Abstract
Synthesis of extracellular sulfated molecules requires active 3′-phosphoadenosine 5′-phosphosulfate (PAPS). For sulfation to occur, PAPS must pass through the Golgi membrane, which is facilitated by Golgi-resident PAPS transporters. Caenorhabditis elegans PAPS transporters are encoded by two genes, pst-1 and pst-2. Using the yeast heterologous expression system, we characterized PST-1 and PST-2 as PAPS transporters. We created deletion mutants to study the importance of PAPS transporter activity. The pst-1 deletion mutant exhibited defects in cuticle formation, post-embryonic seam cell development, vulval morphogenesis, cell migration, and embryogenesis. The pst-2 mutant exhibited a wild-type phenotype. The defects observed in the pst-1 mutant could be rescued by transgenic expression of pst-1 and hPAPST1 but not pst-2 or hPAPST2. Moreover, the phenotype of a pst-1;pst-2 double mutant were similar to those of the pst-1 single mutant, except that larval cuticle formation was more severely defected. Disaccharide analysis revealed that heparan sulfate from these mutants was undersulfated. Gene expression reporter analysis revealed that these PAPS transporters exhibited different tissue distributions and subcellular localizations. These data suggest that pst-1 and pst-2 play different physiological roles in heparan sulfate modification and development.
Keywords: C. elegans, Development, Gene Knock-out, Glycosaminoglycan, Heparan Sulfate, Nucleotide Sugar Transporter/Solute Carrier Family 35 (SLC35), Sulfation
Introduction
Organogenesis, tissue morphogenesis, and cell growth require diverse types of extracellular sulfation. Sulfated molecules are crucial for the establishment of a hydrophilic extracellular environment and for intercellular signaling (1–8). In eukaryotes, Golgi-resident sulfotransferases transfer sulfates from an active sulfate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS),5 to membrane-associated and secreted molecules like glycosaminoglycans (GAGs) (9–11). The sulfation reaction yields 3′-phosphoadenosine 5′-phosphate (PAP) as a byproduct. Biochemical and cytological studies have revealed that PAPS is synthesized by PAPS synthase, a bifunctional enzyme found in the nucleus and/or cytosol but not in the Golgi apparatus (12–15). PAPS transporters transport PAPS from the cytosol into the Golgi lumen (16). Recent reports have demonstrated that PAPS/PAP concentrations in the Golgi apparatus are important for biosynthetic regulation of sulfated molecules, including heparan sulfate (HS) (17–20). Thus, understanding how PAPS metabolism is regulated by PAPS transporters in vivo will provide important insight into the mechanisms underlying developmental control of extracellular sulfation.
PAPS transporter activity was first demonstrated in rat liver Golgi-derived vesicles (16). The characterization of purified proteins involved in PAPS transport activity suggests that they act through an antiport mechanism (13, 21–24). Recently, two human PAPS transporter genes were cloned and named PAPST1 (Slc35b2) (25) and PAPST2 (Slc35b3) (26, 27). The Drosophila slalom gene, which encodes PAPST1, was identified as a segment polarity gene (28, 29). In zebrafish, mutations in pinscher, the gene encoding PAPST1, caused defects in skeletal development and axon sorting (30). Although the gene encoding PAPST2 showed genetic interactions with the genes that encode HS modification enzymes in the fruit fly, its physiological roles are largely unknown (27).
The nematode Caenorhabditis elegans is a model organism that is well suited for developmental genetics because of its simple and well organized organs, including those of the reproductive, digestive, nervous, and epithelial tissue systems. The C. elegans genome contains orthologs of all the known enzymes involved in PAPS metabolism, including PAPS synthase (pps-1, T14G10.1), PAPS reductase (R53.1), Golgi-resident PAP phosphatase (Y6B3B.5), sulfate transporters (sulp-2, F14D12.5; sulp-4, K12G11.1) (31), and PAPS transporters (pst-1, M03F8.2; pst-2, F54E7.1). In addition, C. elegans expresses genes that are involved in the sulfation of tyrosine and HS but not chondroitin (32–36). This represents an advantage of using C. elegans over vertebrate species to investigate the role of PAPS metabolism in development. In vertebrates, chondroitin sulfate is required for chondrogenesis (2, 18, 19, 30), and defects in chondroitin sulfation obscure the importance of other sulfated molecules. Previous studies have demonstrated that extracellular sulfated molecules play pivotal roles in nervous system development, gonadal cell migration, cuticle formation, and embryogenesis in C. elegans (37–42). Most recently, pst-1 alleles were shown to be required for viability and neuronal development (43).
In this study, we aimed to investigate the roles of PAPS transporters in development and morphogenesis. To that end, we isolated C. elegans deletion mutants of the PAPS transporters and analyzed the defects in development and morphogenesis.
EXPERIMENTAL PROCEDURES
Materials
[35S]PAPS (1.59 Ci/mmol), GDP-[U-14C]fucose (271 mCi/mmol), and CMP-[9-3H]sialic acid (33.6 Ci/mmol) were purchased from PerkinElmer Life Sciences. UDP-[6-3H]galactose (20 Ci/mmol), UDP-N-acetyl-d-[6-3H]galactosamine (20 Ci/mmol), UDP-[1-3H]glucose (20 Ci/mmol), GDP-[2-3H]mannose (40 Ci/mmol), UDP-[U-14C]glucuronic acid (300 mCi/mmol), and UDP-[U-14C]xylose (264 mCi/mmol) were purchased from American Radiolabeled Chemicals Inc. (St. Louis, MO). UDP-N-acetyl-d-[U-14C]glucosamine (293 mCi/mmol) was purchased from GE Healthcare.
C. elegans Strains
We used C. elegans N2 as the wild-type strain. Strains were maintained and cultured as described previously (44). We used strains carrying the following alleles and balancer chromosomes: ayIs4 I (45), tm3316 III, syIs80 III (46), jcIs1 IV (47), tm3364 V, and nT1[qIs51] (IV;V). All except pst-1(tm3364) and pst-2(tm3316) were obtained from the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health, National Center for Research Resources (NCRR).
Isolation of Worms with Deleted Alleles
Worms with deletions of the pst-1(tm3364) and pst-2(tm3316) alleles were isolated from pools of worms that had been mutagenized by using the trimethylpsoralen/UV method (48). The pst-1(tm3364) mutant contained an 1186-bp deletion and a 9-bp insertion, which removed the third and fourth exons of the gene and resulted in a frameshift mutation. These deleted exons are identical among the three pst-1 splice variants. The pst-1(tm3364) deletion mutant expressed only one transmembrane region in the N terminus of the protein. The pst-2(tm3316) mutant contained a 296-bp deletion that removed part of the fifth exon, resulting in a frameshift mutation. This deletion mutant expressed only five transmembrane regions in the N terminus of the protein. The pst-1 deletion mutant was balanced and maintained as the pst-1(tm3364)/nT1[qIs51] strain. A double pst-1;pst-2 deletion mutant was maintained as the pst-1(tm3364)/nT1[qIs51], pst-2(tm3316) strain. To eliminate possible additional mutations, the pst-1(tm3364) and pst-2(tm3316) strains were back-crossed four and two times, respectively.
Analysis of GAGs
Freshly cultured nematodes were sonicated with a GE-70 ultrasonic processor (Branson Ultrasonics) and freeze-dried. The dried samples (136 mg of wild type, 72.8 mg of pst-1, and 182 mg of pst-2) were extracted with acetone and then treated with 6 ml of 1.0 m NaBH4/0.05 m NaOH at 4 °C for 20 h. The reaction was stopped with the addition of acetic acid. The samples were adjusted to 5% trichloroacetic acid and centrifuged. The soluble fraction was extracted with ether. As shown previously (34, 49, 50), the amount of HS in C. elegans was extremely small; thus, for further processing, we added a carrier of 100 μg of shark cartilage chondroitin 6-O-sulfate (Seikagaku Corp.), which contained negligible amounts of nonsulfated disaccharides. The aqueous phase was adjusted to 80% ethanol. The resultant precipitate was dissolved in 50 mm pyridine acetate and subjected to gel filtration on a PD-10 column with 50 mm pyridine acetate as an eluent. The flow-through fraction was collected and evaporated. The dried samples were dissolved in water and applied to a column (7 ml) of cation-exchange resin AG 50W-X2 (H+ form, Bio-Rad) pre-equilibrated with water. The unbound fraction, which contained the librated O-linked saccharides, was neutralized with 1 m NH4HCO3. The purified GAG fraction was digested with chondroitinase ABC or a mixture of heparitinases I and II, and then the digests were derivatized with 2-aminobenzamide and analyzed by high performance liquid chromatography as described previously (51).
Analysis of Embryonic Development
Four-dimensional microscopy was performed as described previously (49), except that embryos were mounted on a 2% agarose pad in M9 solution to examine the defects in the ventral cleft enclosure. This is because observation of the ventral cleft was difficult with embryos mounted on an 8-well printed microscopic glass slide (Matsunami Glass Inc.). To evaluate and compare the embryonic phenotypes of the mutants, embryos were dissected from the homozygous adult animals and cultured for 18 h in M9 at 20 °C.
Alkaline Bleach Assay
An alkaline bleach assay was performed basically as described (52). Ten adult hermaphrodites were transferred to a 5-μl M9 drop and placed on an 8-well glass slide followed by the addition of 2× alkaline hypochlorite solution (2 n NaOH, 80% NaOCl solution (10% available chlorine)). For the “time to stop thrashing,” we recorded the time starting from the addition of hypochlorite and ending when all worms stopped thrashing.
Plasmid Construction
We generated plasmids that attached enhanced green fluorescent protein (egfp) or mCherry gene to the ends of different PAPS transporter or reporter genes. Plasmids were constructed on a pFX_EGFPT vector backbone, essentially as described previously (53, 54), and named according to the gene they carried: Ppst-1a::egfp, Ppst-1bc::egfp, pst-1ab(fl)::egfp, pst-1c(fl)::egfp, Ppst-2::egfp, pst-2a(fl)::egfp, pst-2b(fl)::egfp, Punc-119::egfp, and Prgef-1::egfp (Ppst-1a stands for pst-1a promoter and fl stands for full length (supplemental Figs. 1 and 2)). Briefly, cDNAs were amplified by reverse transcription-PCR from total RNA of wild-type C. elegans with specific primers tagged with the NotI restriction site. We inserted the cDNA fragments digested with NotI into the Pdpy-7::egfp or the Pdpy-7::mCherry plasmid to create Pdpy-7::pst-1b::egfp, Pdpy-7::pst-2a::egfp, and Pdpy-7::aman-2::mCherry (15, 55). Similarly, cDNAs were amplified by reverse transcription-PCR from total RNA of human prostate cancer (LNCaP) cells with specific primers tagged with NotI restriction sites, and cDNA fragments digested with NotI were cloned into the Peft-4::Venus plasmid to make Peft-4::hPAPST1::Venus and Peft-4::hPAPST2::Venus (53). To make the tissue-specific pst-1b::egfp and pst-2a::egfp expression constructs, Pdpy-7::pst-1a::egfp and Pdpy-7::pst-2a::egfp were digested with NotI to yield the pst-1a and pst-2a sequences, respectively. These were cloned into Pmyo-3::egfp, Punc-119::egfp, and Prgef-1::egfp plasmids. The plasmids for the yeast expression system were prepared using the GATEWAYTM cloning system (Invitrogen) as described previously (25–27). We used two steps of attB adaptor PCR for preparation of the attB-flanked PCR products. The first PCR step was performed with gene-specific primers, and the second PCR step was performed with attB adaptive primers. The PCR products of pst-1a and pst-2 were cloned into pDONRTM201. Then each construct was converted into a yeast expression vector, YEp352GAP-II, which was inserted into additional attB cassettes sequences and three influenza HA epitope tag sequences at the position of the multicloning site of YEp352GAP-II. YEp352GAP-II-pst1a-HA and YEp352GAP-II-pst2-HA plasmids were transformed into yeast. DNA sequence analysis was performed with the Prism 3130 Genetic Analyzer (Applied Biosystems). The PCR primers used in this section are listed in Table S1.
DNA Microinjection
Microinjections were performed as described by Mello and Fire (56). Expression constructs under the control of the eft-4 promoter or other promoters were injected at 2–10 or 30 ng/μl, respectively, with co-injection of a tissue-specific marker, Ptph-1::dsred or Paman-2::aman-2::mCherry at 20 ng/μl and/or rol-6(gf) at 80 ng/μl.
Subcellular Fractionation of Yeast and Transport Assay
Golgi-rich subcellular fractionation and nucleotide sugar transport assays were performed as described previously (25–27). Each YEp352GAP-II-pst-1a-HA and YEp352GAP-II-pst-2-HA plasmid was transformed into yeast (Saccharomyces cerevisiae) strain W303-1a (MATa, ade2–1, ura3–1, his3–11,15, trp1–1, leu2–3,112, and can1–100) by the lithium acetate procedure. These transformed yeast cells were grown at 30 °C in a synthetic defined medium lacking uracil. Yeast cells were converted into spheroplasts, homogenized, and fractionated to yield a 100,000 × g Golgi-rich subcellular fraction. Then, each Golgi-rich membrane fraction (200 or 100 μg of protein) was incubated in 50 μl of reaction buffer (20 mm Tris-HCl, pH 7.5, 0.25 m sucrose, 5.0 mm MgCl2, 1.0 mm MnCl2, and 10 mm 2-mercaptoethanol) that contained various substrates (mixture of radiolabeled and cold substrate) at 25 °C for 5 min. After incubation, the radioactivity incorporated in the Golgi-rich subcellular fractionation was trapped using a 0.45-μm nitrocellulose filter (Advantec MFS) and measured using liquid scintillation. The amount of radioactivity incorporated was calculated as the difference from the background value obtained from the same assay at 0 min for each sample.
Western Blot Analysis
The Golgi-rich subcellular fraction samples were suspended in 3× SDS sample buffer (New England Biolabs) and incubated 4 °C for 16 h. The samples were subjected to 10% SDS-polyacrylamide gel electrophoresis. The membrane to which the separated proteins were transferred was probed with anti-HA monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and horseradish peroxidase-conjugated anti-mouse IgG antibody and stained with ECL Plus (GE Healthcare).
Analysis of Subcellular Localization of PST-1·EGFP and PST-2·EGFP
To determine subcellular localization of the PST-1·EGFP protein, transgenic animals carrying extrachromosomal arrays of Pdpy-7::pst-1b::egfp and Pdpy-7::aman-2::mCherry were freeze-cracked and fixed in methanol for 5 min at −20 °C (57). After blocking with TBST (Tris-buffered saline containing 0.2% Tween 20), samples were incubated with the anti-GFP monoclonal antibody (1:100 dilution, mFX73; Wako Pure Chemical Industries) and anti-DsRed polyclonal antibody (1:100 dilution, Clontech) at room temperature for 2 h. Then, samples were washed twice with TBST and incubated at room temperature for 2 h with Alexa 488- or 594-conjugated anti-mouse IgG (H+L) (Molecular Probes) and/or Alexa 594-conjugated anti-rabbit IgG secondary antibody (Molecular Probes), diluted 1:200. Subcellular localization of the PST-2·EGFP protein was determined by examining transgenic animals that carried extrachromosomal arrays of pst-2a(fl)::egfp and Paman-2::aman-2::mCherry. The specimens were observed on a Zeiss LSM510 system (Carl Zeiss). The extent of co-localization of PST-1·EGFP and PST-2·EGFP with AMAN-2·mCherry was determined as the percent intensity of the co-localized signal relative to the total GFP-specific signal using the colocalization threshold plug-in within NCBI Image J software.
RESULTS
Identification of C. elegans PAPS Transporter Genes pst-1 and pst-2
Only a single orthologous gene for each human PAPS transporter is found in the C. elegans genome: pst-1 (M03F8.2) and pst-2 (F54E7.1) (54, 33). The amino acid sequence of the human PAPS transporter-1 (hPAPST1) is 40.0, 40.5, and 36.3% identical to that of PST-1a, PST-1b, and PST-1c, respectively (supplemental Fig. S1A). On the other hand, the human PAPS transporter-2 (hPAPST2) amino acid sequence is 40.3 and 30.1% identical to that of PST-2a and PST-2b, respectively (supplemental Fig. S2A). The Web tool Phobius (58, 59) predicted that all the PST-1 variants possess nine transmembrane regions, whereas PST-2a and PST2b possess 10 and six transmembrane regions, respectively (supplemental Figs. S1A and S2A). The C. elegans genome database (Wormbase) indicates that the pst-1 and pst-2 loci encode three and two splice variants, respectively, and both loci are included in operons (OP5092 and OP3316) (supplemental Figs. S1B and S2B).
Substrate Specificities of PST-1 and PST-2 Proteins Expressed in Yeast Cells
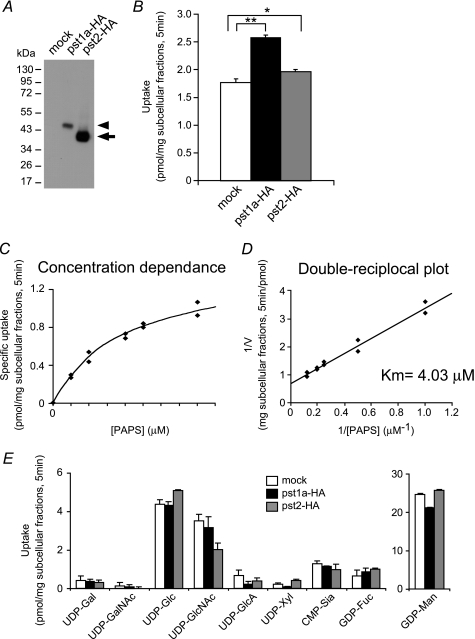
To determine the functional properties of PST-1 and PST-2, a heterologous yeast expression system was used (25–27). Western blot analysis demonstrated that PST-1a and PST-2 proteins were both expressed in the yeast P100 membrane fraction (Fig. 1A). The P100 fraction derived from yeast cells expressing PST-1a exhibited significant PAPS transport activity (Fig. 1B). The substrate concentration dependence on PAPS transport by PST-1a is shown in Fig. 1, C and D. The apparent Km value of PST-1a was estimated to be 4.03 μm. PST-2 protein also exhibited statistically significant transport activity (Fig. 1B). However, the Km value was not determined, because of its small transport activity. No significant difference was observed in transport activity of PST-1a and PST-2 for nucleotide sugars (Fig. 1E). These results suggest that the PST-1 and PST-2 proteins are PAPS-specific transporters.
FIGURE 1.
Transport activities of PST-1a and PST-2 for PAPS and nucleotide sugars by yeast expression. A, Western blot analysis of the Golgi-rich subcellular fraction prepared from yeast cells expressing mock (left lane), HA-tagged PST-1a (center lane), and HA-tagged PST-2 (right lane) using anti-HA monoclonal antibody. The loaded amount of subcellular fraction proteins was 5 μg from the cells expressing mock or HA-tagged PST-1a and 0.5 μg from the cells expressing HA-tagged PST-2. The arrowhead and arrow indicate HA-tagged PST-1a and HA-tagged PST-2, respectively. B, PAPS uptake of PST-1a and PST-2. 200 μg of Golgi-rich subcellular fraction for each sample was incubated in 50 μl of reaction buffer containing 5 μm PAPS (mixture of 2 μm [35S]PAPS and 3 μm PAPS) at 25 °C for 5 min, and the incorporated radioactivity was measured. Values shown are the mean ± S.D. obtained from two independent experiments. Open bar, mock; solid bar, PST-1a-HA; gray bar, PST-2-HA. *, p < 0.05; **, p < 0.001; assessed by two-tailed Student's t test. C, substrate concentration dependence of PST-1a. 200 μg of Golgi-rich subcellular fraction for each sample was incubated in 50 μl of reaction buffer containing different concentrations of [35S]PAPS at 25 °C for 5 min, and the incorporated radioactivity was measured. Specific uptake of PST-1a was calculated by subtracting the value of the mock uptake from the values of PST-1a-HA uptake. D, double-reciprocal plot used to determine the Km value of PST-1a. E, nucleotide sugar uptake of PST-1a and PST-2. 100 μg of Golgi-rich subcellular fraction for each sample was incubated in 50 μl of reaction buffer containing 2 μm various nucleotide sugars at 25 °C for 5 min, and the incorporated radioactivity was measured. Values shown are the mean ± S.D. obtained from two independent experiments. Open bar, mock; solid bar, PST-1a-HA; gray bar, PST-2-HA.
Isolation of pst-1 and pst-2 Deletion Mutants
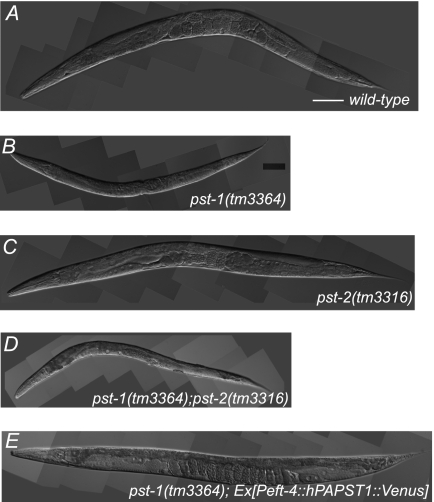
To gain insight into the physiological roles of PAPS transporters in C. elegans, we isolated strains with deletions in each of the pst genes from a library of trimethylpsoralen/UV-mutagenized worms by PCR screening. We isolated mutants that were predicted to have large transmembrane deletions in both alleles and thus likely to lack nucleotide sugar or PAPS transport activity (supplemental Figs. S1A and S2A). The isolated pst-1(tm3364) homozygotes had pleiotropic defects and an embryonic lethal phenotype (Fig. 2B and Table 1; also see below). In contrast, the isolated pst-2(tm3316) homozygotes were viable and fertile (Fig. 2C and Table 1; also see below) but had slightly reduced brood sizes (not shown).
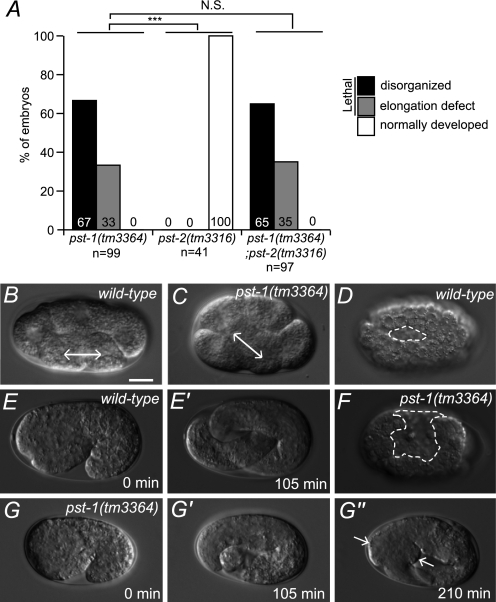
FIGURE 2.
pst-1(tm3364), pst-2(tm3316), and pst-1(tm3364);pst-2(tm3316) C. elegans mutants and transgenic rescue by hPAPST1. A–D, representative DIC images of adult C. elegans wild type (A), pst-1(tm3364) mutant (B), pst-2(tm3316) mutant (C), and pst-1(tm3364);pst-2(tm3316) double mutant (D). The pst-1(tm3364);pst-2(tm3316) double mutant showed morphological features similar to those of the pst-1(tm3364) mutant. E, representative DIC image of an adult pst-1(tm3364) mutant expressing the hPAPST1 transgene under the eft-4 promoter (Peft-4::hPAPST1::Venus construct). Scale bar = 100 μm.
TABLE 1.
Summary of the phenotypes of PAPS transporter mutant animals
+, Abnormality was observed; ++, severe abnormality was observed; −, abnormality was not observed; ND, not determined.
| Phenotype | pst-1(tm3364) | pst-2(tm3316) | pst-1, pst-2 |
|---|---|---|---|
| Embryonic | |||
| AB/P1 asymmetric cell division | − | − | ND |
| Cytokinesis | − | − | ND |
| EMS cell division | + | − | ND |
| Ea and Ep cell ingression | − | − | ND |
| Ventral enclosure | + | − | ND |
| Epidermal elongation | + | − | + |
| Larval | |||
| Cuticle formation | + | − | ++ |
| Vulval morphology | + | − | + |
| Seam cell morphology | + | − | + |
| DTC migration | + | − | + |
Reduced HS Sulfation in pst-1(tm3364) and pst-2(tm3316) Mutant Worms
Next, we examined HS sulfation in the PAPS transporter mutants. Because of the embryonic lethality of the pst-1 mutant, liquid culture samples containing pst-1(tm3364)/nT1[qIs51] heterozygous and pst-1(tm3364)/pst-1(tm3364) homozygous animals were used for biochemical analysis. For the pst-2 mutant, liquid culture samples containing the pst-2(tm3316) homozygous animals were used. In wild-type worms, HS disaccharide analysis revealed the expected profile (Table 2) (15, 34, 35, 41). In both pst-1 and pst-2 mutant worms, the number of nonsulfated units increased, and the number of sulfated units decreased; this suggested that both pst-1 and pst-2 were involved in sulfation of HS in vivo. Notably, the disaccharide profile of the pst-1 mutants was different from that of the pst-2 mutant. The levels of trisulfated disaccharide units (ΔHexA(2S)α1–4GlcN(NS,6S)) were reduced in the pst-1 mutant worms, but they were not affected in the pst-2 mutant worms compared with the controls (N2 worms). Conversely, the levels of ΔHexAα1–4GlcNAc(6S) were not affected in the pst-1 mutant worms, but they were reduced in the pst-2 mutant worms compared with controls. These results suggest that sulfation patterns of HS depend on both PST-1 and PST-2 activity.
TABLE 2.
Disaccharide composition of HS from C. elegans wild-type (N2) and mutant (pst-1 and pst-2) strains
| Disaccharidea | N2 | pst-1b | pst-2c |
|---|---|---|---|
| pmol/mg HS (%) | |||
| ΔHexAα1–4GlcNAcc | 23.6 ± 0.3 (47) | 35.3 ± 0.8 (69) | 22.6 ± 2.7 (71) |
| ΔHexAα1–4GlcNAc(6S) | 4.8 ± 1.2 (10) | 5.3 ± 3.0 (10) | 1.5 ± 0.3 (5) |
| ΔHexAα1–4Glc(NS) | 12.9 ± 1.7 (26) | 8.2 ± 3.2 (17) | 4.9 ± 2.3 (15) |
| ΔHexA(2S)α1–4Glc(NS) | 7.2 ± 1.9 (14) | 1.6 ± 0.5 (3) | 1.7 ± 1.0 (5) |
| ΔHexA(2S)α1–4Glc(NS,6S) | 1.6 ± 0.5 (3) | 0.7 ± 0.1 (1) | 1.0 ± 0.4 (3) |
| Total disaccharide | 50.1 ± 3.5 | 51.1 ± 5.9 | 31.7 ± 3.6 |
| Sulfation degreed | 0.74 | 0.37 | 0.40 |
a ΔHexAα, Glc, and GlcNAc represent unsaturated hexuronic acid, glucosamine, and N-acetylglucosamine, respectively.
b Liquid culture samples containing pst-1(tm3364)/nT1[qIs51] heterozygotes and pst-1(tm3364)/pst-1(tm3364) homozygotes were used.
c Liquid culture samples containing only pst-2(tm3316) homozygous animals were used.
d Sulfation degree is expressed as the average number of sulfate groups/disaccharide unit.
Defects in pst-1 Are Rescued by Expression of Human PAPS Transporter-1 but Not Human PAPS Transporter-2
We performed rescue experiments in pst-1 mutant worms by injecting plasmid constructs that carried wild-type genes under the eft-4 promoter, which is active in almost all cells (41). Transgenic expression of pst-1b::egfp in pst-1(tm3364) animals rescued all the observed defects. This suggested that the eft-4 promoter was sufficient to drive expression of constructs in cells that lacked PST-1 function (not shown). hPAPST1 and hPAPST2 are well characterized human Golgi-resident PAPS transporters (25, 26, 27). Transgenic expression of hPAPST1::Venus also rescued all of the defects observed in pst-1(tm3364) animals; this suggested that PST-1 is the homolog of the human PAPS transporter-1 (Fig. 2E). However, expression of hPAPST2::Venus did not rescue any of the defects, although we confirmed that the hPAPST2 protein was expressed in the transgenic worms (data not shown).
PST-1 Is Required for Cuticle Formation
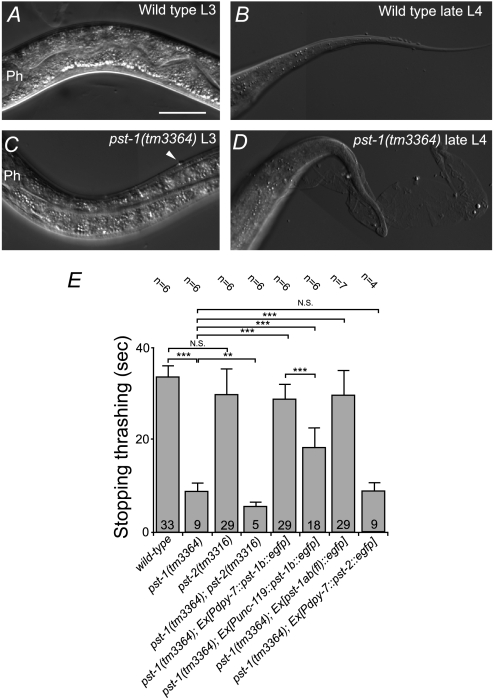
Homozygous pst-1(tm3364) embryos from the pst-1(tm3364) heterozygous hermaphrodites hatched and developed into small, fragile L3 (5%), L4 (44%), or adult (51%) animals with impaired cuticles (n = 82; Fig. 2B). In contrast, development of pst-2(tm3316) homozygotes was similar to that of the wild type (Fig. 2C). Although pst-1(tm3364) mutant larvae exhibited apparently normal locomotion nematode growth medium agar plate during the L3 stage, they exhibited “skiddy” locomotion during the L4 to adult stages (not shown). A previous report showed that skiddy locomotion is correlated with a defect in cuticle formation (52). Differential interference contrast (DIC) microscopy analysis showed that the cuticle started to become irregular in the early L3 stage (compare Fig. 3, A and C). During the L4 to adult stages, the cuticle became more severely abnormal, resulting in defective molting with some blisters (compare Fig. 3, B and D). The old, unshed cuticle remained attached to the body in various places. In contrast, no cuticle abnormality was observed in the pst-2(tm3316) mutant worms. We also examined cuticle fragility by testing alkaline bleach sensitivity (52). Consistently, mutant pst-1(tm3364) worms exhibited very fragile cuticles (rapidly stopped moving in the alkaline solution), whereas mutant pst-2(tm3316) worm cuticle fragility was similar to that of the wild-type animals (Fig. 3E).
FIGURE 3.
Cuticle defect phenotype in pst-1(tm3364), pst-2(tm3316), and pst-1(tm3364);pst-2(tm3316) C. elegans mutant adult animals. A–D, representative DIC images of regions behind the head (A and C) and tail (B and D) of wild-type L3 (A), wild-type late-L4 (B), pst-1(tm3364) mutant L3 (C), and pst-1(tm3364) mutant late-L4 larvae (D). Ph, posterior pharyngeal bulb; arrowhead (in C), in pst-1(tm3364) L3 larvae, some small protuberances of outer cuticle were observed, which were not observed in wild-type animals. During mid-L4 to the young adult stage, pst-1(tm3364) mutant animals showed abnormal molting (D). Bar = 30 μm. E, analysis of alkaline bleach sensitivity. The time to stop thrashing in alkaline bleach solution was an indicator of cuticle fragility. The pst-1(tm3364) mutant animals were more fragile (stopped faster) than the wild-type animals. In contrast, pst-2(tm3316) mutants showed cuticle fragility similar to wild-type animals. The pst-1(tm3364);pst-2(tm3316) double mutant animals were more fragile than the pst-1(tm3364) single mutant animals. In transgenic pst-1(tm3364) mutant animals that carried an extrachromosomal array of Pdpy-7::pst-1b::egfp or Punc-119::pst-1b::egfp, the cuticle fragility was restored to the wild-type level, but the former was slightly more fragile than the latter. In transgenic pst-1(tm3364) mutant animals that carried an extrachromosomal array of Pdpy-7::pst-2::egfp, the cuticle fragility was similar to that of pst-1(tm3364) mutants without the transgene. Ex, extrachromosomal array; N.S. = not significant. **, p < 0.01; ***, p < 0.001; assessed by two-tailed Student's t test.
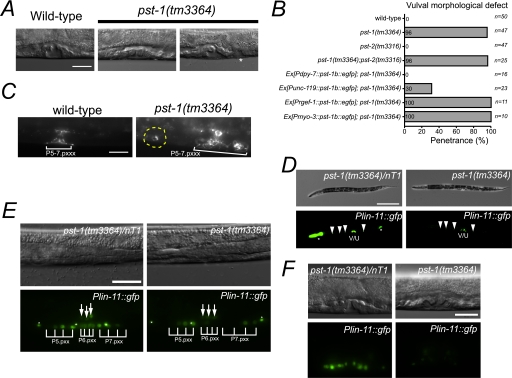
PST-1 Is Required for Vulval Morphogenesis
DIC analysis showed that almost all pst-1 knock-out animals exhibited various forms of disorganized vulval morphology. In 79% (n = 39) of pst-1(tm3364) mutant mid-L4 larvae, the vulva contained some abnormally positioned cells, which resulted in a longer invagination along the anterior-posterior axis or a small, ectopic invagination (Fig. 4, A and B). In 16% (n = 8) of pst-1(tm3364) mid-L4 larvae, vulval cells were arranged relatively normally but exhibited a collapsed invaginated space (Fig. 4A, middle) similar to that observed in the squashed vulva (Sqv) phenotype caused by a deficiency in chondroitin glycosaminoglycan synthesis (44, 49, 60, 61). The vulval architecture in the pst-1(tm3364) mutant was visualized with the transgenic expression of ajm-1::gfp, an adherence junction marker of epithelial and vulval cells. This showed a disturbance during the mid-L4 stage (Pn.pxxx stage) (Fig. 4C). A large fraction (43%) of the pst-1(tm3364) mutant animals exhibited a fragmented vulva, and 17.4% (n = 23) exhibited incorrectly positioned cells. This observation suggested that vulval cell migration and orientation are affected in pst-1(tm3364) mutants. A LIM class homeobox gene, lin-11, which is involved in specific cell fates of vulva cell types, was transgenically expressed with the Plin-11::gfp construct. In wild-type secondary cells, lin-11 typically shows an asymmetric expression pattern (46) (Fig. 4D). In pst-1(tm3364) mutants, the asymmetric lin-11::gfp(syIs80) expression pattern was irregular (Fig. 4E, 33.3%, n = 15) at the L3 stage (Pn.pxx stage), and expression levels were drastically reduced at the mid-L4 stage (Pn.pxxx stage) (Fig. 4F, 100%, n = 20). However, in VC motor neurons, lin-11::gfp expression was undisturbed (Fig. 4D).
FIGURE 4.
Vulval defects of the pst-1(tm3364) mutant. A, wild-type vulva forms an invagination space that looks like a pointed hat during early to mid-L4 stage (left). The pst-1(tm3364) mutation induced a collapse of the vulval invagination space (middle). In the mid-L4 stage, the pst-1(tm3364) vulva exhibited an ectopic invagination space (asterisk, right panel). Scale bar = 20 μm. B, penetrance in several genetic backgrounds of vulval morphological defects, including the squashed vulva and inappropriate vulval cell positioning. Ex, extrachomosomal array. C, the vulval structures of wild-type (left) and pst-1(tm3364) homozygote (right) animals were visualized with AJM-1·GFP. Incorrectly positioned cells are circled with a yellow dashed line. Scale bar = 20 μm. D, DIC and fluorescence images of pst-1(tm3364)/nT1 (left) and pst-1(tm3364) homozygote (right) mid-L4 animals that expressed Plin-11::gfp. The pst-1(tm3364)/nT1 animals with the syIs80 transgene showed Plin-11::gfp expression patterns similar to those of wild-type animals (data not shown). The GFP signals derived from the nT1[qIs51] balancer chromosome are indicated with asterisks. Arrowheads indicate VC motor neurons, and V/U indicates Plin-11::gfp expression in the vulva and uterus. Bar = 200 μm. E, DIC and fluorescence images of pst-1(tm3364)/nT1 (left) and pst-1(tm3364) homozygote (right) vulvas during the L3 stage (Pn.pxx stage). GFP signals from VC motor neurons are indicated with asterisks. GFP signals from the uterus are indicated with arrows. Bar = 25 μm. F, DIC and fluorescence images of pst-1(tm3364)/nT1 (left) and pst-1(tm3364) (right) homozygote vulvas during the mid-L4 stage (Pn.pxxx stage). Bar = 25 μm.
Additionally, the expression levels of GFP were not affected in worms expressing Pegl-17::gfp where egl-17 encodes a fibroblast growth factor family member, which is specifically expressed in secondary cells at the Pn.pxxx stage. However, the patterning of cells that expressed Pegl-17::gfp was affected at this stage (data not shown). These results suggested that secondary cell fate is adopted in pst-1(tm3364) and that PST-1 is involved in vulval morphogenetic events via regulation of lin-11 expression.
PST-1 Is Required for Post-embryonic Seam Cell Development and Distal Tip Cell (DTC) Migration
Disruption of the pst-1 gene resulted in abnormal seam cell development (see supplemental Fig. S3 and the legend for details). The results suggested that pst-1 is involved in post-embryonic seam cell development. In addition to epithelial defects, pst-1 mutant animals were also defective in DTC migration. During the L4 stage in most wild-type animals, two DTCs that reached the dorsal side migrated along the dorsal body wall muscles toward the middle of the animal (supplemental Fig. S3E). In pst-1(tm3364) mutant larvae, 26.4% (n = 49) of anterior and 45.6% (n = 48) of posterior DTCs failed to migrate along the dorsal side and remained on the ventral side in the early to mid-L4 stage (supplemental Fig. S3, F and G). In pst-2(tm3316) mutant larvae, no abnormalities in seam cell development or DTC migration were observed (supplemental Fig. S3G).
PST-1 Is Required during Embryogenesis for Oriented EMS Cell Division, Neuroblast Migration, and Epidermal Elongation
The second generation (F1) of pst-1(tm3364) homozygous embryos, which is predicted to lack both maternal and zygotic pst-1 activity, displayed a fully penetrant embryonic lethal phenotype (Fig. 5A). This suggested that the first generation of pst-1(tm3364) homozygous embryos grew to adults because maternally supplied molecules rescued the deficient embryos. Two-thirds (67%) of the pst-1(tm3364) F1 embryos displayed severe morphogenetic defects that resulted in disorganized morphology before the early stage of elongation; the other one-third (33%) displayed relatively mild morphogenetic defects that resulted in developmental arrest during the elongation stage. This embryonic phenotype was reminiscent of the disruption of HS synthesis by rib-1 and rib-2 knock-outs (40, 41, 62) or the disruption of PAPS synthesis (15). This supports the notion that pst-1 is involved in HS sulfation during embryonic development. In contrast, embryos that lacked pst-2 activity displayed little or no overt embryonic lethality (Fig. 5A). Analysis of pst-1(tm3364) F1 embryos by DIC optics revealed that none (n = 23) were affected in the initial step of gastrulation (ingression of Ea and Ep cells), but a small fraction of them were affected in EMS cell division orientation (6.9%, n = 29; Fig. 5, B and C). In wild-type embryonic morphogenesis, ventral neuroblasts migrated toward the ventral cleft created by the ingression of mesodermal cells, where they formed a substrate for the epidermis during ventral enclosure (63). In pst-1(tm3364) F1 embryos that exhibited severe morphogenetic defects, ventral enclosure failed, resulting in an unenclosed ventral surface (Fig. 5, D and F), suggesting that PST-1 was involved in neuroblast migration in the early morphogenesis phase. The pst-1(tm3364) F1 embryos that underwent normal ventral enclosure elongated to variable lengths ranging from 1.5- to 3-fold increases in size and then retracted, resulting in a swollen body morphology (Fig. 5, E and G).
FIGURE 5.
Embryonic phenotypes of pst-1 and pst-2. A, percentage of embryonic mutant phenotypes in pst-1(tm3364), pst-2(tm3316), and double pst-1(tm3364);pst-2(tm3316) mutants. N.S., not significant. ***, p < 0.001. B and C, EMS cell division axis is affected in pst-1(tm3364) embryos. In wild-type embryos at the 6-cell stage, EMS cells divide along the anterior/posterior axis (bidirectional arrow (B)). In pst-1(tm3364) mutant embryos at the 6-cell stage, the axis of EMS division is skewed (bidirectional arrow (C)). D and F, DIC images of wild-type (D) and pst-1(tm3364) (F) embryos at the ventral cleft enclosure stage. The dashed lines indicate the edges of an unenclosed ventral surface. E–G″, time lapse analysis of wild-type (E and E′) and pst-1(tm3364) (G and G″) embryos during the elongation stage. Arrows indicate swollen body morphology. Scale bar = 10 μm.
Differential Expression Patterns of pst-1 and pst-2
To determine the localization of pst-1 and pst-2, we injected four types of pst-1 and three types of pst-2 transgene expression vectors into wild-type animals. (Constructs and expression patterns summarized in supplemental Figs. S1C and S2C and Table 3.) In animals that carried the pst-1ab(fl)::egfp, pst-1c(fl)::egfp, and Ppst-1bc::egfp vectors, GFP fluorescence was specifically observed in seam cells and amphid sheath cells during embryonic and larval development (Fig. 6, A and B). GFP expression was also detected in the hypodermis of L4 transgenic animals that carried pst-1ab(fl)::egfp (Fig. 6C) and pst-1c(fl)::egfp. Although the pst-1 mutant displayed a vulval defect, no vulval expression was observed in these transgenic worms (Fig. 6C). In contrast, in transgenic animals that carried Ppst-1a::egfp, GFP fluorescence was observed in almost all cells throughout development except germ cells, where extrachromosomal transgenes are generally silenced (Fig. 6D). The pst-1ab(fl)::egfp and pst-1c(fl)::egfp constructs included the entire sequence of pst-1a, but Ppst-1a::egfp carried only the 5′-promoter region of pst-1a. Thus, broad expression of Ppst-1a::egfp might be induced due to the absence of a regulatory region for transgene expression. Additionally, the expression pattern induced by a promoter of the M03F8.3 gene, a gene immediately upstream of pst-1, was similar to that induced by the pst-1a promoter (sEx10297 transgene). Expression data from a genome-wide in situ hybridization analysis indicated that pst-1 mRNA was specifically expressed in lateral seam cells, the adult germ line, and early embryos (a cDNA group, CELK04198, NextDB (nematode expression pattern database)).
TABLE 3.
Summary of the expression patterns of PAPS transporter genes
| Reporter construct | Expression pattern |
|---|---|
| Ppst-1a::egfp | Ubiquitous |
| Ppst-1bc::egfp | Epidermis (amphid sheath cells, hypodermis, seam cells) |
| pst-1-ab(fl)::egfp | Epidermis (amphid sheath cells, hypodermis, seam cells) |
| pst-1c(fl)::egfp | Epidermis (amphid sheath cells, hypodermis, seam cells) |
| Ppst-2::egfp | Intestine, gland cells |
| pst-2a(fl)::egfp | Intestine, gland cells |
| pst-2b(fl)::egfp | No expression |
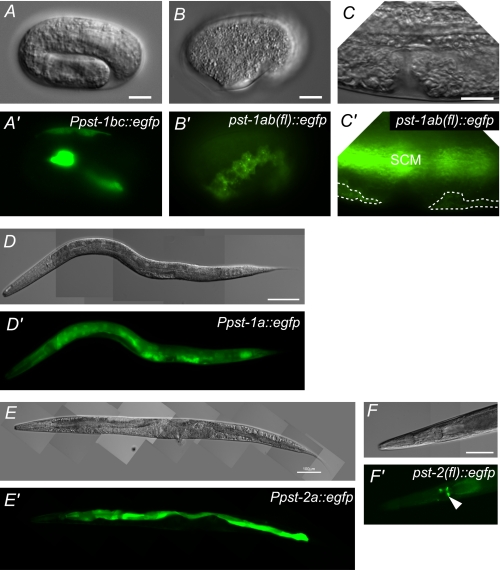
FIGURE 6.
Expression patterns of pst-1 and pst-2 reporter constructs in wild-type animals. Transgene reporter analyses of pst-1 (A–D) and pst-2 (E and F). DIC (A–F) and fluorescent (A′–F′) images are shown. GFP was visualized directly under a fluorescence microscope except in B′ and C′, which were sliced samples immunostained with an anti-GFP antibody. A and A′, expression pattern of Ppst-1bc::egfp in a folded embryo. EGFP is dominantly expressed in seam cells. B, B′, C, and C′, expression pattern of pst-1ab(fl)::egfp in an embryo (B) and the mid-ventral region (C) of a mid-L4 hermaphrodite. EGFP is dominantly expressed in seam cells. EGFP is expressed in seam cells (SCM) and the hypodermis (indicated with dashed lines) but not in the vulva (C′). D and D′, expression pattern of Ppst-1a::egfp in a mid-L4 larva. EGFP is expressed in almost all cells. E and E′, expression pattern of Ppst-2a::egfp in a young adult animal. EGFP is expressed in the intestine and pharyngeal gland cells. GFP expression in pharyngeal gland cells is weaker than in the intestine in this animal. F and F′, expression pattern of pst-2(fl)::egfp in the head region of a young adult animal. The EGFP signal in the pharyngeal gland cells is indicated with an arrowhead. Scale bars = 10 μm (A–C) and 100 μm (E–F).
In transgenic animals that carried the Ppst-2a::egfp and pst-2(fl)::egfp constructs, EGFP fluorescence was specifically observed in intestinal and pharyngeal gland cells (Fig. 6, E and F). However, no GFP signal was observed in worms carrying Ppst-2b::egfp (data not shown). This suggested that the pst-2b promoter does not have inherent activity. Although there are no expression data for pst-2 in the NextDB, pst-2 is among the set of 603 germ line-specific/enriched genes identified by SAGE (serial analysis of gene expression). In the SAGE library, we found 10 tags for pst-2 in the germ line and four tags in the soma, suggesting that pst-2 is expressed predominantly in the germ line (64). Taken together, these results suggest that although both pst-1 and pst-2 are likely to be expressed in the germ line, they have mutually exclusive expression patterns in somatic cells; pst-1 tends to be dominantly expressed in tissues generated from ectodermal cells, and pst-2 tends to be expressed in tissues generated from endomesodermal cells.
PST-1 and PST-2 Are Localized to the Golgi Apparatus
In cultured mammalian cells, PAPS transporters have been shown to localize to the Golgi membrane (20, 25, 26). Our reporter analyses indicated that pst-1 and pst-2 are expressed in the hypodermis and intestine, respectively. Thus, we examined the subcellular localizations of PST-1·EGFP and PST-2·EGFP in those tissues. Consistent with the previous reports, the transporter proteins PST-1·EGFP and PST-2·EGFP co-localized with a marker of the Golgi apparatus, mCherry-tagged alpha-mannosidase II (AMAN-2). This suggested that PST-1 and PST-2 proteins reside in the Golgi (supplemental Fig. S4, A and B). However, in merged images, it was apparent that although PST-2·EGFP almost completely co-localized with AMAN-2, PST-1·EGFP only partly co-localized with AMAN-2 (supplemental Fig. S4C). The localization of AMAN-2 is predicted to be in the cis/medial-Golgi compartments rather than the trans-Golgi compartment. Thus, our results suggested that PST-2, but not PST-1, was distributed primarily in the cis/medial-Golgi compartments.
PST-1 Expression in the Epidermis Is Sufficient for Proper Larval Epithelial Development, and Its Expression in the Neuroectoblast Is Sufficient for Rescuing Embryonic Lethality
To determine which cell types required pst-1 gene function, we examined whether epidermal, neuronal, or muscle expression of pst-1 was sufficient to rescue the embryonic, vulval, and cuticle phenotypes of deletion mutants. Epidermis-specific expression of pst-1b::egfp under the control of the dpy-7 promoter rescued vulval and abnormal cuticle phenotypes (Figs. 3E and 4B) and exhibited weak activity, but it was sufficient to rescue the embryonic phenotype (Table 4). The neuroectoderm- and neuron-specific expression of pst-1b::egfp under the control of the unc-119 promoter only partially rescued vulval and abnormal cuticle phenotypes (Figs. 3E and 4B). However, it rescued the embryonic lethal phenotype more completely than epidermis-specific expression (Table 4). In contrast, differentiated neuron-specific or body wall muscle-specific expression of pst-1b::egfp under the control of rgef-1 or myo-3 promoters did not rescue any of the phenotypes. These results suggested that pst-1 has specific functions in epidermal tissues during larval development and in epidermal precursors and/or neuroblasts during embryogenesis.
TABLE 4.
Either neuroectodermal or epidermal expression of PST-1 was necessary and sufficient for embryogenesis
| Promoter | Target tissue | No. of plates with proliferating progeny/total test platesa | Rescue activityb |
|---|---|---|---|
| 10 larvae/plate | |||
| None | 0/2 | − | |
| pst-1bc | Hypodermis, seam cells, amphid sheath | 2/2, 2/2c | ++ |
| dpy-7 | Hypodermis, seam cells | 3/5, 3/3c | + |
| unc-119 | Neuroectoderm, differentiated neurons | 2/2, 2/2c | ++ |
| rgef-1 | Differentiated neurons | 0/3 | − |
| myo-3 | Body wall muscles | 0/3 | − |
a Ten L4 larvae of mutant or transgenic animals were transferred to a fresh plate seeded with OP50 to determine whether the animals could proliferate after 1 week.
b Lethality in transgenic pst-1(tm3364) mutant embryos that carried Pdpy-7::pst-1b::egfp was higher than in those that carried Punc-119::pst-1b::egfp or pst-1bc(fl)::egfp.
c Two independent transgenic lines were tested.
pst-2 Synergizes with pst-1 in Cuticle Formation but Not in Vulval Development or Embryogenesis
To investigate the genetic relationship between pst-1 and pst-2, we examined the pst-1;pst-2 double mutant phenotypes. The pst-1 mutants exhibited abnormalities in embryogenesis, vulval development, and cuticle formation, but the pst-2 mutants displayed essentially normal phenotypes (Figs. 3E and 4B). These data suggested that pst-1 and pst-2 do not have redundant roles in embryogenesis and vulval development. In contrast, the alkaline bleach sensitivity test revealed that the pst-1;pst-2 double mutant had a more severely fragile cuticle than the pst-1 single mutant (Fig. 3E), despite their similarities in DIC analyses. This suggested that pst-1 and pst-2 had some redundant functions in cuticle formation. To determine whether the redundancy between pst-1 and pst-2 resulted from the molecular function of the PAPS transporter, we ectopically expressed pst-2 in the epidermis of the pst-1(tm3364) mutant and examined the effect on cuticle abnormality. The alkaline bleach sensitivity test revealed that pst-2 expressed in the epidermis under control of the dpy-7 promoter failed to rescue cuticle fragility in pst-1(tm3364) mutants; in contrast, pst-1b expression successfully rescued cuticle fragility (Fig. 3E). These results suggested that PST-1 and PST-2 play differential physiological roles in cuticle formation.
DISCUSSION
This study showed that, in C. elegans, the PAPS transporter pst-1 gene, but not the pst-2 gene, is essential for diverse aspects of epithelial development, somatic gonadal cell migration, and viability. We observed embryonic defects in pst-1 knock-out worms similar to those observed in embryos deficient in pps-1 (15) or rib-1/rib-2, which lacked HS-synthesizing enzymes (41, 62). During larval development, the pst-1 mutant showed defective cuticle formation similar to that observed in larvae depleted of pps-1 or tyrosylprotein sulfotransferase-A (tpst-1) genes (15, 37). Although disaccharide analysis revealed that both pst-1 and pst-2 were involved in HS sulfation, none of the defects caused by the pst-1 mutation could be restored by the heterogeneous expression of hPAPST2 or PST-2. Moreover, HS sulfation patterns isolated from pst-1 mutant animals were clearly different from those isolated from pst-2 mutant animals. Furthermore, pst-1 and pst-2 displayed different expression patterns. These observations indicated that hPAPST1/PST-1 has distinct characteristics from hPAPST2/PST-2 in vivo. Our data also suggested that subcellular localization of the PST-1·EGFP protein was slightly different from that of the PST-2·EGFP protein. Thus, each PAPS transporter may reside in different intracellular compartments; this would allow differential sulfation reactions within a single cell. Intriguingly, differential “Golgi units” are proposed to regulate different glycosylation reactions in Drosophila cells (65).
Analysis using the yeast heterologous system clearly suggested that PST-1 and PST-2 are PAPS-specific transporters. Although PST-2 showed weaker transport activity compared with PST-1, the transporter activity in our assay was statistically significant, and PST-2 showed no transport activity for nucleotide sugars. These results together with the reduced sulfation in the pst-2 null allele HS disaccharide analysis strongly indicate that PST-2 is also a PAPS transporter. It is intriguing that the total amount of HS disaccharide units was decreased in pst-2 mutant but not in the pst-1 null allele. Because sulfation takes place simultaneously with elongation of the GAG chain (66), depletion of PAPS in the intracellular compartment containing PST-2 may specifically influence the synthesis of GAG chains and reduce the heparan sulfate content measured in pst-2 null mutant animals.
The pst-1, pst-2 double mutant showed that PST-1 and PST-2 had synergistic effects in cuticle formation. Precise cuticle formation requires both epithelial cells and pharyngeal gland cells (67). Epithelial cells synthesize the cuticle, and pharyngeal gland cells are thought to secrete a surface coating that covers the cuticle. Our results showed that pst-2 was expressed in pharyngeal gland cells, and thus in these cells, PST-2 may be involved in the secretion or synthesis of components of the cuticle surface coat. Another nucleotide sugar transporter, SRF-3, transports UDP-GlcNAc and UDP-Gal into Golgi apparatus-enriched vesicles from the cytosol. SRF-3 is expressed in pharyngeal gland cells and seam cells. It is involved in the biosynthesis of glycoconjugates for the outer surface and the cuticle (68, 69). Considering the expression patterns of PST-2, it is possible that PST-2 may cooperate with SRF-3 in this process. We also found that PST-2 was strongly expressed in the intestine, consistent with the tissue distribution of hPAPST2 transcripts (26). This implies that PST-2 and hPAPST2 may play a common role in the intestine; for example, they may participate in host-pathogen interactions.
Vulval morphogenesis, including invagination and final cell positioning along the anterior-posterior axis, was affected in pst-1 mutant animals. Our data suggest that pst-1 is required for the precise expression of lin-11::gfp. Similar findings have been reported for worms that expressed mutant lin-17, a gene that encodes the Frizzled Wnt receptor (46, 70, 71). HS proteoglycans are essential for Wnt signaling, both in vertebrates and invertebrates (72), and thus the sulfation of HS proteoglycans could modulate Wnt/Frizzled signaling or lin-11 transcriptional regulation in vulval cells.
Immunostaining experiments have indicated that HS is present in vulval cells and around the vulva (Ref. 73 and our unpublished data). Consistent with this finding, abnormal vulval morphogenesis is thought to result from the loss of rib-2 (a glucosaminyltransferase) function (74). Other glycosyltransferase genes (sqv-6, sqv-2, sqv-8, and sqv-3) involved in establishing proteoglycan linkages are required for synthesis of both HS and chondroitin. Mutations of sqv genes cause the Sqv phenotype, which was also observed in the pst-1 mutant. As ample evidence indicates that chondroitin is not sulfated in this organism (34–36), the Sqv phenotype in pst-1 mutant would be due to undersulfation of HS rather than chondroitin. However, animals with mutations in the sqv genes do not exhibit abnormal anterior-posterior cell positioning or small ectopic invaginations (60), despite their requirement for HS synthesis. This apparent discrepancy might be ascribed to the different half-lives of the different gene products studied (mRNA or proteins) and/or to different metabolic functions of the enzymes studied (PAPS transport versus GAG linkage). Further study of these genes in the vulval development will provide useful information concerning the fundamental machinery of GAG synthesis and metabolism, as well as the regulation of extracellular signaling by GAGs.
Mutation of pst-1 also resulted in abnormal EMS cell division and post-embryonic seam cell development, which are regulated by Wnt signaling (75). This gives rise to the intriguing possibility that sulfation of extracellular molecules could be involved in regulation of proteins associated with Wnt signaling in diverse processes in C. elegans.
The lethality observed in the pst-1 mutants isolated in this study occurred at a different embryonic stage than that observed in the pps-1 null mutant (tm1109) that we isolated previously (15). The animals with pst-1 mutations survived through the embryonic stage of the second generation; in contrast, animals with pps-1 mutations died in L2/L3 of the first generation. This difference in timing can be explained by several possibilities. (i) Cytosolic sulfation is essential for L2/L3 growth. To date, no null mutation of ssu-1, which encodes the only known C. elegans ortholog of cytosolic sulfotransferase, has been isolated (76–78). However, deficiencies in ssu-1, by RNA interference or reduction-of-function mutations, did not result in a lethal phenotype. (ii) Extracellular sulfation is essential for L2/L3 growth, and PST-1 may not be the only Golgi-resident PAPS transporter, or there may be a novel mechanism for providing PAPS to Golgi-resident sulfotransferases. (iii) Neither cytosolic nor extracellular sulfation is required for L2/L3 growth, and PPS-1 has a function distinct from PAPS synthesis in vivo. Understanding how lethality is caused by pps-1 and pst-1 deficiencies will shed light on the mechanisms that underlie the regulation of PAPS metabolism and sulfation in C. elegans development.
While this manuscript was in preparation, Bülow and colleagues (43) published a paper concentrating on the analysis of pst-1 and pst-2 functions in the nervous system of C. elegans. They show that pst-1 is essential in nervous system development and other functions, and our results are complementary to their results indicating the essentiality of PAPS transporters in the organism.
Supplementary Material
Acknowledgments
We thank Dr. K. Fukushima and Prof. K. Yamashita (Tokyo Institute of Technology, Yokohama, Japan) for providing the mammalian total cDNA. We also thank the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, NCRR, for the worms and E. coli strains. We also thank Adam Kleinschmit for useful comments.
This work was supported by a grant-in-aid from the Japan Society for the Promotion of Science Fellows (to K. D.), a grant-in-aid for young scientists (B) (to S. M.), and Grant-in-Aid for Scientific Research B-21390025 (to H. K.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan. This research was also supported by MEXT Grant-in-Aid for Scientific Research B-20370051 (to S. N.) and by the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Corp. (to K. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- PAPS
- 3′-phosphoadenosine 5′-phosphosulfate
- DIC
- differential interference contrast
- DTC
- distal tip cell
- EGFP
- enhanced green fluorescent protein
- GFP
- green fluorescent protein
- GAG
- glycosaminoglycan
- HA
- hemagglutinin
- hPAPST
- human PAPS transporter
- HS
- heparan sulfate
- PAP
- 3′-phosphoadenosine 5′-phosphate
- Sqv
- squashed vulva
- NS
- 2-N-sulfate
- 2S
- 2-O-sulfate
- 6S
- 6-O-sulfate.
REFERENCES
- 1.Kehoe J. W., Bertozzi C. R. (2000) Chem. Biol. 7, R57–R61 [DOI] [PubMed] [Google Scholar]
- 2.Schwartz N. B., Domowicz M. (2002) Glycobiology 12, 57R–68R [DOI] [PubMed] [Google Scholar]
- 3.Honke K., Zhang Y., Cheng X., Kotani N., Taniguchi N. (2004) Glycoconj. J. 21, 59–62 [DOI] [PubMed] [Google Scholar]
- 4.Zhang H., Uchimura K., Kadomatsu K. (2006) Ann. N.Y. Acad. Sci. 1086, 81–90 [DOI] [PubMed] [Google Scholar]
- 5.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 6.Sugahara K., Mikami T. (2007) Curr. Opin. Struct. Biol. 17, 536–545 [DOI] [PubMed] [Google Scholar]
- 7.Nadanaka S., Kitagawa H. (2008) J. Biochem. 144, 7–14 [DOI] [PubMed] [Google Scholar]
- 8.Morita I., Kizuka Y., Kakuda S., Oka S. (2008) J. Biochem. 143, 719–724 [DOI] [PubMed] [Google Scholar]
- 9.Robbins P. W., Lipmann F. (1957) J. Biol. Chem. 229, 837–851 [PubMed] [Google Scholar]
- 10.Klaassen C. D., Boles J. W. (1997) FASEB J. 11, 404–418 [DOI] [PubMed] [Google Scholar]
- 11.Strott C. A. (2002) Endocr. Rev. 23, 703–732 [DOI] [PubMed] [Google Scholar]
- 12.Robbins P. W., Lipmann F. (1958) J. Biol. Chem. 233, 681–685 [PubMed] [Google Scholar]
- 13.Zaruba M. E., Schwartz N. B., Tennekoon G. I. (1988) Biochem. Biophys. Res. Commun. 155, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 14.Besset S., Vincourt J. B., Amalric F., Girard J. P. (2000) FASEB J. 14, 345–354 [DOI] [PubMed] [Google Scholar]
- 15.Dejima K., Seko A., Yamashita K., Gengyo-Ando K., Mitani S., Izumikawa T., Kitagawa H., Sugahara K., Mizuguchi S., Nomura K. (2006) J. Biol. Chem. 281, 11431–11440 [DOI] [PubMed] [Google Scholar]
- 16.Schwarz J. K., Capasso J. M., Hirschberg C. B. (1984) J. Biol. Chem. 259, 3554–3559 [PubMed] [Google Scholar]
- 17.Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellén L. (2008) J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 18.Sohaskey M. L., Yu J., Diaz M. A., Plaas A. H., Harland R. M. (2008) Development 135, 2215–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederick J. P., Tafari A. T., Wu S. M., Megosh L. C., Chiou S. T., Irving R. P., York J. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11605–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dick G., Grøndahl F., Prydz K. (2008) Glycobiology 18, 53–65 [DOI] [PubMed] [Google Scholar]
- 21.Capasso J. M., Hirschberg C. B. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7051–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandon E. C., Milla M. E., Kempner E., Hirschberg C. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10707–10711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozeran J. D., Westley J., Schwartz N. B. (1996) Biochemistry 35, 3695–3703 [DOI] [PubMed] [Google Scholar]
- 24.Ozeran J. D., Westley J., Schwartz N. B. (1996) Biochemistry 35, 3685–3694 [DOI] [PubMed] [Google Scholar]
- 25.Kamiyama S., Suda T., Ueda R., Suzuki M., Okubo R., Kikuchi N., Chiba Y., Goto S., Toyoda H., Saigo K., Watanabe M., Narimatsu H., Jigami Y., Nishihara S. (2003) J. Biol. Chem. 278, 25958–25963 [DOI] [PubMed] [Google Scholar]
- 26.Kamiyama S., Sasaki N., Goda E., Ui-Tei K., Saigo K., Narimatsu H., Jigami Y., Kannagi R., Irimura T., Nishihara S. (2006) J. Biol. Chem. 281, 10945–10953 [DOI] [PubMed] [Google Scholar]
- 27.Goda E., Kamiyama S., Uno T., Yoshida H., Ueyama M., Kinoshita-Toyoda A., Toyoda H., Ueda R., Nishihara S. (2006) J. Biol. Chem. 281, 28508–28517 [DOI] [PubMed] [Google Scholar]
- 28.Lüders F., Segawa H., Stein D., Selva E. M., Perrimon N., Turco S. J., Häcker U. (2003) EMBO J. 22, 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali R. A., Mellenthin K., Fahmy K., Da Rocha S., Baumgartner S. (2005) Dev. Genes. Evol. 215, 537–543 [DOI] [PubMed] [Google Scholar]
- 30.Clément A., Wiweger M., von der Hardt S., Rusch M. A., Selleck S. B., Chien C. B., Roehl H. H. (2008) PLoS Genet. 4, e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman T., Chernova M. N., Clark J. S., Jiang L., Alper S. L., Nehrke K. (2005) Am. J. Physiol. Cell Physiol. 289, C341–C351 [DOI] [PubMed] [Google Scholar]
- 32.Bülow H. E., Hobert O. (2004) Neuron 41, 723–736 [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi S., Dejima D., Nomura K. (2009) Trends Glycosci. Glycotechnol. 21, 179–191 [Google Scholar]
- 34.Yamada S., Van Die I., Van den Eijnden D. H., Yokota A., Kitagawa H., Sugahara K. (1999) FEBS Lett. 459, 327–331 [DOI] [PubMed] [Google Scholar]
- 35.Toyoda H., Kinoshita-Toyoda A., Selleck S. B. (2000) J. Biol. Chem. 275, 2269–2275 [DOI] [PubMed] [Google Scholar]
- 36.Nabetani T., Miyazaki K., Tabuse Y., Tsugita A. (2006) Proteomics 6, 4456–4465 [DOI] [PubMed] [Google Scholar]
- 37.Kim T. H., Hwang S. B., Jeong P. Y., Lee J., Cho J. W. (2005) FEBS Lett. 579, 53–58 [DOI] [PubMed] [Google Scholar]
- 38.Berninsone P. M. (December18, 2006) in WormBook (C. elegans Research Community, ed) doi/10.1895/wormbook.1.125.1, http://www.wormbook.org
- 39.Gumienny T. L., MacNeil L. T., Wang H., de Bono M., Wrana J. L., Padgett R. W. (2007) Curr. Biol. 17, 159–164 [DOI] [PubMed] [Google Scholar]
- 40.Franks D. M., Izumikawa T., Kitagawa H., Sugahara K., Okkema P. G. (2006) Dev. Biol. 296, 409–420 [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa H., Izumikawa T., Mizuguchi S., Dejima K., Nomura K. H., Egusa N., Taniguchi F., Tamura J., Gengyo-Ando K., Mitani S., Nomura K., Sugahara K. (2007) J. Biol. Chem. 282, 8533–8544 [DOI] [PubMed] [Google Scholar]
- 42.Bülow H. E., Tjoe N., Townley R. A., Didiano D., van Kuppevelt T. H., Hobert O. (2008) Curr. Biol. 18, 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharya R., Townley R. A., Berry K. L., Bülow H. E. (2009) J. Cell Sci. 122, 4492–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burdine R. D., Branda C. S., Stern M. J. (1998) Development 125, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 46.Gupta B. P., Sternberg P. W. (2002) Dev. Biol. 247, 102–115 [DOI] [PubMed] [Google Scholar]
- 47.Mohler W. A., Simske J. S., Williams-Masson E. M., Hardin J. D., White J. G. (1998) Curr. Biol. 8, 1087–1090 [DOI] [PubMed] [Google Scholar]
- 48.Gengyo-Ando K., Mitani S. (2000) Biochem. Biophys. Res. Commun. 269, 64–69 [DOI] [PubMed] [Google Scholar]
- 49.Mizuguchi S., Uyama T., Kitagawa H., Nomura K. H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. (2003) Nature 423, 443–448 [DOI] [PubMed] [Google Scholar]
- 50.Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Ando K., Mitani S., Sugahara K. (2004) J. Biol. Chem. 279, 53755–53761 [DOI] [PubMed] [Google Scholar]
- 51.Kinoshita A., Sugahara K. (1999) Anal. Biochem. 269, 367–378 [DOI] [PubMed] [Google Scholar]
- 52.Gravato-Nobre M. J., Nicholas H. R., Nijland R., O'Rourke D., Whittington D. E., Yook K. J., Hodgkin J. (2005) Genetics 171, 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gengyo-Ando K., Yoshina S., Inoue H., Mitani S. (2006) Biochem. Biophys. Res. Commun. 349, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 54.Dejima K., Murata D., Mizuguchi S., Nomura K. H., Gengyo-Ando K., Mitani S., Kamiyama S., Nishihara S., Nomura K. (2009) FASEB J. 23, 2215–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilleard J. S., Barry J. D., Johnstone I. L. (1997) Mol. Cell. Biol. 17, 2301–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mello C., Fire A. (1995) Methods Cell Biol. 48, 451–482 [PubMed] [Google Scholar]
- 57.Miller D. M., Shakes D. C. (1995) Methods Cell Biol. 48, 365–394 [PubMed] [Google Scholar]
- 58.Käll L., Krogh A., Sonnhammer E. L. (2005) Bioinformatics 21, Suppl. 1, i251–i257 [DOI] [PubMed] [Google Scholar]
- 59.Käll L., Krogh A., Sonnhammer E. L. (2007) Nucleic Acids Res. 35, W429–W432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herman T., Hartwieg E., Horvitz H. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hwang H. Y., Olson S. K., Esko J. D., Horvitz H. R. (2003) Nature 423, 439–443 [DOI] [PubMed] [Google Scholar]
- 62.Morio H., Honda Y., Toyoda H., Nakajima M., Kurosawa H., Shirasawa T. (2003) Biochem. Biophys. Res. Commun. 301, 317–323 [DOI] [PubMed] [Google Scholar]
- 63.Chisholm A. D., Hardin J. (December1, 2005) in WormBook (C. elegans Research Community, ed) doi/10.1895/wormbook.1.35.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- 64.Wang X., Zhao Y., Wong K., Ehlers P., Kohara Y., Jones S. J., Marra M. A., Holt R. A., Moerman D. G., Hansen D. (2009) BMC Genomics 10, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yano H., Yamamoto-Hino M., Abe M., Kuwahara R., Haraguchi S., Kusaka I., Awano W., Kinoshita-Toyoda A., Toyoda H., Goto S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silbert J. E., Sugumaran G. (1995) Biochim. Biophys. Acta 1241, 371–384 [DOI] [PubMed] [Google Scholar]
- 67.Nelson F. K., Albert P. S., Riddle D. L. (1983) J. Ultrastruct. Res. 82, 156–171 [DOI] [PubMed] [Google Scholar]
- 68.Höflich J., Berninsone P., Göbel C., Gravato-Nobre M. J., Libby B. J., Darby C., Politz S. M., Hodgkin J., Hirschberg C. B., Baumeister R. (2004) J. Biol. Chem. 279, 30440–30448 [DOI] [PubMed] [Google Scholar]
- 69.Cipollo J. F., Awad A. M., Costello C. E., Hirschberg C. B. (2004) J. Biol. Chem. 279, 52893–52903 [DOI] [PubMed] [Google Scholar]
- 70.Sternberg P. W., Horvitz H. R. (1988) Dev. Biol. 130, 67–73 [DOI] [PubMed] [Google Scholar]
- 71.Sawa H., Lobel L., Horvitz H. R. (1996) Genes Dev. 10, 2189–2197 [DOI] [PubMed] [Google Scholar]
- 72.Lin X. (2004) Development 131, 6009–6021 [DOI] [PubMed] [Google Scholar]
- 73.Minniti A. N., Labarca M., Hurtado C., Brandan E. (2004) J. Cell Sci. 117, 5179–5190 [DOI] [PubMed] [Google Scholar]
- 74.Bender A. M., Kirienko N. V., Olson S. K., Esko J. D., Fay D. S. (2007) Dev. Biol. 302, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eisenmann D. M. (June25, 2005) in WormBook (C. elegans Research Community, ed) doi/10.1895/wormbook.1.7.1, http://www.wormbook.org
- 76.Carroll B. T., Dubyak G. R., Sedensky M. M., Morgan P. G. (2006) J. Biol. Chem. 281, 35989–35996 [DOI] [PubMed] [Google Scholar]
- 77.Hattori K., Inoue M., Inoue T., Arai H., Tamura H. O. (2006) J. Biochem. 139, 355–362 [DOI] [PubMed] [Google Scholar]
- 78.Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., Hewitson M., Holz C., Khan M., Lazik S., Martin C., Nitzsche B., Ruer M., Stamford J., Winzi M., Heinkel R., Röder M., Finell J., Häntsch H., Jones S. J., Jones M., Piano F., Gunsalus K. C., Oegema K., Gönczy P., Coulson A., Hyman A. A., Echeverri C. J. (2005) Nature 434, 462–469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.