Abstract
Plasminogen activator inhibitor-1 (PAI-1) is a multifunctional glycoprotein that plays a critical role in the pathogenesis of chronic kidney and cardiovascular diseases. Although transforming growth factor (TGF)-β1 is a known inducer of PAI-1, how it controls PAI-1 expression remains enigmatic. Here we investigated the mechanism underlying TGF-β1 regulation of PAI-1 in kidney tubular epithelial cells (HKC-8). Surprisingly, overexpression of Smad2 or Smad3 in HKC-8 cells blocked PAI-1 induction by TGF-β1, whereas knockdown of them sensitized the cells to TGF-β1 stimulation, suggesting that Smad signaling is not responsible for PAI-1 induction. Blockade of several TGF-β1 downstream pathways such as p38 MAPK or JNK, but not phosphatidylinositol 3-kinase/Akt and ERK1/2, only partially inhibited PAI-1 expression. TGF-β1 stimulated β-catenin activation in tubular epithelial cells, and ectopic expression of β-catenin induced PAI-1 expression, whereas inhibition of β-catenin abolished its induction. A functional T cell factor/lymphoid enhancer-binding factor-binding site was identified in the promoter region of the PAI-1 gene, which interacted with T cell factor upon β-catenin activation. Deletion or site-directed mutation of this site abolished PAI-1 response to β-catenin or TGF-β1 stimulation. Similarly, ectopic expression of Wnt1 also activated PAI-1 expression and promoter activity. In vivo, PAI-1 was induced in kidney tubular epithelia in obstructive nephropathy. Delivery of Wnt1 gene activated β-catenin and promoted PAI-1 expression after obstructive injury, whereas blockade of Wnt/β-catenin signaling by Dickkopf-1 gene inhibited PAI-1 induction. Collectively, these studies identify PAI-1 as a direct downstream target of Wnt/β-catenin signaling and demonstrate that PAI-1 induction could play a role in mediating the fibrogenic action of this signaling.
Keywords: β-Catenin, Gene Regulation, Plasminogen Inhibitor, Transforming Growth Factor β (TGFβ), Wnt Pathway
Introduction
Plasminogen activator inhibitor-1 (PAI-1)2 is a multifunctional, secreted glycoprotein that plays a critical role in regulating various biological processes involving extracellular proteolysis and tissue remodeling (1, 2). Aside from its classical role in inhibiting the activity of tissue-type and urokinase-type plasminogen activators, PAI-1 influences numerous cellular behaviors via diverse mechanisms. It binds with high affinity to extracellular protein vitronectin, thereby modulating its interaction with αvβ3 integrin. PAI-1 is also known to bind to urokinase-type plasminogen activator and its receptor complex on the cell surface, regulating internalization and intracellular signaling (1). Through a multitude of actions, PAI-1 plays an important role in diverse cellular processes such as cell migration, wound healing, tumor metastasis, angiogenesis, and tissue remodeling. It becomes increasingly clear that the functions of PAI-1 extend far beyond its ability to block plasminogen activation.
PAI-1 is expressed in almost all cell types, predominantly in endothelial cells, adipocytes, and hepatocytes (3). In normal kidney, however, its expression level is extremely low or undetectable. Injury to renal parenchyma in a wide variety of acute and chronic kidney diseases often causes marked induction of PAI-1 expression (4–6). Circulating plasma PAI-1 levels also rise in patients with renal insufficiency, which may contribute to the progression of chronic kidney and cardiovascular lesions (7). In the past several years, extensive experimental studies using genetically modified animal models have revealed that PAI-1 is a powerful fibrogenic factor that promotes inflammatory cell recruitment and extracellular matrix accumulation (1, 2, 8). Mice with genetic ablation of PAI-1 are protected against the development of renal fibrotic lesions after a variety of insults in different models, such as diabetic nephropathy (9), unilateral ureteral obstruction (UUO) (10), anti-glomerular basement membrane nephritis (11), and remnant kidney after five-sixth (5/6) nephrectomy (2). Conversely, transgenic mice with PAI-1 overexpression are more susceptible to renal injury and develop more severe fibrotic lesions than wild-type controls (11, 12). Based on these findings, PAI-1 has been proposed as a promising therapeutic target for developing new strategies for treatment of progressive kidney diseases (1, 13).
The expression of PAI-1 in kidney tissues is tightly controlled by a diverse array of signals, such as transforming growth factor-β1 (TGF-β1), angiotensin II, and other extracellular cues (3, 14). In light of an increased production of these pathogenic factors in diseased kidneys (15, 16), up-regulation of PAI-1 likely represents a convergent pathway that mediates their fibrogenic actions. Of many extracellular cues regulating PAI-1 expression, TGF-β1 is the most studied and is considered a major and potent inducer under pathologic conditions. However, the mechanism governing TGF-β1 regulation of PAI-1 expression remains poorly understood.
In this study, we have investigated the regulation of the PAI-1 gene by TGF-β1 in kidney tubular epithelial cells. Unexpectedly, we show that Smads, the principal downstream mediators of TGF-β1 signaling (17), actually inhibit TGF-β1-mediated PAI-1 induction. We further demonstrate that the canonical pathway of Wnt/β-catenin signaling, which is activated in fibrotic kidneys (18), is responsible, at least partially, for PAI-1 induction. Our studies identify PAI-1 as a direct downstream target of the canonical pathway of Wnt/β-catenin signaling and unravel a potential mechanism underlying this signal pathway in the pathogenesis of renal fibrosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
Human proximal tubular epithelial cell (HKC, clone 8) was provided by Dr. L. Racusen (Johns Hopkins University, Baltimore, MD). The cells were cultured in Dulbecco's modified Eagle's medium-Ham's F12 medium supplemented with 5% fetal bovine serum. The cells were typically seeded at ∼70% confluence in complete medium containing 5% fetal bovine serum for 24 h and then serum-starved for 16 h, followed by incubation with recombinant TGF-β1 (R & D Systems, Minneapolis, MN) for various periods of time at the concentration of 2 ng/ml or different concentration for 48 h except as otherwise indicated. The cells were then collected for Western blot analyses. In some experiments, the cells were pretreated with various chemical inhibitors at the concentrations specified or vehicle (0.1% Me2SO) for 30 min, followed by incubating in the absence or presence of 2 ng/ml TGF-β1. PD98059 (MEK1 inhibitor), wortmannin (PI3K inhibitor), SC68376 (p38 MAPK inhibitor), SP600125 (JNK inhibitor), and Ro31-8220 (pan-specific protein kinase C inhibitor) were purchased from Calbiochem (La Jolla, CA). SB431542 (a specific TGF-β1 type I receptor/activin receptor-like kinase-5 inhibitor) was obtained from Tocris Bioscience (Ellisville, MO). The specific β-catenin small molecule inhibitor ICG-001 was described previously (19).
Plasmid Transient Transfection
For transient transfection, HKC-8 cells were seeded in six-well plates at 5 × 105 cells/well. The cells were then transfected with HA-tagged Smad2 or HA-tagged Smad3 expression vectors (20) (provided by Dr. X. Lin, Baylor College of Medicine, Houston, TX) for 24 h and then treated with various concentrations of TGF-β1 for 48 h. The empty pcDNA3 vector was used as a mock transfection control.
Small Interfering RNA Inhibition
For small interfering RNA (siRNA) inhibition studies, HKC-8 cells were transiently transfected with negative control siRNA (Am4635), Smad2 siRNA (identification number 115715), or Smad3 siRNA (identification number 107877; Ambion, Austin, TX) at the final concentration of 120 nm by using Oligofectamine reagent according to the instructions specified by the manufacturer (Invitrogen), as described previously (20). After transfection for 24 h, the cells were then serum-starved for 16 h, followed by incubation with various concentrations of TGF-β1 for an additional 48 h. Whole cell lysates were then collected for Western blot analyses.
Establishment of Stable Cell Lines
HKC-8 cells were transfected with FLAG-tagged N-terminal truncated, stabilized β-catenin expression vector (pDel-β-cat), or HA-tagged Wnt1 expression vector (pHA-Wnt1; Upstate Biotechnology, Lake Placid, NY), respectively, using the Lipofectamine 2000 according to the instructions specified by the manufacturer (Invitrogen). The empty vector pcDNA3 (Invitrogen) was used as a mock transfection control. Twenty-four hours after transfection and every 3–4 days thereafter, the cells were refed with fresh selective medium containing G418 (Geneticin; Invitrogen) at a final concentration of 0.8 mg/ml. Neomycin-resistant clones were selected and expanded. Ectopic expression of Wnt1 or β-catenin in the stable cell lines was confirmed by Western blot analysis.
Animal Model
Male CD-1 mice that weighed ∼20–22 g were purchased from Harlan Sprague-Dawley (Indianapolis, IN). UUO was performed using an established protocol, as described previously (18). Sham-operated mice were used as normal controls. For delivery of Wnt1 or DKK1 (Dickkopf-1) gene, naked Wnt1 expression plasmid (pHA-Wnt1) or DKK1 expression plasmid (pFLAG-DKK1; provided by Dr. Xi He, Harvard Medical School, Boston, MA) (21) was injected intravenously at 1 mg/kg of body weight 1 day before (day −1) UUO, by use of a hydrodynamics-based in vivo gene transfer approach, as described previously (18, 22). Control UUO mice were injected with empty vector pcDNA3 in an identical manner. Groups of mice (n = 5) were killed at 7 days after UUO, and the kidney tissues were removed for various analyses. The animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh.
Western Blot Analysis
The preparation of whole cell lysates and kidney tissue homogenates and Western blot analysis of protein expression were carried out by using routine procedures as described previously (23). The primary antibodies were obtained from the following sources: anti-PAI-1 (sc-5297; Santa Cruz Biotechnology); anti-β-catenin (catalog number 610154; BD Transduction Laboratories, San Jose, CA); anti-dephosphorylated, active β-catenin (catalog number 05-665; Upstate Biotechnology); anti-FLAG (M2, F3165; Sigma); anti-HA (6E2; Cell Signaling Technology, Danvers, MA); anti-Smad2 (catalog number 51-1300); anti-Smad3 (catalog number 51-1500; Invitrogen); anti-phospho-Smad2 (Ser465/467) (catalog number 3101); anti-phopsho-Smad3 (Ser423/425) (catalog number 9514; Cell Signaling); anti-α-tubulin (Sigma); and anti-glyceraldehyde-3-phosphate dehydrogenase (Ambion, Austin, TX). Quantification was performed by measurement of the intensity of the signals with the use of National Institutes of Health Image analysis software.
RNA Isolation and Reverse Transcription-PCR
Total RNA was extracted using a TRIzol RNA isolation system (Invitrogen). The first strand of cDNA was synthesized using 2 μg of RNA in 20 μl of reaction buffer by reverse transcription using avian myeloblastosis virus reverse transcription (Promega, Madison, WI) and random primers at 42 °C for 30 min. PCR was carried out using a standard PCR kit and a 1-μl aliquot of cDNA, HotStarTaq polymerase (Qiagen) with specific primer pairs, as described previously (18). The sequences of the primer pairs were as follows: PAI-1, 5′-GGC CAT TAC TAC GAC ATC CTG-3′ (sense) and 5′-GGT CAT GTT GCC TTT CCA GT-3′ (antisense); β-actin, 5′-TCA AGA TCA TTG CTC CTC CTG AGC-3′ (sense) and 5′-TGC TGT CAC CTT CAC CGT TCC AGT-3′ (antisense). Relative levels of PAI-1 mRNA were calculated after normalizing with the housekeeping gene β-actin.
Reporter Plasmid Construction and Site-directed Mutagenesis
PAI-1 promoter-luciferase reporters were constructed by cloning various lengths of the PAI-1 promoter region into the pGL3-Basic luciferase vector (Promega). Different lengths of the human PAI-1 gene promoter fragments were generated by using PCR and cloned into the pGL3-Basic luciferase vector using routine cloning procedures. The correct sequences of all PAI-1 promoter-luciferase constructs, as indicated in Fig. 6A, were confirmed by DNA sequencing at the University of Pittsburgh DNA sequencing core facility. For generating mutant PAI-1 promoter-luciferase reporter, a point mutation (A to T) at the core TCF site was introduced in the 0.42PAI-1-Luc reporter plasmid by using site-directed mutagenesis according to the protocol specified by the manufacturer (QuikChange II XL kit; Stratagene, La Jolla, CA). Mutation was confirmed by DNA sequencing.
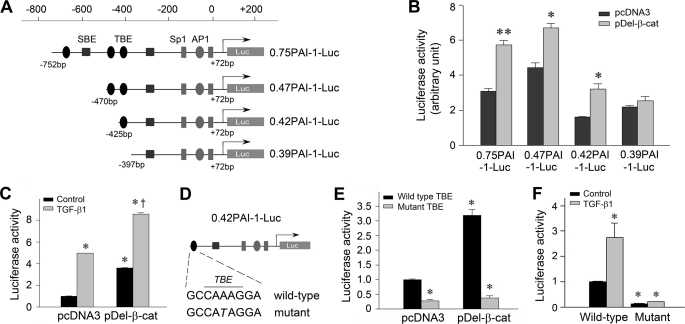
FIGURE 6.
PAI-1 gene promoter harbors a functional TBE that is responsible for β-catenin-mediated PAI-1 induction. A, diagram shows the construction of various luciferase reporter plasmids containing different lengths of the promoter region of human PAI-1 gene linked to the coding sequence of luciferase gene. Putative TBE, SBE, Sp1, and AP1 sites are indicated. B, β-catenin activates PAI-1 promoter in tubular epithelial cells. HKC-8 cells were cotransfected with various PAI-1 promoter-luciferase reporter plasmids as shown in A and stabilized β-catenin expression vector (pDel-β-cat) or empty vector (pcDNA3). R. reniformis luciferase vector (pRL-TK) was used as an internal control for transfection efficiency. Luciferase activities (arbitrary unit) were calculated, reported after normalizing for transfection efficiency, and presented as the means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01 versus pcDNA3 controls. C, PAI-1 promoter harbors a functional TBE (0.42PAI-1-Luc) that is responsive to β-catenin and TGF-β1 stimulation. HKC-8 cells were cotransfected with 0.42PAI-1-Luc reporter plasmid with pcDNA3 or pDel-β-cat, followed by incubation without or with TGF-β1 (2 ng/ml) for 48 h. D–F, site-directed mutation in the TBE of PAI-1 promoter abolishes its responsiveness to β-catenin and TGF-β1 stimulation. The construction of wild-type and mutant TBE reporter plasmids (D) and luciferase activities (E and F) are presented. *, p < 0.05 versus controls; †, p < 0.05 versus pDel-β-cat alone.
Luciferase Assay
HKC-8 cells were cotransfected by using Lipofectamine 2000 reagent with various PAI-1-Luc luciferase reporter constructs (1.0 μg) plus pDel-β-cat (2.0 μg) expression vector or empty pcDNA3 vector, respectively. Luciferase reporter plasmid containing four copies of the Smad-binding elements (SBE4-Luc) was described previously (24) and obtained from Addgene (plasmid 16495; Addgene, Cambridge, MA). A fixed amount (0.1 μg) of internal control reporter Renilla reniformis luciferase driven under a thymidine kinase promoter (pRL-TK; Promega) was also cotransfected for normalizing the transfection efficiency. In some experiments, after transfection for 24 h, the cells were treated without or with TGF-β1 (2 ng/ml) for an additional 24 h. Luciferase assay was performed using a dual luciferase assay system kit according to the manufacturer's protocols (Promega). Relative luciferase activity (arbitrary unit) was reported as fold induction over controls after normalizing for transfection efficiency.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed to analyze in vivo interactions of TCF and putative TCF-binding element (TBE) in human PAI-1 promoter (25). This assay was carried out essentially according to the protocols specified by the manufacturer (ChIP assay kit; Upstate Biotechnology). Briefly, HKC-8 cells after various treatments as indicated were cross-linked with 1% formaldehyde and then resuspended in SDS lysis buffer containing protease inhibitors. The chromatin solution was sonicated, and the supernatant was diluted 10-fold. An aliquot of total diluted lysate was used for total genomic DNA as input DNA control. The anti-TCF-1 antibody (clone 7H13; catalog number 05-510; Upstate Biotechnology) was added and incubated at 4 °C overnight, followed by incubation with protein A-agarose for 1 h. The precipitates were washed, and the chromatin complexes were eluted. After reversal of the cross-linking at 65 °C for 4 h, the DNA was purified, and ChIP samples were used as a template for PCR. Primer sets P1/P2 and P3/P4 encompass different regions of the PAI-1 promoter containing proximal and distal putative TBE, respectively. The sequences of primers used for ChIP assay were given in Fig. 7A. PCR samples were analyzed by electrophoresis on a 2.0% agarose gel.
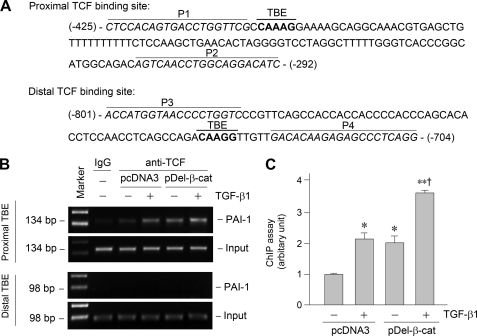
FIGURE 7.
Activation of β-catenin promotes TCF binding to the proximal TBE in PAI-1 promoter. A, partial sequence of PAI-1 gene promoter region. Bold letters indicate the putative TBE. P1 and P2 indicate the primer pair encompassing the proximal TBE, whereas P3 and P4 indicate the prime pair for the distal TBE. B and C, ChIP assay revealed that activation of β-catenin (either by TGF-β1 or by overexpressing stabilized β-catenin) promoted TCF binding to the proximal TBE but not distal TBE. Representative ChIP assay (B) and quantitative ChIP data (C) are presented. *, p < 0.05 versus controls; **, p < 0.01 versus controls; †, p < 0.05 versus TGF-β1 or pDel-β-cat alone (n = 3).
Immunohistochemical Staining
Paraffin-embedded mouse kidney sections (3-μm thickness) were prepared by a routine procedure (18, 26). Immunohistochemical staining for PAI-1 protein was performed using a routine protocol as described previously (18).
Statistical Analysis
Statistical analysis was performed using SigmaStat software (Jandel Scientific Software, San Rafael, CA). Comparisons between groups were made using one-way analysis of variance, followed by the Student-Newman-Keul's test. A p value of < 0.05 was considered significant.
RESULTS
Smad Signaling Does Not Mediate PAI-1 Induction by TGF-β1
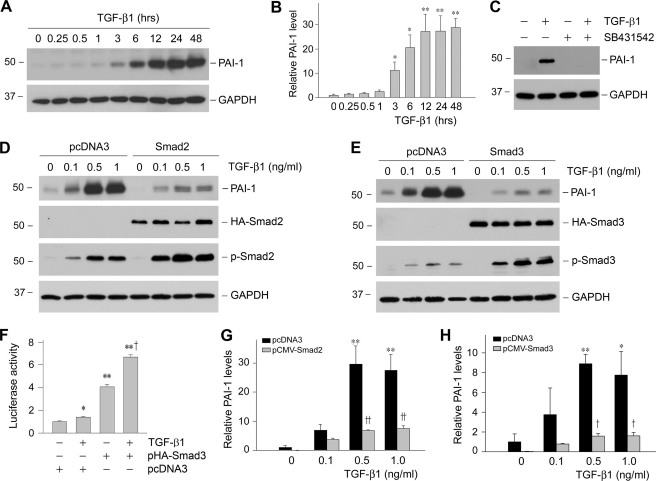
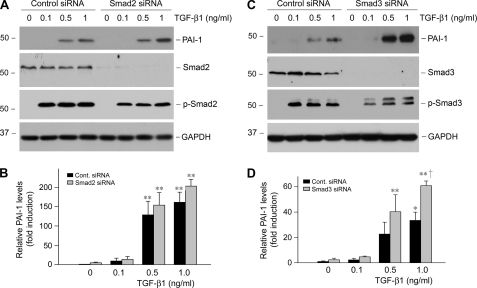
We studied PAI-1 regulation by TGF-β1 in human proximal tubular epithelial cells (HKC-8). As shown in Fig. 1A, TGF-β1 markedly induced PAI-1 expression in a time-dependent manner. Quantitative determination of PAI-1 induction is presented in Fig. 1B. The effect of TGF-β1 on PAI-1 induction was also dose-dependent, starting at a concentration as low as 0.5 ng/ml (data not shown). This induction of PAI-1 expression was dependent on TGF-β receptor signaling, because inhibition of TGF-β type I receptor by SB431542, a potent and specific small molecule activin receptor-like kinase-5 inhibitor (27), completely abolished PAI-1 expression (Fig. 1C). Because Smad signaling is the principal downstream effector of TGF-β1, we investigated the role of Smads in mediating PAI-1 induction. Surprisingly, overexpression of either Smad2 or Smad3 drastically inhibited PAI-1 induction in response to TGF-β1 stimulation (Fig. 1, D, E, G, H). Of note, exogenous Smad was able to respond to TGF-β1 stimulation to promote the SBE-driven luciferase reporter activity (Fig. 1F). To confirm the negative role of Smad in PAI-1 regulation, we utilized an opposite approach by knocking down the endogenous Smad expression, as previously reported (20). As shown in Fig. 2, knockdown of Smad2 or Smad3 by siRNA sensitized HKC-8 cells to TGF-β1 stimulation, resulting in greater induction of PAI-1 expression. Therefore, it appears that Smad signaling actually inhibits, rather than mediates, PAI-1 expression in proximal tubular epithelial cells after TGF-β1 stimulation.
FIGURE 1.
Smad blocks PAI-1 expression induced by TGF-β1 in proximal tubular epithelial cells. A and B, TGF-β1 induced PAI-1 expression in a time-dependent manner. Human proximal tubular epithelial cells (HKC-8) were treated with TGF-β1 (2 ng/ml) for various periods of time as indicated. Shown in A is the representative Western blot result. Quantitative determination of PAI-1 induction after normalization with GAPDH is presented in B. C, TGF-β receptor signaling is required for PAI-1 induction. HKC-8 cells were pretreated with activin receptor-like kinase-5 inhibitor SB431542 (10 μm), followed by incubation with TGF-β1 (2 ng/ml) for 24 h as indicated. D and E, overexpression of Smad2 or Smad3 inhibited PAI-1 induction in response to TGF-β1 stimulation. HKC-8 cells were transiently transfected with HA-tagged Smad2 or Smad3, or empty pcDNA3 vector, followed by incubation with different doses of TGF-β1 as indicated. Whole cell lysates were immunoblotted with specific antibodies against PAI-1, HA, phospho-Smad2, phospho-Smad3, and GAPDH, respectively. F, Smad3 promotes the SBE-driven luciferase reporter activity. HKC-8 cells were cotransfected with SBE4-Luc reporter plasmid and Smad3 expression vector (pHA-Smad3) or empty vector (pcDNA3), followed by incubation with TGF-β1 for 48 h as indicated. Relative luciferase activities (arbitrary unit) were reported. G and H, graphic presentations of PAI-1 abundance after transfection with Smad2 or Smad3 are shown. The data are presented as the means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01 versus controls. †, p < 0.05; ††, p < 0.01 versus pcDNA3.
FIGURE 2.
Knockdown of Smad2 or Smad3 enhances PAI-1 induction after TGF-β1 stimulation. HKC-8 cells were transiently transfected with control (Cont.) siRNA, Smad2, or Smad3 siRNA (120 nm) and then treated with different doses of TGF-β1 as indicated. Whole cell lysates were immunoblotted with specific antibodies against PAI-1, Smad2, Smad3, phospho-Smad2, phospho-Smad3, and GAPDH, respectively. Representative Western blots (A and C) and the relative abundance of PAI-1 after normalization with GAPDH (B and D) are presented. The data (relative to the controls = 1.0) are presented as the means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01 versus controls. †, p < 0.01 versus control siRNA group with TGF-β1.
Activation of MAPK Signaling Is Partially Responsible for PAI-1 Induction
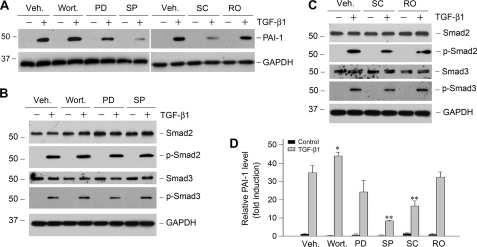
We next investigated any other potential signal pathways downstream of TGF-β1 that are responsible for PAI-1 induction in HKC-8 cells. It is well known that TGF-β1 is capable of activating several signal pathways including PI3K/Akt, ERK-1/2, p38 MAPK, JNK, and protein kinase C in different cells (28–30). To elucidate the potential involvement of these signal pathways in PAI-1 regulation, HKC-8 cells were pretreated with specific inhibitors for 30 min, followed by incubation in the absence or presence of TGF-β1. As shown in Fig. 3, blockade of p38 MAPK and JNK attenuated PAI-1 induction by TGF-β1, whereas inhibition of ERK-1/2 upstream kinase (MEK1) and protein kinase C exhibited little influence on PAI-1 expression. Inhibition of PI3K actually promoted TGF-β1-mediated PAI-1 induction, suggesting that this pathway has negative effects on PAI-1 expression. Of note, these inhibitors had little effect on Smad2/3 abundance and their phosphorylation after TGF-β1 stimulation, except that inhibition of p38 MAPK slightly reduced Smad activation (Fig. 3, B and C). Considered together, two branches of MAPK signaling, namely p38 MAPK and JNK, are partially responsible for TGF-β1-mediated PAI-1 induction in tubular epithelial cells.
FIGURE 3.
Activation of MAPK signaling is partially responsible for PAI-1 induction by TGF-β1. HKC-8 cells were pretreated with either various chemical inhibitors or vehicle (Veh., 0.1% Me2SO) as indicated for 30 min, followed by incubation in the absence or presence of TGF-β1 (2 ng/ml) for 24 h. PD, PD98059 (MEK1 inhibitor) (10 μm); wort., wortmannin (PI3K inhibitor) (10 nm); SC, SC68376 (p38 MAPK inhibitor) (20 μm); SP, SP600125 (JNK inhibitor) (20 μm); RO, Ro31-8220 (pan-specific protein kinase C inhibitor) (10 μm). A, representative Western blots of PAI-1 expression after various treatments. B and C, Smad2/3 abundance and phosphorylation in the presence of various inhibitors as indicated. D, quantitative determination of the relative PAI-1 abundance after normalization with GAPDH. The data (relative to the controls = 1.0) are presented as the means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01 versus vehicle controls.
Activation of β-Catenin Signaling by TGF-β1 Mediates PAI-1 Induction
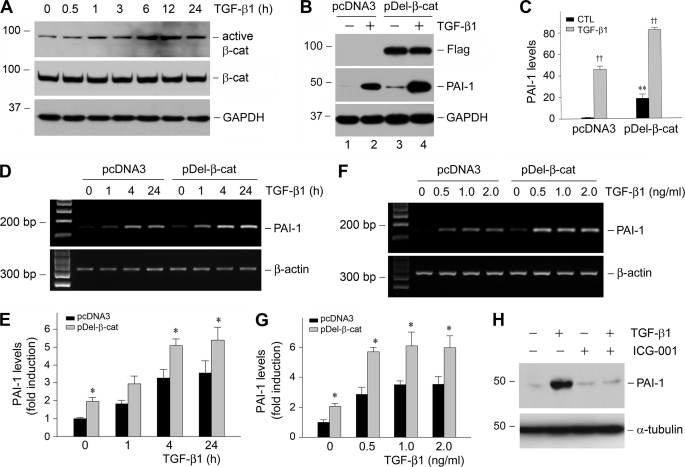
Recent studies show that the canonical pathway of Wnt/β-catenin signaling is involved in the pathogenesis of renal interstitial fibrosis (18, 31), which prompted us to explore whether this signal pathway plays a role in mediating TGF-β1-induced PAI-1 expression. As shown in Fig. 4A, TGF-β1 stimulated β-catenin activation in tubular epithelial cells in a time-dependent manner, as revealed by an increase in dephosphorylated, active β-catenin levels. This activation of β-catenin by TGF-β1 was not affected by the intracellular levels of Smad abundance (data not shown). To determine any involvement of β-catenin activation in PAI-1 regulation, we established stable HKC-8 cell line by transfecting with FLAG-tagged, N-terminal truncated, stabilized β-catenin expression vector (pDel-β-cat). It was revealed that ectopic expression of stabilized β-catenin induced PAI-1 expression at basal conditions (Fig. 4B, lane 1 versus lane 3). Quantitative determination revealed a more than 15-fold induction of PAI-1 expression in the HKC-8 cells transfected with stabilized β-catenin, compared with that transfected with empty vector (pcDNA3). Likewise, exogenous β-catenin also further promoted PAI-1 induction under TGF-β1-stimulated conditions (Fig. 4B, lane 2 versus lane 4). Similar results were obtained when PAI-1 mRNA was detected (Fig. 4, D–G).
FIGURE 4.
Activation of β-catenin signaling by TGF-β1 mediates PAI-1 induction. A, TGF-β1 induces β-catenin activation in tubular epithelial cells. HKC-8 cells were treated with TGF-β1 (2 ng/ml) for various periods of time as indicated, and the cell lysates were immunoblotted with antibodies against active β-catenin, total β-catenin, and GAPDH, respectively. B and C, ectopic expression of stabilized β-catenin promotes PAI-1 induction at basal and TGF-β1-stimulated conditions. HKC-8 cells stably transfected with either empty vector (pcDNA3) or FLAG-tagged, N-terminal truncated β-catenin (pDel-β-cat) were incubated in the absence or presence of TGF-β1 (2 ng/ml) for 48 h. A representative Western blot (B) and the quantitative data (C) are presented. CTL, control. **, p < 0.01 versus pcDNA3 controls. ††, p < 0.01 versus without TGF-β1 (n = 3). D–G, expression of stabilized β-catenin also promotes PAI-1 mRNA expression. HKC-8 cells stably transfected with either empty vector (pcDNA3) or FLAG-tagged, N-terminal truncated β-catenin (pDel-β-cat) were incubated in the absence or presence of TGF-β1 for various periods of time (D and E) at different concentrations (F and G). The quantitative data on PAI-1 mRNA induction (fold over the controls) are presented in E and G. *, p < 0.05 versus pcDNA3 controls. H, inhibition of β-catenin signaling blocked TGF-β1-induced PAI-1 expression. HKC-8 cells were pretreated with a specific small molecule β-catenin inhibitor (ICG-001) (10 μm) for 30 min, followed by incubation in the absence or presence of TGF-β1 (2 ng/ml) for 24 h.
We also assessed the necessity of β-catenin activation for TGF-β1-induced PAI-1 expression. To this end, HKC-8 cells were treated with ICG-001, a highly specific small molecule β-catenin inhibitor (19), followed by incubation with TGF-β1. As shown in Fig. 4H, inhibition of β-catenin signaling abolished PAI-1 induction by TGF-β1 in HKC-8 cells.
Multiple Signal Pathways Are Involved in Regulating PAI-1 Induction
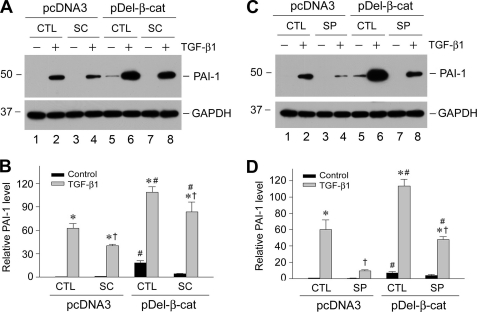
Because p38 MAPK and JNK signaling are partially responsible for PAI-1 induction by TGF-β1 (Fig. 3), we next investigated whether these pathways influence PAI-1 induction by β-catenin. As shown in Fig. 5, inhibition of p38 MAPK attenuated PAI-1 expression induced by exogenous β-catenin in basal conditions (Fig. 5A, lane 5 versus lane 7). Similarly, blockade of p38 MAPK activation also reduced PAI-1 expression in HKC-8 stable cell line overexpressing β-catenin after TGF-β1 stimulation (Fig. 5A, lane 6 versus lane 8). Inhibition of JNK also attenuated PAI-1 expression in cell lines expressing stabilized β-catenin under both basal and TGF-β1-stimulated conditions (Fig. 5, C and D). These observations suggest that multiple pathways downstream of TGF-β1 are involved in regulating PAI-1 expression in tubular epithelial cells.
FIGURE 5.
Multiple signal pathways downstream of TGF-β1 are involved in mediating PAI-1 induction. HKC-8 cells stably transfected with either empty vector (pcDNA3) or FLAG-tagged, N-terminal truncated β-catenin (pDel-β-cat) were incubated with TGF-β1 (2 ng/ml) in the absence or presence of SC68376 (SC, p38 MAPK inhibitor) (A and B) or SP600125 (SP, JNK inhibitor) (C and D). Representative Western blot (A and C) and quantitative data (B and D) on PAI-1 induction (fold over the controls) are presented. *, p < 0.05 versus controls without TGF-β1. †, p < 0.05 versus controls without inhibitors. #, p < 0.05 versus pcDNA3 controls (n = 3). CTL, control.
PAI-1 Gene Promoter Harbors a Functional TCF Site Responsible for β-Catenin Action
To elucidate the mechanism underlying β-catenin regulation of PAI-1 expression, we analyzed the structure and function of human PAI-1 gene promoter. Bioinformatics analysis revealed three putative TCF/lymphoid enhancer-binding factor-binding sites (TBEs), as well as numerous other transcription factor binding sites such as Sp1, AP1, and SBE, in the proximal promoter region of the human PAI-1 gene (Fig. 6A). To test the functionality of these TBEs, we constructed various luciferase reporter plasmids containing different lengths of the promoter region of the human PAI-1 gene linked to the coding sequence of luciferase gene. After these constructs were cotransfected into HKC-8 cells in the presence or absence of the stabilized β-catenin expression vector, luciferase activities were assessed. As shown in Fig. 6B, a functional TBE, located at −425 to −397 relative to transcription initiation site, designated as proximal TBE, was responsive to exogenous β-catenin stimulation. Deletion of this region abolished luciferase induction by stabilized β-catenin (Fig. 6B). Interestingly, luciferase-reporter plasmid containing this proximal TBE (0.42PAI-1-Luc) not only responded to exogenous β-catenin but also was responsive to TGF-β1 stimulation (Fig. 6C). A combination of exogenous β-catenin and TGF-β1 further promoted luciferase-reporter activity (Fig. 6C). To confirm the importance of this TBE in regulating PAI-1 expression, we utilized a site-directed mutagenesis approach to introduce a point mutation (A to T) in the TCF core-binding site (Fig. 6D). As shown in Fig. 6 (E and F), destruction of this TBE completely abolished its responsiveness to β-catenin and TGF-β1 stimulation. Of note, site-directed mutation of this TBE in PAI-1 promoter also resulted in a decreased luciferase-reporter activity under basal conditions (Fig. 6, E and F), suggesting that endogenous β-catenin signaling is also critical for conferring the constitutive expression of the PAI-1 gene.
Activation of β-Catenin Promotes TCF Binding to the Proximal TBE in PAI-1 Promoter
To provide further evidence for a role of β-catenin in regulating PAI-1 expression, we examined the physical and functional interaction between TCF and its cognate TBE in PAI-1 promoter by using an in situ chromatin immunoprecipitation (ChIP) assay. Based on the functional data in Fig. 6, we primarily focused on studying the interaction between TCF and the proximal, functional TBE, whereas we used the distal, nonfunctional TBE as a control (Fig. 7A). We found that activation of β-catenin, either by TGF-β1 or by overexpressing stabilized β-catenin, promoted TCF binding to the proximal TBE but not to distal TBE (Fig. 7, B and C). Moreover, combination of TGF-β1 and stabilized β-catenin further promoted the binding of TCF to the proximal TBE in the PAI-1 promoter (Fig. 7, B and C). Therefore, β-catenin activation is able to facilitate TCF binding to the proximal TBE, leading to PAI-1 transcription.
Ectopic Expression of Wnt1 Promotes PAI-1 Expression
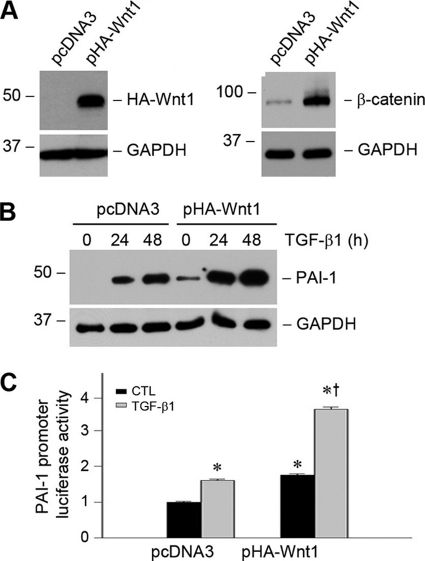
Because Wnt is the upstream activator of β-catenin signaling, we next examined the effect of Wnt1, the prototypic Wnt that transmits its signal via the canonical pathway, on PAI-1 gene expression in tubular epithelial cells. To this end, we established a stable HKC-8 cell line in which HA-tagged Wnt1 was expressed (Fig. 8A). As shown in Fig. 8A, ectopic expression of Wnt1 resulted in stabilization and accumulation of β-catenin in HKC-8 cells. It was revealed that Wnt1 expression alone induced PAI-1 expression in tubular epithelial cells under basal conditions (Fig. 8B). Stimulation with TGF-β1 further promoted PAI-1 induction in HKC-8 cells. Exogenous Wnt1 also stimulated PAI-1 promoter activities in tubular epithelial cells after transfection with PAI-1 promoter-luciferase reporter plasmid (0.42PAI-1-Luc) (Fig. 8C). Therefore, Wnt1, through activation of its downstream mediator β-catenin, is able to activate PAI-1 promoter and induce its gene expression.
FIGURE 8.
Ectopic expression of Wnt1, an upstream activator of β-catenin signaling, promotes PAI-1 expression. A, ectopic expression Wnt1 increases β-catenin protein abundance in tubular epithelial cells. HKC-8 cells were stably transfected with HA-tagged Wnt1, and cell lysates were immunoblotted with antibodies against HA and β-catenin, respectively. B, ectopic expression of Wnt1 promotes PAI-1 induction at basal and TGF-β1-stimulated conditions. HKC-8 cells stably transfected with either pcDNA3 or pHA-Wnt1 were incubated in the absence or presence of TGF-β1 (2 ng/ml) for various periods of time as indicated. C, Wnt1 stimulates PAI-1 promoter activities in tubular epithelial cells. HKC-8 cells were cotransfected with PAI-1 promoter-luciferase reporter plasmid (0.42PAI-1-Luc) and Wnt1 expression vector (pHA-Wnt1) or empty vector (pcDNA3), followed by incubation without or with TGF-β1 as indicated. The luciferase activities (arbitrary unit) were calculated, reported after normalizing for transfection efficiency, and presented as the means ± S.E. of three experiments. *, p < 0.05 versus pcDNA3 controls. †, p < 0.05 versus without TGF-β1 (n = 3). CTL, control.
Wnt/β-Catenin Signaling Modulates PAI-1 Expression in Vivo
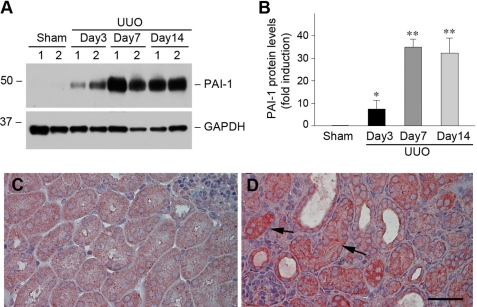
To investigate PAI-1 regulation by Wnt/β-catenin signaling in vivo, we first examined PAI-1 expression in a mouse model of renal fibrosis induced by UUO, in which Wnt/β-catenin signaling is activated (18, 31). As shown in Fig. 9 (A and B), PAI-1 was markedly induced in the obstructed kidney at different time points after UUO, as shown by Western blot analyses. Immunohistochemical staining exhibited that PAI-1 protein was predominantly localized in renal tubular epithelia in obstructive nephropathy (Fig. 9D, arrows). This expression pattern of PAI-1 after UUO is consistent with that of β-catenin in this model (18), suggesting a close correlation between PAI-1 expression and β-catenin activation in vivo.
FIGURE 9.
PAI-1 is induced specifically in renal tubular epithelial cells after obstructive injury. A and B, Western blot analyses show PAI-1 induction in the obstructed kidney at different time points after UUO. Representative Western blot (A) and quantitative data after normalization with GAPDH (B) are presented. The data are presented as the means ± S.E. of five animals (n = 5). *, p < 0.05; **, p < 0.01 versus sham controls. C and D, immunohistochemical staining shows the localization of PAI-1 protein in obstructive nephropathy. Kidney sections from sham (C) and UUO (D) were immunostained with antibody against PAI-1. The arrows indicate PAI-1-positive renal tubular epithelial cells. Scale bar, 50 μm.
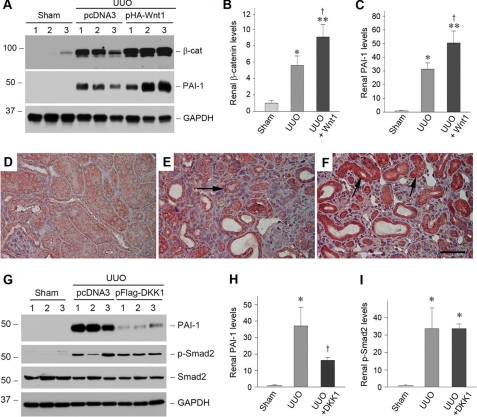
To establish a direct relationship of PAI-1 expression and Wnt/β-catenin signaling in vivo, we manipulated the Wnt/β-catenin signaling by delivery of Wnt1 gene through a hydrodynamics-based strategy (32, 33). As shown in Fig. 10 (A–C), delivery of the exogenous Wnt1 gene induced renal β-catenin and PAI-1 expression after obstructive injury. Immunohistochemical staining demonstrates that exogenous Wnt1 expression significantly enhanced PAI-1 induction in renal tubular epithelial cells after UUO (Fig. 10F), suggesting that Wnt/β-catenin signaling stimulates PAI-1 expression in vivo as well. To confirm this observation, we deliver the DKK1 gene, an endogenous antagonist of the Wnt/β-catenin canonical pathway (21, 34), in mice after UUO. As shown in Fig. 10 (G–I), blockade of Wnt/β-catenin signaling by DKK1 inhibited renal PAI-1 expression, but not Smad2 phosphorylation, after obstructive injury. Of note, DKK1 also inhibited PAI-1 induction by TGF-β1 in HKC-8 cells in vitro (data not shown). Together, it appears clear that PAI-1 is a transcriptional target of Wnt/β-catenin signaling both in vitro and in vivo.
FIGURE 10.
Wnt/β-catenin signaling modulates PAI-1 expression in vivo. A–C, expression of exogenous Wnt1 gene induces renal β-catenin and PAI-1 expression after obstructive injury. Representative Western blots (A) and relative levels (fold induction over sham controls) of β-catenin (B) and PAI-1 (C) are presented. Numbers 1–3 indicate individual animals in a given group. The data are presented as the means ± S.E. of five animals (n = 5). *, p < 0.05; **, p < 0.01 versus sham controls. †, p < 0.05 versus UUO controls. D–F, immunohistochemical staining shows the localization of PAI-1 protein in different groups. Kidney sections from sham (D), UUO (E), and UUO plus Wnt1 (F) were immunostained with antibody against PAI-1. The arrows indicate positive staining. Scale bar, 50 μm. G–I, blockade of Wnt/β-catenin canonical pathway by DKK1 inhibits renal PAI-1 expression but not Smad2 phosphorylation after obstructive injury. Representative Western blots (G) and relative levels (fold induction over sham controls) of PAI-1 (H) and phospho-Smad2 (I) are presented. The data are presented as the means ± S.E. of three to five animals (n = 3–5). *, p < 0.05 versus sham controls. †, p < 0.05 versus UUO controls.
DISCUSSION
PAI-1 is arguably one of the most studied TGF-β-targeted genes (3). However, the molecular mechanism and intracellular mediators dictating PAI-1 gene regulation by TGF-β1 remain incompletely understood. In this study, we have presented compelling evidence suggesting that PAI-1 is a transcriptional target of the canonical pathway of Wnt/β-catenin signaling. This conclusion is supported by several lines of evidence. First, overexpression of stabilized β-catenin in kidney tubular epithelial cells induces PAI-1 expression under both basal and TGF-β1-stimulated conditions. Second, inhibition of β-catenin by small molecule inhibitor abolishes PAI-1 induction by TGF-β1. Third, the PAI-1 gene promoter region harbors a functional TBE, which mediates the interaction with TCF in response to β-catenin activation. Fourth, activation of β-catenin by overexpressing Wnt1 induces PAI-1 expression in vitro and in vivo. Finally, modulation of β-catenin signaling by ectopic expression of DKK1 inhibits PAI-1 gene expression in kidney tubular epithelia in vivo. Given that TGF-β1 activates β-catenin in tubular epithelial cells, this signal pathway likely plays an important role in mediating TGF-β1-triggered PAI-1 induction. Collectively, our present study provides significant and novel insights into the mechanism governing PAI-1 gene regulation. Furthermore, these results suggest that PAI-1 induction could play a critical role in mediating the fibrogenic action of Wnt/β-catenin signaling.
Wnt/β-catenin signaling has been implicated in kidney development and the pathogenesis of various kinds of renal diseases (18, 22, 31, 35, 36). Wnt signal is transmitted through their receptors, the Frizzled (Fzd) family of proteins, and coreceptors, members of the low density lipoprotein receptor-related protein-5 or -6. Upon binding to their plasma membrane receptors, Wnt proteins induce a series of downstream signaling events, resulting in dephosphorylation of β-catenin. This leads to the stabilization of β-catenin, rendering it to translocate into the nuclei, where it binds to TCF/lymphoid enhancer-binding factor and recruits the coactivator and its associated transcriptional machinery to initiate the expression of the Wnt-targeted genes (37–39). We have recently demonstrated that the majority of 19 different Wnt genes are up-regulated with distinct patterns in the kidneys after obstructive injury, which leads to a dramatic accumulation and activation of β-catenin, predominantly in renal tubular epithelia (18). Such an activation of Wnt/β-catenin signaling in diseased kidney is clearly associated with the pathogenesis of renal fibrotic lesions (40), because inhibition of the canonical Wnt signaling by DKK1 reduces renal fibrosis in the mouse model of obstructive nephropathy. This notion is further strengthened by the present study demonstrating that PAI-1 is a transcriptional target of the Wnt/β-catenin signaling.
Our finding that Smad2 and Smad3 inhibit PAI-1 induction is quite surprising, in view of the fact that Smads are the principal downstream effectors of TGF-β1 signaling. Upon binding to its Type II and type I receptors, TGF-β1 triggers Smad2 and Smad3 phosphorylation at serine residues in the C termini, induces their association with common partner Smad4 and subsequently translocation into the nuclei, where they control the transcription of TGF-β1-responsive genes (17, 41). Although there are so-called CAGA boxes, also known as SBE, in the PAI-1 gene promoter region (42), overexpression of either Smad2 or Smad3 in tubular epithelial cells does not stimulate PAI-1 induction in response to TGF-β1. Rather, Smads significantly block endogenous PAI-1 induction (Fig. 1). This unexpected observation is confirmed by knocking down of endogenous Smads using siRNA strategy (Fig. 2). The exact reasons behind the discrepancy between this study and previous reports regarding the role of Smads in PAI-1 expression are unknown. It could simply reflect the differences in cell types and cellular contexts, because Smad signaling is well known for its ability to cross-talk with other signal inputs (43, 44). It should be pointed out that many previous studies on PAI-1 gene regulation solely rely on a highly artificial system to perform functional analysis of its promoter, in which multiple (often 9–12) copies of SBE were linked to a heterologous promoter (42, 45). It remains an open question whether this kind of artificial promoter-reporter assay truly imitates the endogenous gene regulation. On the contrary, the present study demonstrates that endogenous PAI-1 gene expression in tubular epithelial cells is suppressed by Smad signaling.
TGF-β1 is capable of activating multiple downstream signal pathways, including Smad, PI3K, MAPK, and β-catenin. Besides Smad signaling, PI3K also negatively regulates PAI-1 expression, because its inhibition leads to an exaggerated PAI-1 induction in response to TGF-β1 stimulation (Fig. 3). This is consistent with earlier reports showing that PI3K inhibits PAI-1 gene expression in vascular endothelial cells and adipocytes (46, 47). Therefore, it appears clear that some branches of the TGF-β1 downstream signals, such as Smad and PI3K, actually inhibit PAI-1 induction in tubular epithelial cells. Of note, in addition to β-catenin signaling, p38 MAPK and JNK, but not ERK-1/2, are imperative in stimulating PAI-1 expression (Fig. 3). Because the PAI-1 gene promoter also harbors a functional AP1 site, it is not surprising that MAPK signal pathway also plays a role in regulating PAI-1 induction after TGF-β1 stimulation. Taken together, multiple negative and positive signal pathways downstream of TGF-β1 participate in PAI-1 gene regulation, creating a multifaceted regulatory system with great complexity and versatility.
Despite a negative regulation by Smad and PI3K signals, the net effect of TGF-β1 stimulation in tubular epithelial cells is a dramatic induction of PAI-1 expression, underscoring the strength of the positive signals elicited by β-catenin, p38 MAPK, and JNK for PAI-1 induction. In tubular epithelial cells overexpressing β-catenin, inhibition of p38 MAPK or JNK also repressed PAI-1 expression under both basal and TGF-β1-stimulated conditions, suggesting that β-catenin and these MAPK signals may cross-talk and work in concert to up-regulate PAI expression (48). In this regard, it is of interest to point out that p38 MAPK has been shown to directly phosphorylate GSK-3β at its C terminus, leading to its inactivation and subsequent β-catenin stabilization and accumulation (49, 50). Therefore, β-catenin could theoretically mediate the action of p38 MAPK in inducing PAI-1 expression after TGF-β1 stimulation. Of note, we cannot exclude the possibility that a noncanonical pathway of Wnt may also play a role in regulating PAI-1 expression through activation of p38 MAPK and JNK signaling (51–53).
In summary, we have shown herein that PAI-1 is a direct transcriptional target of the canonical pathway of Wnt/β-catenin signaling. By inducing this powerful fibrogenic factor, Wnt/β-catenin signaling plays a critical role in promoting the development and progression of renal fibrotic lesions. In addition, this signaling is known to induce the expression of numerous fibrosis-related genes such as fibronectin, Snail, and Twist in kidney cells (18, 22). In this context, it is desirable to develop feasible and specific strategies to constrain Wnt/β-catenin signaling in vivo and to test their ability to inhibit the progression of chronic kidney diseases.
This work was supported, in whole or in part, by National Institutes of Health Grants DK061408, DK064005, and DK071040.
- PAI-1
- plasminogen activator inhibitor-1
- TGF
- transforming growth factor
- ChIP
- chromatin immunoprecipitation
- UUO
- unilateral ureter obstruction
- TCF
- T cell factor
- TBE
- TCF-binding element
- SBE
- Smad-binding element
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphatidylinositol 3-kinase
- ERK
- extracellular signal-regulated protein kinase
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- MEK
- MAPK/ERK kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Eddy A. A., Fogo A. B. (2006) J. Am. Soc. Nephrol. 17, 2999–3012 [DOI] [PubMed] [Google Scholar]
- 2.Ma L. J., Fogo A. B. (2009) Front. Biosci. 14, 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagamine Y. (2008) Thromb. Haemost. 100, 1007–1013 [PubMed] [Google Scholar]
- 4.Hamano K., Iwano M., Akai Y., Sato H., Kubo A., Nishitani Y., Uyama H., Yoshida Y., Miyazaki M., Shiiki H., Kohno S., Dohi K. (2002) Am. J. Kidney Dis. 39, 695–705 [DOI] [PubMed] [Google Scholar]
- 5.Paueksakon P., Revelo M. P., Ma L. J., Marcantoni C., Fogo A. B. (2002) Kidney Int. 61, 2142–2148 [DOI] [PubMed] [Google Scholar]
- 6.Eddy A. A. (2002) Am. J. Physiol. Renal. Physiol. 283, F209–F220 [DOI] [PubMed] [Google Scholar]
- 7.Segarra A., Chacón P., Martinez-Eyarre C., Argelaguer X., Vila J., Ruiz P., Fort J., Bartolomé J., Camps J., Moliner E., Pelegrí A., Marco F., Olmos A., Piera L. (2001) J. Am. Soc. Nephrol. 12, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 8.Eddy A. A. (2009) Thromb. Haemost. 101, 656–664 [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholas S. B., Aguiniga E., Ren Y., Kim J., Wong J., Govindarajan N., Noda M., Wang W., Kawano Y., Collins A., Hsueh W. A. (2005) Kidney Int. 67, 1297–1307 [DOI] [PubMed] [Google Scholar]
- 10.Oda T., Jung Y. O., Kim H. S., Cai X., López-Guisa J. M., Ikeda Y., Eddy A. A. (2001) Kidney Int. 60, 587–596 [DOI] [PubMed] [Google Scholar]
- 11.Kitching A. R., Kong Y. Z., Huang X. R., Davenport P., Edgtton K. L., Carmeliet P., Holdsworth S. R., Tipping P. G. (2003) J. Am. Soc. Nephrol. 14, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 12.Matsuo S., López-Guisa J. M., Cai X., Okamura D. M., Alpers C. E., Bumgarner R. E., Peters M. A., Zhang G., Eddy A. A. (2005) Kidney Int. 67, 2221–2238 [DOI] [PubMed] [Google Scholar]
- 13.Huang Y., Border W. A., Lawrence D. A., Noble N. A. (2009) Am. J. Physiol. Renal Physiol 297, F1045–F1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura S., Nakamura I., Ma L., Vaughan D. E., Fogo A. B. (2000) Kidney Int. 58, 251–259 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y. (2006) Kidney Int. 69, 213–217 [DOI] [PubMed] [Google Scholar]
- 16.Böttinger E. P. (2007) Semin. Nephrol. 27, 309–320 [DOI] [PubMed] [Google Scholar]
- 17.Massagué J., Gomis R. R. (2006) FEBS Lett. 580, 2811–2820 [DOI] [PubMed] [Google Scholar]
- 18.He W., Dai C., Li Y., Zeng G., Monga S. P., Liu Y. (2009) J. Am. Soc. Nephrol. 20, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eguchi M., Nguyen C., Lee S. C., Kahn M. (2005) Med. Chem. 1, 467–472 [DOI] [PubMed] [Google Scholar]
- 20.Tan R., He W., Lin X., Kiss L. P., Liu Y. (2008) Am. J. Physiol. Renal. Physiol. 294, F1076–F1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semënov M. V., Tamai K., Brott B. K., Kühl M., Sokol S., He X. (2001) Curr. Biol. 11, 951–961 [DOI] [PubMed] [Google Scholar]
- 22.Dai C., Stolz D. B., Kiss L. P., Monga S. P., Holzman L. B., Liu Y. (2009) J. Am. Soc. Nephrol. 20, 1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Kang Y. S., Dai C., Kiss L. P., Wen X., Liu Y. (2008) Am. J. Pathol. 172, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zawel L., Dai J. L., Buckhaults P., Zhou S., Kinzler K. W., Vogelstein B., Kern S. E. (1998) Mol. Cell 1, 611–617 [DOI] [PubMed] [Google Scholar]
- 25.Tan X., Wen X., Liu Y. (2008) J. Am. Soc. Nephrol. 19, 1741–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Tan X., Dai C., Stolz D. B., Wang D., Liu Y. (2009) J. Am. Soc. Nephrol. 20, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., Hill C. S. (2002) Mol. Pharmacol. 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 28.Yang J., Dai C., Liu Y. (2005) J. Am. Soc. Nephrol. 16, 68–78 [DOI] [PubMed] [Google Scholar]
- 29.Schnaper H. W., Jandeska S., Runyan C. E., Hubchak S. C., Basu R. K., Curley J. F., Smith R. D., Hayashida T. (2009) Front. Biosci. 14, 2448–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai C., Yang J., Liu Y. (2003) J. Biol. Chem. 278, 12537–12545 [DOI] [PubMed] [Google Scholar]
- 31.Surendran K., Schiavi S., Hruska K. A. (2005) J. Am. Soc. Nephrol. 16, 2373–2384 [DOI] [PubMed] [Google Scholar]
- 32.Dai C., Yang J., Bastacky S., Xia J., Li Y., Liu Y. (2004) J. Am. Soc. Nephrol. 15, 2637–2647 [DOI] [PubMed] [Google Scholar]
- 33.Dai C., Yang J., Liu Y. (2002) J. Am. Soc. Nephrol. 13, 411–422 [DOI] [PubMed] [Google Scholar]
- 34.Niehrs C. (2006) Oncogene 25, 7469–7481 [DOI] [PubMed] [Google Scholar]
- 35.Surendran K., McCaul S. P., Simon T. C. (2002) Am. J. Physiol. Renal. Physiol. 282, F431–F441 [DOI] [PubMed] [Google Scholar]
- 36.von Toerne C., Schmidt C., Adams J., Kiss E., Bedke J., Porubsky S., Gretz N., Lindenmeyer M. T., Cohen C. D., Gröne H. J., Nelson P. J. (2009) Am. J. Transplant 9, 2223–2239 [DOI] [PubMed] [Google Scholar]
- 37.Huang H., He X. (2008) Curr. Opin. Cell Biol. 20, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White B. D., Nguyen N. K., Moon R. T. (2007) Curr. Biol. 17, R923–R925 [DOI] [PubMed] [Google Scholar]
- 39.Angers S., Moon R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 40.Liu Y. (2010) J. Am. Soc. Nephrol. 21, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 42.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Yang J., Li Y., Liu Y. (2005) Am. J. Physiol. Renal. Physiol. 288, F16–F26 [DOI] [PubMed] [Google Scholar]
- 44.Hua X., Liu X., Ansari D. O., Lodish H. F. (1998) Genes Dev. 12, 3084–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Z., Lam K. S., Wang N., Xu X., Cortes M., Andersen B. (2006) Oncogene 25, 2920–2930 [DOI] [PubMed] [Google Scholar]
- 46.Mukai Y., Wang C. Y., Rikitake Y., Liao J. K. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H1937–H1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venugopal J., Hanashiro K., Nagamine Y. (2007) J. Cell. Biochem. 101, 369–380 [DOI] [PubMed] [Google Scholar]
- 48.Bikkavilli R. K., Malbon C. C. (2009) Commun. Integr. Biol. 2, 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thornton T. M., Pedraza-Alva G., Deng B., Wood C. D., Aronshtam A., Clements J. L., Sabio G., Davis R. J., Matthews D. E., Doble B., Rincon M. (2008) Science 320, 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bikkavilli R. K., Feigin M. E., Malbon C. C. (2008) J. Cell Sci. 121, 3598–3607 [DOI] [PubMed] [Google Scholar]
- 51.Caverzasio J., Manen D. (2007) Endocrinology 148, 5323–5330 [DOI] [PubMed] [Google Scholar]
- 52.Saadeddin A., Babaei-Jadidi R., Spencer-Dene B., Nateri A. S. (2009) Mol. Cancer Res. 7, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 53.Fukukawa C., Nagayama S., Tsunoda T., Toguchida J., Nakamura Y., Katagiri T. (2009) Oncogene 28, 1110–1120 [DOI] [PubMed] [Google Scholar]