Abstract
Orchestrated regulation of neuronal migration and morphogenesis is critical for neuronal development and establishment of functional circuits, but its regulatory mechanism is incompletely defined. We established and analyzed mice with neural-specific knock-out of Trio, a guanine nucleotide exchange factor with multiple guanine nucleotide exchange factor domains. Knock-out mice showed defective cerebella and severe signs of ataxia. Mutant cerebella had no granule cells in the internal granule cell layer due to aberrant granule cell migration as well as abnormal neurite growth. Trio-deficient granule cells showed reduced extension of neurites and highly branched and misguided processes with perturbed stabilization of actin and microtubules. Trio deletion caused down-regulation of the activation of Rac1, RhoA, and Cdc42, and mutant granule cells appeared to be unresponsive to neurite growth-promoting molecules such as Netrin-1 and Semaphorin 6A. These results suggest that Trio may be a key signal module for the orchestrated regulation of neuronal migration and morphogenesis during cerebellar development. Trio may serve as a signal integrator decoding extrinsic signals to Rho GTPases for cytoskeleton organization.
Keywords: Cell Migration, Cytoskeleton, Gene Knockout, Neurodevelopment, Signal Transduction, Guanine Nucleotide Exchange Factor, Parallel Fiber, Small GTPase
Introduction
Cerebellar development is an exquisitely orchestrated process that produces a layered structure of the cerebellar cortex containing granule cells, Purkinje cells, and cerebellar interneurons (1). Granule cell precursors arising from a germinal zone in the rhombic lip migrate tangentially over the cerebellar anlage to form an external granule cell layer (EGL)3 and continue to proliferate in the outer EGL (2). In the inner part of the EGL, post-mitotic granule cells extend bipolar axons parallel to the pial surface and move tangentially along the axis of the developing axons, forming the characteristic early parallel fibers. At the border of the inner EGL, granule neurons extend a third process perpendicular to the surface. After this leading process extension, cell bodies of granule neurons migrate radially along Bergmann glia to position past the Purkinje cell layer to form the IGL (1, 3). Coordinated axonal guidance and neuronal migration are essential steps in this complex developmental process. It is known that various guidance molecules converge on cytoskeleton regulation (4–6). How extracellular signals are transduced from cell-surface receptors to the migration machinery is incompletely understood (7).
Rho-family small GTPases, which are activated by guanine nucleotide exchange factors (GEFs), are key regulators of cytoskeleton dynamics (8–11). Extensive studies suggest that Rho GTPases has an important role in neuronal development, including axon guidance, migration, morphogenesis, and plasticity of neurons (12–14). How migrating neurons interpret guidance cues to GTPase activity and directional movement is a major challenge for understanding neural development.
As a founding member of an intriguing family of Dbl GEFs, Trio contains two GEF domains (15, 16). Several studies using Caenorhabditis elegans and Drosophila show that UNC-73/Trio regulates axon guidance through regulating the activities of Rac GTPases in vivo (17–20). In mammals, Trio is expressed ubiquitously in various tissues, including the central nervous system (15, 21), and Trio complete knock-out mice displayed disorganized hippocampus as well as olfactory bulb and neuronal clusters in the hindbrain (22, 23). It is therefore suggested that Trio may play an important part in the neuronal development. These Trio knock-out mice are embryonic-lethal (22), which prevents analysis of the role of Trio in postnatal development of the brain (particularly the cerebellum).
To address the role of Trio in postnatal development of the brain, we generated mice with brain-specific knock-out of Trio. Our results uncover Trio function and its importance in the developing cerebellum. We also propose that Trio may potentially act as an integrator decoding such signals as Netrin-1 and Semaphorin 6A to small GTPases during cerebellar development.
EXPERIMENTAL PROCEDURES
Generation of Floxed Trio Mice and Nervous System-specific Knock-out Mice Named Trioflox/flox; Nestin-Cre (TrioNKO)
Experiments were conducted in accordance with the Animal Care and Use Committee of Model Animal Research Center of Nanjing University, Nanjing, China.
Mice used in this study were of a mixed 129/B6 background. Bacterial artificial chromosome (BAC) retrieval methods were used for constructing the targeting vector (24). In brief, the Trio locus encoding the GEF1 domain was retrieved from a 129/sv BAC clone (bMQ 214m08, provided by the Sanger Institute) by a retrieval vector containing two homologous arms. Exons 22–25, which encode a portion of the GEF1, were flanked by two loxP sites, and an frt-Neo-frt cassette was used as a positive selection marker. This deletion causes an out-of-frame reading shift, thereby generating a premature stop codon and a loss-of-function allele. Embryonic stem W4 (ES) cells were electroporated with the linearized targeting vector, screened for homologous recombination by PCR and Southern blot assay. Chimeric mice were generated by injecting ES cells into C57BL/6 blastocysts, followed by transfer to pseudo-pregnant mice. The floxed mice with germ line transmission were confirmed by genotyping analysis. To generate Trioflox/flox; Nestin-Cre (TrioNKO) mice, the floxed mice were crossed with Nestin-Cre mice expressing a Cre recombinase specifically in the nervous system (strain B6.Cg (SJL)-TgN (Nes-Cre)Kln, Jackson Laboratory).
Generation of GFAP-Cre Mice
A transgenic expressive vector contains a Cre gene driven by a 2.2-kb human gfap promoter. The vector linearized with BglII and HindIII was introduced into the male pronuclei of fertilized eggs by microinjection. The eggs were then implanted into the oviduct of a pseudopregnant foster mother. The founders and their offspring are identified by PCR genotyping. Transgenic expression patterns of Cre were examined with a R26R reporter line (129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym, (Jackson Laboratory) carrying a lacZ gene whose expression requires excision of loxP-flanked stop sequences.
Southern Blot Analysis
Genomic DNA was digested with EcoRV and transferred onto a nylon membrane followed by probing with a 700-bp 32P-labeled probe outside the 5′-end or the 3′-end of the targeting construct as indicated in Fig. 1A. In wild-type DNA, only an 18-kb band was detected, whereas the floxed allele showed an additional 8.5-kb band with the 5′-probe, and an additional 11.5-kb band with the 3′-probe. A 9.5-kb band was generated by the knock-out allele with the 3′-probe.
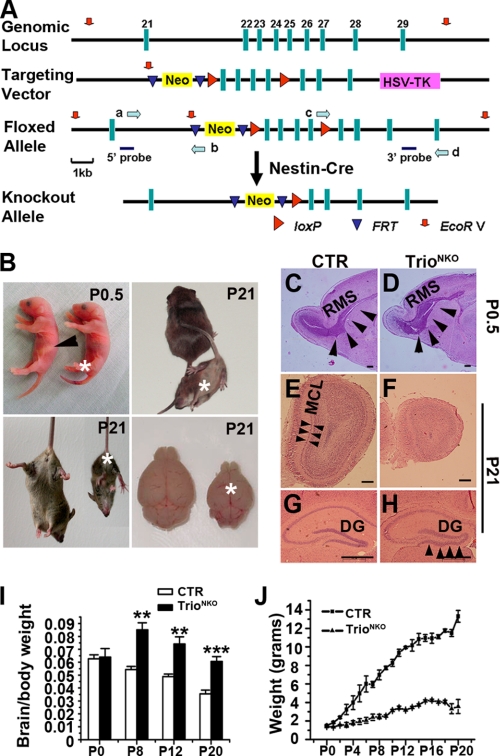
FIGURE 1.
Targeted disruption of the Trio gene in the nervous system. A, schematic representation of Trio knock-out strategy specific to the nervous system. The 11.5-kb genomic DNA fragment containing Trio exons 22–25 was subcloned from 129/sv BAC. The first loxP site (red arrowheads) was targeted downstream of exon 25. Mice containing the floxed allele were crossed with Nestin-Cre (tg) mice to generate Trio+/flox; Nestin-Cre and Trioflox/flox; Nestin-Cre mice. The probes used for Southern blot analysis are shown as solid blue bars, and the locations of PCR primers (a–d) are for screening homologous recombination (green arrows). B, 90% of mutant mice died within 24 h after birth without sucking milk (arrowhead, control stomach filled with milk). Few mutants could survive for 3 weeks with severe ataxia, retarded growth, and smaller brain size. Asterisks indicate the knock-out mice. C and D, Nissl-stained sagittal sections show a reduced size of the mutant olfactory bulb and an enlarged rostral migratory stream (RMS, arrowheads) at P0.5. E and F, hematoxylin and eosin-stained coronal sections of the olfactory bulbs of P21 mice. The mitral cell layer (MCL) is clearly visible in the control mice (arrowheads) but cannot be distinguished from the internal granule cell layer in the mutant mice. G and H, lamination defects of the hippocampus in Trio mutant mice. At P21, granule cells of dentate gyrus (DG) are tightly arranged in the control mice, but not in the mutant (arrowheads). I, the ratio of brain/body weight of Trio-deficient and control mice have no obvious differences at P0, but the ratio of mutant mice increased after P8 compared with the control; **, p < 0.01, ***, p < 0.001. J, the growth curves of CTR and TrioNKO mice. From P2, the weight of mutant mice is significantly lower than control (n = 3–5). Scale bars in C–H, 150 μm.
Western Blot Analysis
Western blot analyses were done for measurement of the expression of Trio and other proteins. Protein samples were prepared according to a method described elsewhere (25). Equal amounts of protein were loaded for 12% SDS-PAGE followed by protein transfer to a nitrocellulose membrane. For Trio analysis, 5% SDS-PAGE was used. The membrane was probed with antibodies specific to Trio (1:200, Santa Cruz Biotechnology, Santa Cruz, CA), Rac1 (1:1000, Upstate), RhoA (1:500, Cell Signaling), and Cdc42 (1:250, Cell Signaling) followed by horseradish peroxidase-conjugated secondary antibody. The membrane was incubated in SuperSignal West Dura Substrate (Pierce) before exposure to film.
Histological and Immunohistochemical Analyses
Histological and immunohistochemical analyses were done on 4% paraformaldehyde-fixed paraffin (8 μm) or frozen sections (20 μm). For the histopathology assay, paraffin-embedded sections were stained with hematoxylin and eosin. Frozen sections were stained for Nissl substance with cresyl violet (Sigma). For immunohistochemical analysis, brain sections were incubated with antibodies to Pax6 (1:1000, kindly provided originally by Dr. Grant Mastick, University of Michigan), calbindin D-28k (1:500, Chemicon), RC2 (1:4, DHSB), GFAP (1:500, Chemicon), Cre (1:500, Novagen), NeuN (1:200, Chemicon), BrdUrd (1:5, Upstate), Tuj1 (1:100, Chemicon), TAG-1 (1:5, DHSB), Tau (1:200, Bioworld), phospho-histone-H3 (1:100, Cell Signaling), and cleaved caspase-3 (1:200, Cell Signaling) overnight. After washing the sections, immunofluorescent dye-conjugated secondary antibodies (1:100 (Invitrogen) or 1:200 (Sigma)) were used. To visualize the cell nucleus, sections were counterstained with propidium iodide (1.5 μm, Invitrogen) or TOPRO-3 iodide (1 mm, Invitrogen). All sections with fluorescent staining were examined with a fluorescence confocal microscope (DMIRE2, Leica).
BrdUrd Labeling
To examine the proliferation of granule cell precursors, P0.5 mice were injected (intraperitoneally) with 5-bromo-2-deoxyuridine (BrdUrd, 50 mg/kg body weight, Sigma) diluted in saline solution. Mice were killed 1 h later. To follow migrating cells in vivo, P8 mice were injected (intraperitoneally) with BrdUrd (50 mg/kg body weight). Mice were killed at the indicated time after injection and fixed with 4% paraformaldehyde in phosphate-buffered solution. Brain sections (20 μm) were stained with antibody to BrdUrd (1:5, Upstate) and visualized by incubation with fluorescein isothiocyanate-labeled secondary antibody.
DiI Labeling of EGL Cells
P0.5 cerebella were perfused and fixed with 4% paraformaldehyde. Small crystals of the lipophilic dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate (DiI) (Invitrogen) were placed on the EGL for 6 days at 4 °C. They were then embedded in 4% agar and cut in the coronal direction at a thickness of 200 μm using a vibratome. After washing with phosphate-buffered solution, sections were analyzed using a fluorescent microscope (DMIRE2, Leica).
Neuronal Cultures
Cerebellar EGL cells isolated from P0.5 animals were digested with trypsin-EDTA (Amersham Biosciences-BRL) for 10 min at 37 °C. They were triturated into a single-cell suspension with fire-polished glass pipettes. Dissociated cells were plated onto coverslips pre-coated with poly-d-lysine (100 μg/ml, Sigma) at a density of 150 neurons per mm2. They were cultured in the plating medium (Dulbecco's modified Eagle's medium with 10% fetal bovine serum). Four hours after culture, plating media were changed into a neurobasal medium supplemented with 2% B27 (Amersham Biosciences). Cells were treated with the following reagents: glutamate (50 μm, Sigma), recombinant Netrin-1 (250 ng/ml, R&D Systems), recombinant nerve growth factor (50 ng/ml, R&D Systems), and Semaphorin 6A/Fc (1000 ng/ml, R&D Systems). For cytoskeleton analysis and Rho GTPases activity assay, dissociated cerebellar neurons were treated with Cytochalasin D, Taxol, Y27632, and NSC23766 24 h after culture. For the in vitro migration assay, microexplant cultures were prepared as described (26). Briefly, P4 cerebellar EGL was cut into pieces (300–400 μm) followed by plating on dishes coated with laminin (25 μg/ml, Sigma). Cultures were maintained in 5% CO2 at 37 °C and analyzed 1–2 days after incubation. For immunocytochemical analysis, cultures were fixed with 4% paraformaldehyde and immunostained with antibodies against Tuj1 (1:100, Chemicon), TAG-1 (1:5, DHSB), Tau (1:200, Bioworld), and tetramethyl rhodamine isothiocyanate (TRITC)-labeled phalloidin (0.5 μg/ml, Sigma).
Time-lapse Videomicroscopy
Cerebellar EGL microexplants cultured for 30 h were placed in a chamber containing 5% CO2 at 37 °C. Images were captured by a motorized inverted research microscope (I × 81, Olympus) equipped with a digital camera (QImaging) driven with Image-Pro Plus 6.0 software. Time-series images were acquired every 10 min using a ×10 dry objective lens equipped with phase optics.
Analyses of Endogenous Activity for Rac1, Cdc42, and RhoA
The constructs PBD-GST and RBD-GST, which contain the GTPase binding domains of PAK and Rhotekin, respectively, were provided by Hollis T. Cline (27). Whole brains of P0.5 mice were lysed in buffer A (25 mm HEPES, pH 7.5, 1% Nonidet P-40, 10 mm MgCl2, 100 mm NaCl, 5% glycerol, 1 mm phenylmethylsulfonyl fluoride). The lysates clarified by centrifugation at 10,000 × g for 2 min were incubated with GST fusion proteins immobilized on glutathione-Sepharose beads for 2 h at 4 °C. Total cell lysates and GST pulldown-associated proteins were subjected to Western blot analysis with antibodies against Rac1 (1:1000, Upstate), RhoA (1:500, Cell Signaling), and Cdc42 (1:250, Cell Signaling).
Statistical Analyses
Data are mean ± S.E. Differences between groups were determined by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Generation and Characterization of TrioNKO Mice
We established a mouse strain with loxP sites flanking exons 22–25 of the coding region for the GEF1 domain of Trio (Trioflox/+). Trioflox/flox mice were fertile and healthy without obvious morphological or behavioral abnormalities. Mice containing the floxed Trio gene were crossed with Nestin-Cre transgenic mice expressing a Cre recombinase to specifically ablate Trio expression in nerve tissues (28) (Fig. 1A). Birth of pups, including Trio-floxed mice with (Trioflox/flox; Nestin-Cre and Trioflox/+; Nestin-Cre) or without (Trioflox/flox and Trioflox/+) Cre occurred in the expected Mendelian ratio. Southern blot assays confirmed the successful floxed alleles (supplemental Fig. S1A) and Cre-mediated specific deletions (supplemental Fig. S1B) in the nervous system. In Trio null mice, the floxed region of Trio was almost completely deleted in the brain, whereas it remained intact in the kidney, lung, and intestine (supplemental Fig. S1B). Western blot assay showed efficient ablation of Trio in the brain at different ages (E14.5 to P0) (supplemental Fig. S1C). We then quantified the relative amounts of Rac1, Cdc42, and RhoA in knock-out nerve tissues to assess the compensatory expressions of the Trio-related Rho GTPases; there was no significant change in the expression of these proteins (supplemental Fig. S1D). In subsequent experiments, we used Trioflox/+; Nestin-Cre mice as controls (CTR); and Trioflox/flox; Nestin-Cre mice as TrioNKO.
We examined the phenotypes at embryonic stages from E15.5 to E18.5. Histological results showed that, except olfactory bulb at E18.5 showing disorganized lamination, the neuronal tissues from mutant embryos, including cerebral cortex and hippocampus, showed comparable histology to CTR (supplemental Fig. S2, A–J). The histology of heart and kidney from P0 TrioNKO mice is also normal (supplemental Fig. S2, K–N).
About 90% of TrioNKO mice died within 1 day after birth without sucking milk (Fig. 1B). The remaining TrioNKO mice could survive for a further 5–22 days. Surviving mice displayed severe signs of ataxia: they had an ataxic gait, and clasped the hind limbs when subjected to tail suspension, whereas the CTR mice extended and shook their hind limbs (Fig. 1B). These features strongly suggested an abnormality in the cerebellum of TrioNKO mice.
Macrophenotypic examination of surviving TrioNKO mice showed a smaller brain and reduced body weight (Fig. 1, B and J). The ratio of brain/body weight at P0.5 was comparable to CTR mice (p = 0.75; n = 3), but it then increased significantly from P8 to P20 (p < 0.01–0.001) (Fig. 1I). This result suggested growth retardation in mutant mice. Histological examination showed a smaller olfactory bulb with an enlarged rostral migratory stream, and defective mitral cell layer formation in mutant mice (Fig. 1, C–F). This defect may account for the failure to suckle milk (29). The hippocampus also showed a disorganized structure in the dentate gyrus (Fig. 1, G and H). Such histological changes in the mutant olfactory bulb and dentate gyrus are consistent with that of embryonic lethal mice with complete knock-out of Trio (22).
Multiple Developmental Defects of Cerebellum of TrioNKO Mice
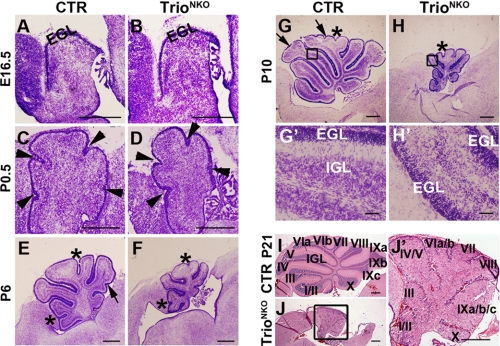
At the early development stage (E16.5), the TrioNKO cerebellum had a similar morphology and histological structure to CTR, with comparable thickness of the EGL (Fig. 2, A and B) and a similar staining pattern of the EGL marker Pax6 (supplemental Fig. S3, A and B), indicating that the early migration of granule cell precursors from the rombic lip was normal. During postnatal development, the TrioNKO cerebellum was profoundly defective as reflected by its small size and lack of granule cells in the IGL (Fig. 2, C–J′). At P0.5, the mutant cerebellum had five typical cardinal lobes formed by four principal fissures and comparable thickness of the EGL, but the size of the mutant cerebellum was reduced (∼80% of the CTR littermates) (Fig. 2, C and D); at later development stages (P6 to P21), the cardinal lobes of the mutant cerebellum developed corresponding lobules (Fig. 2, E–J′), even though some sub-lobules were not formed (e.g. IV and V; VIa and VIb; and IXa, IXb, and IXc) (Fig. 2, I–J′). TrioNKO cerebellar size did not increase after P6. At P21, their sizes were as small as 15% of CTR cerebella (Fig. 2, E–J′).
FIGURE 2.
Nestin-Cre-mediated Trio deletion leads to developmental defects of the cerebellum. A–J′, sagittal sections of cerebella at different developmental stages examined by Nissl staining (A–H′) or hematoxylin and eosin staining (I–J′). A and B, at E16.5, control and mutant cerebella are relatively similar in size and structure. C–J′, from P0.5, mutant cerebella are gradually smaller than control, and a clear IGL is not observed. The overall anteroposterior pattern of foliation is intact in the mutant, although several fissures forming sub-lobules (arrows in E and G) are not developed. Arrowheads represent principal fissures forming cardinal lobes, asterisks indicate fissures forming lobules, and arrows represent fissures forming sub-lobules. Roman numerals represent the corresponding lobules. G′, H′, and J′ are magnifications of G, H, and J, respectively, emphasizing the lack of IGL in the mutant cerebella. Scale bars: A–H and I–J′), 200 μm; G′ and H′, 20 μm.
Glia-specific Knock-out of Trio Has No Effect on Cerebellum Development
We examined Bergmann glial cells of the TrioNKO cerebellum by staining with antibodies to RC2 (marker for early glial cells) and glial fibrillary acidic protein (GFAP) (specific marker for Bergmann glia). There was no obvious morphological difference of early glia (RC2 staining) in early cerebella from P1 CTR mice and TrioNKO mice (supplemental Fig. S4, A and B), but Trio-deleted glial fibers (GFAP staining) became progressively sparse at later stages (P8 to P21) (supplemental Fig. S4, C–F). This observation showed impaired development of Bergmann glial fibers in TrioNKO.
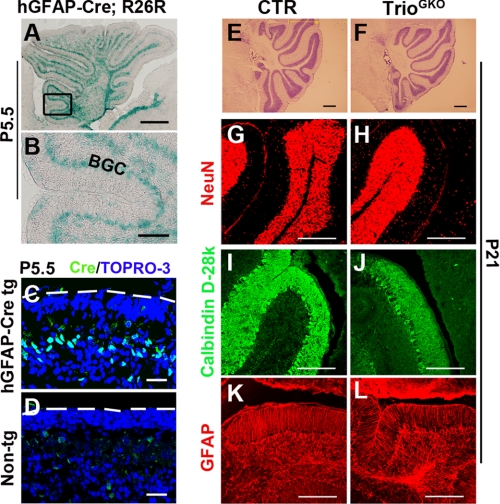
Cre expression driven by a nestin promoter can occur in neuronal and glia progenitors. To determine if the failure of cerebellar development in TrioNKO mice was caused by Trio deletion in the glia, we made an hGFAP-Cre transgenic line expressing Cre in Bergmann glial cells. Results from Cre and lacZ staining showed a Bergmann glia-specific expression of Cre at P5.5 in the cerebellum (Fig. 3, A–D). We then crossed it with the Trioflox/flox line to knock-out Trio in the Bergmann glia cells. The resultant mice Trioflox/flox; GFAP-Cre (referred to as TrioGKO in this report) displayed normal behaviors and normal cerebellar development (Fig. 3, E and F). The mutant cerebellum exhibited normal differentiation of granule cells, normal development of Purkinje cells, and normal formation of Bergmann glial fibers as evidenced by NeuN, calbindin, and GFAP staining, respectively (Fig. 3, G–L). We concluded that specific deletion of Trio in the Bergmann glia has no effect on cerebellar development. The defect in TrioNKO cerebellum may be caused by the impaired development of granule cells.
FIGURE 3.
Normal cerebellar development in Trioflox/flox; GFAP-Cre mice. A and B, Cre activity of the hGFAP-Cre transgene shown by the R26R reporter mouse assay. In hGFAP-Cre; R26R mice, reporter-positive cells were detected at the Bergmann glial cell (BGC) body layer at P5.5. C and D, sagittal sections of the cerebellum from an hGFAP-Cre transgenic mouse and a non-transgene mouse (5.5 days old), immunostained with Cre antibody. Dashed lines indicate the pial surface of the cerebella. E and F, Nissl staining shows the normal structure of the Trioflox/flox; GFAP-Cre cerebella at P21. G and H, post-mitotic granule cells (NeuN), Purkinje cells (Calbindin D-28k), and glial cells (GFAP) are normally located in the mutant as well as the control at P21. Scale bars: A and E–L, 200 μm; B, 40 μm; and C and D, 20 μm.
TrioNKO Cerebellum Exhibits Neuronal Migration Defects
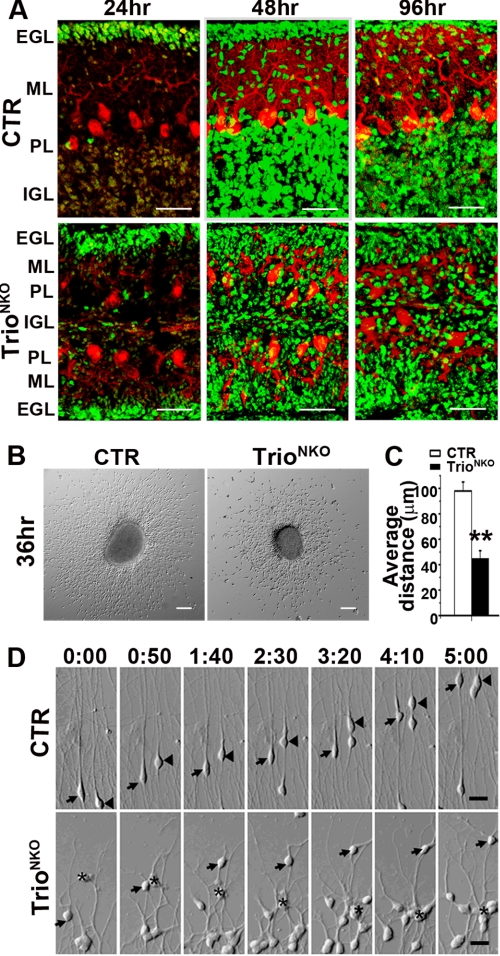
Granule cells are the most abundant neurons in the cerebellum, and their appropriate migration is crucial for IGL formation. We examined cerebellar granule neuron (CGN) migration in vivo and in vitro. For in vivo assay, we labeled granule cells with BrdUrd and determined their migration by localizing labeled cells with an immunofluorescence assay (30). In CTR mice, abundant BrdUrd-positive cells were detected in the EGL 24 h after labeling; labeled cells at the EGL, molecular layer, and IGL showed a typical migration pattern 48 h after labeling; and 85% of labeled cells had completed radial migration and were in the IGL 96 h after labeling (Fig. 4A). In the mutant cerebellum, although a comparable number of labeled cells could be detected in the EGL at 24 h, ∼80% of labeled cells were located outside of the Purkinje cell layer and were distributed randomly at the EGL and molecular layer at 96 h, showing an abnormal pattern of migration (Fig. 4A). In the in vitro assay, granule cells migrated away from the EGL explants independent of glial cells and followed the intrinsic program of differentiation (26, 31). Thirty-six hours after culture, the mean spreading distance of migrating granule cells of Trio-deficient explants were reduced by 2.1-fold compared with that of the control explants (98.15 ± 7.04 μm versus 45.11 ± 6.23 μm, p < 0.01) (Fig. 4, B and C). The mutant granule cells soma was located in random orientations, unlike the CTR cells, which were located in a radial direction (Fig. 4B). To examine the dynamic migration of the cells, we carried out time-lapse videomicroscopy on cerebellar EGL explants 30 h after culture. The CTR granule cells migrated in the opposite direction to the explants along the long leading process bearing typical growth cones (Fig. 4D and supplemental Video S1), whereas TrioNKO granule cells moved in random orientations along their abnormal processes (Fig. 4D and supplemental Video S2). Taken together, we conclude that deletion of Trio caused impaired migration of CGN.
FIGURE 4.
Defective migration of granule cells in TrioNKO cerebella in vivo and in vitro. A, dividing granule cells of the cerebella were pulse-labeled at P9 by BrdUrd and then visualized by BrdUrd immunofluorescent staining at indicated times after injection. B, microexplants of P4 EGL tissues 36 h after culture. Disordered migration of granule cells occurred in the mutant explant. C, decrease in mean migration distance of granule cells from the edge of aggregates in the Trio mutant; **, p < 0.01. D, microexplants were prepared from P4 EGL and cultured for 30 h before imaging. Explants are at the bottom of the frame. Intervals between pictures were 50 min. Control granule cells migrated in the opposite direction to the explant (arrows and arrowheads), whereas most Trio-deficient granule cells moved randomly (arrows) or backwards to the explant (asterisk). Scale bars: A, 25 μm; B, 100 μm; and D, 50 μm.
Neurite Growth and the Cytoskeleton of Trio-deficient CGN Are Impaired
Formation of neuronal neurites is important for cell migration. During postnatal development, post-mitotic neurons start to extrude bipolar axons to form parallel fibers and move tangentially along the fibers in the inner EGL (2, 32). To evaluate the neurite-forming properties of Trio-deficient CGN, we examined parallel fiber formation in vivo and neurite formation in vitro. At P8, the deeper portion of the mutant EGL delineated by expression of the transient marker for early parallel fiber TAG-1 appeared thick and non-compact in contrast with CTR (Fig. 5, A and B). This indicated abnormal development of parallel fibers in the inner EGL of mutant mice. We then studied the parallel fibers in newborn mutant mice when the parallel fibers just began to form. At P0.5, the inner EGL in CTR mice showed well organized spindle-shaped cell bodies along the horizontal axis, displaying typical tangential migration of granule cells. Granule cells in the mutant EGL showed disorganized cell bodies (Fig. 5, C and D), indicating abnormal organization of parallel fibers. To observe parallel fibers directly, we implanted DiI particles in fixed cerebella and examined fiber morphology under a microscope. Long and parallel arranged-shaped fibers can be observed in the CTR cerebellum, whereas the fibers of the mutant cerebellum appear short and disordered (Fig. 5, E and F). In vitro observation of culture explants showed processes growing from the mutant explant (shown by staining for neuronal-specific beta III tubulin Tuj1) to be greatly decreased in contrast to CTR (Fig. 6, A and B). CTR axons had a straight and long morphology, whereas Trio-deleted explants displayed tortuous and short processes (Fig. 6, A and B). A similar observation was obtained by staining explants with the axonal marker Tau (a natural microtubule-stabilizing protein (Fig. 6, C and D′). To examine the parallel fiber-forming ability of granule neurons in vitro, we stained the explants with antibody to TAG-1. In the CTR explant, long and even parallel fibers were detected, whereas the mutant exhibited short and fragmented TAG-1-positive fibers (Fig. 6, E and F′). Observation of the dynamic formation of processes showed that the mutant granule cells extended short and highly branched processed that frequently retracted and changed orientation, indicating that the processes lost guidance (Fig. 6G and supplemental Video S4), whereas the CTR cells displayed typical morphology and behaviors of a growing process (Fig. 6G and supplemental Video S3).
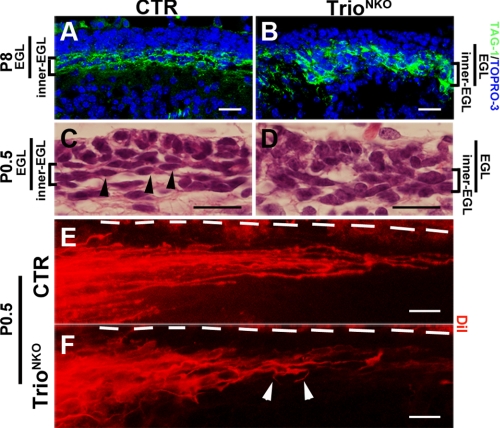
FIGURE 5.
Disrupted formation of parallel fibers in Trioflox/flox; Nestin-Cre mice. A and B, early parallel fibers in the inner EGL labeled with TAG-1 are thick and non-compact in Trio-deficient mice compared with control littermates. C and D, hematoxylin and eosin stained of coronal sections of P0.5 cerebella. Spindle-shaped cells (arrowheads) are horizontally located in the inner EGL of control mice. Mutant cell bodies in the inner EGL are disordered. E and F, DiI-labeled parallel fibers. Horizontal beams of parallel fibers in the inner EGL of CTR cerebellum (E) are not formed in the Trio mutant (F). Disorganized fibers are indicated by white arrowheads. Dashed lines indicate the pial surface of the cerebella. Scale bars in A–F: 20 μm.
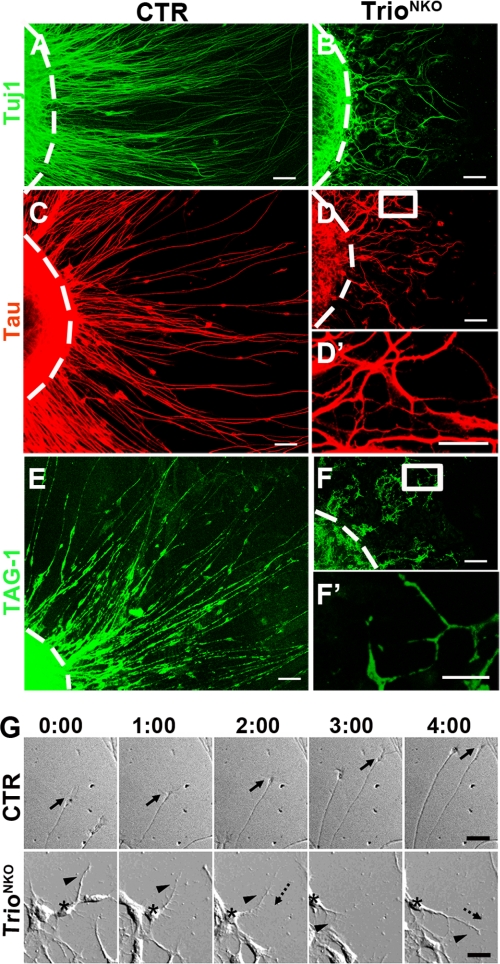
FIGURE 6.
Trio-deleted cerebellar EGL microexplant in culture displayed impaired formation of neurites. Expression of Tuj1 (A and B), Tau (C, D, and D′), and TAG-1 (E, F, and F′) in P4 EGL microexplants 36 h after culture. The dashed lines indicate the original border of the explants. D′ and F′ are magnifications of D and F, respectively. TrioNKO microexplant had short, disoriented, and fewer axons (B, D, and D′) and virtually no long parallel fiber formation (TAG-1 staining) (F and F′). G, microexplants were prepared from P4 EGL and cultured for 30 h before imaging. Explants are at the bottom of the frame. Intervals between pictures were 60 min. The control neuron extended long radial axons under the guidance of growth cones (arrows). The mutant neuron (asterisk) extended short axons with many branches (arrowhead). Frequently, the extended axon may retract (long dashed arrow) and then re-extend again (short dashed arrow). Scale bars: A–D, E, and F, 40 μm; D′ and F′, 10 μm; and G, 50 μm.
To characterize the properties of process formation of mutant CGN, we examined the neurite outgrowth in dissociated CGNs. Sixty hours after culture, the granule neurons underwent a relative stationary phase (5). We stained granule neurons for actin and neuronal-specific Tuj1 (Fig. 7, A and B). CTR granule cells exhibited typical neurite morphologies: long and smooth processes with fewer branches, the microtubule extending to the middle of the growth cone, and a regular growth cone delineated with actin staining. Trio-deficient granule cells clearly showed different morphologies: short and highly branched processes, irregular growth cone at the leading processes with highly branched terminals, and strong filipodia in the axon shaft and the tips of leading process (Fig. 7A). Quantification of the mean number of primary neurites extending from the cell body for each neuron showed no significant difference between CTR and TrioNKO (2.19 ± 0.42 of CTR versus 1.99 ± 0.15 of TrioNKO, p = 0.58; n = 60) (Fig. 7D). TrioNKO neurons had significantly shorter axon length than CTR (68.25 ± 4.26 μm versus 106.75 ± 8.45 μm, p = 0.005; n = 60) (Fig. 7E). Compared with CTR granule cells, branch number of the mutant neurites per 100 μm of axon increased from 1.77 ± 0.56 to 5.43 ± 0.10 (p = 0.01; n = 60) (Fig. 7F), and the mutant branch total length per 100 μm of axon also increased from 29.16 ± 4.42 μm to 58.91 ± 9.33 μm (p = 0.01; n = 60) (Fig. 7G). Thus, deletion of Trio led to inhibition of neurite growth and highly branched neurites.
FIGURE 7.
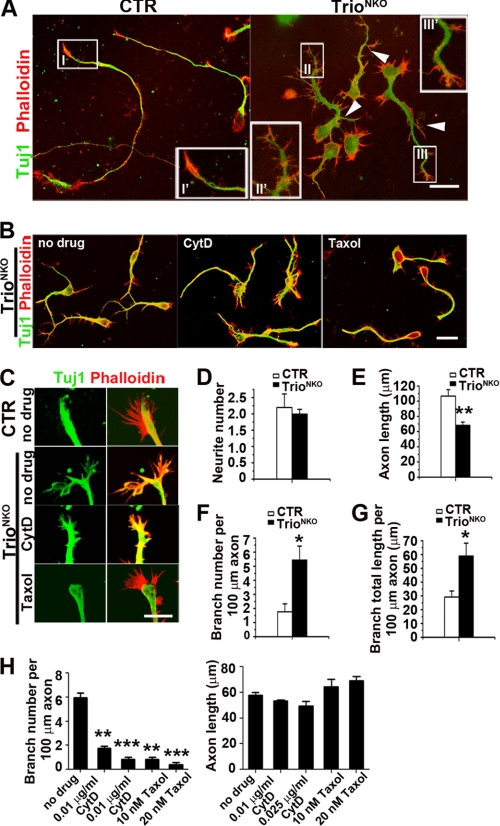
Abnormal neurite morphogenesis of TrioNKO cerebellar granule cells in culture. A, representative granule cells 2.5 days after plating. Neurons were double-labeled with antibody against Tuj1 and TRITC-labeled phalloidin. White arrowheads indicate mutant axon branches. I′, II′, and III′ are magnifications of I, II, and III, respectively, emphasizing irregular growth cone formation in the mutant. B, 24 h after culture, cerebellar granule cells were treated with cytochalasin D and taxol for 36 h. C, 24 h after culture, cerebellar granule cells were treated with cytochalasin D and taxol for 4 h. D–G, statistical analysis for the number and length of neurites; n = 20 neurons from three independent cultures. D, control and mutant granule cells had an equivalent number of neurites arising from the cell body. E, axon length was shorter in the mutant than in the control; *, p < 0.05; **, p < 0.01. F and G, branching density (branches per unit) for axon and total branch length per unit length of axon were increased in Trio-deficient granule cells compared with control. H, branch density and axon length of mutant granule cells treated with cytochalasin D and taxol; **, p < 0.05; ***, p < 0.001. Scale bars: A and B, 20 μm; C, 10 μm.
Neurite is primarily regulated through a dynamic cytoskeleton (33), in which appropriate stabilization of F-actin and dynamic microtubules is an important determinant of its morphogenesis (34). TrioNKO neurons displayed strong filopodia, branched neurites and short processes, so increased stabilization of actin polymerization and reduced stabilization of microtubules in the mutant cells may be expected (35, 36). We measured the effect of the actin-depolymerizing reagent cytochalasin D and the microtubule stabilizer taxol on the morphogenesis and extension of neurites. As expected, treatment with cytochalasin D significantly reduced the number of filopodia in Trio-deficient neurons, and the branch number per unit neurite was also significantly reduced (0.48 ± 0.04 versus 5.23 ± 0.05 of no drug; p < 0.01), and the neurite length was reduced significantly (43.3 ± 5.0 versus 60.9 ± 10 of no drug, p < 0.05) (Fig. 7, B, C, and H). In light of the notion that filopodia are required for neuritogenesis (37, 38), this result could be explained: cytochalasin D depolymerizes the highly stabilized F-actin of mutant granule cells, thereby inhibiting filopodia formation and hence inhibiting neuritogenesis or branch formation. When mutant neurons were treated with taxol, the branch number per unit axon was reduced from 6.16 ± 0.6 to 1.08 ± 0.3 (p < 0.01), but the axon length did not increase significantly (56.8 ± 7.0 versus 69.3 ± 12; p > 0.05) (Fig. 7, B, C, and H). Thus, in addition to stabilized actin polymerization, the destabilized microtubule caused by Trio deletion may also contribute to highly branched neurites, probably through promoting branch formation of growth cones (39).
Characterization of the Physiological Properties of Trio-deficient Granule Cells
GEFs and their Rho small GTPase effectors have diverse functions in multiple physiological processes of cells. To investigate the other potential physiological changes caused by Trio deletion, we characterized their properties by proliferation, differentiation, and apoptosis.
Using the BrdUrd incorporation assay, we found that the proliferative cells located at the outer EGL of P0.5 mutant cerebella had a similar pattern of CTR (supplemental Fig. S3, C and D). To analyze dynamic proliferation of the granule cells, we quantified phospho-histone-H3-positive cells in the cerebella from mice of different ages (Fig. 8, A and B). The number of positive cells in the outer EGL of the mutant cerebella was similar to those of CTR littermates from P0.5 to P10 (p > 0.05) (Fig. 8J). This suggested that the proliferation of granule cells was not affected in Trio knock-out mice.
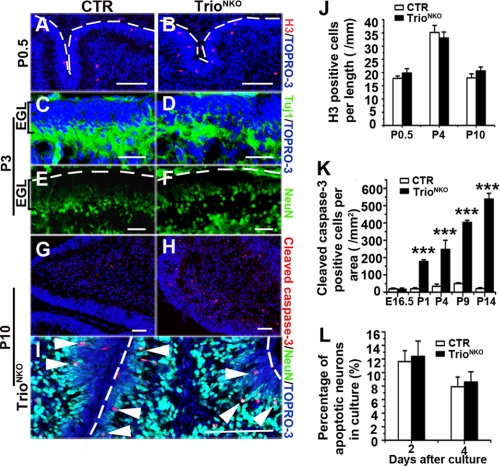
FIGURE 8.
Normal proliferation and differentiation determination but increased apoptosis of Trioflox/flox; Nestin-Cre granule cells. A and B, phospho-histone-H3 (H3) immunostaining shows normal proliferation ability of mutant EGL. Dashed lines indicate the pial surface. C and D, the expression pattern for process extension can be detected by Tuj1 in the mutant inner EGL at P3. E and F, the differentiated neuron maker NeuN exists in the inner EGL in the mutant mice as the control at P3. Dashed lines indicate the pial surface. G and H, apoptosis, characterized by cleaved caspase-3, is greatly aggravated in mutant cerebella. I, most of the apoptotic cells in the mutant are NeuN-positive (white arrowheads) in the inner EGL or molecular layer. Dashed lines indicate the pial surface. J, H3-positive cells at the outer EGL per unit length of EGL. K, quantification of apoptotic cerebellar cells per area of cerebella during different developmental stages (n = 3); ***, p < 0.001. L, quantification of apoptotic granule cells in vitro by propidium iodide staining. Cerebellar neurons were from P4 mice and cultured at high density (600 neurons per mm2). The histogram indicates that the percentage of apoptotic cells in Trio mutant neurons was comparable to CTR cultured for 2 and 4 days. Scale bars in A–I, 100 μm.
We then examined the differentiation of granule cells by staining with antibody to the early post-mitotic immature neuron marker Tuj1 and the differentiated neuron marker NeuN (40). In the cerebella of P3 TrioNKO mice, the inner EGL showed staining of Tuj1 and NeuN with a similar pattern of CTR (Fig. 8, C–F). This suggested that the precursors can enter the differentiation program.
We next examined the level of apoptosis in the developing cerebellum with antibody to cleaved caspase-3. Apoptotic cells in the knock-out cerebellum increased greatly during the postnatal period (all p < 0.001) (Fig. 8, G, H, and K). More than 70% of the apoptotic cells were NeuN-positive in the inner EGL or in the molecular layer (Fig. 8I). To determine if the apoptosis was caused by an intracellular mechanism or by secondary effects of developmental defects, we cultured CGNs in vitro and quantified apoptotic cells by morphological examination. Compared with CTR, Trio-deleted CGNs did not show significantly increased apoptosis after culture for 2 and 4 days (p = 0.66; n = 1000–1500 for 2 days and p = 0.58; n = 1000–1200 for 4 days, respectively) (Fig. 8L). The apoptosis of Trio-deficient CGNs in vivo appears to be a secondary effect to cerebellar defects. Taken together, our observations suggest that deletion of Trio may not directly affect the intracellular physiology of cerebellar cells, including proliferation, differentiation, and apoptosis.
Down-regulation of the Activity of Rho GTPases in Trio-deficient Neurons
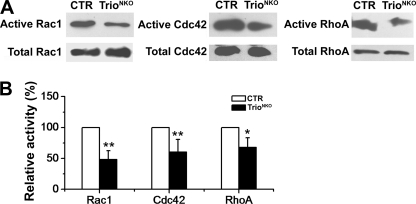
To assess the regulatory ability of Trio on small GTPases, the active Rac1, RhoA, and Cdc42 (direct downstream target of RhoG) were quantified. Early cerebella are too small to provide sufficient protein samples, so we used whole brain tissue to measure the proteins. Western blot assay showed that the total amount of each protein in the mutant brain was unchanged, as mentioned above (supplemental Fig. S1D). Glutathione S-transferase (GST) pulldown assay showed that the amounts of active RhoA, Rac1, and Cdc42 were significantly reduced in the Trio-deficient brain (Fig. 9A). Quantitative analysis showed that the brain tissue from TrioNKO mice (P0.5) displayed only 48.1 ± 14.3% of active Rac1, 60.4 ± 2.0% of active Cdc42, and 66.7 ± 15.6% of active RhoA compared with CTR tissues (Fig. 9B). This result suggests that Trio can activate Rac1, RhoA, and Cdc42 under physiological conditions.
FIGURE 9.
Quantification of active Rho small GTPases. A, GTP-bound Rac1, Cdc42, and RhoA were pulled down using PBD-GST (Rac1 and Cdc42) and RBD-GST (RhoA) from lysates of CTR and mutant total brain at P0.5. Active Rac1, Cdc42, and RhoA pulldown assays were detected by Western blot using antibodies against Rac1, Cdc42, and RhoA, respectively. Total Rac1, Cdc42, and RhoA in cell lysates indicated equal amounts of the GTPases (bottom). B, the histogram shows that the rate of active Rac1, RhoA, and Cdc42 was significantly reduced in mutant mice (p = 0.001 for Rac1, n = 5; p = 0.009 for RhoA, n = 5; p = 0.03 for Cdc42, n = 4); *, p < 0.05; **, p < 0.01.
It is known that RhoA inhibits neurite growth and that Rac1 and Cdc42 promote neurite growth (10). In Trio-deficient CGN, neurite growth is significantly inhibited. To investigate the function of RhoA in mutant CGN, we applied the RhoA-specific inhibitor Y27632 to CGN and analyzed neurite growth behaviors. Adding Y27632 significantly promoted neurite growth in CTR (93.1 ± 3.7 μm versus 109.5 ± 4.4 μm, p < 0.05) and mutant CGN (72.2 ± 4.0 μm versus 88.9 ± 6.1 μm, p < 0.05). The percentages of increased neurite length were comparable (17.6% of CTR and 23.1% of mutant CGN, respectively). Such unaffected response to further inhibition of RhoA implied that the neurite growth inhibition of Trio deficiency CGN might be not attributable to RhoA activity. Addition of the Rac1 inhibitor NSC23766 reduced the neurite length of CTR CGN from 93.1 ± 3.7 μm to 59.6 ± 2.3 μm (p < 0.001), but it slightly inhibited neurite growth of the mutant CGN (72.2 ± 4.0 μm versus 64.5 ± 2.4 μm). Thus, Rac1 might be, more or less, functionally involved in the neurite extension.
Unresponsiveness of Netrin-1 and Semaphorin 6A in Trio-deficient Neurons
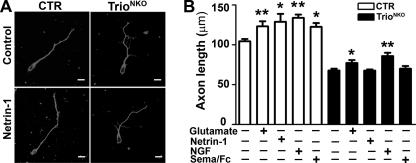
It has been reported that several extracellular cues regulate cerebellar development (41, 42). For example, Netrin-1 repels neurons of the external germinal layer of the postnatal cerebellum (43); Semaphorin 6A controls the initiation of granule cell radial migration (44); nerve growth factor and excitatory neurotransmitters regulate morphogenesis of cultured cerebellar cells (45). To determine if these signals regulate neurite growth through Trio-mediated activation of small GTPases in the cerebellum, we tested the effect of different extracellular cues in CTR or mutant dissociated cerebellar neurons (Fig. 10, A and B). Addition of Netrin-1 and Semaphorin 6A showed an increased neurite growth of CTR neurons (1.24 ± 0.15-fold, p = 0.04; n = 60 and 1.18 ± 0.04-fold, p = 0.01; n = 60, respectively), but no significant changes in Trio-deficient neurons (0.98 ± 0.08-fold, p = 0.67; n = 60 and 1.05 ± 0.20-fold, p = 0.64; n = 60, respectively) (Fig. 10B). Nevertheless, the ratio of increased neurite growth by adding glutamate (1.20 ± 0.05-fold, p = 0.008; n = 60 versus 1.17 ± 0.05-fold, p = 0.02; n = 60) or nerve growth factor (1.28 ± 0.09-fold, p = 0.001 versus 1.30 ± 0.07-fold, p = 0.004) in CTR and mutant neurons was comparable (Fig. 10B). These results suggested that Trio deletion may block Netrin-1 and Semaphorin 6A signals for neurite growth of cerebellar neurons.
FIGURE 10.
Neurite outgrowth in response to exogenous cues. A, neurite outgrowth of control and TrioNKO cerebellar neurons cultured for 2.5 days in vitro with control buffer and the buffer with Netrin-1. B, quantification of mean axon length of cerebellar granule cells 2.5 days after plating with control buffer, glutamate, Netrin-1, nerve growth factor, and Semaphorin 6A/Fc; *, p < 0.05; **, p < 0.01. Scale bar in A, 20 μm.
DISCUSSION
Trio Is Required for Cerebellar Development
The Rho family of small GTPases acts as “intracellular molecular switches” that transduce signals from extracellular stimuli to the cytoskeleton, thereby regulating neuronal migration-related processes via multiple signal pathways (24, 46–52). The GEFs far outnumber the Rho GTPases themselves in a given organism, thereby enabling signals to function in a specific manner. The relative physiological importance of each specific GEF along with downstream effectors is difficult to appreciate. Genetic approaches offer tools to unravel complexities in defining signal modules in the physiological response. We therefore used a conditional knock-out of Trio GEF in neuronal tissue to define its function in cerebellar development.
As a key GEF regulator of Rho small GTPases, Trio was noted for its role in axon guidance and axonagenesis in Drosophila and C. elegans (17, 53). The function and physiological importance of Trio in the cerebellum have not been defined. We show here that Trio-deficient mice display severe phenotypes, including low survival rate, severe ataxia, and multiple developmental defects of the cerebellum. These results suggest that Trio is required for cerebellar development. Various GEFs have been found to be essential for the function or development of neurons (3, 54–57), but only a few GEFs involved in cerebellum development have been reported (58, 59). In light of the severe phenotypes of the Trio-deficient cerebellum, Trio may be defined as a crucial GEF for cerebellar development.
The importance of Trio in the cerebellum may be also supported by observations from human diseases. The GEF1 domain of Trio targets filamin to remodel cytoskeletal actin, and filamin is required for the effect of Trio on cytoskeletal organization (60). The mutation of filamin in humans may give rise to impaired migration of neurons, and hence results in several neuronal defects (including cerebellar abnormalities) (61–63). Trio signaling may thus function essentially in filamin-associated cerebellum diseases. Huntingtin-associated protein 1, a protein closely related to cerebellar degeneration in Huntington disease, can interact with Trio-like peptide directly (64). This implies an important functional role of Trio in the cerebellum during the pathogenesis of Huntington disease.
Trio Coordinately Regulates Neurite Morphogenesis and Migration of CGN through Regulation of the Cytoskeleton
During the processes of IGL formation, cerebellar granule cells undergo profound morphological changes, including neurite generation, neurite extension, and directed turning, which are tightly coupled with guided migration of granule cells (4, 5). Trio deletion caused extensive defects among these processes. However, the present study suggests that the impairment of neurite morphogenesis as well as cell migration is the primary defects in Trio-deficient CGN. Other defects are attributable to the secondary effects of defective cerebellar development. Appropriate regulation of neurite morphogenesis and migration of CGNs appears to be necessary for the multiple developmental processes of cerebellum, in which the Trio signal pathway is required.
The common feature among signaling pathways regulating neuronal migration is eventual involvement of the cytoskeleton, in which microtubule and actin networks are believed to be the most important targets (4). Abnormal dynamics of microtubules and actin result in the altered behavior and morphology of neurites (34). In Trio mutant granule cells, neurites showed retarded extension, enriched branches, and more lamellipodia and filopodia. Addition of reagents interfering with F-actin and microtubule stabilization can alter neurite phenotypes. We suggest that the regulation of the morphogenesis of CGN neurites by Trio signaling is implemented through the organization of cytoskeleton.
A series of studies suggest that Rho GTPases function in neuronal development: Rac1 regulates axon guidance and neurite complexity, Cdc42 regulates axon specification and growth, and RhoA inhibits neurite extension (13, 65–67). Because all these proteins in active forms were reduced in Trio-deficient mice, we speculated that the increased neurite complexity and abnormal migration in Trio-null granule cells might be attributed to reduced Rac1 activity, and retarded neurite extension might be caused by reduced Cdc42 and/or Rac1 activity, although we did not have a proper explanation for RhoA function in this context. This hypothesis is also supported by phenotypic comparison with the knock-out mice of Rac1, Cdc42, and other genes (65, 67–69). In particular, the phenotypes of Rac1 knock-out mice, including impairment of neuronal migration, disorganization of cortical lamination, and increased complexity of neurite, are consistent with Trio knock-out phenotypes (67, 68), supporting an important role of Rac1 in Trio's function. Because Rac3-null mice had no gross anatomical defects in brain structure and neuron organization, and Rac3 had no obvious expression in cerebellar granule cells (69), we didn't emphasize the involvement of Rac3 in the Trio knock-out phenotype.
Trio May Act as a Signal Integrator for CGN Migration
External guidance cues are required to accomplish orchestral regulation of neuron migration (43, 70–73). For example, Netrin-1 is expressed in the EGL and in the molecular layer. Netrin-1 receptor Dcc, which may associate with Trio indirectly via its interaction with PAK1, is expressed in the inner EGL. The transmembrane Semaphorin 6A is expressed in the tangentially migrating granule cells of inner EGL and controls the extension and turning of CGN processes, which couples guided cell migration (43, 44, 74–76). An internal mechanism integrating extracellular cues to migration machinery in the developing cerebellum should be established. Trio may serve as a signal integrator establishing such a mechanism for the developmental program in the cerebellum. Based on observations in the present study, Trio may integrate Netrin-1 and Semaphorin 6A signals to Rho GTPases with unknown signal cascades. This speculation is also supported by observations from the spinal cord and cortical neurons (76). The multiple GEF domains of Trio enable Trio to differentially respond to cues by combination of activation of small GTPases (53, 76, 77) and facilitate an efficient and/or orchestrated means to interconnect the different signal pathways.
Supplementary Material
Acknowledgments
We thank Dr. H. Jiang (University of Texas Southwestern Medical Center at Dallas) for valuable discussions and generous help. We thank Dr. N. Copeland (NCI, National Institutes of Health) for providing materials for the BAC-retrieval system and Dr. David Adams (Sanger Institute, UK) for providing 129 BAC clones.
This work was supported by the National Basic Research Program of China (973 Program) (Grants 2009CB941602, 2007CB947100, and 2005CB522501), National Natural Science Funding of China (Grant 30570911), and the Scientific Research Foundation of Graduate School of Nanjing University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Videos S1–S4.
- EGL
- external granule cell layer
- IGL
- internal granule cell layer
- GEF
- guanine nucleotide exchange factors
- BAC
- bacterial artificial chromosome
- CGN
- cerebellar granule neuron
- BrdUrd
- bromodeoxyuridine
- TRITC
- tetramethylrhodamine isothiocyanate
- ES
- embryonic stem W4 cell
- TrioNKO
- Trioflox/flox; Nestin-Cre
- GST
- glutathione S-transferase
- CTR
- control
- DiI
- 1,1'-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate
- GFAP
- glial fibrillary acidic protein.
REFERENCES
- 1.Hatten M. E. (1999) Annu. Rev. Neurosci. 22, 511–539 [DOI] [PubMed] [Google Scholar]
- 2.Wingate R. J. (2001) Curr. Opin. Neurobiol. 11, 82–88 [DOI] [PubMed] [Google Scholar]
- 3.Wang V. Y., Zoghbi H. Y. (2001) Nat. Rev. Neurosci. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 4.Ayala R., Shu T., Tsai L. H. (2007) Cell 128, 29–43 [DOI] [PubMed] [Google Scholar]
- 5.Komuro H., Yacubova E. (2003) Cell Mol. Life Sci. 60, 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y., Yu T., Zhang X. C., Nagasawa T., Wu J. Y., Rao Y. (2002) Nat. Neurosci. 5, 719–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai L. H., Gleeson J. G. (2005) Neuron 46, 383–388 [DOI] [PubMed] [Google Scholar]
- 8.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 9.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 10.Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 11.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 12.Luo L. (2000) Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 13.Govek E. E., Newey S. E., Van Aelst L. (2005) Genes Dev. 19, 1–49 [DOI] [PubMed] [Google Scholar]
- 14.Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., Wu J. Y., Xiong W. C., Rao Y. (2001) Cell 107, 209–221 [DOI] [PubMed] [Google Scholar]
- 15.Debant A., Serra-Pagès C., Seipel K., O'Brien S., Tang M., Park S. H., Streuli M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5466–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng J., Luo L. (2004) Neuron 44, 779–793 [DOI] [PubMed] [Google Scholar]
- 17.Steven R., Kubiseski T. J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C. W., Pawson T., Culotti J. (1998) Cell 92, 785–795 [DOI] [PubMed] [Google Scholar]
- 18.Awasaki T., Saito M., Sone M., Suzuki E., Sakai R., Ito K., Hama C. (2000) Neuron 26, 119–131 [DOI] [PubMed] [Google Scholar]
- 19.Bateman J., Shu H., Van Vactor D. (2000) Neuron 26, 93–106 [DOI] [PubMed] [Google Scholar]
- 20.Bateman J., Van Vactor D. (2001) J. Cell Sci. 114, 1973–1980 [DOI] [PubMed] [Google Scholar]
- 21.Ma X. M., Huang J. P., Eipper B. A., Mains R. E. (2005) J. Comp. Neurol. 482, 333–348 [DOI] [PubMed] [Google Scholar]
- 22.O'Brien S. P., Seipel K., Medley Q. G., Bronson R., Segal R., Streuli M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12074–12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backer S., Hidalgo-Sáchez M., Offner N., Portales-Casamar E., Debant A., Fort P., Gauthier-Rouvière C., Bloch-Gallego E. (2007) J. Neurosci. 27, 10323–10332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P., Jenkins N. A., Copeland N. G. (2003) Genome. Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He W. Q., Peng Y. J., Zhang W. C., Lv N., Tang J., Chen C., Zhang C. H., Gao S., Chen H. Q., Zhi G., Feil R., Kamm K. E., Stull J. T., Gao X., Zhu M. S. (2008) Gastroenterology 135, 610–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagata I., Nakatsuji N. (1990) Brain Res. Dev. Brain Res. 52, 63–73 [DOI] [PubMed] [Google Scholar]
- 27.Li Z., Aizenman C. D., Cline H. T. (2002) Neuron 33, 741–750 [DOI] [PubMed] [Google Scholar]
- 28.Graus-Porta D., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J. C., Müller U. (2001) Neuron 31, 367–379 [DOI] [PubMed] [Google Scholar]
- 29.Hongo T., Hakuba A., Shiota K., Naruse I. (2000) Biol. Neonate 78, 293–299 [DOI] [PubMed] [Google Scholar]
- 30.Yue Q., Groszer M., Gil J. S., Berk A. J., Messing A., Wu H., Liu X. (2005) Development 132, 3281–3291 [DOI] [PubMed] [Google Scholar]
- 31.Yacubova E., Komuro H. (2002) J. Neurosci. 22, 5966–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatten M. E., Heintz N. (1995) Annu. Rev. Neurosci. 18, 385–408 [DOI] [PubMed] [Google Scholar]
- 33.Dent E. W., Gertler F. B. (2003) Neuron 40, 209–227 [DOI] [PubMed] [Google Scholar]
- 34.Sanes D. H., Reh T. A., Harris W. A. (2006) Development of the Nervous System, Elsevier Inc. [Google Scholar]
- 35.Gallo G., Letourneau P. C. (2004) J. Neurobiol. 58, 92–102 [DOI] [PubMed] [Google Scholar]
- 36.Bielas S. L., Serneo F. F., Chechlacz M., Deerinck T. J., Perkins G. A., Allen P. B., Ellisman M. H., Gleeson J. G. (2007) Cell 129, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalli G., Hall A. (2005) J. Cell Biol. 171, 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalil K., Dent E. W. (2005) Curr. Opin. Neurobiol. 15, 521–526 [DOI] [PubMed] [Google Scholar]
- 39.Homma N., Takei Y., Tanaka Y., Nakata T., Terada S., Kikkawa M., Noda Y., Hirokawa N. (2003) Cell 114, 229–239 [DOI] [PubMed] [Google Scholar]
- 40.Lee M. K., Tuttle J. B., Rebhun L. I., Cleveland D. W., Frankfurter A. (1990) Cell Motil. Cytoskeleton 17, 118–132 [DOI] [PubMed] [Google Scholar]
- 41.Yacubova E., Komuro H. (2003) Cell Biochem. Biophys. 37, 213–234 [DOI] [PubMed] [Google Scholar]
- 42.Park H. T., Wu J., Rao Y. (2002) Bioessays. 24, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alcántara S., Ruiz M., De Castro F., Soriano E., Sotelo C. (2000) Development 127, 1359–1372 [DOI] [PubMed] [Google Scholar]
- 44.Kerjan G., Dolan J., Haumaitre C., Schneider-Maunoury S., Fujisawa H., Mitchell K. J., Chédotal A. (2005) Nat. Neurosci. 8, 1516–1524 [DOI] [PubMed] [Google Scholar]
- 45.Cohen-Cory S., Dreyfus C. F., Black I. B. (1991) J. Neurosci. 11, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Västrik I., Eickholt B. J., Walsh F. S., Ridley A., Doherty P. (1999) Curr. Biol. 9, 991–998 [DOI] [PubMed] [Google Scholar]
- 47.Shamah S. M., Lin M. Z., Goldberg J. L., Estrach S., Sahin M., Hu L., Bazalakova M., Neve R. L., Corfas G., Debant A., Greenberg M. E. (2001) Cell 105, 233–244 [DOI] [PubMed] [Google Scholar]
- 48.Shekarabi M., Kennedy T. E. (2002) Mol. Cell Neurosci. 19, 1–17 [DOI] [PubMed] [Google Scholar]
- 49.Swiercz J. M., Kuner R., Behrens J., Offermanns S. (2002) Neuron 35, 51–63 [DOI] [PubMed] [Google Scholar]
- 50.Guan K. L., Rao Y. (2003) Nat. Rev. Neurosci. 4, 941–956 [DOI] [PubMed] [Google Scholar]
- 51.Huber A. B., Kolodkin A. L., Ginty D. D., Cloutier J. F. (2003) Annu. Rev. Neurosci. 26, 509–563 [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson D. G. (2001) Nat. Rev. Neurosci. 2, 155–164 [DOI] [PubMed] [Google Scholar]
- 53.Newsome T. P., Schmidt S., Dietzl G., Keleman K., Asling B., Debant A., Dickson B. J. (2000) Cell 101, 283–294 [DOI] [PubMed] [Google Scholar]
- 54.Watabe-Uchida M., Govek E. E., Van Aelst L. (2006) J. Neurosci. 26, 10633–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 56.Ma X. M., Kiraly D. D., Gaier E. D., Wang Y., Kim E. J., Levine E. S., Eipper B. A., Mains R. E. (2008) J. Neurosci. 28, 12368–12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tolias K. F., Bikoff J. B., Burette A., Paradis S., Harrar D., Tavazoie S., Weinberg R. J., Greenberg M. E. (2005) Neuron 45, 525–538 [DOI] [PubMed] [Google Scholar]
- 58.Donald S., Humby T., Fyfe I., Segonds-Pichon A., Walker S. A., Andrews S. R., Coadwell W. J., Emson P., Wilkinson L. S., Welch H. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4483–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J., Furuya A., Furuichi T. (2007) J. Cell Biol. 179, 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellanger J. M., Astier C., Sardet C., Ohta Y., Stossel T. P., Debant A. (2000) Nat. Cell Biol. 2, 888–892 [DOI] [PubMed] [Google Scholar]
- 61.Fox J. W., Lamperti E. D., Ekşioğlu Y. Z., Hong S. E., Feng Y., Graham D. A., Scheffer I. E., Dobyns W. B., Hirsch B. A., Radtke R. A., Berkovic S. F., Huttenlocher P. R., Walsh C. A. (1998) Neuron 21, 1315–1325 [DOI] [PubMed] [Google Scholar]
- 62.Robertson S. P. (2004) Clin. Dysmorphol. 13, 123–131 [DOI] [PubMed] [Google Scholar]
- 63.Feng Y., Walsh C. A. (2004) Nat. Cell Biol. 6, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 64.Colomer V., Engelender S., Sharp A. H., Duan K., Cooper J. K., Lanahan A., Lyford G., Worley P., Ross C. A. (1997) Hum. Mol. Genet. 6, 1519–1525 [DOI] [PubMed] [Google Scholar]
- 65.Garvalov B. K., Flynn K. C., Neukirchen D., Meyn L., Teusch N., Wu X., Brakebusch C., Bamburg J. R., Bradke F. (2007) J. Neurosci. 27, 13117–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heasman S. J., Ridley A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 67.Chen L., Liao G., Waclaw R. R., Burns K. A., Linquist D., Campbell K., Zheng Y., Kuan C. Y. (2007) J. Neurosci. 27, 3884–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kassai H., Terashima T., Fukaya M., Nakao K., Sakahara M., Watanabe M., Aiba A. (2008) Eur. J. Neurosci. 28, 257–267 [DOI] [PubMed] [Google Scholar]
- 69.Corbetta S., Gualdoni S., Albertinazzi C., Paris S., Croci L., Consalez G. G., de Curtis I. (2005) Mol. Cell. Biol. 25, 5763–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rakic P., Cameron R. S., Komuro H. (1994) Curr. Opin. Neurobiol. 4, 63–69 [DOI] [PubMed] [Google Scholar]
- 71.Wu W., Wong K., Chen J., Jiang Z., Dupuis S., Wu J. Y., Rao Y. (1999) Nature 400, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bagri A., Tessier-Lavigne M. (2002) Adv. Exp. Med. Biol. 515, 13–31 [PubMed] [Google Scholar]
- 73.Marín O., Plump A. S., Flames N., Sánchez-Camacho C., Tessier-Lavigne M., Rubenstein J. L. (2003) Development 130, 1889–1901 [DOI] [PubMed] [Google Scholar]
- 74.Barallobre M. J., Pascual M., Del Río J. A., Soriano E. (2005) Brain Res. Brain Res. Rev. 49, 22–47 [DOI] [PubMed] [Google Scholar]
- 75.Renaud J., Kerjan G., Sumita I., Zagar Y., Georget V., Kim D., Fouquet C., Suda K., Sanbo M., Suto F., Ackerman S. L., Mitchell K. J., Fujisawa H., Chédotal A. (2008) Nat. Neurosci. 11, 440–449 [DOI] [PubMed] [Google Scholar]
- 76.Briançon-Marjollet A., Ghogha A., Nawabi H., Triki I., Auziol C., Fromont S., Piché C., Enslen H., Chebli K., Cloutier J. F., Castellani V., Debant A., Lamarche-Vane N. (2008) Mol. Cell. Biol. 28, 2314–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Estrach S., Schmidt S., Diriong S., Penna A., Blangy A., Fort P., Debant A. (2002) Curr. Biol. 12, 307–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.