with subunit C, Cfoot, and Chead were examined using a Microcal VP-ITC isothermal titration calorimeter. Each experiment was carried out in 25 mm sodium phosphate, 0.5 mm EDTA, pH 7.0, at 10 °C with EG in the cell and subunit C, Cfoot, or Chead in the syringe. Briefly, a known concentration of ligand (C, Cfoot, or Chead) was titrated into a fixed concentration of EG. Blank experiments (C, Cfoot, and Chead into buffer) were subtracted from titration experiments to negate any heats of dilution. The data were analyzed using the VP-ITC programs in Originlab.
Analytical Gel Filtration Chromatography
Purified EG, C, Cfoot, and Chead were run individually on an H/R S75 1.6 × 50 cm column attached to an AKTA fast protein liquid chromatograph (GE Healthcare). The column was calibrated using blue dextran, albumin, green fluorescent protein, RNase A, and insulin. The ability of EG to form a complex with Chead and Cfoot was examined by mixing the purified proteins and passing them over the column. Complex formation was assessed by a shift in elution volume and confirmed by SDS-PAGE.
Native Agarose Gel Electrophoresis
Native agarose gel electrophoresis was used to examine the interactions between the EG heterodimer and subunit C, Chead, and Cfoot (30 μg each). A 2% (w/v) agarose gel (20 mm acetic acid, 40 mm Tris, pH 8.25) was run at 100 V for 2 h at 4 °C. The gel was fixed for 30 min (25% isopropanol, 10% acetic acid) and subsequently washed three times in 95% ethanol. The gel was flattened on a slab dryer set to 80 °C (gradient heat) for 2 h and stained for 30 min in Coomassie G and destained in fixing solution. For second dimension SDS-PAGE analysis, the bands were excised, soaked in SDS gel loading buffer, and resolved on SDS-PAGE gels.
Limited Proteolysis
Limited trypsin proteolysis was carried out with the EG heterodimer in the absence or presence of subunit C or Chead domain. Trypsin (5 μg/ml) was added to EG, EGC, and EGChead (20 μm) incubated at room temperature, and time points were taken at 0, 5, 10, 20, 30, and 50 min. At each time point, the reaction was stopped by the addition of 1 mm phenylmethylsulfonyl fluoride and placed immediately on dry ice.
Mass Spectrometry
Electrospray Ionization Time-of-Flight Mass Spectrometry was used to verify the masses of expressed subunits and subunit domains. The proteins were dissolved in 50/50/0.1 acetonitrile:water:formic acid and manually injected into the Z-spray source of a Waters Q-TOF Micro mass spectrometer. Mass calibration was performed with 1% phosphoric acid.
Analytical Ultracentrifugation
Because Chead eluted as twice its molecular mass using analytical gel filtration, sedimentation velocity analytical ultracentrifugation was employed to determine the molecular mass of Chead in solution. Three different concentrations were used for measurements to determine whether any concentration-dependent oligomerization would occur. Samples of Chead (156, 95.6, and 26.4 μm) in 25 mm sodium phosphate, pH 7.0, were run on a Beckman XL-A analytical ultracentrifuge at 20 °C. The samples were run in a 12-mm-path length cell and measured from 200 to 310 nm at 50,000 rpm. The data were analyzed and fit to a continuous (c(s)) distribution model using the SEDNTERP and SEDFIT software.
RESULTS
Previously, Féthière et al. (28) have shown that subunits E, G, and C are able to form a 1:1:1 complex in vitro. In vivo cross-linking on the other hand showed that in the intact V-ATPase complex, both Chead and Cfoot were in proximity to EG subunits (24), an arrangement subsequently confirmed and further defined by electron microscopy and three-dimensional image reconstruction (11–13) (a schematic of the subunit arrangement in the V-ATPase is shown in Fig. 1). Based on EM and small angle x-ray scattering studies, we (11) and others (12) speculated that binding of EG to C is likely via Cfoot. Because the C subunit dissociates from the complex during controlled enzyme dissociation, the affinity and nature of its interactions with other subunits in the holoenzyme may give us insight into the mechanism of activity regulation by reversible dissociation. Here we have constructed the isolated C subunit head and foot domains to more closely examine the interactions between subunit C and the EG heterodimer.
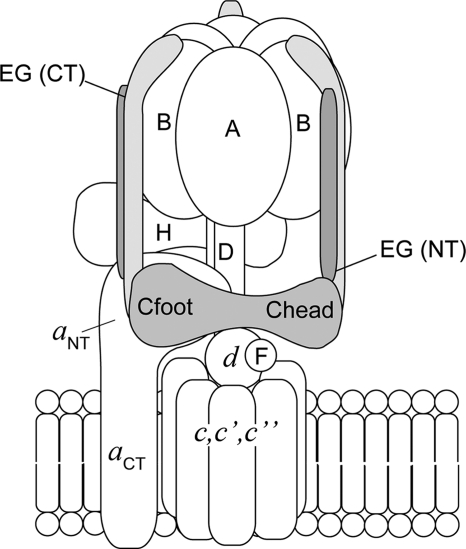
FIGURE 1.
Schematic of the yeast V-ATPase subunit arrangement. The subunits labeled in capital letters belong to the catalytic V1 sector, whereas those in lowercase letters are part of the membrane integral Vo. Details of the functional roles of each subunit can be found in the Introduction. The position and interactions within the holoenzyme between the C subunit (Cfoot and Chead) and the EG heterodimers are shown. As discussed throughout the text, one of these two EG stator contacts is lost in vitro. In addition to their role of holding the catalytic hexamer static during enzyme rotation, the stator subunits act as a structural bridge between V1 and Vo. The C termini of the three peripheral stalks (subunits EG) make contacts with each of the three B subunits, whereas contacts with C, H, and aNT are mediated by the N termini. Two of the three EG heterodimers are in contact with the C subunit, with one EG heterodimer in proximity to both Cfoot and the soluble N terminus of subunit a (aNT) and another in contact with Chead.
Construction, Expression, and Purification of Subunits and Domains
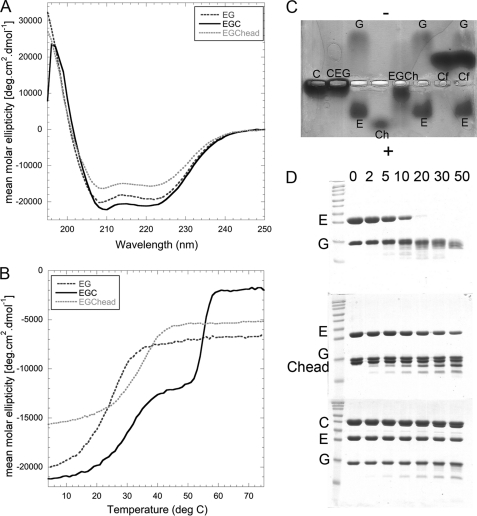
All of the V-ATPase subunits and subunit domains used in this work were expressed in E. coli as N-terminal MBP fusions. The full-length C subunit, in a modified pMAL vector (Vma5 pMAL-PPase), was used as a template for construction of the head and foot domains. The crystal structure of the yeast C subunit (Protein Data Bank entry 1u7l) (23) shows that the molecule is folded as an elongated hairpin with a long helical region (neck), flanked by two globular domains (Cfoot and Chead; Fig. 2, A and B). Cfoot is formed by the N and C termini, which encompasses amino acids 1–151 and 287–392. A distance of ∼10.4 Å was measured between the β carbons of Asn151 and Gln287, the ends of the two polypeptides that make up the foot domain. Fusion PCR was employed to fuse these two polypeptides together via a four-amino acid flexible linker (GAAA). The head domain (Chead) was previously designated to be composed of amino acids 166–263 (23). However, the C subunit crystal structure shows that this domain is connected to the neck domain via two long loops with a measured distance of ∼19 Å between α carbons. To ensure that the head domain would be folded as in the full-length subunit, part of the neck domain was incorporated into the construct used here. Briefly, the distance between β carbons of the amino acids of the neck domain α helices, nearest the head domain, was measured to find the shortest distance between residues. This approach was used to define a site at which to truncate the C subunit for construction of Chead. The shortest distance (∼5 Å) was measured between Asn158 and Ala277, and these two amino acids were thus chosen as the cut site. Briefly, site-directed mutagenesis was employed to delete the first 157 amino acids from the C subunit pMAL-PPase as well as to introduce a stop codon in position 278. The C subunit primary amino acid sequence labeled with the constructs described here and shaded to match the corresponding residues in the crystal structure is shown in Fig. 2 (A and B). Full-length subunit C, Cfoot (amino acids 1–151 and 287–392) and Chead (amino acids 158–277) were expressed in E. coli strain Rosetta2, which carries a pRARE plasmid under chloramphenicol selection. For removal of the cleaved tag, all of three proteins were brought to their predicted isoelectric points (pI) and passed over anion exchange resin. The predicted pI of both C and Cfoot is 6.5 and that of Chead is 6.0. At pH 6–6.5, MBP is negatively charged (predicted charge −6.5 to −8) and binds to DEAE, whereas subunit C and its domains can be collected from the flow through and wash steps. All three were subjected to gel filtration as a final purification step.
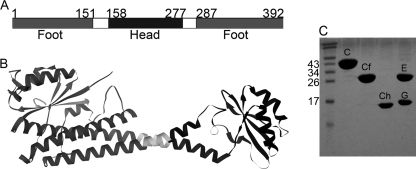
FIGURE 2.
C subunit domain constructs used in this study. A, schematic representation of the primary amino acid sequence of the V-ATPase C subunit with the head (Chead) and foot (Cfoot) domains indicated. Cfoot encompasses the N and C termini of the subunit, whereas Chead is located in the center of the primary sequence. The Chead assignment used here differs from that in Drory et al. (23) in that it includes residues of the so-called neck because of concerns over proper folding of the isolated domain. B, the 1.75 Å resolution crystal structure of the C subunit (Protein Data Bank entry 1u7l) with domains shaded to match the primary sequence in A. C, SDS-PAGE showing the purified proteins used in this study. All of the proteins were expressed as MBP fusions followed by cleavage of the tag and purified as described under “Experimental Procedures.” From left to right, molecular mass standards, C subunit (C), Cfoot (Cf), Chead (Ch), and EG complex (E and G).
The E and G subunits have been shown to form stable heterodimers in vitro when co-expressed in E. coli from a bicistronic vector (32). In the current study, we used a similar approach to generate heterodimeric EG complex. The coding sequences for both subunits, separated by a Shine-Dalgarno sequence, were introduced into the pMAL-PPase vector, with N-terminally MBP tagged subunit G first, followed by untagged E. After protease cleavage, the complex was applied to a cation exchanger to remove the MBP tag with subsequent anion exchange for removal of any excess free G subunit. Gel filtration was used for further purification of the heterodimer.
PreScission protease cleavage of MBP tags resulted in the N-terminal linker sequence GPKVP (plus ESGS from the vector in the case of C and Chead). A representative SDS-PAGE gel showing purified C, Cfoot, Chead, and EG is shown in Fig. 2C (for gel filtration elution profiles, see Figs. 3A and 5).
FIGURE 3.
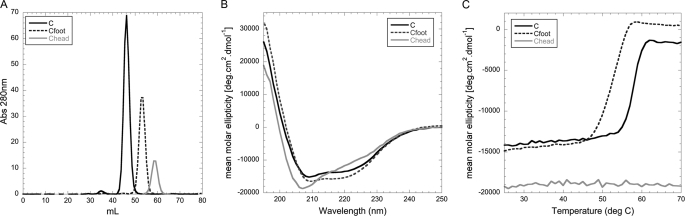
C subunit domain characterization. A, gel filtration elution profiles of C, Cfoot, and Chead on a Superdex S75 1.6 × 50-cm column. All of the proteins elute as single, symmetrical peaks in the order of decreasing molecular mass (C subunit, 44.9 kDa (49.2 kDa observed); Cfoot, 29.6 kDa (34.6 kDa observed); and Chead, 14.5 kDa (26.5 kDa observed)). The slightly larger apparent molecular masses are likely due to the elongated shape of the proteins. B, CD wavelength scans of C, Cfoot, and Chead recorded from 250 to 195 nm in 25 mm sodium phosphate, pH 7, at 25 °C (pH 7.5 and 20 °C in the case of Chead). As can be seen from the spectra, C and Cfoot have roughly the same secondary structure content per residue with the spectra dominated by the characteristic minima at 208 and 222 nm, indicating the presence of α helix. Chead, however, has more β sheet character as seen in the minimum at 208 nm. C, thermal melting of C, Cfoot, and Chead as monitored by recording the CD signal at 222 nm (same buffer conditions as in B) from 25 to 70 °C. Both C and Cfoot exhibit steep transitions at 58 and 53 °C, respectively. Chead, however, is extremely thermally stable such that a transition is never observed, even when measured to 95 °C (data not shown).
FIGURE 5.
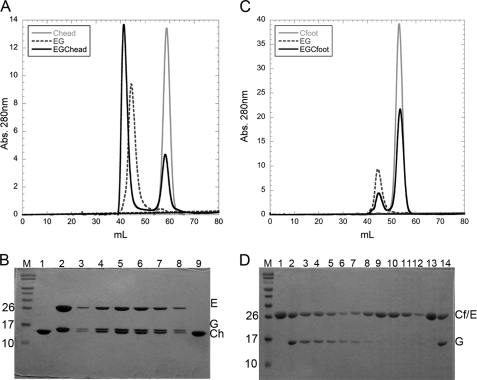
C subunit domain interaction with the peripheral stalk heterodimer, EG. A, gel filtration elution profiles of isolated EG (dashed line) and Chead (gray line) as well as premixed EG and Chead (black line). Purified EG heterodimer was mixed with a molar excess of Chead and subjected to gel filtration under the same conditions as the isolated components. A clear shift in elution volume can be seen for the premixed EG-Chead (black line) with excess Chead eluting at a volume identical to isolated Chead (gray line). B, Coomassie-stained 15% SDS-PAGE of the column fractions in A. M, prestained molecular mass marker; lane 1, Chead control; lane 2, EG heterodimer control; lanes 3–8, fractions under the shifted peak (black solid line) in A; lane 9, peak fraction at 58 ml (excess Chead). The shifted elution peak in A corresponds to complex formation between EG and Chead. C, gel filtration elution profiles of isolated EG and Cfoot as well as premixed EG and Cfoot. Purified EG heterodimer was mixed with a molar excess of Cfoot and subjected to gel filtration as for the isolated components. No complex formation between EG and Cfoot was observed because the two components eluted as separate entities (black solid line). Chromatography of isolated EG and Cfoot are shown for comparison (dotted and gray lines for EG and Cfoot, respectively). D, SDS-PAGE of the chromatogram in C. M, prestained molecular mass marker; lanes 1 and 13, Cfoot; lanes 2 and 14, EG heterodimer control; lanes 3–7, EG elution peak (40–50 ml); lanes 8–12, Cfoot elution peak (50–60 ml). Although E and Cfoot run at approximately the same apparent molecular mass on the gel, both the intensity of the band (at ∼29 kDa) paired with G and the lack of a shift in elution volume confirm the absence of complex formation between EG and Cfoot.
Characterization of C Subunit Head and Foot Domains
The isolated domains of the C subunit, Cfoot and Chead, were characterized with respect to the full-length subunit. Apparent molecular masses were estimated by size exclusion chromatography (see below) and corroborated by Quadrupole TOF mass spectrometry (spectra not shown). According to the mass spectrometry data, the predicted molecular masses correspond well with those observed; for the full-length C subunit 44,955 Da was observed (44,896 Da predicted), for Cfoot 29615 Da was observed (29611 Da predicted), and for Chead 14,564 Da was observed (14,537 Da predicted). All of the predicted molecular masses include the protease linker sequence. Analytical gel filtration gave molecular masses very close to those predicted for both Cfoot (29.6 kDa predicted, 34.6 kDa observed) and the full-length subunit (44.8 kDa predicted, 49.2 kDa observed) (Fig. 3A). Possibly because of its elongated nature, Chead eluted at a molecular mass close to twice its expected value (14.5 kDa predicted, 26.5 kDa observed). All three proteins, however, elute as symmetrical peaks, indicative of monodisperse preparations. To determine whether Chead existed as a monomer or dimer, sedimentation velocity analytical ultracentrifugation was performed (data not shown). At the three concentrations tested, Chead sedimented as a single species with a calculated molecular mass of ∼15.5 kDa, thus confirming that the protein was indeed monomeric in solution.
Far UV Circular Dichroism Spectroscopy
Far UV circular dichroism spectroscopy was employed to confirm that the C subunit domains are properly folded and contain the appropriate secondary structural elements. Fig. 3B shows CD spectra of full-length C subunit as well as Chead and Cfoot. Overall, the CD data for the full-length C subunit agree well with the previously published spectra (33). Secondary structure content per amino acid was counted from the C subunit crystal structure for C, Chead, and Cfoot (23). Both the full-length protein and Cfoot have approximately the same secondary structure content per amino acid, whereas Chead has considerably more β sheet character. This is reflected in the CD spectra (Fig. 3B), where minima at 208 and 222 nm are indicative of α helices (C and Cfoot), and a strong minima at 208 nm reflects high β sheet content (Chead). All three proteins were also examined for thermal stability using CD (Fig. 3C). The full-length C subunit and Cfoot had sharp, steep two-state melting transitions, indicating that the preparations are homogeneous and that thermal unfolding is highly cooperative. Both were found to be reasonably thermally stable with apparent melting temperatures (Tm) of 58 and 53 °C for the full-length C subunit and Cfoot, respectively. In contrast, no transition was observed for Chead up to a temperature of 95 °C (data only shown to 75 °C), indicating that the protein has high thermal stability.
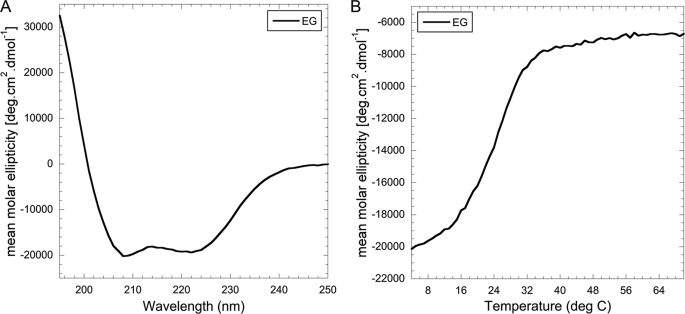
Proper folding of the EG heterodimer was also assessed using CD spectroscopy (Fig. 4A). As can be seen, the spectrum has the appearance typical for highly α helical proteins with the characteristic minima at 222 and 208 nm. Consistent with previously published CD spectra for the EG heterodimer (32), the ratio of the signal at wavelength 208 and 222 nm is close to unity (θ222/θ208 = 0.96), indicative of the presence of α helical coiled coil secondary structure (34). Thermal unfolding experiments conducted with EG heterodimer showed that the complex has a Tm of 25.4 °C (Fig. 4B), a value significantly lower compared with the full-length C subunit and its two domains (Fig. 3C). Interestingly, the mean molar ellipticity of EG complex at higher temperatures does not reach zero, and it remains unclear at this point whether the melting transition observed for EG represents a complete breaking up of the heterodimer or a partial unfolding of, for example, the N-terminal α helical coiled coil domain. Cooling the sample back to 10 °C after heating to 70 °C revealed that the thermal unfolding of EG is largely (70–80%) reversible (data not shown). One possible explanation is that the β sheet containing the C-terminal domain of the complex retains some secondary structure, possibly serving as a starting point for the cooling induced refolding of the N-terminal domains. Another noteworthy feature of the thermal unfolding curve shown in Fig. 4B is the lack of a low temperature plateau, suggesting that there will be some fraction of (partially) unfolded EG complex even at temperatures significantly below the transition temperature.
FIGURE 4.
Secondary structure and thermal stability of the heterodimeric EG complex. A, CD wavelength scan of the EG heterodimer measured from 250 to 195 nm at 10 °C in 25 mm sodium phosphate, pH 7. The wavelength scan of EG (as previously shown) indicates coiled coil content with the ratio of the minima at 208 and 222 nm approaching unity. B, CD thermal melting curve of EG measured at 222 nm from 4 to 75 °C in 25 mm sodium phosphate, pH 7. Thermal melting of the EG heterodimer displays a single cooperative transition with a Tm of ∼25 °C.
In summary, CD spectroscopy showed that the proteins used in this study are folded and contain the expected secondary structure content. Thermal unfolding experiments indicated that the subunit EG heterodimer is significantly less stable compared with the subunit C constructs.
Binding Studies of EG and Chead and Cfoot
In the following sections, the interaction of EG heterodimer with Chead and Cfoot was investigated using size exclusion chromatography, native gel electrophoresis, CD spectroscopy, limited proteolysis, and isothermal titration calorimetry.
Analytical Gel Filtration
It has been shown that although the EG heterodimer cross-links to both the head and foot domains of the C subunit in vivo (24), only one EG binds to the full-length C subunit in vitro (12, 28). To establish the polarity of the EGC interaction, purified EG heterodimer was mixed with a molar excess of Chead or Cfoot and subjected to analytical size exclusion chromatography as described under “Experimental Procedures.” Complex formation was assessed by SDS-PAGE of the fractions under each peak in the chromatogram. As can be seen in Fig. 5A, when EG and Chead are analyzed separately, they elute at 45 and 58 ml, respectively. A mixture of the two, on the other hand, results in peaks eluting at 41 and 58 ml, and SDS-PAGE reveals that the peak at 41 ml (Fig. 5B, lanes 3–8) contains subunits E, G, and Chead, whereas the peak at 58 ml contains the excess Chead (Fig. 5B, lane 9). In contrast, and in good agreement with the idea that subunit C has only one high affinity site for EG, Fig. 5 (C and D) shows that EG and Cfoot do not form a complex with sufficient affinity to co-elute from the column (no peak shift and no co-elution). In summary, the data show that Chead contains a high affinity binding site for EG heterodimer, whereas Cfoot shows no interaction with EG under the conditions of the experiment. This is consistent with earlier size exclusion chromatography experiments that showed that full-length subunit C forms a 1:1:1 complex with subunits EG (12, 28).
Electrophoretic Mobility Shift Assay
The analytical gel filtration data above indicate that the high affinity site for the EGC interaction is on Chead. Because only relatively high affinity interactions are detected by gel filtration, we used electrophoretic mobility shift assays on agarose gels under native conditions to assess the ability of Cfoot to bind EG. Briefly, EG, C, Chead, and Cfoot were run both individually and with EG preincubated with subunit C and subunit C domains. A representative gel is shown in Fig. 6C. Surprisingly, although E and G have high enough affinity to co-elute from anion exchange, cation exchange, and gel filtration columns, the two subunits run in opposite directions on the native agarose gel under the conditions tested (Fig. 6C, third and sixth lanes; identity of bands confirmed by second dimension SDS-PAGE (not shown)). This behavior can be explained by the fact that subunit G is a basic protein with a predicted isoelectric point (pI) of ∼9, whereas subunit E is acidic with a pI of ∼5 (35, 36). As shown above, EG heterodimer has limited thermal stability, and it is possible that under the conditions of the native gel, the binding energy of the two subunits is not sufficient to withstand the opposing forces of the electric field. The situation changes, however, when subunits EG are electrophoresed in presence of subunit C or Chead. With subunit C, no free E or G is observed, but the resulting EGC complex possesses insufficient charge and/or is too large to migrate significantly (Fig. 6C, second lane). With Chead, no band corresponding to free E, G, or Chead is observed, and instead, all three proteins co-migrate toward the positive electrode. Curiously, Cfoot is migrating toward the cathode even though its predicted isoelectric point is lower than the pH of the buffer used for the gel. However, no complex formation with EG is observed, and all three proteins migrate as the individual species. In conclusion, the electrophoretic mobility shift assay experiment confirms the results obtained from the analytical size exclusion chromatography in that EG is able to form a complex with Chead but not Cfoot.
FIGURE 6.
Characterization of the EGC and EGChead complexes: enhanced stability of EG in the presence of C and Chead. A, CD wavelength scans of EG, EGChead, and EGC measured from 250 to 195 nm at 10 °C in 25 mm sodium phosphate, pH 7. The wavelength scan of EG (dark gray dotted line) displays coiled coil content (see Fig. 4A). The wavelength scan of EGChead (light gray dotted line) displays the same characteristic, but there is some decrease in signal, presumably because of the high β sheet character of Chead. Interestingly, the signal at 208 nm compared with 222 nm does not increase in the EGChead spectrum (see Fig. 3B for comparison). The shape of the EGC spectrum (black solid line) does not change appreciably with only a slight increase in overall ellipticity. B, CD thermal melting curves of EG, EGChead, and EGC measured at 222 nm from 4 to 75 °C in 25 mm sodium phosphate, pH 7. The EG heterodimer is sensitive to thermal denaturation with a Tm of ∼25 °C (see Fig. 4B.). In complex with Chead, however, the melting temperature is increased by 10 °C. This spectrum represents the melting of EG only, because Chead does not melt (see Fig. 3C). The melting curve for EGC shows two transitions: one is at 31 °C (presumably EG bound to C) and the next is the thermal denaturation of C (55 °C, see Fig. 3C). This indicates that EG binding to C or Chead leads to a significant increase of EG thermal stability. C, electrophoretic mobility shift assay. C, Chead (Ch), Cfoot (Cf), and EG were run individually and premixed on a 2% native agarose gel, pH 8.25, at 4 °C. The identities of the various bands (labeled on the gel) were verified by cutting out the bands and running them in a second dimension on an SDS-PAGE gel. Surprisingly, E and G do not co-migrate on the native gel. Co-migration of EG was only observed in the presence of C or Chead, further indication that the EG interaction is greatly stabilized by binding to C or Chead. D, Coomassie-stained 15% SDS-PAGE gel of limited trypsin proteolysis of EG, EGChead, and EGC. The tops of the gels are labeled according to the time (in min) post-addition of trypsin, and subunit identities are indicated on the left. Although subunit E is completely degraded in the isolated EG complex after 30 min, significant protection to the heterodimer is conferred upon binding C or Chead.
CD Spectroscopy of the EGC(head) Complex
The data above clearly indicate that EG and C(head) are able to form a stable complex, whereas EG and Cfoot do not. We therefore tested whether complex formation changes the thermal stability or secondary structure content of EG (Fig. 6, A and B). The thermal stability of the EG complex alone is limited, with a melting temperature of 25.4 °C (see section “Far UV Circular Dichroism Spectroscopy” above). In complex with C or Chead, however, the temperature of the first unfolding transition increases by 5–10 °C to 30–34.5 °C for EGC and EGChead, respectively (Fig. 6B). At the same time, complex formation leads to a slightly more linear signal in the low temperature region of the CD melting curves. As already noted above, the CD signal does not reach zero at temperatures beyond thermal unfolding, indicating some residual secondary structure in EG C-terminal domain and/or C(head). The wavelength scan in Fig. 6A shows that the overall secondary structure does not change appreciably for EG upon binding Chead. However, this lack of an apparent change in the shape of the spectrum upon EG binding Chead is particularly striking when compared with Fig. 3B, which shows the same wavelength range for the free Chead.
Limited Proteolysis
The electrophoretic mobility shift assay and thermal unfolding data above demonstrated that EG is stabilized by its interaction with C and Chead. To further examine this increase in stability, we carried out limited proteolysis on EG in the absence and presence of subunit C and Chead. As can be seen from Fig. 6D (top gel), in the absence of C or Chead, subunit EG is rapidly degraded over the 50-min time course. The situation is changed dramatically in the presence of subunit C (bottom gel) or Chead (middle gel). Here, much less degradation of EG is observed after 50 min, especially in the presence of intact subunit C.
Isothermal Titration Calorimetry
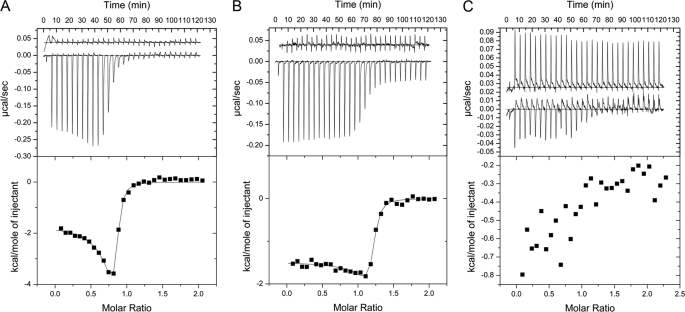
To quantitate the thermodynamics of the interactions between EG and C, Chead and Cfoot, we used isothermal titration calorimetry. As can be seen in Fig. 7 (A and B), the EGC and EGChead interactions are both exothermic, and saturation of binding occurs at a ∼1:1 molar ratio. The data for EGC (Fig. 7A) was fit to a two-site model, with only one of those sites being occupied (N1 = ∼0.02 and N2 = ∼0.80). The fit allowed for calculation of the binding constant and thermodynamics of the interaction. The Kd was found to be 42 nm, ΔH = −1664 cal/mol, and ΔS = +28 cal/(mol·K). These parameters were then used for calculation of the ΔG of association of EG and C at a temperature of 283 K. A value of −40 kJ/mol was calculated. The same experiment was carried out using EG and Chead (Fig. 7B), and the data were fit to the same two-site model, again with only one site occupied (N1 = ∼0.0164 and N2 = ∼1.16). The data show that the thermodynamics of the EGChead interaction are strikingly similar to those of the EGC interaction with a Kd of 45 nm, ΔH = −1480 cal/mol, ΔS = +28 cal/(mol·K), and ΔG = −39.8 kJ/mol (283 K). In the top panel of Fig. 7B, a continued, constant heat release beyond EGChead saturation occurs that is not due to heat of dilution of Chead. When performing the titration using EG and Cfoot, no significant heat release was measured, and the data could not be reliably fit (Fig. 7C). This last observation is again consistent with the results from the gel filtration and native gel electrophoresis that failed to show a high affinity interaction between EG and Cfoot.
FIGURE 7.
Affinity of the interaction between EG and C, Chead, and Cfoot. Isothermal titration calorimetry was used to determine the binding affinities between EG heterodimer and the C subunit constructs. A, purified C subunit was titrated into EG, and heats of binding were measured at 10 °C in 25 mm sodium phosphate, 0.5 mm EDTA, 1 mm tris(2-carboxy) ethyl phosphine, pH 7. The base-line data (heat of C subunit dilution) and binding isotherm are displayed in the top panel, and the enthalpy change is plotted as a function of molar ratio in the bottom panel. The change in enthalpy versus molar ratio data were fit to a two-site model allowing for calculation of Kd and ΔG. The Kd was found to be ∼42 nm with a ΔG of association of ∼−40 kJ/mol. B, the base-line data (heat of Chead dilution) and binding isotherm (top panel) as well as enthalpy change versus molar ratio (bottom panel) for the EG Chead interaction. Purified Chead was titrated into EG, and heats of binding were measured at 10 °C in 25 mm sodium phosphate, 0.5 mm EDTA, 50 mm NaCl, pH 7. The enthalpy change versus molar ratio data were fit to a two-site model, allowing for calculation of Kd and ΔG. The Kd was found to be ∼45 nm with a ΔG of association ∼−39.8 kJ/mol, similar to the data for the EG-C interaction. C, the base-line data (heat of Cfoot dilution) and binding isotherm (top panel) as well as enthalpy change versus molar ratio (bottom panel) for the EG Cfoot interaction. Purified Cfoot was titrated into EG, and heats of binding were measured at 10 °C in 25 mm sodium phosphate, 0.5 mm EDTA, 1 mm TCEP, pH 7. The heats measured for the EG Cfoot titration were too close to the noise level to be reliably fit. These data not only confirm that the high affinity EG binding site on C is Chead but also validate our Chead construct because the thermodynamics of the EGC interaction are nearly identical to those of EGChead.
It can be seen in Fig. 7 (A and B) that the heat of EG-C(head) binding increases transiently before saturation. This heat increase may be explained, in part, by the limited thermal stability of EG and its reversible nature (discussed in the section “Far UV Circular Dichroism Spectroscopy” above) as well as the increase in stability of EG when bound to C(head). Even at the temperature of the isothermal titration calorimetry experiment (10 °C), a subpopulation of EG may exist, in which the α helical N termini are partially “open” (unfolded). We propose that at the beginning of the titration, C(head) preferentially binds to “closed” (fully folded) EG, but upon progression of the titration, total heat release from complex formation between EG and C(head) will also include heat release from closing (refolding) of the partially unfolded EG complexes. Thus, the heat release toward saturation will increase as we would expect to detect both a heat of EG folding and a heat of binding to C(head). Evidence to support this explanation comes from experiments where EG complex was injected into the cell. Under these conditions, a large endothermic effect (heat uptake) was observed, and we speculate that this heat uptake is due to partial unfolding of the N-terminal domains of EG, which in turn may be caused by the large dilution of the complex into the cell (data not shown).
DISCUSSION
The eukaryotic V-ATPase is unique among rotary ATPases in its subunit composition and its mode of regulation by reversible dissociation. The mechanism of reversible dissociation, or regulated association, had been first observed for the yeast (16) and insect (17) V-ATPase but has since been confirmed also for the enzyme from other animals including mammals (18–20). Regulated dissociation involves controlled breaking and reformation of the protein-protein interactions that constitute the ATPase-ion channel interface (Fig. 1 for a schematic of the complex). This interface is formed by a central rotating stalk (subunits DF from the V1 binding to subunit d from the Vo) and three peripheral stalks (subunit EG heterodimers), which are connected to the membrane-bound a subunit either via direct interaction with the a subunit N-terminal domain (aNT) or indirectly via the C subunit (subunit H has been shown not to be required for enzyme assembly and may therefore not be involved in regulated dissociation/association).
Subunit C, found exclusively in the eukaryotic V-type motor, is the only subunit in the complex that can be found in isolation from both V1 and Vo upon enzyme dissociation. In addition, subunit C has been shown to interact with a variety of cellular components including the cytoskeleton (37–39) and the RAVE complex (40), possibly suggesting some form of sequestration during controlled dissociation. Interestingly, whereas deletion of the gene encoding subunit C prevents assembly of V1 with Vo (41), this mutation has little affect on assembly of the remainder of the V1 and Vo subunits (42). These features indicate that C subunit likely plays an important role in the mechanism of disassembly/reassembly. Based upon recent EM reconstructions, subunit C appears to bind to two of the three peripheral stators (11–13), consistent with earlier cross-linking data that showed subunits EG to be in proximity to both the head and foot domains of the C subunit (24). Data from in vitro studies on the other hand indicated that the subunit EG heterodimer was able to bind subunit C with a 1:1:1 ratio (28). These data indicate that the head and foot domains of the C subunit have different affinities for EG.
Until now, the question of which subunit C domain contains the high affinity site for binding EG has not been addressed directly, although previous indirect evidence has suggested that it is Cfoot. For example, two different C subunit crystal structures were solved revealing different conformations of the head domain (23). In the lower resolution conformer, the head domain was shown to be bent at the head-neck interface, demonstrating potential flexibility in this region. Interestingly, this lower resolution structure corresponds well with the density belonging to the C subunit in a recent EM reconstruction from our laboratory; however, the connection between Chead and EG was not well resolved (11). This lack of density for the EGChead interaction lead us to speculate that it is likely the EGCfoot interaction that is with high affinity. A similar conclusion was reached based on a recent small angle x-ray scattering model of the isolated EGC complex, which appeared to fit well with Cfoot forming the connection to EG (12). Although these data seemed to suggest that the Cfoot domain contained the high affinity binding site for the EG heterodimer, the flexibility of the neck domain of subunit C as seen in the two crystal forms (23), and the limited resolution of the EM and small angle x-ray scattering models (11, 12) makes the assignment of Cfoot as the EG binding site appear somewhat ambiguous.
In an effort to identify the high affinity EG binding site on subunit C and characterize this interaction, we have constructed the isolated domains of the C subunit, Chead and Cfoot. Analytical gel filtration and CD experiments indicate that the two domains are properly folded and monodisperse. Using these techniques coupled with native agarose electrophoresis, limited trypsin proteolysis, and isothermal titration calorimetry, we present evidence that it is the head domain of subunit C that forms a stable complex with the EG heterodimer, whereas under the conditions tested, no complex formation between EG and the foot domain of subunit C was observed. The thermodynamics of the EGC and EGChead interactions were examined using isothermal titration calorimetry and were found to be nearly identical, with ΔG of association (ΔGa) = ∼−40 kJ/mol and dissociation constants of ∼40 nm. In the same experiments conducted with Cfoot, no significant heat release was observed upon titration into EG. Interestingly, the in vivo cross-linking study mentioned above demonstrated that although Chead cross-links to both E and G, Cfoot cross-links to E and the soluble N terminus of the a subunit (24). It is therefore possible that the interaction between Cfoot and a second EG heterodimer, as seen in EM reconstructions of the intact complex, requires the presence of the N-terminal domain of the a subunit. It should be noted that although the eukaryotic V-ATPase contains three EG peripheral stators (43), only two are present in the related archaeal A- and bacterial A/V-ATPase (44–46). Although both of these stators interact with the N-terminal domain of the archaeal and bacterial subunit a homologue (subunit I), only one binds the soluble domain of subunit I with high affinity in vitro (47). Together, these observations indicate that some of the interactions at the ATPase-ion channel interface are only formed in the intact V-, A-, or A/V-ATPase complex. In this context it is interesting to note that photochemical cross-linking in yeast V-ATPase identified two regions in the N-terminal domain of subunit a that were in close proximity to E and G subunits (48).
As mentioned above, the unique mechanism of activity regulation in the V-ATPase involves the breaking and reformation of the protein-protein interactions that constitute the V1-Vo interface. This dramatic rearrangement as well as the interactions between EG and C in the holoenzyme have been well documented, but the details of controlled dissociation are poorly understood. The interactions between EG and the C subunit have been shown to be mediated by the N termini of the parallel EG heterodimer with the C termini distal to the membrane (Fig. 1 and Ref. 11). Mutagenesis of the G subunit N terminus has been shown to inhibit enzyme dissociation, disrupt assembly, and interrupt its interactions with subunit E (25). In addition, E subunit N-terminal residues have been shown to be necessary for the interaction with both subunits G and C (26, 49). As we have shown here, the EGC(head) complex is more stable than the EG heterodimer alone. For example, in the presence of C(head), EG is more thermally stable, migrates as a complex on native agarose gel, and is protected significantly from protease digestion. Because both C and Chead appear to have a positive effect on EG stability, we hypothesize that the C subunit stabilizes the N termini of EG.
Although there is no structure available for the eukaryotic EG heterodimer, the proteins comprising the peripheral stalk (EH)4 in the related archaeal A- and bacterial A/V-ATPase are more stable and thus have been more amenable to structural characterization than their eukaryotic counterparts. Although the archaeal EG heterodimer is shorter in length and more stable than yeast EG, their secondary structure predictions and biological functions are similar (50, 51), indicating that they may be used as a structural model for eukaryotic EG. NMR spectroscopy and domain mapping experiments (50) showed that although subunit G is entirely composed of α helix, subunit E contains a globular C-terminal domain of mixed αβ character as seen earlier in the crystal structure of the isolated domain (52). Overall, the EG heterodimer is elongated with the N termini involved in a coiled coil, whereas the C-terminal globular domain of the E subunit is in contact only with the very C-terminal α helix of the G subunit (50). The recent crystal structure of the Thermus thermophilus EG heterodimer (53) (Protein Data Bank entry 3k5b) confirmed the domain mapping analysis of the archaeal EG heterodimer (50) and showed for the first time that the N-terminal domains of E and G are folded in a right-handed coiled coil, a fold predicted earlier for the b subunit dimer of the E. coli F-ATP synthase (54). Interestingly, the crystal structure of the T. thermophilus EG begins with a coiled coil starting from residue 2 of the E subunit, but the N-terminal 21 residues of the G subunit are not present in the structure (53). Although the S. cerevisiae G subunit is approximately the same size as that of T. thermophilus, the yeast subunit E is 48 amino acids longer, suggesting the presence of some additional structural element in the N-terminal region of the eukaryotic EG heterodimer.
It was shown previously that the thermal stability of the coiled coil domain of archaeal EG is reasonably high and at 60 °C mirrors the optimal growth temperature of the organism (51). In contrast, our data show that the yeast EG heterodimer is very sensitive to thermal denaturation and that unless bound to the C subunit, the Tm is below the optimal growth temperature for yeast. Because the S. cerevisiae EGC interaction has been mapped to the N termini of the EG heterodimer (25, 26), our data suggest that although eukaryotic E and G are a high affinity heterodimer, their extreme N termini are vulnerable, thus limiting the thermal and chemical stability of the complex in vitro. The predicted coiled coil domain is likely responsible for much of the high affinity EG interaction. As we have shown here, in the presence of C(head), the stability of EG is greatly enhanced, likely through protection of the more labile extreme N termini. Thermal stability of the interaction of subunits E, G, and C may also play a role in the regulation of the V-ATPase complex in vivo. Recently, a subunit E isoform (E1) was identified in mouse testis that is specifically localized to the V-ATPase in the acrosome of developing and mature sperm (55). Although the E1 isoform is able to complement yeast Vma4p at normal growth temperatures, uncoupling of ATPase and proton pumping was observed at elevated temperature, indicating limited thermal stability of the interaction of subunit E1 with the other subunits of the complex (subunits G and C and the N-terminal domain of subunit a).
To summarize, the data presented here indicate that in vitro the high affinity site for EGC complex formation is located on the head domain of the C subunit, and we speculate that the differences in affinities are likely involved in the unique mode of regulation by reversible dissociation. We further speculate that regulated disassembly occurs when an unknown environmental signal leads to a conformational change in the C subunit as seen in two crystal forms found by Drory et al. (23). This conformational change then leads to the release of Cfoot (low affinity site) and upon continued rotation of the enzyme the high affinity interaction, made by Chead, is broken, thereby releasing the C subunit into the cytosol and promoting enzyme dissociation. Interestingly, the free energy change (ΔGa) of the EGC(head) interaction is with −40 kJ/mol close to the free energy change of ATP hydrolysis under conditions found in the cytoplasm (−57 kJ/mol) (56). Although many interactions must be broken during the process of controlled dissociation, it is interesting that the energetic cost of breaking the high affinity EGC(head) interaction could be balanced by hydrolysis of one ATP molecule. This is an especially attractive hypothesis in that it has been shown that the enzyme must be active for dissociation to occur (21, 57).
The work presented here demonstrates that the isolated domains of the C subunit are properly folded and can be used for studying the molecular aspects of the many documented interactions of this subunit with other subunits of the complex in more detail. Further work is required to elucidate the interaction between EG and Cfoot because this interaction may be dependent upon the presence of additional V-ATPase subunits such as the N-terminal domain of the a subunit. In addition, the increase in stability as seen for EG in complex with C(head) may also be found for other V-ATPase subcomplexes, a property that may assist in future studies of V-ATPase structure and function. Such studies are ongoing in our laboratory.
Acknowledgments
We thank Dr. Patricia Kane and Sheena Claire Li for the Vma5 pMALPPase and Vma4-Vma10 pBAD constructs. Lee Parsons is gratefully acknowledged for the contribution of the EG pMALPPase co-expression construct as well as assistance with mass spectrometry. In addition, we thank Dr. Erik Kish-Trier for critical reading of this manuscript as well as advice and expertise in protein purification and data analysis, Brandy Verhalen for technical advice and assistance, and Dr. Stewart Loh for helpful advice and guidance in CD data analysis. Dr. Michael Cosgrove is acknowledged for help with the analytical ultracentrifugation analysis of Chead.
This work was supported, in whole or in part, by National Institutes of Health Grant GM58600.
L. S. Parsons and S. Wilkens, unpublished observations.
To avoid confusion with the eukaryotic H subunit, archaeal H subunit will hereon be called as its eukaryotic counterpart, subunit G.
- V-ATPase
- vacuolar ATPase
- EM
- electron microscopy
- MBP
- maltose-binding protein.
REFERENCES
- 1.Ohsumi Y., Anraku Y. (1983) J. Biol. Chem. 258, 5614–5617 [PubMed] [Google Scholar]
- 2.Forgac M. (2007) Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 3.Kane P. M. (2006) Microbiol. Mol. Biol. Rev. 70, 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saroussi S., Nelson N. (2009) J. Exp. Biol. 212, 1604–1610 [DOI] [PubMed] [Google Scholar]
- 5.Marshansky V., Futai M. (2008) Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinton A., Sennoune S. R., Bond S., Fang M., Reuveni M., Sahagian G. G., Jay D., Martinez-Zaguilan R., Forgac M. (2009) J. Biol. Chem. 284, 16400–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair H. C., Teitelbaum S. L., Ghiselli R., Gluck S. (1989) Science 245, 855–857 [DOI] [PubMed] [Google Scholar]
- 8.Sun-Wada G. H., Toyomura T., Murata Y., Yamamoto A., Futai M., Wada Y. (2006) J. Cell Sci. 119, 4531–4540 [DOI] [PubMed] [Google Scholar]
- 9.Alper S. L. (2002) Annu. Rev. Physiol. 64, 899–923 [DOI] [PubMed] [Google Scholar]
- 10.Wilkens S., Vasilyeva E., Forgac M. (1999) J. Biol. Chem. 274, 31804–31810 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Zheng Y., Mazon H., Milgrom E., Kitagawa N., Kish-Trier E., Heck A. J., Kane P. M., Wilkens S. (2008) J. Biol. Chem. 283, 35983–35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diepholz M., Venzke D., Prinz S., Batisse C., Flörchinger B., Rössle M., Svergun D. I., Böttcher B., Féthière J. (2008) Structure 16, 1789–1798 [DOI] [PubMed] [Google Scholar]
- 13.Muench S. P., Huss M., Song C. F., Phillips C., Wieczorek H., Trinick J., Harrison M. A. (2009) J. Mol. Biol. 386, 989–999 [DOI] [PubMed] [Google Scholar]
- 14.Maher M. J., Akimoto S., Iwata M., Nagata K., Hori Y., Yoshida M., Yokoyama S., Iwata S., Yokoyama K. (2009) EMBO J. 28, 3771–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Numoto N., Hasegawa Y., Takeda K., Miki K. (2009) EMBO Rep. 10, 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane P. M. (1995) J. Biol. Chem. 270, 17025–17032 [PubMed] [Google Scholar]
- 17.Sumner J. P., Dow J. A., Earley F. G., Klein U., Jäger D., Wieczorek H. (1995) J. Biol. Chem. 270, 5649–5653 [DOI] [PubMed] [Google Scholar]
- 18.Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., van der Goot F. G. (2008) PLoS One 3, e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trombetta E. S., Ebersold M., Garrett W., Pypaert M., Mellman I. (2003) Science 299, 1400–1403 [DOI] [PubMed] [Google Scholar]
- 20.Voss M., Vitavska O., Walz B., Wieczorek H., Baumann O. (2007) J. Biol. Chem. 282, 33735–33742 [DOI] [PubMed] [Google Scholar]
- 21.Parra K. J., Kane P. M. (1998) Mol. Cell Biol. 18, 7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane P. M., Parra K. J. (2000) J. Exp. Biol. 203, 81–87 [DOI] [PubMed] [Google Scholar]
- 23.Drory O., Frolow F., Nelson N. (2004) EMBO Rep. 5, 1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue T., Forgac M. (2005) J. Biol. Chem. 280, 27896–27903 [DOI] [PubMed] [Google Scholar]
- 25.Charsky C. M., Schumann N. J., Kane P. M. (2000) J. Biol. Chem. 275, 37232–37239 [DOI] [PubMed] [Google Scholar]
- 26.Jones R. P., Durose L. J., Findlay J. B., Harrison M. A. (2005) Biochemistry 44, 3933–3941 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z., Inoue T., Forgac M., Wilkens S. (2006) FEBS Lett. 580, 2006–2010 [DOI] [PubMed] [Google Scholar]
- 28.Féthière J., Venzke D., Madden D. R., Böttcher B. (2005) Biochemistry 44, 15906–15914 [DOI] [PubMed] [Google Scholar]
- 29.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 32.Féthière J., Venzke D., Diepholz M., Seybert A., Geerlof A., Gentzel M., Wilm M., Böttcher B. (2004) J. Biol. Chem. 279, 40670–40676 [DOI] [PubMed] [Google Scholar]
- 33.Armbrüster A., Svergun D. I., Coskun U., Juliano S., Bailer S. M., Grüber G. (2004) FEBS Lett. 570, 119–125 [DOI] [PubMed] [Google Scholar]
- 34.Lau S. Y., Taneja A. K., Hodges R. S. (1984) J. Biol. Chem. 259, 13253–13261 [PubMed] [Google Scholar]
- 35.Supeková L., Supek F., Nelson N. (1995) J. Biol. Chem. 270, 13726–13732 [DOI] [PubMed] [Google Scholar]
- 36.Foury F. (1990) J. Biol. Chem. 265, 18554–18560 [PubMed] [Google Scholar]
- 37.Xu T., Forgac M. (2001) J. Biol. Chem. 276, 24855–24861 [DOI] [PubMed] [Google Scholar]
- 38.Vitavska O., Merzendorfer H., Wieczorek H. (2005) J. Biol. Chem. 280, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 39.Vitavska O., Wieczorek H., Merzendorfer H. (2003) J. Biol. Chem. 278, 18499–18505 [DOI] [PubMed] [Google Scholar]
- 40.Smardon A. M., Kane P. M. (2007) J. Biol. Chem. 282, 26185–26194 [DOI] [PubMed] [Google Scholar]
- 41.Ho M. N., Hill K. J., Lindorfer M. A., Stevens T. H. (1993) J. Biol. Chem. 268, 221–227 [PubMed] [Google Scholar]
- 42.Doherty R. D., Kane P. M. (1993) J. Biol. Chem. 268, 16845–16851 [PubMed] [Google Scholar]
- 43.Kitagawa N., Mazon H., Heck A. J., Wilkens S. (2008) J. Biol. Chem. 283, 3329–3337 [DOI] [PubMed] [Google Scholar]
- 44.Esteban O., Bernal R. A., Donohoe M., Videler H., Sharon M., Robinson C. V., Stock D. (2008) J. Biol. Chem. 283, 2595–2603 [DOI] [PubMed] [Google Scholar]
- 45.Vonck J., Pisa K. Y., Morgner N., Brutschy B., Müller V. (2009) J. Biol. Chem. 284, 10110–10119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau W. C., Rubinstein J. L.Proc. Natl. Acad. Sci. U.S.A. 107, 1367–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto M., Unzai S., Saijo S., Ito K., Mizutani K., Suno-Ikeda C., Yabuki-Miyata Y., Terada T., Toyama M., Shirouzu M., Kobayashi T., Kakinuma Y., Yamato I., Yokoyama S., Iwata S., Murata T. (2008) J. Biol. Chem. 283, 19422–19431 [DOI] [PubMed] [Google Scholar]
- 48.Qi J., Forgac M. (2008) J. Biol. Chem. 283, 19274–19282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohira M., Smardon A. M., Charsky C. M., Liu J., Tarsio M., Kane P. M. (2006) J. Biol. Chem. 281, 22752–22760 [DOI] [PubMed] [Google Scholar]
- 50.Kish-Trier E., Wilkens S. (2009) J. Biol. Chem. 284, 12031–12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kish-Trier E., Briere L. K., Dunn S. D., Wilkens S. (2008) J. Mol. Biol. 375, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lokanath N. K., Matsuura Y., Kuroishi C., Takahashi N., Kunishima N. (2007) J. Mol. Biol. 366, 933–944 [DOI] [PubMed] [Google Scholar]
- 53.Lee L. K., Stewart A. G., Donohoe M., Bernal R. A., Stock D. (2010) Nat. Struct. Mol. Biol. 17, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Rizzo P. A., Bi Y., Dunn S. D. (2006) J. Mol. Biol. 364, 735–746 [DOI] [PubMed] [Google Scholar]
- 55.Sun-Wada G. H., Imai-Senga Y., Yamamoto A., Murata Y., Hirata T., Wada Y., Futai M. (2002) J. Biol. Chem. 277, 18098–18105 [DOI] [PubMed] [Google Scholar]
- 56.Nicholls D., Ferguson S. J. (2002) Bioenergetics 3, Adademic Press, London [Google Scholar]
- 57.Huss M., Wieczorek H. (2007) FEBS Lett. 581, 5566–5572 [DOI] [PubMed] [Google Scholar]