Abstract
Integrin αvβ8 is a critical regulator of transforming growth factor β activation in vasculogenesis during development, immune regulation, and endothelial/epithelial-mesenchymal homeostasis. Recent studies have suggested roles for integrin β8 in the pathogenesis of chronic obstructive pulmonary disease, brain arteriovenous malformations, and select cancers (Araya, J., Cambier, S., Markovics, J. A., Wolters, P., Jablons, D., Hill, A., Finkbeiner, W., Jones, K., Broaddus, V. C., Sheppard, D., Barzcak, A., Xiao, Y., Erle, D. J., and Nishimura, S. L. (2007) J. Clin. Invest. 117, 3551–3562; Su, H., Kim, H., Pawlikowska, L., Kitamura, H., Shen, F., Cambier, S., Markovics, J., Lawton, M. T., Sidney, S., Bollen, A. W., Kwok, P. Y., Reichardt, L., Young, W. L., Yang, G. Y., and Nishimura, S. L. (2010) Am. J. Pathol. 176, 1018–1027; Culhane, A. C., and Quackenbush, J. (2009) Cancer Res. 69, 7480–7485; Cambier, S., Mu, D. Z., O'Connell, D., Boylen, K., Travis, W., Liu, W. H., Broaddus, V. C., and Nishimura, S. L. (2000) Cancer Res. 60, 7084–7093). Here we report the first identification and characterization of the promoter for ITGB8. We show that a SP binding site and a cyclic AMP response element (CRE) in the ITGB8 core promoter are required for its expression and that Sp1, Sp3, and several AP-1 transcription factors form a complex that binds to these sites in a p38-dependent manner. Furthermore, we demonstrate the requirement for Sp3, ATF-2, and p38 for the transcription and protein expression of integrin β8. Additionally, reduction of SP3 or inhibition of p38 blocks αvβ8-mediated transforming growth factor β activation. These results place integrin β8 expression and activity under the control of ubiquitous transcription factors in a stress-activated and pro-inflammatory pathway.
Keywords: AP-1 Transcription Factor, Integrin, p38, Promoters, Sp1, Transforming Growth Factor β (TGFβ)
Introduction
Integrins are a large family of cell surface molecules mediating diverse biologic roles in angiogenesis, vasculogenesis, lymphoid trafficking, immune cell function, and cancer cell growth and metastasis (5, 6). Integrins consist of a single α and a single β subunit, forming 24 known heterodimers and can be subclassified according to subunit preference (7). The αv subunit-containing subfamily consists of five members: αvβ1, αvβ3, αvβ5, αvβ6, and αvβ8. The integrin αvβ8 is of particular interest because it is the only αv subunit-containing integrin that has been shown to be essential for development (8–14). The integrin β8 subunit is expressed in various epithelial (squamous mucosa, airway epithelium, renal tubular epithelium, and neuroepithelium) (1, 15–17), mesenchymal (fibroblasts, kidney mesengial cells, and astrocytes) (18, 19), and select immune cell types (splenocytes, CD44+ T-cells, and dendritic cells) (20). Integrin β8 functions to mediate cross-talk between immune cell types or mesenchymal cells with epithelial or endothelial cells (15, 17, 20, 21).
In adult tissues, increased expression of β8 has been shown in airway fibroblasts in lung tissue from chronic obstructive pulmonary disease patients and its expression is reduced in adenocarcinomas of the lung and in perivascular astrocytes in brain arteriovenous malformations (1, 2, 4). ITGB8 has been found to be increased in BRCA-1 positive breast cancers and has been identified as part of a six-gene expression signature predicting lung metastasis from breast cancer (3, 22).
The main, if not the only, physiologically relevant ligand for integrin αvβ8 is transforming growth factor β (TGF-β),3 a multifunctional cytokine that regulates cell proliferation, differentiation, survival, matrix synthesis, and immune cell function (23). TGF-β is ubiquitously expressed in three isoforms in mammals (TGF-β 1–3), but is maintained in an inactive form by its non-covalent interaction with its propeptide, the latency associated peptide of TGF-β (24). The integrin αvβ8 binds to the integrin-recognition motif (RGD) of the latency associated peptide of TGF-β and mediates the activation of TGF-β1 and -3 (24). Overlapping phenotypes of mice deficient for itgb8 with a variety of TGF-β deficiency models have revealed that integrin αvβ8 is essential for the in vivo activation of TGF-β1 and TGF-β3 during development (9, 20, 25–28). Thus, αvβ8 is a major “gatekeeper” of TGF-β function. Most of the functions of αvβ8, in vivo, including vascular differentiation (17), airway fibrosis in chronic obstructive pulmonary disease (1, 23, 29), cancer cell proliferation (4), and metastasis (19) have been attributed to αvβ8-mediated activation of TGFβ.
The mechanistic basis underlying the regulation of β8 expression during development and in normal and adult pathologic tissue remains largely unknown, as the genomic regions regulating ITGB8 have not been defined. Interestingly, a genetic variant in the ITGB8 locus has recently been found to correlate with risk of brain arteriovenous malformation development and the at-risk brain arteriovenous malformation variant was associated with reduced β8 protein expression (2). Here we describe the first characterization of the promoter and elements that regulate ITGB8 expression and place it under the control of p38, a stress-activated and pro-inflammatory protein.
EXPERIMENTAL PROCEDURES
Tissue Culture and Reagents
HeLa, H1264, U373, 293 cells, primary human lung fibroblasts, and human astrocytes were cultured in 10% fetal bovine serum (Invitrogen) in Dulbecco's modified Eagle's medium (Cellgro®, Mediatech, Inc., Manassas, VA) plus penicillin and streptomycin (University of California San Francisco Cell Culture Facility). Primary human bronchial epithelial cells (HBEC) were maintained in Bronchial Epithelial Growth Media (Lonza, Allendale, NJ) on collagen-coated dishes as previously described (1). HBEC, lung fibroblasts, and astrocytes were obtained using leftover tissues from human surgical specimens, in accordance with an approved IRB protocol, as described (30). The TGF-β reporter cell line, TMLC (gift from John Munger and Dan Rifkin, New York University, New York), was cultured as previously described (31). The chemical inhibitors of the p38 (SB202190) and extracellular-signaling response kinase (ERK) (PD98059) MAP kinase pathways were obtained from Calbiochem (EMD4 Biosciences, Gibbstown, NJ), whereas the inhibitor of the Jun N-terminal kinase (JNK) pathway (SP600125) was obtained from A.G. Scientific, Inc. (San Diego, CA). DTBP (dimethyl-3,3′-dithiobispropionimidate·2HCl) and 16% formaldehyde were purchased from Pierce (Rockford, IL).
5′ Rapid Amplification of cDNA Ends (RACE)
RNA was extracted from U373 and 293 cell lines using the RNeasy kit (Qiagen, Valencia, CA). 5′ RACE was performed using RNA from the above cells and human placental RNA from the SMARTTM RACE kit (Clontech, Mountain View, CA) according to the manufacturer's protocol. The ITGB8-specific primer used for second-strand synthesis and subsequent re-amplifications of fragments was GSP2j3 (5′-tgttgctggcatcccgagcccgagcttcctcccttgcc-3′). Amplified fragments were cloned using the TOPO-TA cloning kit (Invitrogen) and sequenced.
Construction of ITGB8 Promoter Reporter Plasmids
Genomic DNA from U373 cells was used as template for the PCR amplification of genomic sequences 5′ and into the first exon of ITGB8 and cloned into the secreted alkaline phosphatase (SEAP) reporter vector (Clontech). All other deletion constructs (−1280/+69; −973/+69; −694/+69; −623/+69; −491/+69; −330/+69; −1275/−494; −623/−494; −623/−336) were created utilizing restriction enzyme sites. Constructs were purified for transfection using the Qiagen endotoxin-free maxi-prep kit.
Transcription factor binding site mutant reporter constructs were made by site-directed PCR mutagenesis using the high fidelity Phusion DNA polymerase (Finnzymes USA, Woburn, MA) from −1280/+69 as template. A fragment spanning from the origin to the AflII site was amplified using the primers, 5′-ggtaccgagctcttacgcgtg-3′ and 5′-tttgatcttaagGctAaGcttcccagtaaacggaacaaaaagttacag-3′, which mutated the upstream putative CRE site to a HindIII site (mutations indicated by uppercase), then cloned into −1280/+69 to create CRE(−41)mt. DMRT5mt was created by amplifying a fragment between the AflII and BspEI sites using primers, 5′-cgcgggcttaagatcaaaagacccactAtGCAttgcaaaagccc-3′ and 5′-tcccggagcgactggccgagatttctcgctg-3′, which mutated the putative DMRT5 site to a NsiI site, then cloned into −1280/+69. E2Fmt was created by a similar amplification of the same region and cloning as for DMRT5mt using AflII and BspEI, but using the primers, 5′-gggcgccgcttaagatcaaaagacccactgtaacttgcaaaagc-3′ and 5′-tcccggagcgactggccgagattActAgTtgctcc-3′, which mutated the putative E2F site to a SpeI site. NFκBmt was created by amplifying a fragment between the BspE1 and BstAP1 sites in −1280/+69 using primers 5′-gagaaatctcggccagtcgctccggaaacagcccctg-3′ and 5′-ccccgcagctgctgcaGacaaagtggagtcaagggacctcGActgcccccaacgccc-3′, which mutated the putative NFκB/c-Rel site and created a PstI site upstream for identification purposes by changing a single nucleotide, then cloned into −1280/+69. ΔAP4 was created by a single cleavage of the BstAP1 site, Klenow-filled, then re-ligated, resulting in the deletion of “cagc” from the putative AP4 site. The downstream CRE(−626)mt was created by amplifying a fragment between the two SacII sites in −1280/+69 using primers, 5′-ttaaccgcggATatctcatgcctcaccaatgtcccgcccacgctgct-3′ and 5′-atataccgcgggggttcctgctcagcaggcgcagggcagcctctgtcat-3′, which mutated this putative CRE site to an EcoRV site, then cloned into −1280/+69. The SPmt was created using this same method as for CRE(626)mt, but using an alternative forward primer, 5′-atatccgcggtgacttcatgcctcaccaatgtcGAATTcacgctgctccgagctgt-3′, which mutated the putative SP site to an EcoRI site. All constructs were confirmed by sequencing.
Reporter Assays
SEAP reporter constructs were transfected into all cell types using the Lipofectamine 2000 reagent (Invitrogen). A Renilla luciferase reporter construct (Promega, Madison, WI) driven by the TK1 (thymidine kinase) promoter was co-transfected at a 1:50 molar ratio to the SEAP constructs to normalize for transfection efficiency. The medium was replaced on the cells 24 h after transfection then at 48 h both reporter assays were performed. SEAP (Phospha-Light detection system, Applied Biosystems, Foster City, CA) and luciferase (Renilla luciferase assay kit, Promega) were detected using the manufacturer's protocols. Raw data were normalized by subtracting the background signal from cells treated with Lipofectamine 2000 alone, then the resulting SEAP signal was divided by the Renilla luciferase signal. Each experiment was normalized to each other by dividing by the activity of pSEAP basic, which does not possess a promoter driving SEAP expression.
Nuclear Extraction and Electromobility Shift Assays (EMSA)
Nuclear extracts were obtained using the NE-PER kit (Pierce) with protease inhibitors (Protease Inhibitor Mixture Set 1, Calbiochem) and phosphatase inhibitors, 1 mm Na3VO4 and 10 mm NaF. Prior to the final step of the kit protocol, the nuclear extracts were subjected to three flash freeze-thaw cycles.
Both strands of each of the oligonucleotide probes (+ strands only: wild type, 5′-agtgtgggccgcggtgacttcatgcctcaccaatgtcccgcccacgct-3′; CRE/CAAT/SP triple mutant, 5′-agtgtgggccgcgCATGGCAGTtgcctcaGGCGACCAGACGTTacgct-3′; CRE/CAAT double mutant, 5′-agtgtgggccgcgCATGGCAGTtgcctcaGGCGAgtcccgcccacgct-3′; SP mutant, 5′-agtgtgggccgcggtgacttcatgcctcaccaatCCAGACGTTacgct-3′; wild type, consensus CRE (32), 5′-gcattacctcatcccgtgagccttcg-3′) were synthesized and PAGE-purified (Operon, Huntsville, AL), then annealed using a standard protocol (Sigma). Additionally, the wild type probe was biotinylated at the 5′ end of each strand prior to purification (Operon). Antibodies used for supershift analysis were all from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), anti-Sp1 (sc-59) or (sc-420X), anti-Sp3 (sc-644), anti-CREB-binding protein (sc-369X), anti-ATF-2 (sc-6233X), anti-C/EBPβ (sc-150X), anti-c-Jun (sc-1694X), anti-JunD (sc-74X), anti-JunB (sc-46X), anti-c-Fos (sc-52X), anti-Fra-1 (sc-605X), and anti-Fra-2 (sc-171X). Additional antibodies against ATF-2 (numbers 9226 and 9225, Cell Signaling, Danvers, MA) were used for confirmation.
EMSAs were performed using the non-radioactive, LightShift Chemiluminescent EMSA kit (Pierce) with slight modifications. 50 μg of nuclear extracts were first incubated with competitor probes (100 fmol) and/or antibodies (0.8–6 μg) in binding buffer (32) including 10 μg of bovine serum albumin for 45 min at room temperature. Then the wild type, biotinylated probe was added to the reactions (1 fmol) and incubated for an additional 15 min at room temperature. The reactions were subsequently electrophoresed through a 4% polyacrylamide, non-denaturing gel. The rest of the assay was performed according to the manufacturer's protocol (Pierce).
Plasmid and siRNA Transfections and RT-PCR
Control (catalog numberAM4641), SP1 (ID number 143158), and SP3 (ID number 115338) siRNAs were purchased from Ambion, Inc. (Applied Biosystems). The ATF2 (sc-29205) siRNA and control (sc-37007) were purchased from Santa Cruz Biotechnology, Inc. Additional siRNAs against SP3 (J-023096-09), ATF2 (J-009871-06), and Control (D-001810-10) were purchased from Dharmacon, Inc. (Lafayette, CO). These siRNAs were transfected into primary lung fibroblasts using the electroporation technique by Amaxa Nucleofector II (Amaxa Biosystems, Lonza). For transcript analysis, medium was replaced after 24 h and RNA was extracted at 48 h post-transfection. RNA extraction and cDNA synthesis were performed using the RNeasy and Quantitect kits (Qiagen), respectively. For surface protein extraction, medium was replaced after 24 h, then after 72 h the cells were stained for integrin β8 and analyzed by flow cytometry. The p38αDN and pcDNA constructs (gifts from Jiahuai Han, The Scripps Research Institute, La Jolla, CA) were transfected into lung fibroblasts using Amaxa Nucleofector II as described above. RNA was extracted from these cells 24 h post-transfection.
Quantitative RT-PCR was performed using SYBR Green, as described (33). Primer sequences for ACTB, GAPDH, SP1, SP3, and MAPK14 (p38α) were as described (1, 34–36). Two sets of primers were used to amplify ITGB8 (ITGB8, 5′-tggtcgaggagtttgtgtttg-3′ and 5′-agccactgaagcattggca-3′; hβ8, 5′-aggatcttctacccctcttgc-3′ and 5′-atctggacagatggcggtaatg-3′) and their results were averaged.
Standard RT-PCR was performed using the Taq Core PCR Kit (Qiagen) as previously described (21). The primers for ATF2 were 5′-gcatgttccagctgcagtccc-3′ and cttctgtatgtggactactcgg-3′ and ACTB (β-actin) were as described (15).
Flow Cytometry
Flow cytometry was performed as previously described (4) using antibodies against integrin β8 (clone 14E5 (21)).
Immunoblots
45 μg of nuclear extracts (isolated as described above) were immunoblotted as previously described (15). Commercially purchased antibodies were used against ATF-2 (dual-phospho-Thr69/71) (number 9225, Cell Signaling), HSP 27 (phospho-Ser78) (SPA-523, Assay Designs, Ann Arbor, MI), and Lamins A/C (sc-7292, Santa Cruz Biotechnology).
TGF-β Activation Assay
Fibroblasts were treated with SB202190 or transfected with p38αDN, as described above. After 48 h, the cells were washed, detached, and co-cultured with TGF-β reporter cells (TMLC) as previously described (15, 21) in the presence of control (clone W6/32, ATCC, Manassas, VA), pan-TGF-β (clone 1D11, ATCC), or integrin β8 antibodies (clone 37e1 (21)).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation assays were performed using the ChIP Assay Kit (17-295, Upstate/Millipore, Temecula, CA) according to the manufacturer's protocol, with modifications using the DTBP bifunctional cross-linker, essentially as described (37). Briefly, primary lung fibroblasts were treated with or without the p38 inhibitor, SB202190, for 2 h prior to cross-linking with 5 mm DTBP in cold phosphate-buffered saline for 15 min at 4 °C and inactivation (100 mm Tris, pH 8.0, 150 mm NaCl), followed by a second cross-linking step with 1% formaldehyde in phosphate-buffered saline for 15 min at 4 °C. Samples were immunoprecipitated with 10 μg of primary antibodies, as above, in addition to anti-Sp3 (07-107) antibody or normal rabbit IgG (PP64B) from Upstate/Millipore, overnight at 4 °C. After de-cross-linking, DNA was extracted (QIAquick PCR purification kit, Qiagen) and quantitative PCR was performed, as described above. Association of amplified fragments was normalized to input chromatin, then expressed relative to IgG. Primer pairs used were: P3 (5′-cccgtttactgggaaggt-3′, 5′-ccttaaccctggactgga-3′), P2 (5′-actggtccaactctccaa-3′, 5′-gcaaacccgaatctctca-3′), and P1 (5′-cgatgggaatggcaactat-3′, 5′-caggagcagcagtaacag-3′).
Statistical Analysis
All statistics performed were by comparing two sets of values using the Students' t test in the graphing and statistical analysis software program, PrismTM 4 (GraphPad Software, Inc., La Jolla, CA); * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
RESULTS
Characterization of the Transcriptional Start Site of ITGB8
To determine the transcriptional start site (TSS) of ITGB8, the 5′ end of the transcript was mapped by 5′ RACE (Fig. 1A). These experiments indicate that the four most 5′ TSS are −410, −404, −236, and −217, from the published TSS for ITGB8 (located on the plus strand of human chromosome 7 at nucleotide 20,337,250) (38). This result extends the published 5′-untranslated region to a total length of 1,115 nucleotides. Sequence analysis indicates that no TATA box is present in 1,213 bp of the 5′-flanking sequence to the most 5′ TSS. A CCAAT site is located 28 bp upstream of the most 5′ TSS. This newly identified TSS includes a close match to the consensus initiator (INR) element YYANWYY, containing the critical adenosine as the +1 nucleotide (39, 40). Hereto forward, the newly identified TSS will be designated as +1. ESTs (DA238257.1 and DA994035.1) representing 5′ oligo-capped mRNA sequences from a 5′ end-enriched cDNA library from brain tissue show transcripts that map +14 and +43 relative to our newly identified TSS, respectively.
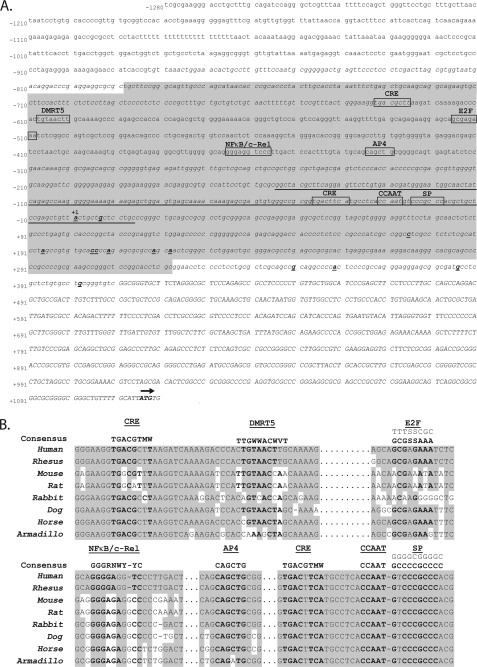
FIGURE 1.
Characterization of the 5′ TSS of the ITGB8 gene with surrounding sequence analysis. A, location of the TSS and other sequence features 5′ of the ITGB8 gene. The published ITGB8 5′ untranslated region is indicated in capital letters. The start codon is labeled in bold with an arrow above. The 5′ RACE results from the TSS are labeled in bold and underlined. The most 5′ of these results is labeled as +1 (above), determining the numbering for the presented sequence. Predicted CpG islands are represented in italics. The region of significant homology across 28 mammalian species is underlined. The predicted, overlapping promoter regions are highlighted in light gray. Putative transcription factor binding sites are boxed and labeled above. B, homology of putative transcription factor binding sites of interest in the sequences 5′ of the ITGB8 gene across eight placental mammalian species. Consensus binding sites for the transcription factors are indicated above, the E2F consensus includes both strands in the 5′-3′ orientation. Homologous bases are highlighted in gray.
Characterization of the ITGB8 Core Promoter
In silico analysis of sequences 5′ of the TSS for ITGB8 identified two overlapping promoter regions, one from −779 to −635 (McPromoter (41)) and one from −725 to +322 (Dragon Genome Explorer (42)) (Fig. 1A). Two separate CpG islands, which are commonly found in promoters, also overlap with these predicted promoter regions (−882 to −625; −311 to +1115) (UCSC Genome Browser (43)).
Sequence analysis for homology across mammalian species (rankVista (44)) indicates that a region in the 5′ flanking sequence of ITGB8 (−154 to +13, p = 0.00032) shows significant interspecies homology. Putative transcription factor binding sites (MatInspector and Genomatix (45)) were found in and around these areas of conservation including putative cyclic AMP responsive elements (CRE; −626 and −41), doublesex-mab3-related transcription factor-5 (DMRT5) (−601), E2F (−509), NFκB/c-Rel (−357), activator protein-4 (AP4) (−335), and the Sp1 family (−19) (Fig. 1A). Homology across eight placental mammalian species is illustrated in Fig. 1B. The putative CRE, CCAAT, and SP binding sites close to the TSS exhibit the region of highest interspecies homology and are absolutely conserved among these eight species (Fig. 1B).
The Core Promoter of the ITGB8 Gene Is from −493 to +69 bp Relative to the TSS
Promoter reporter assays were performed using deletion constructs containing 5′ sequences from −1280 to +69 relative to the ITGB8 TSS (Fig. 2A). Cell lines derived from cell types that normally express integrin αvβ8 initially used for promoter assays were U373 (glioblastoma), H1264 (lung carcinoma), and HeLa (cervical carcinoma) (data not shown) (1). All three cell lines demonstrated expression of the longest construct, indicating that the −1280 to +69 region contains promoter activity (Fig. 2A). This construct was also expressed in cells that normally do not express integrin β8, human microvascular endothelial cells-1, or primary dermal fibroblasts, suggesting that this region does not contain elements for cell type-specific expression of integrin β8 (data not shown). In H1264, U373, and HeLa cells, the 5′-deletion construct encompassing −491 to +69 had similar activity as the full-length construct, demonstrating that this region contains the core ITGB8 promoter (Fig. 2A). In HeLa cells, the construct −330 to +69 maintained the same low level of activity as the longer 5′-deletion constructs but showed no activity in H1264 or U373 cells, indicating that this region of the core promoter has activity in specific cell types (Fig. 2A).
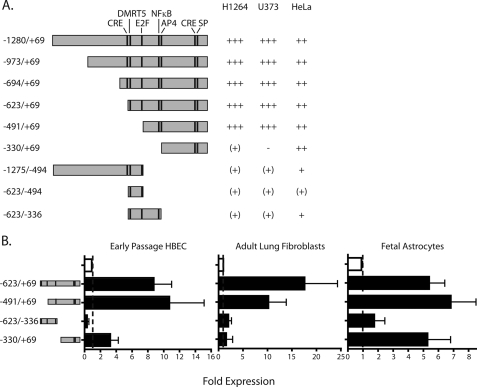
FIGURE 2.
Identification of the ITGB8 core promoter in cell lines and select primary cells that endogenously express integrin β8. A, ITGB8 reporter constructs and activities in H1264, U373, and HeLa cell lines. −, zero; (+), <2-fold; +, 2–3-fold; ++, >3–8-fold; +++, >8-fold increase in activity, relative to background of the basic reporter construct containing no promoter elements. The constructs are drawn to scale with the location of putative transcription factor binding sites of interest indicated with a line. B, ITGB8 reporter assay results in primary cells, HBEC, adult lung fibroblasts, and fetal astrocytes using select constructs are indicated to the left (± S.E.). Baseline is indicated by a line. The empty bar is baseline, pSEAP basic, whereas filled bars represent relative activity of reporter constructs indicated at the left.
When the activities of four constructs, −623/+69, −491/+69, −623/−336, and −330/+69, were tested in primary cells that normally express αvβ8, HBEC, adult lung fibroblasts, and fetal astrocytes, the core promoter activity of construct, −491/+69, was confirmed (Fig. 2C). The smaller 5′-deletion construct, −330/+69, maintained high expression in fetal astrocytes, supporting that this smaller segment of the core promoter is sufficient to drive expression in specific cell types (Fig. 2C).
The CRE and SP Binding Site in the ITGB8 Core Promoter Are Required to Drive Expression
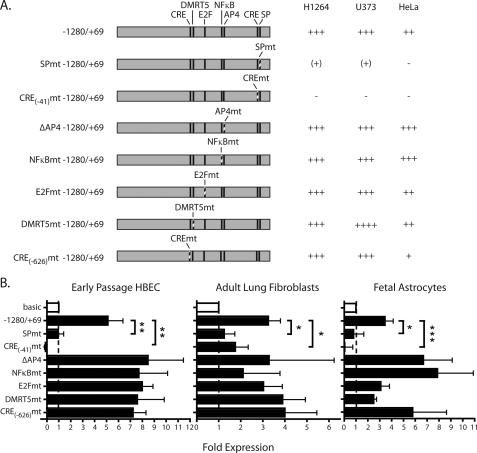
There are four putative transcription factor binding sites with high degrees of interspecies conservation within the ITGB8 core promoter, NFκB/c-Rel, AP4, CRE, and SP (Fig. 1B). We made reporter constructs with mutations in each of these sites (Fig. 3A). Mutations in the CRE(−41) or SP binding site significantly reduced or completely abrogated expression in both cell lines and primary cells (Fig. 3, A and B). Mutation in the NFκB/c-Rel site or deletion of the AP4 site, individually or combined, did not alter reporter expression (Fig. 3, A and B, data not shown). We also performed mutational analysis of CRE(−626), DMRT5, and E2F putative transcription factor binding sites upstream of the core promoter and were predicted by in silico analysis (Fig. 1). Mutations in the CRE(−626), DMRT5, or E2F binding sites did not significantly change reporter expression (Fig. 3). These data indicate that the CRE(−41) and SP binding site located in the core promoter (−491/+69) are required for ITGB8 promoter activity.
FIGURE 3.
The SP and CRE binding sites are required for ITGB8 promoter activity. A, mutant ITGB8 reporter constructs and activities in H1264, U373, and HeLa cell lines. All constructs are drawn to scale with mutated sites indicated by dashed lines. Fold-induction is as defined as described in the legend to Fig. 2. B, mutant ITGB8 reporter assays in primary HBEC, adult lung fibroblasts, and fetal astrocytes (±S.E.). Baseline is indicated by a line. The empty bar is baseline, pSEAP basic, whereas filled bars represent relative activity of mutant reporter constructs indicated at the left. * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
Specific Complexes Form at the CRE and SP Sites in the Core Promoter of ITGB8
EMSAs using a probe corresponding to the sequence containing the CRE, CCAAT, and SP binding sites in the ITGB8 core promoter were performed in nuclear extracts from primary adult lung fibroblasts (Fig. 4A). Similar binding patterns were observed in nuclear extracts from primary HBEC (data not shown). At least six migrating bands were identified with this probe (Fig. 4A, lane 1). We determined that the slowest migrating five bands are specific protein-DNA complexes, because the wild type competitor completely displaced all five bands (Fig. 4A, lane 2, and Fig. 5B, lane 2). A competitor mutated in the CRE and CCAAT binding sites displaced most of the complexes from bands 1, 2, and all from band 5, but did not disrupt complexes in bands 3 and 4 (Fig. 4A, lane 3). In contrast, a competitor with a wild type consensus CRE (32) displaced bands 3 and 4, but not bands 1, 2, and 5 (Fig. 4A, lane 4). Similarly, a competitor mutated in the SP site also displaced bands 3 and 4, but not bands 1, 2, and 5 (Fig. 4A, lane 5). A competitor mutated for all three sites, CRE, CCAAT, and SP did not displace any bands, confirming that complexes in bands 1–5 are specific to these sites (Fig. 4A, lane 6). These results demonstrate that specific complexes form at the CRE site (bands 3 and 4) and SP site (bands 1, 2, and 5) in the ITGB8 core promoter.
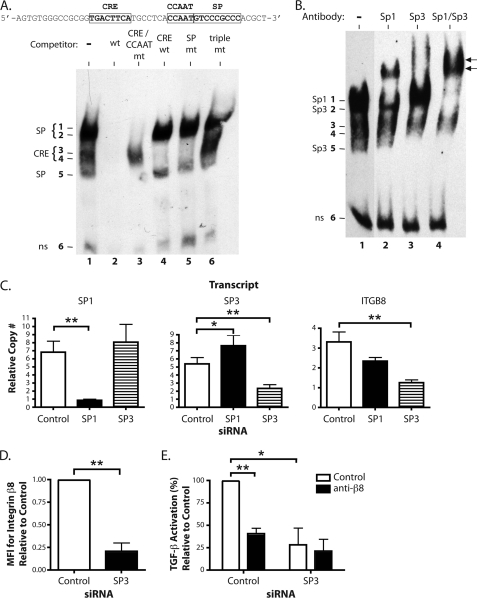
FIGURE 4.
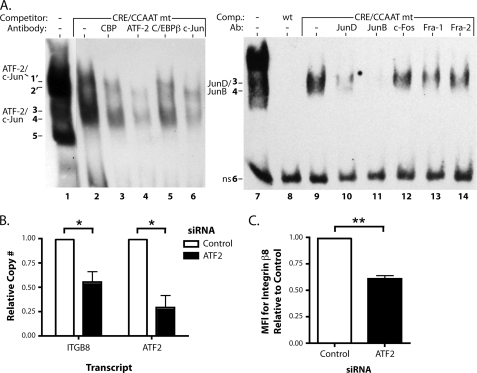
Sp3 is required for ITGB8 expression and αvβ8-mediated TGF-β activation. A, EMSA in adult lung fibroblasts using a biotinylated probe (sequence indicated above) covering the CRE, CCAAT, and SP transcription factor binding sites in the core promoter with or without the indicated unlabeled competitors. Wt is the unmutated competitor. CRE/CCAAT mt, SP, or triple mt indicates mutations in each or all of the indicated sites. CRE wt is an unrelated CRE consensus site competitor (32). The figure is a composite from a single representative gel. B, EMSA supershift analysis using polyclonal antibodies against Sp1, Sp3, or Sp1 and Sp3 combined. Supershifted complexes are indicated by arrows. The figure is a composite from a single representative gel. C, quantitative RT-PCR results for SP1, SP3, and the ITGB8 in adult lung fibroblasts transfected with siRNA against SP1, SP3, or control (± S.E.). The measured transcript is labeled above each respective graph. D, flow cytometry for surface expression of the integrin β8 subunit on adult lung fibroblasts transfected with siRNA against SP3 or control (± S.E.). MFI, mean fluorescence intensity. E, TGF-β activation assays of adult lung fibroblasts treated with siRNA against SP3 or control using control or anti-β8 blocking antibodies (± S.E.). * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
FIGURE 5.
AP-1 complexes interact with the ITGB8 promoter and ATF-2 regulates ITGB8 expression. A, EMSA supershift analysis using polyclonal antibodies against CRE binding and AP-1 transcription factors. Addition of the CRE/CCAAT double mutant competitor was necessary to displace the Sp1/Sp3 complexes for easier visualization of bands 3 and 4. Bands co-migrating and not displaced by the CRE/CCAT competitor are indicated by 1′ and 2′. Supershifts in the left panel were performed with antibodies against CBP, ATF-2, C/EBPβ, and c-Jun and on the right panel with antibodies against JunD, JunB, c-Fos, Fra-1, and Fra-2. Each panel is a composite from a single representative gel. B, quantitative RT-PCR for ITGB8 and ATF2 from adult lung fibroblasts transfected with an siRNA against ATF2 (± S.E.). C, flow cytometry for surface levels of integrin β8 on adult lung fibroblasts transfected with the same siRNA against ATF2 as in B (± S.E.). MFI, mean fluorescence intensity. * = p ≤ 0.05; ** = p ≤ 0.01.
Sp1 and Sp3 Bind to the Putative SP Binding Site in the Core Promoter of ITGB8
The most abundant DNA binding complexes in the EMSAs using the CRE/CCAAT/SP probe are bands 1 and 2, which are specific to the SP site. Antibodies against Sp1 and Sp3 were used to supershift these complexes (Fig. 4B). Sp1 is contained in the complex in band 1 as shown by its supershift using antibodies against Sp1 (Fig. 4B, lane 2). Sp3 is contained in complexes in bands 2 and 5 as shown by their supershifts using antibodies against Sp3 (Fig. 4B, lane 3). Three bands, 1, 2, and 5, were supershifted using antibodies against both Sp1 and Sp3 together, confirming that these complexes contain Sp1 and Sp3 (Fig. 4B, lane 4). These data show that Sp1 and Sp3 from nuclear extracts of lung fibroblasts can bind to the SP binding site in the core promoter of ITGB8.
Sp3 Is Required for Basal Expression of ITGB8
Primary human adult lung fibroblasts were treated with siRNAs against SP1, SP3, or a control, and analyzed by quantitative RT-PCR for integrin β8 transcript levels. As shown in Fig. 4C, significant reductions of SP1 or SP3 transcripts were obtained by treatment with their respective siRNAs. For cells treated with siRNA against SP1, SP3 transcript levels were significantly increased, whereas treatment with siRNA against SP3 did not alter SP1 levels (Fig. 4C, left and middle panels). Although siRNA against SP1 showed a trend toward reduction in ITGB8 transcript levels, only siRNA against SP3 significantly reduced ITGB8 levels (Fig. 4C, right panel). This result was confirmed using a second siRNA against SP3, which resulted in a 61% reduction in SP3 transcript levels (p = 0.001) and a 31.5% reduction in ITGB8 transcript levels (p < 0.0001). This reduction in ITGB8 transcript levels by siRNA against SP3 was also observed at the protein level as determined by flow cytometry (Fig. 4D). These data show that Sp3 is required for basal expression of integrin β8 in primary adult lung fibroblasts.
Sp3 Is Required for Integrin αvβ8-mediated TGF-β Activation by Lung Fibroblasts
Primary adult lung fibroblasts were transfected with siRNAs against SP3 or control, then subjected to a TGF-β activation assay using blocking antibodies against integrin β8 (Fig. 4E). As shown in Fig. 4E, TGF-β activation was reduced by 59% in the presence of blocking antibodies against integrin β8 using control siRNA-transfected cells. Total TGF-β activation was significantly reduced by 72% in cells transfected with siRNA against SP3. Addition of integrin β8 blocking antibodies had only a negligible effect on SP3 siRNA-transfected cells. These results indicate that Sp3 is required for transcriptional regulation of integrin β8 expression, which in turn regulates αvβ8-mediated TGF-β activation by lung fibroblasts.
AP-1 Complexes Bind to the Putative CRE Binding Site in the Core Promoter of ITGB8
EMSA competition experiments using the CRE/CCAAT mutant competitor with the CRE/CCAAT/SP probe revealed two faint bands that co-migrate with bands 1 and 2, suggesting that these bands represent complexes that are also specific to CRE or CCAAT, labeled 1′ and 2′ (Fig. 5A, lane 2). Thus, supershift analysis for CRE binding and AP-1 transcription factors was performed in the presence of the CRE/CCAAT mutant competitor for better visualization of bands 1′ and 2′ (Fig. 5A, lanes 2-6). Antibodies against either ATF-2 or c-Jun displaced band 1′ and partially displaced bands 3 and 4 (Fig. 5A, lanes 4 and 6). We hypothesize that the antibodies are binding to their protein targets in such a way that it causes their dissociation from the DNA, although it is also possible that these complexes are being supershifted, but are not detected in this non-radioactive EMSA. Longer exposures show a very faint, supershifted band in the lane with the extract containing c-Jun antibodies (data not shown). These data suggest that an AP-1 complex containing ATF-2 and c-Jun in nuclear extracts of lung fibroblasts bind to the CRE site in the core promoter of ITGB8.
Other AP-1 transcription factors were analyzed for their ability to bind to the CRE site by EMSA supershift analysis. Antibodies against JunD and JunB showed a significant reduction in the intensity of DNA-binding complexes in bands 3 and 4 (Fig. 5A, lanes 10 and 11). However, antibodies against c-Fos, Fra-1, and Fra-2 did not cause differences in the binding pattern (Fig. 5A, lanes 12–14). A slower migrating complex could be seen at longer exposures in some supershift experiments using antibodies against JunD or JunB (data not shown). In summary, these data suggest that AP-1 complexes containing JunD and JunB from nuclear extracts of lung fibroblasts bind to the CRE site in the core promoter of ITGB8.
ITGB8 Expression Is Regulated by ATF-2 and p38
ATF2 siRNA significantly reduced ATF2 levels (Fig. 5B), which led to the reduction of ITGB8 transcript levels by ∼45% (Fig. 5B). A second siRNA against ATF2 confirmed this result, with a 45% reduction in ATF2 transcript levels (p = 0.0048) and a 36% reduction in ITGB8 transcript levels (p < 0.0001). The reduction in ITGB8 transcript levels was functionally significant because a 40% reduction in integrin β8 protein levels was also observed in cells treated with siRNAs against ATF2 (Fig. 5C). These results suggest a role for an AP-1 complex containing ATF-2 in regulating integrin β8 expression.
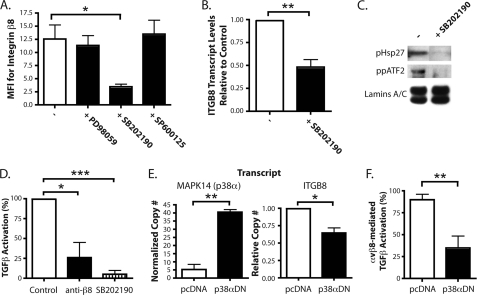
AP-1 is primarily regulated by the MAPK signaling pathways (46, 47). The MAPK pathways are classically divided into the ERK, JNK, and p38 pathways (48). To determine which MAPK signaling pathway may be involved in the regulation of integrin β8 expression through AP-1, primary adult lung fibroblasts were treated with chemical inhibitors to each of these pathways then analyzed for surface expression of integrin β8 using flow cytometry (Fig. 6A). Only the p38 inhibitor, SB202190, significantly reduced integrin β8 surface expression (Fig. 6A). A similar reduction by this inhibitor was also seen at the transcript level (Fig. 6B). SB202190 inhibited phosphorylation of a canonical target of p38, HSP 27, and also inhibited the dual phosphorylation of ATF-2, another known target (49) (Fig. 6C). These data suggest a role for p38 in regulating ITGB8 expression through activation of ATF-2 in lung fibroblasts.
FIGURE 6.
p38 regulates ITGB8 expression and αvβ8-mediated TGF-β activation. A, flow cytometry for integrin β8 on adult lung fibroblasts treated with MAPK inhibitors, PD98059 (ERK), SB202190 (p38), and SP600125 (JNK) (± S.E.). MFI, mean fluorescence intensity. B, quantitative RT-PCR results for ITGB8 expression in adult lung fibroblasts treated with SB202190, normalized to GAPDH and β-actin and relative to control (± S.E.). C, immunoblot for phosphorylated HSP 27 and dual-phosphorylated ATF-2 from nuclear extracts from adult lung fibroblasts treated ± SB202190. Immunoblot for the nuclear localized proteins, lamins A and C, was used as a loading control. D, TGF-β activation assays of adult lung fibroblasts treated with anti-β8 blocking antibodies or SB202190 (± S.E.). E, quantitative RT-PCR results for MAPK14 (p38α) and ITGB8 in adult lung fibroblasts transfected with plasmids expressing a p38α dominant-negative isoform (p38αDN) or the empty vector control, pcDNA (± S.E.). The measured transcript is labeled above each respective graph. F, TGF-β activation assays of adult lung fibroblasts transfected with plasmids expressing a p38α dominant-negative isoform (p38αDN) or the empty vector control, pcDNA. Percentage (%) of αvβ8-mediated TGF-β activation shown (± S.E.). * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
Integrin αvβ8-mediated TGF-β Activation by Lung Fibroblasts Is Regulated by the p38 Pathway
Primary adult lung fibroblasts were treated with SB202190 and subjected to a TGF-β activation assay (Fig. 6D). As shown in Fig. 6D, treatment with either the blocking antibody against αvβ8 or SB202190 significantly reduced TGF-β activation by 74 and 95%, respectively. Although the p38 inhibitor essentially abolished all of the TGF-β activation by these cells, this effect was not significantly different from the effect of integrin β8 blocking antibodies. To determine the specificity of the inhibitor, a plasmid expressing a dominant-negative isoform of p38α (p38αDN) was transfected into lung fibroblasts (50). Overexpression of p38αDN (Fig. 6E, left panel) caused a significant decrease in ITGB8 levels by ∼35% (Fig. 6E, right panel). Furthermore, overexpression of p38αDN also caused a 65% decrease in αvβ8-mediated TGF-β activation (Fig. 6F). These data suggest that p38α, through the regulation of integrin β8 expression, regulates αvβ8-mediated TGF-β activation by lung fibroblasts.
Interaction of an AP-1 Complex Containing ATF-2, c-Jun, and Sp3 with the ITGB8 Promoter Requires p38 Signaling
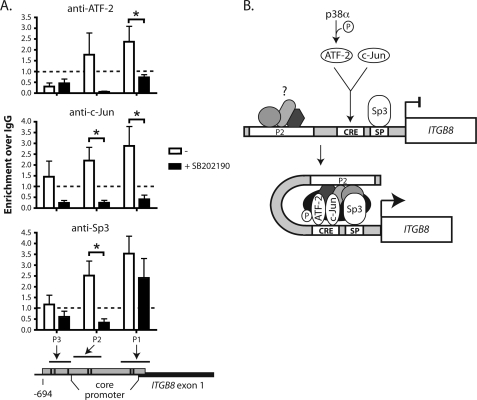
Chromatin immunoprecipitation assays using antibodies specific for the phosphorylated form of ATF-2 (pATF-2) or all forms of ATF-2, c-Jun, and Sp3 were performed on adult lung fibroblasts treated with the p38 inhibitor, SB202190, to assess their association with the ITGB8 promoter (Fig. 7A). Chromatin immunoprecipitates (pATF-2, ATF-2, c-Jun, or Sp3) from untreated cells increased PCR amplicons from the core promoter regions, P1 and P2, but not from P3, which is outside of the ITGB8 core promoter (Fig. 7A). Chromatin immunoprecipitates (pATF-2, ATF-2, or c-Jun) of fibroblasts treated with SB202190 had a reduced PCR signal, relative to untreated fibroblasts, at these ITGB8 promoter sites (Fig. 7A). These results demonstrate that ATF-2, c-Jun, and Sp3 are associated with the ITGB8 core promoter and form a higher order complex that is dependent on p38 signaling.
FIGURE 7.
ATF-2, c-Jun, and Sp3 association with the ITGB8 core promoter requires p38 signaling. A, chromatin immunoprecipitation of ATF-2, c-Jun, or Sp3 in adult lung fibroblasts treated with the p38 inhibitor, SB202190, with PCR amplification of regions from the ITGB8 promoter (± S.E.). Genomic locations of the amplicated regions are indicated in the schematic below the graphs. Essentially identical results were obtained using antibodies against phospho-ATF-2 and total ATF-2 and thus, results were pooled. B, hypothetical model for regulation of the ITGB8 promoter by p38, ATF-2, c-Jun, and Sp3. p38 phosphorylates ATF-2, which heterodimerizes with c-Jun to bind to the CRE site in the ITGB8 core promoter. Along with Sp3, which is already bound to its cognate SP site adjacent to the CRE site in the ITGB8 core promoter, these transcription factors form a higher-order chromatin complex that interacts either directly with the P2 region or indirectly through yet unidentified transcription factors to activate transcription of ITGB8. * = p ≤ 0.05.
DISCUSSION
Integrin β8 Expression and αvβ8-mediated TGF-β Activation Are Regulated by Sp3, AP-1, and the p38 Pathway
Here we report the first identification and characterization of the promoter for human ITGB8, a gene that is crucial for the regulation of TGF-β function in vivo. We identify and characterize two essential cis-acting promoter elements, one that binds Sp1 and Sp3 and one that binds an AP-1 transcriptional complex containing ATF-2. Both Sp3 and ATF-2 are required for efficient ITGB8 transcription and protein expression. We show that the regulation of ITGB8 expression is dependent on p38, a MAPK upstream of ATF-2. Finally, we demonstrate that Sp3 or p38, through regulation of integrin β8 expression, regulates αvβ8-mediated TGF-β activation. Altogether, these results suggest a mechanistic basis for the regulation of αvβ8-mediated TGF-β activation by stress-mediated and pro-inflammatory signaling pathways.
The Identification of the ITGB8 Core Promoter
The ITGB8 core promoter contains, in close proximity, a CCAAT, CRE, and SP binding site, and an INR consensus. These elements are found commonly in eukaryotic core promoters usually residing within 100 bp of the TSS (51, 52). Indeed, the ITGB8 core promoter (−491 to +69 relative to the TSS), which contains the minimal sequence required to drive gene expression in all cell lines in this study, contains all of these elements. However, other yet unidentified regulatory elements reside in the sequence between −330 and −491 because a smaller core promoter construct (−330 to +69), which contains the CCAAT, CRE, SP1, and INR, drives efficient expression only in specific cell types. Thus, cell type-specific regulatory elements are required for efficient core promoter activity in the region between −330 and −491. We tested putative NFκB or AP4 binding sites, which are found in the −330 to −491 sequence, singularly or combined, and found neither was required for core promoter activity. However, we cannot exclude the possibility that either site, or both, in combination with another currently unidentified site within the −330 to −491 region, is required. Transcription factor matrices do not reveal additional highly conserved consensus transcription factor binding sites in the −330 to −491 region and thus, the exact functional sequence(s) and cognate transcription factor(s) remain to be elucidated.
The Sp3 Transcription Factor Regulates ITGB8 Expression
The SP consensus sequence located at −19 in the core promoter is required for activity, because a deletion mutant at this site completely abolishes the activity of the promoter construct and knock-down of SP1 and SP3 reduce expression of β8 by 29 and 62%, respectively. Although Sp1 and Sp3 are thought to be functionally redundant, SP3 knock-down is more effective in reducing β8 expression than SP1, possibly due to a compensatory increase in SP3 expression with SP1 knock-down, which likely masks any effect on β8 expression.
The SP family of transcription factors is highly conserved, consists of Sp1–9, and binds with high selectivity to the SP consensus binding sequence, the GC box (53). Sp1–4 form a subgroup of the SP family that contain homologous activation domains and C-terminal zinc finger regions, which mediate DNA binding specificity (54). We limited our investigation to Sp1 and Sp3 because they are expressed in tissues where β8 is known to be expressed and have been shown to regulate expression of a wide array of integrin genes (55–65). Their functions partially overlap as demonstrated by their knock-out phenotypes in mice (66–68). Sp1 and Sp3 are required for survival and are generally involved in cellular differentiation, proliferation, and organogenesis (69).
An AP1 Complex Is Required for ITGB8 Core Promoter Activity
The CRE site located at −40 is also required for ITGB8 core promoter function. Many different transcription factors and complexes are capable of interacting with CRE, such as CRE-binding protein (CREB), CREB-binding partner (CBP), and a multitude of AP-1 factors (70). AP-1 is a transcription factor complex typically composed of a homo- or heterodimer of a member of the Jun and Fos or ATF transcription factor families including c-Jun, JunB, JunD, c-Fos, FosB, Fra-1, Fra-2, ATF-1–4, and JDP (Jun dimerization partner) (46). In fact, several different AP-1 factors such as ATF-2, c-Jun, JunD, and JunB bound to the CRE site in the ITGB8 promoter.
ATF-2 bound to the consensus CRE site in the core promoter of ITGB8 in a complex with c-Jun, probably in a heterodimer, because antibodies against ATF-2 and c-Jun supershifted the same complex in EMSA and both ATF-2 and c-Jun bound to the core promoter region in chromatin immunoprecipitations. Additionally, some, but not all, of the components of one of the ATF-2 complexes could be supershifted with antibodies to JunB and JunD, suggesting that a multimeric AP-1 complex can form around the CRE on the ITGB8 core promoter. However, it is likely that the functional complex at the CRE is one that contains ATF-2 and c-Jun, because Jun-Jun dimers have a lower affinity for CRE than ATFs/CREBs, whereas c-Jun is a potent transcriptional activator (47).
We confirmed that ATF-2 regulates integrin β8 expression because knock-down of ATF2 caused a significant reduction in integrin β8 levels. Both transcript and surface levels were reduced by ∼40–45%, which could either be due to residual ATF-2 protein levels after ATF2 knock-down, incomplete knock-down of ATF2, and/or compensation by other AP-1 factors in the absence of ATF-2.
ATF-2 is part of a subfamily of AP-1 transcription factors, the ATF/CREB family, which contains a homologous DNA binding domain that mediates interaction with consensus CRE or 12-O-tetradecanoylphorbol-13-acetate-response elements (TRE) and are regulated by cAMP-dependent and stress-activated kinase pathways (70, 71). ATF-2 is the most extensively characterized and is ubiquitously expressed (71). ATF2−/− mice die either perinatally due to severe respiratory distress due to deficient pulmonary differentiation, or succumb later to defective immune responses to microbial infection (72, 73). These data and the discovery that ATF-2 regulates ITGB8 expression also support a role for integrin β8 in the lung and the immune response. ATF2 is mapped to a region on chromosome 2q that is subject to loss of heterozygosity in multiple cancer types and specific alleles of ATF2 are linked to a subset of lung cancers suggesting that ATF-2 may operate as a tumor suppressor in these cancers (74). We have previously shown that integrin β8 behaves as a tumor suppressor in lung cancer cells in vitro and in vivo (4). Therefore, ATF-2 may act as a tumor suppressor in lung cancers at least in part via regulation of integrin β8 expression.
Sp1/Sp3 and AP-1 are often found as components in large multimeric transcription factor complexes that can interact with each other (75, 76). The close proximity of the SP and CRE consensus sequences in the ITGB8 promoter suggest these complexes may interact to regulate β8 expression. Although we did not identify any complexes that contained both Sp1 or Sp3 and AP-1 factors by EMSA analysis, it remains plausible that these adjacent complexes interact to drive expression of integrin β8, but remain either transient or with insufficient affinity to be detected by EMSA. In fact, the results from the chromatin immunoprecipitation experiments suggest that these transcription factors do interact in a higher order chromatin complex at the ITGB8 core promoter because they associate with the same regions of DNA.
Sp1, Sp3, and AP-1 factors are common transcription factors found to regulate integrin genes. Promoter studies of other integrins, such as α2, α3, α5, α6, α11, αIIb, β4, β5, CD11β-δ, and CD18, have shown requirements for Sp1/Sp3 and/or AP-1 transcription factors and their cognate sites for expression (55–64, 77–79). In particular, the promoter of ITGAV (integrin αv), the gene that encodes the heterodimerization partner of integrin β8, is also regulated by Sp1 and Sp3 (65). These data suggest that the regulatory regions have duplicated and co-evolved with the integrin genes.
The p38 Pathway Regulates β8 Expression
We demonstrate that β8 expression is regulated by p38 and that αvβ8-mediated TGF-β activation is p38-dependent, linking, for the first time, the p38 pathway to the regulation of TGF-β activation through integrin β8 expression. In β8 expressing cells, ATF-2, a known target of the p38 MAPK pathway, is phosphorylated, and therefore activated, in a p38-dependent manner. Phosphorylated ATF-2 and c-Jun both bind to the ITGB8 core promoter in a p38-dependent manner. In contrast, Sp3 binds to its cognate site in the ITGB8 core promoter in a p38-independent manner.
Taken together, we hypothesize a model for the regulation of integrin β8 expression through p38, phospho-ATF-2 and Sp3 (Fig. 7B). By EMSA, we showed that Sp3, ATF-2, and c-Jun directly bind to their cognate sites in the core promoter and that these sites are required for promoter expression. We showed that both the “P1” region, which contains these sites, and an adjacent region, “P2,” of the ITGB8 promoter are required for full promoter expression as determined by reporter assays. However, we were unable to determine which putative transcription factor binding sites within the P2 region were required for promoter expression. As expected, chromatin immunoprecipitation of Sp3, ATF-2, or c-Jun resulted in their enrichment at the P1 and P2 regions. This enrichment at the P2 region was unexpected because we were unable to show direct binding of Sp3, ATF-2, or c-Jun to sequences in the P2 region by EMSA. This result could either be due to larger DNA fragments that span the directly adjacent “P1 and P2” regions present in the nuclear sonicates or to a direct or indirect association of Sp3, ATF-2, or c-Jun with the P2 region. Evidence in support of the later is that Sp3 dissociates with P2 but maintains its association with the P1 region in cells treated with the p38 inhibitor. Therefore we hypothesize a model where p38 phosphorylates ATF-2, which binds to c-Jun, and facilitates interaction with the CRE site in the ITGB8 P1 region of the core promoter, which is in close proximity to Sp3 constitutively bound to its SP site. A higher-order chromatin complex forms containing Sp3 and AP-1 that interacts with an adjacent (P2) region in the core promoter either directly or through as of yet unknown factors, which activates the basal transcriptional machinery and transcription of the ITGB8 gene.
The p38 pathway is activated by a wide range of cellular stresses and inflammatory cytokines and also regulates the expression of many pro-inflammatory cytokines (49). Hence, p38 is critical for normal immune and inflammatory responses. Many of the studies defining p38 function rely on relatively specific p38 pyridinylimidazole inhibitors such as SB202190 (80). There are four mammalian isoforms of p38, and these compounds selectively inhibit both p38α and p38β isoforms, but not p38γ or p38δ (81). Knock-out studies indicate a dominant role for p38α in vivo (82, 83). Thus, p38α is the most likely isoform to be involved in regulation of β8 expression, which we confirmed using a dominant-negative isoform of p38α and causing a reduction in ITGB8 expression by ∼35%. This modest reduction in ITGB8 levels is similar to the reduction caused by knock-down of ATF-2, suggesting that p38 has its effect primarily through its activation of ATF-2. Although we cannot exclude the involvement of the other p38 isoforms in regulating ITGB8 expression, p38β is the only other isoform that is reportedly inhibited by SB202190, suggesting that it might also contribute to the regulation of ITGB8 (81).
TGF-β is a crucial mediator of immune and epithelial-mesenchymal homeostasis through diverse effects on cellular differentiation, proliferation, and apoptosis (23). Previous studies have demonstrated important roles for AP-1, p38, and TGF-β in normal epithelial-mesenchymal homeostasis and wound healing of the skin and lung (23, 47, 84, 85). We have also shown that integrin β8 regulates epithelial-mesenchymal and endothelial-mesenchymal homeostasis in the airway and brain, respectively (1, 2, 15, 17). Here we show a direct link between p38, AP-1, and TGF-β in these processes via regulation of integrin β8 expression.
Acknowledgment
We are grateful for the generous gift of the p38α dominant-negative vector from Jiahuai Han of the Scripps Research Institute, La Jolla, CA.
This work was supported, in whole or in part, by National Institutes of Health Grants HL090662, HL63993, and NS04415 (to S. L. N.).
- TGF-β
- transforming growth factor β
- HBEC
- human bronchial epithelial cell
- ERK
- extracellular-signaling response kinase
- JNK
- Jun N-terminal kinase
- DTBP
- dimethyl-3,3′-dithiobispropionimidate
- SEAP
- secreted alkaline phosphatase
- EMSA
- electromobility shift assays
- CREB
- cAMP-response element-binding protein
- TSS
- transcriptional start site
- CRE
- cyclic AMP responsive elements
- DMRT
- doublesex-mab3-related transcription factor
- AP4
- activator protein-4
- MAPK
- mitogen-activated protein kinase
- siRNA
- small interfering RNA
- RT
- reverse transcription.
REFERENCES
- 1.Araya J., Cambier S., Markovics J. A., Wolters P., Jablons D., Hill A., Finkbeiner W., Jones K., Broaddus V. C., Sheppard D., Barzcak A., Xiao Y., Erle D. J., Nishimura S. L. (2007) J. Clin. Invest. 117, 3551–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su H., Kim H., Pawlikowska L., Kitamura H., Shen F., Cambier S., Markovics J., Lawton M. T., Sidney S., Bollen A. W., Kwok P. Y., Reichardt L., Young W. L., Yang G. Y., Nishimura S. L. (2010) Am. J. Pathol. 176, 1018–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culhane A. C., Quackenbush J. (2009) Cancer Res. 69, 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambier S., Mu D. Z., O'Connell D., Boylen K., Travis W., Liu W. H., Broaddus V. C., Nishimura S. L. (2000) Cancer Res. 60, 7084–7093 [PubMed] [Google Scholar]
- 5.Danen E. H., Sonnenberg A. (2003) J. Pathol. 201, 632–641 [DOI] [PubMed] [Google Scholar]
- 6.Hynes R. O. (2004) Matrix Biol. 23, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takada Y., Ye X., Simon S. (2007) Genome Biol. 8, 215.211–215.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X., Griffiths M., Wu J., Farese R. V., Jr., Sheppard D. (2000) Mol. Cell. Biol. 20, 755–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J., Motejlek K., Wang D., Zang K., Schmidt A., Reichardt L. F. (2002) Development 129, 2891–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader B. L., Rayburn H., Crowley D., Hynes R. O. (1998) Cell 95, 507–519 [DOI] [PubMed] [Google Scholar]
- 11.Hodivala-Dilke K. M., McHugh K. P., Tsakiris D. A., Rayburn H., Crowley D., Ullman-Culleré M., Ross F. P., Coller B. S., Teitelbaum S., Hynes R. O. (1999) J. Clin. Invest. 103, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens L. E., Sutherland A. E., Klimanskaya I. V., Andrieux A., Meneses J., Pedersen R. A., Damsky C. H. (1995) Genes Dev. 9, 1883–1895 [DOI] [PubMed] [Google Scholar]
- 13.Huang X. Z., Wu J. F., Cass D., Erle D. J., Corry D., Young S. G., Farese R. V., Jr., Sheppard D. (1996) J. Cell Biol. 133, 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty J. H., Monahan-Earley R. A., Brown L. F., Keller M., Gerhardt H., Rubin K., Shani M., Dvorak H. F., Wolburg H., Bader B. L., Dvorak A. M., Hynes R. O. (2002) Mol. Cell. Biol. 22, 7667–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araya J., Cambier S., Morris A., Finkbeiner W., Nishimura S. L. (2006) Am. J. Pathol. 169, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarad G., Wang B., Khan S., DeVore J., Miao H., Wu K., Nishimura S. L., Wible B. A., Konieczkowski M., Sedor J. R., Schelling J. R. (2002) J. Biol. Chem. 277, 47826–47833 [DOI] [PubMed] [Google Scholar]
- 17.Proctor J. M., Zang K., Wang D., Wang R., Reichardt L. F. (2005) J. Neurosci. 25, 9940–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhe-Reddy S., Khan S., Konieczkowski M., Jarad G., Wu K. L., Reichardt L. F., Takai Y., Bruggeman L. A., Wang B., Sedor J. R., Schelling J. R. (2006) J. Biol. Chem. 281, 19688–19699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cambier S., Gline S., Mu D., Collins R., Araya J., Dolganov G., Einheber S., Boudreau N., Nishimura S. L. (2005) Am. J. Pathol. 166, 1883–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Travis M. A., Reizis B., Melton A. C., Masteller E., Tang Q., Proctor J. M., Wang Y., Bernstein X., Huang X., Reichardt L. F., Bluestone J. A., Sheppard D. (2007) Nature 449, 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mu D., Cambier S., Fjellbirkeland L., Baron J. L., Munger J. S., Kawakatsu H., Sheppard D., Broaddus V. C., Nishimura S. L. (2002) J. Cell Biol. 157, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedenfalk I., Duggan D., Chen Y., Radmacher M., Bittner M., Simon R., Meltzer P., Gusterson B., Esteller M., Kallioniemi O. P., Wilfond B., Borg A., Trent J., Raffeld M., Yakhini Z., Ben-Dor A., Dougherty E., Kononen J., Bubendorf L., Fehrle W., Pittaluga S., Gruvberger S., Loman N., Johannsson O., Olsson H., Sauter G. (2001) N. Engl. J. Med. 344, 539–548 [DOI] [PubMed] [Google Scholar]
- 23.Nishimura S. L. (2009) Am. J. Pathol. 175, 1362–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Annes J. P., Munger J. S., Rifkin D. B. (2003) J. Cell Sci. 116, 217–224 [DOI] [PubMed] [Google Scholar]
- 25.Yang Z., Mu Z., Dabovic B., Jurukovski V., Yu D., Sung J., Xiong X., Munger J. S. (2007) J. Cell Biol. 176, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shull M. M., Ormsby I., Kier A. B., Pawlowski S., Diebold R. J., Yin M., Allen R., Sidman C., Proetzel G., Calvin D., Annunziata N., Doetschman T. (1992) Nature 359, 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aluwihare P., Mu Z., Zhao Z., Yu D., Weinreb P. H., Horan G. S., Violette S. M., Munger J. S. (2009) J. Cell Sci. 122, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu Z., Yang Z., Yu D., Zhao Z., Munger J. S. (2008) Mech. Dev. 125, 508–516 [DOI] [PubMed] [Google Scholar]
- 29.Goodwin A., Jenkins G. (2009) Biochem. Soc. Trans. 37, 849–854 [DOI] [PubMed] [Google Scholar]
- 30.Finkbeiner W. E. (1997) in The Lung: Scientific Foundations (Crystal R. G. ed) pp. 415–433, Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 31.Abe M., Harpel J. G., Metz C. N., Nunes I., Loskutoff D. J., Rifkin D. B. (1994) Anal. Biochem. 216, 276–284 [DOI] [PubMed] [Google Scholar]
- 32.Kitabayashi I., Eckner R., Arany Z., Chiu R., Gachelin G., Livingston D. M., Yokoyama K. K. (1995) EMBO J. 14, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhen G., Park S. W., Nguyenvu L. T., Rodriguez M. W., Barbeau R., Paquet A. C., Erle D. J. (2007) Am. J. Respir. Cell Mol. Biol. 36, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borovecki F., Lovrecic L., Zhou J., Jeong H., Then F., Rosas H. D., Hersch S. M., Hogarth P., Bouzou B., Jensen R. V., Krainc D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11023–11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Citron B. A., Dennis J. S., Zeitlin R. S., Echeverria V. (2008) J. Neurosci. Res. 86, 2499–2504 [DOI] [PubMed] [Google Scholar]
- 36.Hale K. K., Trollinger D., Rihanek M., Manthey C. L. (1999) J. Immunol. 162, 4246–4252 [PubMed] [Google Scholar]
- 37.Fujita N., Wade P. A. (2004) Methods 33, 81–85 [DOI] [PubMed] [Google Scholar]
- 38.Moyle M., Napier M. A., McLean J. W. (1991) J. Biol. Chem. 266, 19650–19658 [PubMed] [Google Scholar]
- 39.Smale S. T. (1997) Biochim. Biophys. Acta 1351, 73–88 [DOI] [PubMed] [Google Scholar]
- 40.Heintzman N. D., Ren B. (2007) Cell. Mol. Life Sci. 64, 386–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohler U., Stemmer G., Harbeck S., Niemann H. (2000) Pac. Symp. Biocomput. 5, 380–391 [DOI] [PubMed] [Google Scholar]
- 42.Bajic V. B., Seah S. H., Chong A., Zhang G., Koh J. L., Brusic V. (2002) Bioinformatics 18, 198–199 [DOI] [PubMed] [Google Scholar]
- 43.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brudno M., Poliakov A., Minovitsky S., Ratnere I., Dubchak I. (2007) Nucleic Acids Res. 35, W669–W674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hess J., Angel P., Schorpp-Kistner M. (2004) J. Cell Sci. 117, 5965–5973 [DOI] [PubMed] [Google Scholar]
- 47.Reddy S. P., Mossman B. T. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1161–L1178 [DOI] [PubMed] [Google Scholar]
- 48.Krishna M., Narang H. (2008) Cell. Mol. Life Sci. 65, 3525–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuenda A., Rousseau S. (2007) Biochim. Biophys. Acta 1773, 1358–1375 [DOI] [PubMed] [Google Scholar]
- 50.Huang S., Jiang Y., Li Z., Nishida E., Mathias P., Lin S., Ulevitch R. J., Nemerow G. R., Han J. (1997) Immunity 6, 739–749 [DOI] [PubMed] [Google Scholar]
- 51.FitzGerald P. C., Shlyakhtenko A., Mir A. A., Vinson C. (2004) Genome Res. 14, 1562–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi W., Zhou W., Xu D. (2007) Physiol. Genomics 31, 374–384 [DOI] [PubMed] [Google Scholar]
- 53.Suske G., Bruford E., Philipsen S. (2005) Genomics 85, 551–556 [DOI] [PubMed] [Google Scholar]
- 54.Bouwman P., Philipsen S. (2002) Mol. Cell. Endocrinol. 195, 27–38 [DOI] [PubMed] [Google Scholar]
- 55.Lu N., Carracedo S., Ranta J., Heuchel R., Soininen R., Gullberg D. (2010) Matrix Biol. 3, 166–176 [DOI] [PubMed] [Google Scholar]
- 56.Gingras M. E., Masson-Gadais B., Zaniolo K., Leclerc S., Drouin R., Germain L., Guérin S. L. (2009) Invest. Ophthalmol. Vis. Sci. 50, 57–67 [DOI] [PubMed] [Google Scholar]
- 57.Katabami K., Kato T., Sano R., Ogura M., Mizuno H., Itoh S., Tsuji T. (2006) J. Cell Biochem. 97, 530–543 [DOI] [PubMed] [Google Scholar]
- 58.Lai C. F., Feng X., Nishimura R., Teitelbaum S. L., Avioli L. V., Ross F. P., Cheng S. L. (2000) J. Biol. Chem. 275, 36400–36406 [DOI] [PubMed] [Google Scholar]
- 59.Ye J., Xu R. H., Taylor-Papadimitriou J., Pitha P. M. (1996) Mol. Cell. Biol. 16, 6178–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosmarin A. G., Luo M., Caprio D. G., Shang J., Simkevich C. P. (1998) J. Biol. Chem. 273, 13097–13103 [DOI] [PubMed] [Google Scholar]
- 61.Noti J. D. (1997) J. Biol. Chem. 272, 24038–24045 [DOI] [PubMed] [Google Scholar]
- 62.Block K. L., Shou Y., Poncz M. (1996) Blood 88, 2071–2080 [PubMed] [Google Scholar]
- 63.Onishi T., Yamakawa K., Franco O. E., Kawamura J., Watanabe M., Shiraishi T., Kitazawa S. (2001) Biochim. Biophys. Acta 1538, 218–227 [DOI] [PubMed] [Google Scholar]
- 64.Noti J. D., Johnson A. K., Dillon J. D. (2000) J. Biol. Chem. 275, 8959–8969 [DOI] [PubMed] [Google Scholar]
- 65.Czyz M., Cierniewski C. S. (1999) Eur. J. Biochem. 265, 638–644 [DOI] [PubMed] [Google Scholar]
- 66.Bouwman P., Göllner H., Elsässer H. P., Eckhoff G., Karis A., Grosveld F., Philipsen S., Suske G. (2000) EMBO J. 19, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krüger I., Vollmer M., Simmons D. G., Simmons D., Elsässer H. P., Philipsen S., Suske G. (2007) Dev. Dyn. 236, 2235–2244 [DOI] [PubMed] [Google Scholar]
- 68.Marin M., Karis A., Visser P., Grosveld F., Philipsen S. (1997) Cell 89, 619–628 [DOI] [PubMed] [Google Scholar]
- 69.Li L., He S., Sun J. M., Davie J. R. (2004) Biochem. Cell Biol. 82, 460–471 [DOI] [PubMed] [Google Scholar]
- 70.Hai T., Hartman M. G. (2001) Gene 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 71.Bhoumik A., Ronai Z. (2008) Cell Cycle 7, 2341–2345 [DOI] [PubMed] [Google Scholar]
- 72.Maekawa T., Bernier F., Sato M., Nomura S., Singh M., Inoue Y., Tokunaga T., Imai H., Yokoyama M., Reimold A., Glimcher L. H., Ishii S. (1999) J. Biol. Chem. 274, 17813–17819 [DOI] [PubMed] [Google Scholar]
- 73.Reimold A. M., Kim J., Finberg R., Glimcher L. H. (2001) Int. Immunol. 13, 241–248 [DOI] [PubMed] [Google Scholar]
- 74.Woo I. S., Kohno T., Inoue K., Ishii S., Yokota J. (2002) Int. J. Oncol. 20, 527–531 [PubMed] [Google Scholar]
- 75.Kardassis D., Papakosta P., Pardali K., Moustakas A. (1999) J. Biol. Chem. 274, 29572–29581 [DOI] [PubMed] [Google Scholar]
- 76.Eckert R. L., Crish J. F., Efimova T., Dashti S. R., Deucher A., Bone F., Adhikary G., Huang G., Gopalakrishnan R., Balasubramanian S. (2004) J. Invest. Dermatol. 123, 13–22 [DOI] [PubMed] [Google Scholar]
- 77.Zutter M. M., Painter A. D., Yang X. (1999) Blood 93, 1600–1611 [PubMed] [Google Scholar]
- 78.Nishida K., Kitazawa R., Mizuno K., Maeda S., Kitazawa S. (1997) Biochem. Biophys. Res. Commun. 241, 258–263 [DOI] [PubMed] [Google Scholar]
- 79.Takaoka A. S., Yamada T., Gotoh M., Kanai Y., Imai K., Hirohashi S. (1998) J. Biol. Chem. 273, 33848–33855 [DOI] [PubMed] [Google Scholar]
- 80.Jackson P. F., Bullington J. L. (2002) Curr. Top. Med. Chem. 2, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 81.LoGrasso P. V., Frantz B., Rolando A. M., O'Keefe S. J., Hermes J. D., O'Neill E. A. (1997) Biochemistry 36, 10422–10427 [DOI] [PubMed] [Google Scholar]
- 82.Aouadi M., Binetruy B., Caron L., Le Marchand-Brustel Y., Bost F. (2006) Biochimie 88, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 83.Hui L., Bakiri L., Stepniak E., Wagner E. F. (2007) Cell Cycle 6, 2429–2433 [DOI] [PubMed] [Google Scholar]
- 84.Angel P., Szabowski A., Schorpp-Kistner M. (2001) Oncogene 20, 2413–2423 [DOI] [PubMed] [Google Scholar]
- 85.Angel P., Szabowski A. (2002) Biochem. Pharmacol. 64, 949–956 [DOI] [PubMed] [Google Scholar]