Abstract
Microvirin (MVN), a recently isolated lectin from the cyanobacterium Microcystis aeruginosa PCC7806, shares 33% identity with the potent anti-human immunodeficiency virus (HIV) protein cyanovirin-N (CV-N) isolated from Nostoc ellipsosporum, and both lectins bind to similar carbohydrate structures. MVN is able to inhibit infection by a wide variety of HIV-1 laboratory-adapted strains and clinical isolates of different tropisms and subtypes in peripheral blood mononuclear cells. MVN also inhibits syncytium formation between persistently HIV-1-infected T cells and uninfected CD4+ T cells and inhibits DC-SIGN-mediated HIV-1 binding and transmission to CD4+ T cells. Long term passaging of HIV-1 exposed to dose-escalating concentrations of MVN resulted in the selection of a mutant virus with four deleted high mannose-type glycans in the envelope gp120. The MVN-resistant virus was still highly sensitive to various other carbohydrate binding lectins (e.g. CV-N, HHA, GNA, and UDA) but not anymore to the carbohydrate-specific 2G12 monoclonal antibody. Importantly, MVN is more than 50-fold less cytotoxic than CV-N. Also in sharp contrast to CV-N, MVN did not increase the level of the activation markers CD25, CD69, and HLA-DR in CD4+ T lymphocytes, and subsequently, MVN did not enhance viral replication in pretreated peripheral blood mononuclear cells. Therefore, MVN may qualify as a useful lectin for potential microbicidal use based on its broad and potent antiviral activity and virtual lack of any stimulatory properties and cellular toxicity.
Keywords: AIDS, Antiviral Agents, Human Immunodeficiency Virus, Lectin, Virus Entry, Microbicide
Introduction
The human immunodeficiency virus type 1 (HIV-1)2 is a global health problem of unprecedented dimensions and has already caused an estimated 25 million deaths worldwide. Sub-Saharan Africa is the region that is affected the most. In this region, women account for ∼60% of all HIV-1 infections (44). Today's prevention options (condoms, mutual monogamy, and sexually transmitted disease treatment) are not feasible for millions of people around the world, especially for women. Therefore, there is an urgent need for the development of other prevention methods so that women have opportunities to protect themselves. Microbicides are self-administering prophylactic agents, impeding transmission of HIV-1 or other sexually transmitted diseases. HIV-1 transmission can be defined by virus crossing the epithelial barrier and productively infecting a CD4+ target cell. The entry of HIV-1 into its target cell is mediated by the glycoproteins gp120 and gp41, which are present on the viral envelope. These glycoproteins bind first to the cellular receptor CD4 and then to a so-called HIV co-receptor, CCR5 and/or CXCR4. Both gp120 and gp41 are heavily glycosylated proteins (1–3).
Carbohydrate binding agents (CBAs), such as the plant lectins Hippeastrum hybrid agglutinin (HHA), Galanthus nivalis agglutinin (GNA), Urtica dioica agglutinin (UDA) and BanLec, or the prokaryotic cyanovirin-N (CV-N) and griffithsin bind to multiple glycans that are present on the envelope of HIV and subsequently inhibit the viral entry process (4–8). CV-N is an 11-kDa protein derived from the cyanobacterium (blue-green alga) Nostoc ellipsosporum. It has attracted worldwide interest because of its ability to neutralize a very broad spectrum of HIV-1 strains and primary virus isolates and to protect macaques after vaginal or rectal chimeric simian HIV infection (9–15). CV-N exhibits a high specificity for terminal Manα(1–2)Man moieties on high mannose (Man-8 or Man-9) glycans (9, 11–13, 16–18). However, incubation of peripheral blood mononuclear cells (PBMCs) with CV-N very often also enhanced viral replication levels at suboptimal concentrations (19), and CV-N had pronounced mitogenic/stimulatory effects on PBMCs (20). The mAb 2G12, a potent neutralizing anti-HIV-1 IgG, binds to a constellation of high mannose-type carbohydrates on gp120 (21) and protects macaques against vaginal transmission upon chimeric simian HIV challenge (22, 23).
Recently, microvirin (MVN) was discovered as a novel lectin from the cyanobacterium Microcystis aeruginosa PCC7806 and shows 33% identity at the amino acid level with CV-N (24). MVN has a molecular mass of 14.3 kDa and shows high specificity for high mannose structures containing terminal Manα(1–2)Manα moieties (24).
EXPERIMENTAL PROCEDURES
Test Compounds and Monoclonal Antibodies
The mannose-specific lectin MVN (14.3 kDa) from the microcystin-producing strain M. aeruginosa PCC7806 was expressed in Escherichia coli and purified as described previously (24). CV-N (11 kDa) was a kind gift from Dr. C. A. Bewley (17). Batches of the stock solutions of MVN and CV-N were tested for endotoxin content with the Limulus Amebocyte Lysate assay (Cambrex Bioscience, Verviers, Belgium), and at the highest concentrations tested, the stock solutions were found to contain less than 1 ng/ml endotoxin. The plant lectins GNA (50 kDa), HHA (50 kDa), and UDA (8.5 kDa) were kindly provided by Dr. E. Van Damme (University Ghent, Belgium). Phytohemagglutinin (PHA) (128 kDa) was purchased from Sigma. Pradimicin S (PRM-S) (910 Da) was produced by the strain Actinomadura sp. TP-A0020 and isolated and purified from the culture supernatants as described previously (25). PRO 2000 (∼5 kDa) was provided by Indevus Pharmaceuticals Inc. (Lexington, MA). AMD3100 (mozobil; CXCR4 antagonist; 794 Da) was kindly provided by Dr. Gary Bridger (at that time at AnorMED (Langley, Canada)) (26).
The 2G12 mAb and the b12 mAb were purchased from Polymun Scientific (Vienna, Austria). The mAbs labeled with phycoerythrin or fluorescein isothiocyanate (FITC) used were CD25, CD69, HLA-DR, and CD4 and were purchased from BD Biosciences (Erembodegem, Belgium). The rabbit anti-human IgG-FITC was purchased from DakoCytomation.
Cells, Cell Cultures, and Viruses
The MT-4 T cell line was a kind gift of Dr. L. Montagnier (at that time at the Pasteur Institute (Paris, France)), and the Raji/DC-SIGN+ B cells were kindly provided by Dr. L. Burleigh (Pasteur Institute). Human T-lymphocytic C8166, HUT-78, and SupT1 cells were obtained from the American Type Culture Collection (Manassas, VA). These cell lines were cultivated in RPMI 1640 medium supplemented with 10% fetal bovine serum (BioWittaker Europe, Verviers, Belgium) and 2 mm l-glutamine (Invitrogen) and maintained at 37 °C in a humidified CO2-controlled atmosphere. Buffy coat preparations from healthy donors were obtained from the Blood Transfusion Center (Leuven, Belgium). PBMCs were cultured in cell culture medium (RPMI 1640) containing 10% fetal bovine serum and 2 mm l-glutamine or were activated with PHA (2 μg/ml) for 3 days and cultured in cell culture medium in the presence of 2 ng/ml interleukin (IL)-2 (Roche Applied Science).
HIV-1 NL4.3 (X4), IIIB (X4), and BaL (R5) were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, National Institutes of Health). HIV-1 HE (R5/X4) was isolated from a Belgian AIDS patient in 1987 (27). Primary clinical isolates representing different HIV-1 clades and a HIV-2 isolate were all kindly provided by Dr. L. Lathey from BBI Biotech Research Laboratories, Inc. (Gaithersburg, MD), and their coreceptor use (R5 of X4) was determined in our laboratory in the astroglioma U87.CD4 cell line transfected with either CCR5 (U87.CD4.CCR5) or CXCR4 (U87.CD4.CXCR4). The HIV-1 NL4.3 strain was made resistant to 2G12 mAb as described previously (28).
Antiviral Replication Assay in the MT-4 T Cell Line
MT-4 cells were infected with the X4 HIV-1 strain NL4.3. Briefly, 5-fold dilutions of the compounds were added to 96-well flat-bottomed plates (International Medical, Brussels, Belgium). Then, to each well, 7.5 × 104 MT-4 cells were added, and the cells were infected with ∼100 tissue culture infective dose 50% (TCID50) of the viruses HIV-1 NL4.3, NL4.3 2G12-res, and NL4.3 MVN-res. Cytopathic effect (CPE) induced by the virus was checked microscopically at regular times. When strong CPE was observed in untreated HIV-infected cells (mostly after 4 or 5 days of culture), the cell viability was assessed spectrophotometrically via the in situ reduction of the tetrazolium dye MTS, using the CellTiter 96 AQueous One Solution Cell proliferation assay (Promega, Madison, WI). The absorbance was then recorded at 490 nm with a 96-well plate reader and compared with four cell control wells (cells without virus and drugs) and four virus control wells (cells with virus but without drugs). The IC50 (the concentration that inhibited HIV-1-induced cell death by 50%) was calculated from each dose-response curve.
The CC50 (50% cytotoxic concentration) of compounds was determined from the reduction of viability of uninfected MT-4 cells exposed to the compounds, as measured by the MTS method described above.
Antiviral Testing of MVN against HIV-1 Isolates in PBMCs
PHA-stimulated blasts were seeded at 0.5 × 106 cells/well into a 48-well plate containing varying concentrations of compound in medium containing IL-2. The virus stocks were added at a final dose of ∼100 TCID50 of HIV-1, HIV-2, or SIV. Cell supernatant was collected at days 9–12, and HIV-1 core antigen (Ag) in the culture supernatant was analyzed by a p24 Ag ELISA kit (PerkinElmer Life Sciences). For HIV-2 and SIV p27 Ag, detection the INNOTEST from Innogenetics (Temse, Belgium) was used.
Giant Cell Formation in Co-cultures of CD4+ SupT1 Cells with Persistently HIV-1-infected HUT-78 Cells
Persistently HIV-1(IIIB)-infected HUT-78 cells (HUT-78/IIIB) were generated by infection of HUT-78 cells with HIV-1 IIIB. The cells were subcultured every 3–4 days, and persistent virus infection was monitored in the culture supernatants using HIV-1 p24 Ag ELISA.
For the co-cultivation assay, different concentrations of the test compound along with 1 × 105 SupT1 cells/50 μl were added to 96-well plates. Then a similar amount of HUT-78/IIIB cells (50 μl) was added. After 1 day, the IC50 values were determined microscopically based on the appearance of giant cells or syncytia in the co-cultures.
Short Exposure of HIV-1 to MVN and Virus Capture by Raji/DC-SIGN Cells
High amounts of HIV-1 HE (100 μl; ∼3.2 × 106 pg p24/ml) were exposed to serial dilutions of the test compounds (200 μl) for 30 min. Then, MVN-exposed virus suspensions were mixed with Raji/DC-SIGN cell suspensions (200 μl; 5 × 105 cells) for 1 h at 37 °C, after which the cells were thoroughly washed with culture medium. The Raji/DC-SIGN cell cultures were then analyzed for HIV-1 p24 Ag content by a p24 Ag ELISA kit.
Exposure of Raji/DC-SIGN Cells to HIV-1 and Subsequent Co-cultivation with CD4+ C8166 Cells in the Presence of MVN
Raji/DC-SIGN cells were exposed to HIV-1 HE for 1 h at 37 °C and thoroughly washed. Various concentrations of the test compound (100 μl) were added in a 96-well plate, along with 1 × 105 C8166 CD4+ T cells (50 μl). Then a similar amount of HIV-1-exposed Raji/DC-SIGN cells were added, and giant cell formation was evaluated microscopically 20–24 h later.
Selection of MVN-resistant HIV-1 NL4.3
HIV-1 NL4.3 was added to MT-4 cells (0.4 × 106 cells/ml) in 24-well plates in the presence of 40 ng/ml MVN. Every 5 days, the replicating virus was passaged in fresh uninfected cells in the presence of MVN at the same concentration as in the previous passage or at a 2-fold higher concentration when extensive CPE was obtained in the previous cell culture. After 41 passages (∼205 days of culture), virus was recovered that was able to replicate in the presence of 700 nm MVN (HIV-1 NL4.3-MVN-res). The induced CPE and p24 Ag production level of this virus was comparable with that of the wild-type NL4.3 virus (in the absence of MVN).
Genotyping of the HIV-1 Env Region
Viral RNA was extracted from culture supernatants using the QIAamp Viral RNA minikit (Qiagen, Hilden, Germany). The genotyping of both the gp120 and gp41 genes was determined in this assay as described before (29).
Flow Cytometric Analyses
MT-4 cells were infected with NL4.3 wild-type and NL4.3-MVN-res virus and analyzed when CPE started to occur (3–4 days after infection). Briefly, after washing with phosphate-buffered saline containing 2% fetal bovine serum, cells were preincubated with or without MVN or CV-N at different concentrations for 30 min, washed, and then incubated with 2G12 mAb or b12 mAb for 30 min at 4 °C. Then the cells were washed and incubated with rabbit anti-human IgG-FITC for 30 min at 4 °C. As a control for aspecific background staining, cells were stained in parallel with rabbit anti-human IgG-FITC only. Then the cells were washed and fixed with 1% aqueous formaldehyde solution, and data were acquired with a FACSCalibur flow cytometer (BD Biosciences) and CellQuest software (BD Biosciences). Data were analyzed with FLOWJO software (Tree Star, San Carlos, CA). For the calculations of the mAb binding, the mean fluorescence intensity (MFI) of each sample was expressed as a percentage of the MFI of control cells (after subtracting the MFI of the background staining). Finally, the IC50 values of the compounds were calculated.
The expression of cellular activation markers was measured after a 3-day incubation of PBMCs with varying concentrations of MVN or CV-N at 37 °C. Briefly, after washing with phosphate-buffered saline containing 2% fetal bovine serum, cells were incubated with FITC-conjugated anti-CD4 mAb in combination with phycoerythrin-conjugated anti-CD25, anti-CD69, or anti-HLA-DR mAbs for 30 min at 4 °C. For aspecific background staining, cells were stained in parallel with Simultest Control IgG γ1/γ2a (BD Biosciences). Finally, the cells were washed, fixed with 1% formaldehyde solution, and analyzed with a FACSCalibur, and data were acquired with CellQuest software and analyzed with FLOWJO software.
Bio-Plex Cytokine Assay
PBMCs were cultured in the presence of several concentrations of MVN and CV-N, and culture supernatant was collected after 72 h. The cytokine production profile was determined by the Bio-Plex 200 system (Bio-Rad) and Bio-Plex Human Cytokine 27-plex assay according to the manufacturer's instructions. The 27-plex assay kit contains beads conjugated with mAbs specific for IL-1α, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, eotaxin, fibroblast growth factor (FGF), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage-CSF (GM-CSF), interferon-γ (IFN-γ), IP-10 (interferon-inducible protein-10), MCP-1 (monocyte chemoattractant protein-1), MIP-1α (macrophage inflammatory protein-1α), MIP-1β, platelet-derived growth factor-BB (PDGF-BB), regulated on activation normal T-cell expressed and secreted (RANTES), tumor necrosis factor-α (TNF-α), and vascular endothelial growth factor (VEGF). For each cytokine, nine standards ranging from 0.5 pg/ml to 32,000 pg/ml were constructed, and the minimum detectable dose was between 0.5 and 5 pg/ml. Standard curves and the concentrations of the cytokines within the samples were generated with Bio-Plex Manager 4.1 software.
Susceptibility of PBMCs for HIV-1
Freshly isolated PBMCs were cultured in the presence of MVN, CV-N, HHA, and PHA for 24 h. Then the cells were collected, washed in culture medium, suspended in RPMI medium with 2 ng/ml IL-2, and seeded in a 48-well flat bottom plate (5 × 105 cells in 450 μl), and 50 μl of the CCR5-tropic clade B HIV-1 BaL stock was added at ∼100 TCID50. The supernatant of each sample was collected after 7 days, and viral replication was measured by p24 Ag ELISA.
Statistical Analysis
Statistical analysis performed on the results included the calculation of the mean, S.E., and p values by use of Student's t test. The significance level was set as p = 0.05. Statistical analysis was performed with GraphPad Prism 5 statistical software (GraphPad Software Inc., San Diego, CA).
RESULTS
Antiviral Activity Profile of MVN in PBMCs
The anti-HIV activity of the new mannose-specific CBA MVN was compared with that of the other mannose-specific CBAs CV-N, HHA, and 2G12 mAb (Table 1). MVN was evaluated in PBMCs against a wide variety of HIV-1 clades and showed antiviral activity against all HIV-1 group M clades with IC50 values ranging between 2.1 and 167 nm. In fact, MVN was more active than CV-N against HIV-1 isolates A and B but was 10-fold less active against a clade C isolate. Apparently, MVN exerted no activity against HIV-1 group O, HIV-2 (Table 1), and SIVmac251 (data not shown). In addition, MVN proved to be equally active against laboratory-adapted strains irrespective of their co-receptor use as evidenced by the comparable IC50 values for X4 NL4.3 and R5 BaL (Table 1). In addition, MVN showed generally a better anti-HIV-1 activity profile than HHA. This is in sharp contrast with the data of the 2G12 mAb, which is active against a limited set of isolates (mainly clade A and B) (Table 1), as is also described by others (30).
TABLE 1.
Anti-HIV activity profile of MVN and a few selected CBAs
Viral co-receptor usage (R5 or X4) was determined in U87.CD4.CCR5 and U87.CD4.CXCR4 cells and is indicated in parentheses.
| Agenta | HIV-1 |
HIV-2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Laboratory strain |
Group M |
Group O |
|||||||
| NL4.3 (X4) | BaL (R5) | A; UG273 (R5) | B; US2 (R5) | C; DJ259 (R5) | D; UG270 (X4) | A/E; ID12 (R5) | BCF06 (X4) | BV-5061W (X4) | |
| MVN | 8 | 22 | 9 | 2 | 167 | 48 | 4 | >350 | >350 |
| CV-N | 38 | 22 | 127 | 16 | 19 | 29 | 5 | 14 | 33 |
| HHA | 2 | 120 | 580 | 108 | >400 | 98 | 32 | 24 | 178 |
| 2G12 mAbb | 140 | 3710 | 18 | 40 | >50,000 | >20,000 | >50,000 | >20,000 | >20,000 |
a IC50 (compound concentration in nm required to inhibit viral p24 (for HIV-1) or p27 (for HIV-2) production 50% in PBMCs).
b IC50 (compound concentration in ng/ml required to inhibit viral p24 (for HIV-1) or p27 (for HIV-2) production with 50% in PBMCs).
Next, the cytotoxicity of MVN was evaluated because this can be a major concern for practical use of the CBAs, since cytotoxicity was observed for CV-N (CC50 of 191 nm and 900 nm in MT-4 cells and PBMCs, respectively) (19, 20). However, we could not observe any toxic effect of MVN in MT-4 cells (CC50 > 35 μm) and PBMCs (CC50 > 7 μm).
Antiviral Activity of MVN against High Inputs of HIV-1
In this set of experiments, the antiviral activity of MVN against a series of NL4.3 virus inputs starting at ∼100 TCID50 (normally used in our experiments) up to 5000 TCID50 was evaluated and compared with the activity of the 2G12 mAb. MVN remained active with slightly increasing IC50 values of 9.0, 29.5, 27.3, 42.6, and 41.5 nm for virus inputs of 100, 500, 1000, 2500, and 5000 TCID50, respectively. Also for HHA, slightly increasing IC50 values of 0.6, 2.3, 3.1, 3.0, and 8.7 nm for virus inputs of 100, 500, 1000, 2500, and 5000 TCID50, respectively, were observed. In contrast, for the 2G12 mAb, there was a strong dose-dependent loss of activity with IC50 values of 0.56, 2.4, and 3.5 μg/ml for virus inputs of 100, 500, and 1000 TCID50, and at 2500 and 5000 TCID50, 2G12 mAb was completely inactive (IC50 > 20 μg/ml).
Effect of MVN on Giant Cell Formation in Co-cultures of HUT-78/IIIB and SupT1 Cells
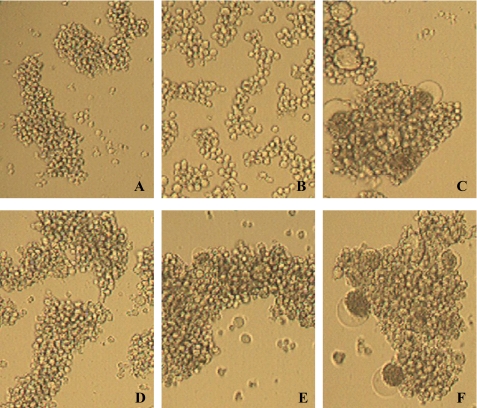
Because the CBAs are known to bind to gp120, we investigated whether MVN could prevent virus-cell contact and fusion. Therefore, the inhibitory activity against syncytium (giant cell) formation between persistently HIV-1-infected HUT-78 T cells and uninfected CD4+ SupT1 cells was determined for MVN. As shown in Fig. 1, MVN dose-dependently inhibited syncytium formation (124 nm ± 38 nm, n = 2).
FIGURE 1.
MVN inhibits giant cell formation. Shown are light microscopic pictures of the following cell cultures: SupT1 cells (A); HUT-78 cells persistently infected with HIV-1 IIIB (B); co-culture of SupT1 cells and HUT-78/IIIB cells (C); and co-culture of SupT1 cells and HUT-78/IIIB cells in the presence of MVN at 140 nm (D), 28 nm (E), and 6 nm (F).
Effect of MVN on the Capture of HIV-1 by Raji/DC-SIGN Cells and on Subsequent Virus Transmission to Uninfected CD4+ T Cells
A possible HIV mucosal infection pathway is the transmission of DC-SIGN captured virus to CD4+ T cells. We investigated the potential of MVN to prevent capture of the HIV-1 strain HE (R5/X4) by DC-SIGN using Raji cells transfected with DC-SIGN (31). Short exposure of these cells to HIV-1 results in a significant capture of virus to their cell membrane (32). Initially, HIV-1 was pre-exposed to MVN for 30 min before the virus was administered to the Raji/DC-SIGN cells. After 1 h, free virus and MVN were carefully removed by serial washing steps, and the amount of captured virus was determined by measurement of the p24 Ag content of the cells. MVN was able to efficiently inhibit binding of HIV-1 to the Raji/DC-SIGN cells (IC50 = 189 nm).
Next, the potential of MVN to prevent the transmission of HIV-1 captured by DC-SIGN to CD4+ T cells was examined. First, HIV-1 strain HE was given the opportunity to be captured by Raji/DC-SIGN cells. Then the HIV-1-captured Raji/DC-SIGN cells were washed and co-cultured with C8166 cells in the presence of various concentrations of MVN. Giant cell formation (appearing upon transmission of HIV-1 from the Raji/DC-SIGN cells to the C8166 CD4+ T cells) was recorded as a parameter of efficiency of virus transmission. MVN prevented the transmission of DC-SIGN-captured virus at an IC50 value of 168 nm.
Determination of Mutations in the Genome of the HIV-1 NL4.3 MVN-resistant Virus Isolate
A major problem in HIV treatment is the emergence of resistant virus mutants. Therefore, the development of resistance of HIV to MVN was investigated. The NL4.3 virus was cultured in MT-4 cells in the presence of suboptimal concentrations of MVN, and the drug concentration was increased when full CPE was observed. After 41 passages (∼205 days), a virus was isolated that was able to replicate in the presence of 700 nm MVN (>100-fold resistance). This NL4.3-MVN-resistant (MVN-res) virus was further analyzed for amino acid changes in its env genome. The data were compared with the sequence of wild-type HIV-1 NL4.3, which was subcultured in parallel in control medium. Interestingly, in the MVN-res virus, up to four (two pure and two mixed) mutations in gp120 were observed in N-glycosylation motifs for high mannose type glycans: Asn295, Asn339, Asn386, and Asn392 (Table 2). Additional mutations were found at K132E and A336T.
TABLE 2.
Amino acid mutations in the HIV-1 NL4.3 strain under sustained MVN pressure (up to 700 nm)
| High mannose type N-glycan sitea | Mutations in gp120 in the NL4.3-MVNres strain |
|---|---|
| 230NKT232 | |
| 234NGT236 | |
| 241NVS243 | |
| 262NGS264 | |
| 289NQS291 | |
| 295NCT297 | T297I |
| 332NIS334 | |
| 339NNT341 | T341(T,I)b |
| 386NST388 | N386(K,N) |
| 392NST394 | N392D |
| 448NIT450 |
a Position of the amino acids determined according to Leonard et al. (1).
b Amino acids in parentheses indicate a mixture of the two amino acids picked up by sequencing in the resistant virus.
Cross-resistance of the MVN-resistant Virus to Other CBAs
The sensitivity of the MVN-res virus against different CBAs and several other entry inhibitors was evaluated and compared with the sensitivity of the wild-type virus and a 2G12-res virus (Table 3). Although four N-glycan deletions were observed in the MVN-res virus, only minor changes in the antiviral activity of CV-N, HHA, GNA, and UDA against MVN-res virus were noted (<5-fold resistance). Also, the antiviral activities of the polyanion PRO 2000 and the CXCR4 antagonist AMD3100 were only slightly different between wild type (WT) and MVN-res viruses. In contrast, the mAb 2G12 was completely inactive against the MVN-res virus, and for PRM-S, a 5-fold decrease in activity was observed. Subcultivation of virus without compound for 41 passages did not affect its sensitivity to MVN, as evidenced by its comparable IC50 value (8 ± 1 nm, n = 3). Of note, whereas MVN-res virus showed cross-resistance to 2G12 mAb, 2G12-res virus kept full sensitivity to MVN.
TABLE 3.
Sensitivity profile of wild-type, MVN-resistant and 2G12-resistant HIV-1 NL4.3 virus
| Agenta | WT | MVN-res | Resistance/sensitivityb | 2G12-res | Resistance/sensitivityb |
|---|---|---|---|---|---|
| -fold | -fold | ||||
| MVN | 6 ± 1 | 576 ± 84 | [96] | 10 ± 4 | [2] |
| CV-N | 16 ± 10 | 60 ± 18 | [4] | 45 ± 35 | [3] |
| HHA | 4 ± 0.8 | 12 ± 4 | [3] | 0.5 ± 0.2 | [8]* |
| GNA | 15 ± 8 | 7 ± 1 | [2]* | 2 ± 1 | [7]* |
| UDA | 284 ± 79 | 718 ± 340 | [3] | 36 ± 9 | [8]* |
| PRM-S | 1,901 ± 11 | 9,363 ± 1,165 | [5] | 4,615 ± 2,714 | [2] |
| 2G12 mAbc | 1,310 ± 530 | >50,000 | [>38] | >50,000 | [>38] |
| b12 mAbc | 380 ± 110 | 2,740 ± 50 | [7] | 640 ± 220 | [2] |
| PRO 2000 | 342 ± 2 | 298 ± 216 | [1] | 196 ± 96 | [2]* |
| AMD3100 | 19 ± 5 | 8 ± 3 | [2]* | 5 ± 0.1 | [4]* |
a IC50 (drug concentration in nm required to inhibit virus replication in MT-4 cells by 50%).
b Values in brackets represent the degree (-fold) of resistance or sensitivity (indicated by an asterisk) of the test agents, compared with wild type virus. Values are mean ± S.E. from 2–7 separate experiments.
c IC50 (drug concentration in ng/ml required to inhibit virus replication in MT-4 cells by 50%).
Anti-gp120 mAb Staining Profile of HIV-1 MVN-res-infected Cells
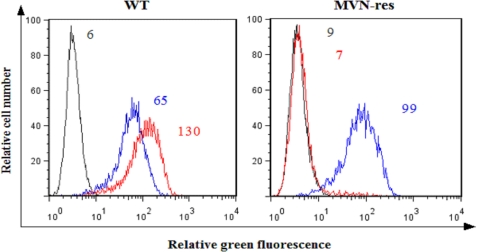
In order to further investigate the cross-resistance of MVN-res virus to 2G12 mAb, we evaluated the binding ability of 2G12 mAb to the resistant virus. MT-4 cells were infected with WT NL4.3 and MVN-res virus and stained for gp120 by means of the anti-gp120 mAb b12, which recognizes a specific epitope in the CD4-binding site of gp120 (33, 34) or the mannose-specific 2G12 mAb (Fig. 2). WT-infected cells stained positive for both mAbs b12 and 2G12 (histogram peaks in the left panel: MFI values were 65 and 130, respectively). Also, cells infected with MVN-res virus did express gp120 at their cell surface, as evidenced by the positive b12 mAb staining (MFI = 99). However, 2G12 mAb was no longer able to bind to the MVN-res gp120, as evidenced by the histogram peak that coincides with the background staining (MFI = 7).
FIGURE 2.
No staining of 2G12 mAb on MVN-res-infected MT-4 cells. MT-4 cells were infected with WT (left) and MVN-res (right) NL4.3, and after 3–4 days, an incubation was done with the 2G12 mAb (red histograms) and the b12 mAb (blue histograms), followed by a staining with rabbit anti-human IgG-FITC. The MFI values of the background fluorescence (gray histograms) for the 2G12 mAb (red histograms) and for the b12 mAb (blue histograms) are indicated.
Effect of MVN on the Binding of 2G12 mAb to gp120 on HIV-1-infected MT-4 Cells
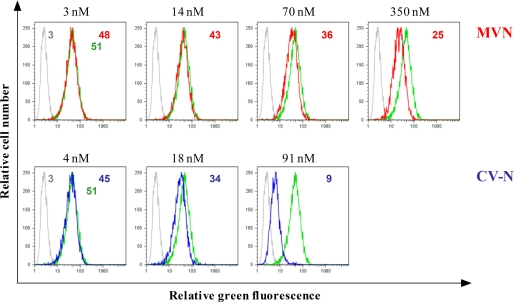
A microarray with eight different carbohydrates, of which most of the structures are derivatives of the high mannose structure displayed by the HIV envelope protein gp120, showed that MVN, just like CV-N, binds to carbohydrates. Because the strongest signals came from structures that contain α-1,2-mannose residues (24), and based on the fact that the neutralizing anti-HIV mAb 2G12 binds to high mannose carbohydrates of gp120, we investigated whether MVN interferes with 2G12 mAb binding on HIV-1-infected T cells. Therefore, HIV-1 NL4.3-infected MT-4 cells were preincubated with several concentrations of MVN, and subsequently, binding was performed with the 2G12 mAb (Fig. 3). As a control, CV-N was included. CV-N inhibited the binding of 2G12 mAb in a dose-dependent manner (IC50 = 28 nm). This is in agreement with our previous report showing a comparable level of inhibition of CV-N for 2G12 mAb binding on MT-4 cells infected with HIV-1 IIIB, NDK, and MN (28). MVN also inhibited 2G12 mAb binding, although to a lesser extent (IC50 = 259 nm), resulting in a ∼10-fold lower inhibiting activity for 2G12 mAb binding as compared with CV-N.
FIGURE 3.
Inhibition of the binding of 2G12 mAb to HIV-1 NL4.3-infected MT-4 cells by MVN and CV-N. MT-4 cells infected with HIV-1 strain NL4.3 were incubated with 2G12 mAb in the absence (green histograms) or presence of various concentrations of MVN (red histograms, top) and CV-N (blue histograms, bottom). The light gray histograms show the background fluorescence. The MFI values of the different incubation conditions are indicated in each histogram.
Evaluation of Potential Stimulatory Effects of MVN on PBMCs
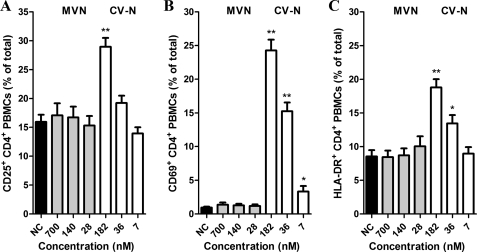
One of the major drawbacks of lectins is their generally assumed potential mitogenic properties. Recently, we described that CV-N markedly stimulated PBMCs (20). Thus, MVN was subjected to a thorough investigation of its potential cell-activating properties. In all of our experiments, CV-N was included as a positive control. Freshly isolated PBMCs were cultured in the presence of MVN or CV-N for 72 h, and the expression of the activation markers CD25, CD69, and HLA-DR of the CD4+ T cells was analyzed by flow cytometry. In unstimulated PBMCs, the mean ± S.E. of the percentages of CD25+ CD4+ cells was 15.9 ± 1.3%. Treatment of PBMCs with 700 nm MVN did not significantly increase the amount of CD4+ CD25+ cells (17.1% ± 2.1, p = 0.66; Fig. 4A). In contrast, treatment of PBMCs with 182 nm CV-N increased the number of CD4+ CD25+ cells dramatically (28.9 ± 1.5%, p < 0.0001; Fig. 4A). The MVN treatment also did not affect the expression of the early activation marker CD69 (mean ± S.E. for control and 700 nm MVN were 0.93 ± 0.16 and 1.4 ± 0.34%, respectively), whereas CV-N elicited a spectacular increase of 26-fold in CD69+ cells (24.3 ± 1.6%; p < 0.0001) (Fig. 4B). Finally, for the late activation marker HLA-DR, again no increase in CD4+ HLA-DR+ cells was measured in the MVN-treated cells (p = 0.96 for the highest concentrations of MVN; Fig. 4C). This is in sharp contrast with the data from treatment with 182 nm CV-N because in this condition, a 2-fold increase in CD4+ HLA-DR+ cells was observed (p < 0.0001; Fig. 4C).
FIGURE 4.
MVN does not increase the expression of cellular activation markers. PBMCs were cultured in the presence of varying concentrations of the lectins MVN and CV-N and incubated at 37 °C. At day 3, the PBMCs were analyzed by flow cytometry for their expression of cellular activation markers with phycoerythrin-conjugated anti-CD25 (A), anti-CD69 (B), or anti-HLA-DR (C) mAbs in combination with FITC-conjugated anti-CD4 mAb. Data represent mean percentage ± S.E. for 4–16 independent experiments. *, p < 0.05; **, p < 0.0001 for comparison with untreated PBMCs (Student t test).
Cytokine and Chemokine Profile of MVN-incubated PBMCs
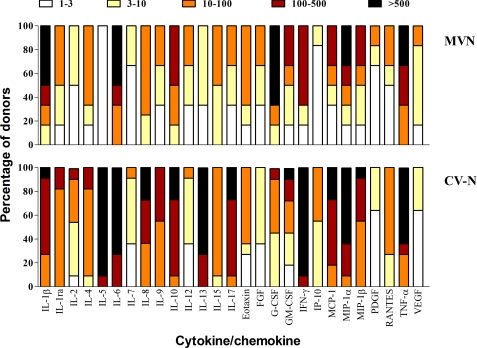
Potential activation of PBMCs may also be reflected by the production of cytokines and chemokines. Therefore, the supernatant of PBMCs was analyzed for its cytokine content by means of the Bio-Plex array system. To investigate potential activating properties of the lectins more thoroughly, PBMCs were cultured in the presence of 140 nm MVN or 182 nm CV-N for 72 h. In the culture supernatant, the concentrations of IL-1α, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, eotaxin, FGF, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, and VEGF were vascular endothelial growth factor were determined. A detailed overview of the cytokine profiles of MVN- and CV-N-treated PBMCs from multiple blood donors is given in Fig. 5. For each lectin, the concentration of the separate cytokines was compared with that of the untreated control and calculated as a -fold increase value. We observed considerable variability in the lectin-induced cytokine profile between the different PBMC donors. Therefore, the -fold increase values obtained from the different donors were divided over different ranking groups (i.e. 1–3-, 3–10-, 10–100-, 100–500-, and >500-fold increase), and the number in each rank is given as a percentage of the total and indicated by a specific color (Fig. 5). Despite the fact that MVN had no significant effect on the expression of the cellular activation markers, we observed that it had a stimulatory effect on the production of several cytokines. In fact, the levels of IL-1β, IL-6, IL-10, G-CSF, IFN-γ, MIP-1α, and TNF-α increased more than 100-fold for a minimum of 50% of the donors. However, overall, the cytokine response of MVN-treated PBMCs was less pronounced as compared with that of CV-N, with the exceptions of IL-1β and G-CSF.
FIGURE 5.
Induction of cytokines/chemokines by MVN. PBMCs from healthy donors were incubated for 72 h with medium only or MVN or CV-N at 140 and 182 nm, respectively. Supernatants were collected, and cytokine levels were measured by the Bio-Plex array system. The -fold increase values of the cytokine concentrations in the supernatant of stimulated PBMCs with respect to the concentrations in the supernatant of untreated PBMCs were determined from six and 11 different donors for MVN and CV-N, respectively. The -fold increase values are divided into subgroups: 1–3-fold increase (white squares), 3–10-fold increase (yellow squares), 10–100-fold increase (orange squares), 100–500-fold increase (dark red squares), and >500-fold increase (black squares). The amount of -fold increase values for each cytokine is given as a percentage in the total amount of donors (y axis).
Susceptibility of MVN-treated PBMCs for the R5 HIV-1 BaL Strain
Activation of PBMCs can ultimately result in higher susceptibility to infection with HIV-1, so next, we determined whether treatment of PBMCs with the CBAs influenced the infectivity of HIV-1. PBMCs from two different donors were treated with 700 nm MVN, 182 nm CV-N, 400 nm HHA, and 16 nm PHA. After 24 h, the cells were washed and exposed to the CCR5-tropic HIV-1 BaL strain. After 7 days, the p24 Ag production levels were measured in the supernatants as a parameter for viral infectivity and replication. As shown in Fig. 6, the p24 Ag production for the untreated cells was 10.2 and 38.1 ng/ml for donor 1 (A) and donor 2 (B), respectively. Pretreatment of PBMCs with 700 nm MVN had almost no effect on the infectivity of the R5 HIV-1 BaL strain: 3.3 and 30.1 ng/ml for donors 1 and 2, respectively. In contrast, pretreatment of PBMCs with 182 nm CV-N resulted in a spectacular increase in virus production; the p24 Ag levels rose to 354 and 443 ng/ml for donors 1 and 2, respectively. Remarkably, CV-N made the cells even more susceptible for infection with HIV-1 BaL than the mitogenic lectin PHA. Finally, pretreatment of PBMCs with 400 nm HHA had a minor, if any, stimulatory effect on the viral replication level (Fig. 6).
FIGURE 6.
Susceptibility of MVN-pretreated PBMCs for R5 HIV-1 infection. Freshly isolated PBMCs from two donors (A and B) were pretreated for 24 h with MVN, CV-N, HHA, and PHA and then washed and infected with the R5 HIV-1 BaL strain for 7 days. No compound was added during the infection, and viral replication was measured by a p24 Ag ELISA in the collected supernatants.
DISCUSSION
The focus of this work is on MVN, a novel cyanobacterial lectin. MVN, isolated from cyanobacterium M. aeruginosa PCC7806, shows 33% identity to CV-N, a lectin isolated from the cyanobacterium N. ellipsosporum. Both lectins show specificity for high mannose structures containing terminal α(1,2)-mannose moieties (24). CV-N is extensively studied, and it was shown that it possesses two carbohydrate binding sites. Because it predominantly occurs in a dimer state, it should be tetravalent. Both carbohydrate sites are conserved in MVN, and it should therefore at least act in a divalent manner. On SDS-PAGE, MVN runs as a dimer, and this probably also reflects the native state of the protein, which supports multivalence (24). In a recent paper on CV-N (35), it was shown that an engineered monovalent mutant variant of CV-N is devoid of any anti-HIV-1 activity. This is strong, albeit indirect, evidence for multivalence in MVN. Recently, it also has been shown that the 2G12 mAb exists in monomer form but also in dimer form and that this latter has increased neutralizing anti-HIV-1 activity (30). Although CV-N has been considered as a potential anti-HIV microbicide, we previously demonstrated that its antiviral activity was overshadowed by accompanied mitogenic side effects, and very often an enhancement of viral replication levels at suboptimal concentrations was observed (19, 20).
If the ultimate goal was the use of MVN as a microbicide, then this compound should inhibit transmission of (i) cell-free viruses, (ii) donor-infected T cells, and (iii) HIV-1 captured by DC-SIGN to CD4+ T cells. In this study, MVN inhibited cell-free virus infection of a broad range of HIV-1 clades in PBMCs, demonstrating a broad spectrum anti-HIV activity. All of the investigated HIV-1 M clades were sensitive to MVN. Many of these isolates have a preference for the co-receptor CCR5, which is important because HIV transmission occurs mainly through CCR5-using (R5) viruses. MVN was also able to inhibit syncytium formation between persistently HIV-1-infected T cells and uninfected CD4+ T cells, thus suggesting an inhibitory role of MVN during attachment of gp120 to cellular receptors and subsequent fusion steps. In addition, we demonstrated a clear inhibitory activity of MVN for transmission of DC-SIGN-captured virus to CD4+ T cells. This is a major asset for MVN because DC-SIGN-directed entry and transmission to CD4+ T-lymphocytes is considered as an important avenue of primary infection of women exposed to HIV-1 through sexual intercourse (36). Also, to block the multiple pathways of HIV mucosal transmission, it is likely that a combination of several microbicide molecules will lead to the best protection against HIV transmission. The combination of MVN with the FDA-approved gp41 fusion inhibitor enfuvirtide (T-20) resulted in strong synergistic inhibition of HIV-1 NL4.3 replication in MT-4 cells, as we also recently showed with the combination of HHA with T-20 (37). Also, in combination with CADA, a CD4 receptor down-regulating HIV inhibitor (38), antiviral synergy was observed (data not shown).
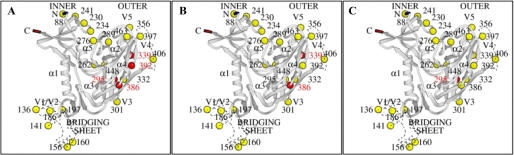
Despite the partial homology between MVN and CV-N, there were also remarkable differences observed between these two cyanobacterial lectins. In contrast to CV-N, MVN was not active against a HIV-1 group O isolate and against HIV-2 and SIV. The glycosylation shield on gp120 of these peculiar lentivirus types should be quite different. It is plausible that MVN and CV-N recognize the several mannose moieties on gp120 in a different way. Indeed, from Fig. 3, it is clear that CV-N binds to the 2G12 mAb epitope on gp120 more firmly than MVN, resulting in a stronger competition for 2G12 mAb binding. The different preference for mannose sugars on gp120 between MVN and CV-N can also be deducted from their resistance profile. The MVN-res virus was selected after 41 passages, and two pure mutations were detected that deleted the glycans at positions Asn295 and Asn392 (Fig. 7A). In addition, two mixed mutations were observed that affected the glycans at position Asn339 and Asn386. In comparison, the CV-N-res virus was selected after 60 passages, with two pure mutations that deleted the glycans at positions Asn339 and Asn386 (19) (Fig. 7B). The fact that the glycans on Asn339 and Asn386 of the MVN-res virus were only partially deleted can explain why the virus showed some cross-resistance to CV-N (4-fold resistance). The α(1,2)-mannose-specific PRM-S appeared to be 5-fold less active against MVN-res virus, and it is reported that a PRM-S-resistant IIIB virus has, among other mutations, also a mutation deleting the glycan on Asn295 (39). Thus, it seems that Asn295 might be an important glycan for the activity of PRM-S, and this can explain its reduced activity against the MVN-res virus. Interestingly, MVN kept its activity against the HIV-1 NL4.3 2G12-res virus. This 2G12-res virus was selected already after six passages and has only one pure mutation that deletes the glycan at Asn295 (28) (Fig. 7C). The mutations in gp120 of MVN-res virus suggest that MVN can use additional glycans to interact with gp120 and exert its antiviral activity. As thus expected, the 2G12 mAb was not able to bind to the MVN-res virus having a mutation at Asn295, whereas the binding of the b12 mAb to the MVN-res virus was not affected (Fig. 2). Importantly, the MVN-res virus was also still sensitive to various other CBAs (HHA, GNA, and UDA), demonstrating that cross-resistance between the different CBAs is not a general phenomenon.
FIGURE 7.
Ribbon diagrams showing the N-glycosylation site mutations (red circles) in gp120 of the MVN-res HIV-1 (A), the CV-N-res HIV-1 (B), and the 2G12-res HIV-1 (C). The 24 putative glycosylation sites are represented by colored circles and their accompanying amino acid number. The red circles indicate the deleted N-glycosylation sites that appear under MVN (A), CV-N (B), and 2G12 mAb (C) pressure in NL4.3 HIV-1.
A major advantage of MVN compared with CV-N is its much better safety profile. In MT-4 cells and PBMCs, MVN did not exert any cellular toxicity at a dose up to 7 μm, which is in sharp contrast to CV-N (the CC50 value in MT-4 cells and PBMCs was 190 and 900 nm, respectively). Thus, the toxicity/activity window is much wider for MVN compared with CV-N, making MVN a much more interesting candidate in a clinical setting. Also, MVN did not activate the cells as evidenced by the weak increase in expression of the cellular activation markers, whereas CV-N increased by 2-, 26-, and 2-fold the expression of the activation markers CD25, CD69, and HLA-DR, respectively. A recently published lectin, BanLec, that is isolated from bananas, Musa acuminata, possessed potent anti-HIV activity (7). However, BanLec showed strong mitogenic activity in human and murine T cells (40–42). These data suggest that MVN might be more interesting than BanLec as a potential component for an antiviral microbicide. MVN had, however, a substantial effect on PBMCs in the release of several cytokines, although this effect was clearly less pronounced than for CV-N. It appears that the induction of activation markers has more serious implications compared with the release of cytokines in terms of susceptibility to HIV infection and replication. Indeed, when PBMCs were pretreated with MVN for 24 h before exposure to R5 HIV-1, no enhancement in viral replication could be observed. On the contrary, cells pretreated with CV-N became profoundly activated and showed a much higher level of viral replication compared with untreated cells. Our statement that for microbicidal gel applications, the cellular activation has more dangerous consequences for enhanced viral susceptibility than general cytokine release is in agreement with the safety data obtained for PRO 2000. This compound was tested in a clinical phase III trial (MDP301) and was found to be safe and well tolerated. However, it induced the release of several cytokines but had only minimal effect on the expression of the cellular activation markers CD25 and CD69 (43). Also, with the plant lectins HHA, GNA, and UDA, several cytokines, such as IL-β, IL-6, G-CSF, and MCP-1, were induced, but only weak, if any, cellular activation was observed, and these lectins also did not enhance viral replication in T cells pretreated with HHA, GNA, or UDA (20). Finally, the effect of cytokine release from activated cells on viral replication will also strongly depend on the net balance of pro- and anti-inflammatory cytokines.
In conclusion, we demonstrated that MVN has anti-HIV-1 activity comparable with that of CV-N but a much higher safety profile. However, it would be important to investigate the potential side effects of MVN on the normal flora of the cervicovaginal tract before further development as a potential microbicidal agent.
Acknowledgments
We are grateful to Becky Provinciael, Sandra Claes, Eric Fonteyn, and Yoeri Schrooten for excellent technical assistance and especially thank Dr. Kristel Van Laethem for sequencing and Dr. Mathy Froeyen for valuable help in drawing Fig. 7.
This work was supported by the Foundation Dormeur, Concerted Actions of the K.U. Leuven Grant GOA 10/014, Center of Excellence Grant EF/05/15, Foundation of Scientific Research Grant FWO G-485-08, and the CHAARM project of the European Commission.
- HIV-1
- human immunodeficiency virus type 1
- Ag
- antigen
- CC50
- 50% cytotoxic concentration
- CBA
- carbohydrate binding agent
- CPE
- cytopathic effect
- CV-N
- cyanovirin-N
- ELISA
- enzyme-linked immunosorbent assay
- FITC
- fluorescein isothiocyanate
- GNA
- G. nivalis agglutinin
- HHA
- Hippeastrum hybrid agglutinin
- HUT-78/IIIB
- persistently HIV-1(IIIB)-infected HUT-78 cells
- mAb
- monoclonal antibody
- MFI
- mean fluorescence intensity
- MVN
- microvirin
- MVN-res
- NL4.3-MVN-resistant
- PBMC
- peripheral blood mononuclear cell
- PHA
- phytohemagglutinin
- PRM-S
- pradimicin S
- TCID50
- tissue culture infective dose 50%
- UDA
- U. dioica agglutinin
- WT
- wild type
- IL
- interleukin
- G-CSF
- granulocyte colony-stimulating factor
- FGF
- fibroblast growth factor
- GM-CSF
- granulocyte-macrophage-CSF
- IFN-γ
- interferon-γ
- IP-10
- interferon-inducible protein-10
- MCP-1
- monocyte chemoattractant protein-1
- MIP
- macrophage inflammatory protein
- PDGF-BB
- platelet-derived growth factor-BB
- TNF-α
- tumor necrosis factor-α
- VEGF
- vascular endothelial growth factor
- RANTES
- regulated on activation normal T cell expressed and secreted.
REFERENCES
- 1.Leonard C. K., Spellman M. W., Riddle L., Harris R. J., Thomas J. N., Gregory T. J. (1990) J. Biol. Chem. 265, 10373–10382 [PubMed] [Google Scholar]
- 2.Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyatt R., Kwong P. D., Desjardins E., Sweet R. W., Robinson J., Hendrickson W. A., Sodroski J. G. (1998) Nature 393, 705–711 [DOI] [PubMed] [Google Scholar]
- 4.O'Keefe B. R., Vojdani F., Buffa V., Shattock R. J., Montefiori D. C., Bakke J., Mirsalis J., d'Andrea A. L., Hume S. D., Bratcher B., Saucedo C. J., McMahon J. B., Pogue G. P., Palmer K. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6099–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini J. (2007) Nat. Rev. Microbiol. 5, 583–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzarini J., Van Laethem K., Hatse S., Froeyen M., Peumans W., Van Damme E., Schols D. (2005) J. Biol. Chem. 280, 41005–41014 [DOI] [PubMed] [Google Scholar]
- 7.Swanson M. D., Winter H. C., Goldstein I. J., Markovitz D. M. (2010) J. Biol. Chem. 285, 8646–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori T., O'Keefe B. R., Sowder R. C., 2nd, Bringans S., Gardella R., Berg S., Cochran P., Turpin J. A., Buckheit R. W., Jr., McMahon J. B., Boyd M. R. (2005) J. Biol. Chem. 280, 9345–9353 [DOI] [PubMed] [Google Scholar]
- 9.Bewley C. A., Gustafson K. R., Boyd M. R., Covell D. G., Bax A., Clore G. M., Gronenborn A. M. (1998) Nat. Struct. Biol. 5, 571–578 [DOI] [PubMed] [Google Scholar]
- 10.Bewley C. A., Kiyonaka S., Hamachi I. (2002) J. Mol. Biol. 322, 881–889 [DOI] [PubMed] [Google Scholar]
- 11.Bolmstedt A. J., O'Keefe B. R., Shenoy S. R., McMahon J. B., Boyd M. R. (2001) Mol. Pharmacol. 59, 949–954 [DOI] [PubMed] [Google Scholar]
- 12.Botos I., O'Keefe B. R., Shenoy S. R., Cartner L. K., Ratner D. M., Seeberger P. H., Boyd M. R., Wlodawer A. (2002) J. Biol. Chem. 277, 34336–34342 [DOI] [PubMed] [Google Scholar]
- 13.Boyd M. R., Gustafson K. R., McMahon J. B., Shoemaker R. H., O'Keefe B. R., Mori T., Gulakowski R. J., Wu L., Rivera M. I., Laurencot C. M., Currens M. J., Cardellina J. H., 2nd, Buckheit R. W., Jr., Nara P. L., Pannell L. K., Sowder R. C., 2nd, Henderson L. E. (1997) Antimicrob. Agents Chemother. 41, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori T., Shoemaker R. H., Gulakowski R. J., Krepps B. L., McMahon J. B., Gustafson K. R., Pannell L. K., Boyd M. R. (1997) Biochem. Biophys. Res. Commun. 238, 218–222 [DOI] [PubMed] [Google Scholar]
- 15.Witvrouw M., Fikkert V., Hantson A., Pannecouque C., O'keefe B. R., McMahon J., Stamatatos L., de Clercq E., Bolmstedt A. (2005) J. Virol. 79, 7777–7784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esser M. T., Mori T., Mondor I., Sattentau Q. J., Dey B., Berger E. A., Boyd M. R., Lifson J. D. (1999) J. Virol. 73, 4360–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bewley C. A., Otero-Quintero S. (2001) J. Am. Chem. Soc. 123, 3892–3902 [DOI] [PubMed] [Google Scholar]
- 18.Shenoy S. R., O'Keefe B. R., Bolmstedt A. J., Cartner L. K., Boyd M. R. (2001) J. Pharmacol. Exp. Ther. 297, 704–710 [PubMed] [Google Scholar]
- 19.Balzarini J., Van Laethem K., Peumans W. J., Van Damme E. J., Bolmstedt A., Gago F., Schols D. (2006) J. Virol. 80, 8411–8421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huskens D., Vermeire K., Vandemeulebroucke E., Balzarini J., Schols D. (2008) Int. J. Biochem. Cell Biol. 40, 2802–2814 [DOI] [PubMed] [Google Scholar]
- 21.Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A. (2003) Science 300, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 22.Hessell A. J., Rakasz E. G., Poignard P., Hangartner L., Landucci G., Forthal D. N., Koff W. C., Watkins D. I., Burton D. R. (2009) PLoS Pathog. 5, e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mascola J. R., Stiegler G., VanCott T. C., Katinger H., Carpenter C. B., Hanson C. E., Beary H., Hayes D., Frankel S. S., Birx D. L., Lewis M. G. (2000) Nat. Med. 6, 207–210 [DOI] [PubMed] [Google Scholar]
- 24.Kehr J. C., Zilliges Y., Springer A., Disney M. D., Ratner D. D., Bouchier C., Seeberger P. H., de Marsac N. T., Dittmann E. (2006) Mol. Microbiol. 59, 893–906 [DOI] [PubMed] [Google Scholar]
- 25.Saitoh K., Tenmyo O., Yamamoto S., Furumai T., Oki T. (1993) J. Antibiot. 46, 580–588 [DOI] [PubMed] [Google Scholar]
- 26.Schols D., Struyf S., Van Damme J., Esté J. A., Henson G., De Clercq E. (1997) J. Exp. Med. 186, 1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauwels R., Andries K., Desmyter J., Schols D., Kukla M. J., Breslin H. J., Raeymaeckers A., Van Gelder J., Woestenborghs R., Heykants J. (1990) Nature 343, 470–474 [DOI] [PubMed] [Google Scholar]
- 28.Huskens D., Van Laethem K., Vermeire K., Balzarini J., Schols D. (2007) Virology 360, 294–304 [DOI] [PubMed] [Google Scholar]
- 29.Van Laethem K., Schrooten Y., Lemey P., Van Wijngaerden E., De Wit S., Van Ranst M., Vandamme A. M. (2005) J. Virol. Methods 123, 25–34 [DOI] [PubMed] [Google Scholar]
- 30.West A. P., Jr., Galimidi R. P., Foglesong C. P., Gnanapragasam P. N., Huey-Tubman K. E., Klein J. S., Suzuki M. D., Tiangco N. E., Vielmetter J., Bjorkman P. J. (2009) J. Virol. 83, 98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geijtenbeek T. B., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y. (2000) Cell 100, 587–597 [DOI] [PubMed] [Google Scholar]
- 32.Balzarini J., Van Herrewege Y., Vermeire K., Vanham G., Schols D. (2007) Mol. Pharmacol. 71, 3–11 [DOI] [PubMed] [Google Scholar]
- 33.Burton D. R., Pyati J., Koduri R., Sharp S. J., Thornton G. B., Parren P. W., Sawyer L. S., Hendry R. M., Dunlop N., Nara P. L. (1994) Science 266, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 34.Roben P., Moore J. P., Thali M., Sodroski J., Barbas C. F., 3rd, Burton D. R. (1994) J. Virol. 68, 4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matei E., Zheng A., Furey W., Rose J., Aiken C., Gronenborn A. M. (2010) J. Biol. Chem. 285, 13057–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hladik F., McElrath M. J. (2008) Nat. Rev. Immunol. 8, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenabian M. A., Saïdi H., Charpentier C., Van Herrewege Y., Son J. C., Schols D., Balzarini J., Vanham G., Bélec L. (2009) J. Antimicrob. Chemother. 64, 1192–1195 [DOI] [PubMed] [Google Scholar]
- 38.Vermeire K., Princen K., Hatse S., De Clercq E., Dey K., Bell T. W., Schols D. (2004) AIDS 18, 2115–2125 [DOI] [PubMed] [Google Scholar]
- 39.Balzarini J., François K. O., Van Laethem K., Hoorelbeke B., Renders M., Auwerx J., Liekens S., Oki T., Igarashi Y., Schols D. (2010) Antimicrob. Agents Chemother. 54, 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peumans W. J., Zhang W., Barre A., Houlès Astoul C., Balint-Kurti P. J., Rovira P., Rougé P., May G. D., Van Leuven F., Truffa-Bachi P., Van Damme E. J. (2000) Planta 211, 546–554 [DOI] [PubMed] [Google Scholar]
- 41.Gavrovic-Jankulovic M., Poulsen K., Brckalo T., Bobic S., Lindner B., Petersen A. (2008) Int. J. Biochem. Cell Biol. 40, 929–941 [DOI] [PubMed] [Google Scholar]
- 42.Koshte V. L., van Dijk W., van der Stelt M. E., Aalberse R. C. (1990) Biochem. J. 272, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huskens D., Vermeire K., Profy A. T., Schols D. (2009) Antiviral Res. 84, 38–47 [DOI] [PubMed] [Google Scholar]
- 44.Joint United Nations Program on HIV/AIDS (2008) Report on the Global AIDS Epidemic, UNAIDS, Geneva, Switzerland [Google Scholar]