Abstract
We identified three heterozygous nonsynonymous single nucleotide polymorphisms in the small heterodimer partner (SHP, NROB2) gene in normal subjects and CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy)-like patients, including two novel missense mutations (p.R38H, p.K170N) and one of the previously reported polymorphism (p.G171A). Four novel heterozygous mutations were also identified in the intron (Intron1265T→A), 3′-untranslated region (3′-UTR101C→G, 3′-UTR186T→C), and promoter (Pro-423C→T) of the SHP gene. The exonic R38H and K170N mutants exhibited impaired nuclear translocation. K170N made SHP more susceptible to ubiquitination mediated degradation and blocked SHP acetylation, which displayed lost repressive activity on its interacting partners ERRγ and HNF4α but not LRH-1. In contrast, G171A increased SHP mRNA and protein expression and maintained normal function. In general, the interaction of SHP mutants with LRH-1 and EID1 was enhanced. K170N also markedly impaired the recruitment of SHP, HNF4α, HDAC1, and HDAC3 to the apoCIII promoter. Molecular dynamics simulations of SHP showed that G171A stabilized the nuclear receptor boxes, whereas K170N promoted the conformational destabilization of all the structural elements of the receptor. This study suggests that genetic variations in SHP are common among human subjects and the Lys-170 residue plays a key role in controlling SHP ubiquitination and acetylation associated with SHP protein stability and repressive function.
Keywords: Gene Regulation, Gene Transcription, Genetic Polymorphism, Nuclear Receptors, Protein Degradation, Protein-DNA Interaction, CADASIL, Function, Polymorphisms, Structure
Introduction
Genetic variations can cause disease by affecting important biological processes. Genetic variations include deletions, insertions, repetitive elements, and large chromosomal rearrangements. Genetic variations can also occur at the level of a single base (termed a single nucleotide polymorphism (SNP)2). SNPs occur at a specific site in the DNA sequence and are the most common type of genetic variation among people. Polymorphisms of genes may influence gene transcription, mRNA stability, and protein activity. SNP variations in the DNA sequences of humans may help predict an individual's response to certain drugs, susceptibility to environmental factors, and risk of developing particular diseases. Thus, SNPs are thought to be critical enablers in realizing the concept of personalized medicine and can be highly useful in diagnostics and drug development (1).
The human nuclear receptor small heterodimer partner (SHP, NROB2) gene is located on chromosome 1p36.1, which consists of two exons separated by an intron (2). SHP is a 257-amino acid protein that contains ligand binding and dimerization domains but lacks a DNA binding domain (3). It plays important roles in regulating metabolic diseases (4, 5), liver cancer progression (6–8), and microRNAs gene transcription and function (9–13). SNPs in the human SHP gene have been identified in normal subjects and patients with diabetes, obesity, and lipodystrophy syndromes (14–19). Some genetic variants in SHP are associated with disease conditions in these studies.
SHP is expressed in the cerebellum and other regions of the brain; however, the function of SHP in brain remains undetermined. In this study we screened germinal SHP gene polymorphisms in normal subjects and CADASIL-like patients. CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) is caused by mutations in the Notch3 receptor (20). We identified six novel variants in the SHP gene that have not been identified before in other diseases. Interestingly, the three exonic SHP mutants exhibited differential functional alterations with regard to their nuclear translocation, gene expression, repressive activity, protein-protein interaction, stability, and degradation. The later was associated with post-translational modification of SHP ubiquitination and acetylation. This report is the first study that identified novel SHP variants in humans and elucidated the molecular and structural basis for the altered functions of SHP mutants.
EXPERIMENTAL PROCEDURES
Study Subjects
Genomic DNAs (provided by Dr. Eliecer Coto) were obtained from leukocytes of a total of 84 subjects (34 healthy and 50 CADASIL-like patients, recruited through the Neurology and the Molecular Genetics Departments of Hospital University Central Asturias, Spain). A total of 14 patients had a NOTCH3 gene-causative mutation, and 36 had no observed Notch3 mutations. The normal subjects were collected at two different times (10 and 24 cases each time). Informed consent was obtained from the patients and controls to participate in a study of the genetic bases of stroke, approved by the Ethical Committee of Hospital UnivCentral Asturias (Spain). Case numbers and Notch3 mutations in CADASIL patients are provided in supplemental Tables S1 and S2.
Mutation Analysis by PCR and Direct Sequencing
Two sets of primers that cross the intron-exon boundaries, SHPe1 forward (5′-GCCACTTCATTGACTGAGGTGATA-3′) and reverse (5′-AATGAGGACCCAATGAGATAACAGA-3′) and SHPe2 forward (5′-CTCTAAAGGAGGAGGTAGTAGTGGGAC-3′) and reverse (5′-GGTGATGGGATTGTGAACTGATAC-3′), were used in a PCR to amplify the exon1 and exon2 from genomic DNAs. The primers SHPin forward (5′-GGGACCATCCTCTTCAACCC) and SHPin reverse (5′-CTTGGAGGCCTGGCACATCT) were used to amplify the intron, and primers SHPpro forward (5′-TAGATTGGACAGTGGGCAAAGT-3′) and SHPpro reverse (5′-CAGCTCTCTGGCTCTGTGTTCT-3′) were used to amplify the promoter region of the SHP gene. PCR was carried out using high fidelity Tag DNA polymerase. PCR products were purified and then sequenced and analyzed on an ABI 377 automated DNA sequencer (PerkinElmer Life Sciences). Sequencing was completed by the Sequencing and Genomic Core laboratory, which is one of the HSC Core Research Facilities at the University of Utah. The identified SHP variants were deposited to the NCBI data base, and the accession numbers are 142101870 (c.113G→A), 142101873 (c.510A→T), 142101875 (c.512G→C), 142101877 (3′-UTR101C→G), 142101879 (3′-UTR186T→C), 142101881 (Intron1265T→A), and 142101883 (Pro-423C→T).
Mutation Nomenclature
The cDNA NM_021969.2 and protein NP_068804.1 sequences were used for mutation nomenclature, with DNA +1 corresponding to the A of the ATG start codon. All sequence variants were verified using the Mutalyzer program.
Plasmid Construction and Transient Transfection
The cDNA of the wild-type human SHP gene was cloned in the expression plasmids pcDNA3-GFP and pCMV-FLAG, respectively. SHP genes carrying the identified mutations were generated by PCR-based site-directed mutagenesis. All SHP constructs including SHPR38H, SHPK170N, and SHPG171A were subcloned into the pcDNA3-GFP and pCMV-FLAG vectors and verified by sequencing. The primers used were G113A forward (5′-cgaccccgtagccactgcctatgtagg-3′) and G113A reverse (5′-cctacataggcagtggctacggggtcg-3′), A510T forward (5′-ggaatatgcctgcctgaatgggaccatcctcttc-3′) and A510T reverse (5′-gaagaggatggtcccattcaggcaggcatattcc-3′), G512C forward (5′-tatgcctgcctgaaagggaccatcctcttcaac-3′) and G512C reverse (5′-gttgaagaggatggtccctttcaggcaggcata-3′). For luciferase reporter assay, YY1 promoter reporter was used to determine SHP repression of ERRγ transactivation using HeLa cells (11). SHP promoter was used to determine SHP repression of LRH-1 transactivation (8). ApoCIII and apoB promoters (21) from Dr. Frances Sladek were used to determine SHP repression of HNF4a transactivation. The luciferase reporter activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer's instructions. Renilla luciferase activity was used to normalize transfection efficiency.
Ubiquitination Assays
HeLa cells were transfected with the FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, and FLAG-SHPG171A and HA-ubiquitin plasmids by Lipofectamine 2000 (Invitrogen) and treated with 5 μm MG132 (Cayman) after transfection. Twenty-four hours later cells were harvested and lysed in 500 μl of lysis buffer (50 mm Tris at pH 8.0, 250 mm NaCl, 0.5% Nonidet P-40, 1 mm dithiothreitol, and protease inhibitors) and immunoprecipitated with anti-FLAG M2 magnetic beads (Sigma) for 4 h at 4 °C. The beads were washed three times with the lysis buffer and analyzed with anti-HA (Sigma) and anti-FLAG (Sigma) by Western blots.
Acetylation Assays
HeLa cells were transfected with FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, and FLAG-SHPG171A for 24 h or co-transfected with CBP, P300, PCAF, Tip60, and males absent on the first (MOF) expression vectors (22), then treated with 0.2 μm trichostatin A for an additional 2 h. Whole cell extracts were prepared in lysis buffer (50 mm Tris at pH 8.0, 125 mm NaCl, 0.125% Nonidet P-40, 1 mm dithiothreitol, and protease inhibitors) and immunoprecipitated with anti-FLAG M2 magnetic beads (Sigma) as described in the ubiquitination assays. Western blots were performed, and immunoprecipitated proteins were detected with antibodies against acetylated lysine (Cell Signaling) and anti-FLAG M2 antibodies (Sigma).
Glutathione S-Transferase (GST) Pulldown Assays
GST-ERRγ (Dr. Uwe Borgmeyer), GST-LRH-1 (Dr. Timothy Osborne), GST-HNF4α (Dr. Akiyoshi Fukamizu), and GST-EID1 (Dr. Eckardt Treuter) were generously provided as indicated (23–26). MagneGSTTM Pull-Down System (catalog #V8872, Promega) was used in this experiment. In vitro protein synthesis of FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, and FLAG-SHPG171A was performed using the TNT- T7 quick coupled transcription/translation system utilizing the T7 promoter (Promega) according to the manufacturer's instructions. To express GST fusion proteins, GST, GST-ERRγ, GST-LRH1, GST-HNF4α, or GST-EID1 were expressed in Escherichia coli BL21/DE3/RIL. The cells containing the transformed plasmids were cultured overnight and inoculated into 4 ml of LB at 37 °C for 3 h (A600 0.6–0.8), and isopropyl 1-thio-β-d-galactopyranoside was then added to the culture to a final concentration of 1 mm. The culture was allowed to grow at 30 °C with vigorous shaking for about 4 h. Cells were harvested, then washed once in ice-cold phosphate-buffered saline and resuspended in 0.4 ml of cell lysis buffer provided by the kit. For pulldown experiments, bacterially expressed GST fusion proteins were bound to glutathione-containing MagneGSTTM particles, and the binding of the in vitro translated FLAG-SHP protein with GST fusion protein was conducted using the MagneGSTTM pulldown system as described in the manufacturer's manual with some modification. Briefly, 5 μl of particles carrying bait proteins (GST fusion protein or GST alone) were combined with 20 μl of prey protein (the TNT-T7 quick-coupled transcription/translation reaction). After capturing, the particles were washed, and the prey proteins were then eluted by suspending in 1× SDS sample buffer. The released proteins were displayed through SDS-PAGE and transferred to nitrocellulose membranes according to standard procedures. Membranes were washed in Tris-buffered saline containing 0.05% Tween 20 (TBST) and blocked for 1 h with TBST containing 5% nonfat milk, then incubated with anti-FLAG M2-peroxidase (horseradish peroxidase) antibody (catalog #A8592, Sigma) at 1:2000 dilution in TBST containing 5% nonfat milk overnight at 4 °C. Membranes were washed four times with TBST before antibody binding was visualized by SuperSignal West Pico chemiluminescent substrate (catalog #34080, Thermo Fisher Scientific Inc., Rockford, IL) according to the manufacturer's protocol.
Molecular Dynamics Simulations of SHP Wild Type and Mutants
The homology model of SHPWT was constructed on the basis of the sequence similarity with the Ultraspiracle protein (pdb code 1g2n) as previously reported (27). SHPK170Nand SHPG171A models were generated replacing Lys-170 and Gly-171 with asparagine and alanine residues in the structure of SHPWT, respectively. Hydrogen atoms were added using the Protein Preparation Wizard workflow as implemented in the Schrödinger Suite 2009 (28). Each of the three models was solvated in an orthorhombic box using TIP3P water molecules (29). Sodium and chlorine atoms were also placed to counterbalance the net charges of the receptor. After the construction of the solvent environment, each system consisted of ∼36,000 atoms in a volume of 374,000 Å3 (see Table 3).
TABLE 3.
RMSD values of the backbone atoms in different structural motifs of SHP wild type, SHPK170N, and SHPG171N mutants as calculated during the 2–10 ns of the molecular dynamic simulations
| Structural motif | SHPWT |
SHPK170N |
SHPG171A |
|||
|---|---|---|---|---|---|---|
| Average | ±S.D | Average | ±S.D. | Average | ±S.D. | |
| Backbone | 3.56 | 0.46 | 4.59 | 0.32 | 3.67 | 0.26 |
| NRbox1 (residues 21–25) | 2.5 | 0.72 | 3.78 | 0.91 | 2.13 | 0.95 |
| NRbox2 (residues 118–123) | 8.25 | 2.35 | 8.61 | 1.36 | 3.3 | 0.75 |
| NRbox3 (residues 214–219) | 1.55 | 0.43 | 1.81 | 0.59 | 1.09 | 0.25 |
| PSXXLP (residues 74–80) | 1.13 | 0.27 | 2.82 | 0.35 | 2.47 | 0.46 |
| H12 (residues 243–257) | 2.57 | 0.3 | 3.42 | 0.53 | 2.88 | 0.3 |
All the molecular dynamic simulations (MDs) were performed using Desmond 2.2 software (31) and the force field OPLS-2005 as implemented in the Schrödinger Suite 2009. In particular, the simulation protocol consisted of the desmond default relaxation steps, a 2-ns equilibration phase, and a final production phase of 8 ns.
In particular, desmond default relaxation steps were (i) the energetic minimization of the system over a maximum of 2000 steps, with the first 10 steps using the steepest descendent and the convergence criterion set to 50 kcal/mol Å−1 and harmonic restraints placed on the solute atoms (force constant = 50.0 kcal/mol), (ii) an analogous energetic refinement of the system without restraints, (iii) MDs of 12 ps at 10 K with harmonic restraints on the solute heavy atoms (force constant = 50.0 kcal/mol), NVT Berendesen ensemble, (iv) MDs of 12 ps at 10 K retaining the harmonic restrains, NPT Berendesen ensemble; (v) MDs of 24 ps of heating phase to 300 K retaining the harmonic restrains, NPT Berendesen ensemble, (vi) MDs of 24 ps at 300 K without harmonic restrains, NPT Berendesen ensemble. Both equilibration and production phases were run using the NPT Berendesen ensemble at temperature 300 K. During all the simulations, a time step of 2 fs was used. Other not specified parameters were set to the default values. The atomic coordinates of each system were saved every 4.8 ps.
Statistical Analysis
Data are expressed as the mean ± S.D. Statistical analyses were carried out using one-way analysis of variance followed by Student's t test; p < 0.01 was considered significant.
RESULTS
Identifying Novel SHP Gene Mutations in Normal and CADASIL-like Subjects
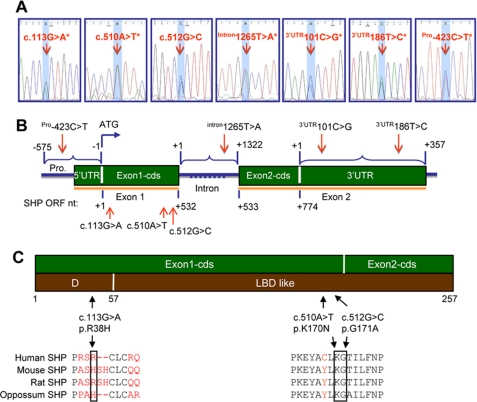
To identify gene polymorphisms of SHP, we collected germinal genomic DNAs of leukocytes from 34 normal and 50 CADASIL-like subjects (supplemental Tables S1 and S2). Promoter, coding region, intron, and 5′- and 3′-UTRs of the SHP gene were screened using PCR and direct sequencing. As shown in Fig. 1, A–C and Tables 1 and 2, novel exonic variants p.R38H (c.113G→A) and p.K170N (c.510A→T) were found in exon 1 of the SHP gene in normal and CADASIL-like subjects. A novel variant Intron1265T→A, 1265 bp downstream of the exon 1/intron boundary, was found in the intron of the SHP gene. Additionally, two novel variants 3′-UTR101C→G and 3′-UTR186T→C, 101 bp and 186 bp downstream of the translation stop site, were found in the 3′-UTR region of the SHP gene, respectively. A novel variant, Pro-423C→T, was also identified in the promoter of the SHP gene. The nucleotide change p.G171A (c.512G→C), a previously identified common SNP in obesity, diabetes, and lipodystrophy subjects (Table 2), was also found in normal and CADASIL-like subjects. All the identified SHP SNPs were in heterozygous forms. Several subjects carried more than one variant, including two normal (#8, #2) and two CADASIL-like (#69, #116) subjects (Table 1). In general, SHP gene exonic mutations are less commonly seen in patients with CADASIL-like that have Notch3 mutations as compared with the controls. On the other hand, SNPs in intron and 3′-UTR of the SHP gene occurred more frequently in CADASIL-like patients than in normal subjects.
FIGURE 1.
Polymorphisms in the human SHP gene in normal and CADASIL subjects. A, electropherograms show the six novel and one previously identified heterozygous sequence variants. All variants were identified in the forward and reverse genomic DNA strands. The SNP of interest is highlighted in light blue. The asterisks indicate novel variants identified in this study which have not been reported before. B, a diagram of the SHP gene shows the location of the variants. Polymorphisms were identified in exon 1 (c.113G→A*, c.510A→T*, c.512G→C), intron (intron1265T→A*), 3′-UTR (3′-UTR101C→G*, 3′-UTR186T→C*), and promoter (Pro-423C→T) of the SHP gene. Mutation nomenclature was numbered based on GenBankTM cDNA NM_021969.2 and protein NM_068804.1 sequences. In the open reading frame (ORF) of the coding region, nucleotide +1 corresponds to the A of the ATG start codon. In the intron region, nucleotide +1 corresponds to the 1st nucleotide of intron. In the 3′-UTR region, nucleotide +1 corresponds to the 1st nucleotide downstream of the stop codon. In the 5′-UTR and promoter region, nucleotide −1 corresponds to the 1st nucleotide upstream of the start codon. C, a diagram of the SHP gene shows the location of variants that result in substitution of histidine for arginine at position 38 in the SHP hinge region, asparagine for lysine at position 170, and alanine for glycine at position 171 in the SHP ligand-binding domain-like domain.
TABLE 1.
Mutations identified in the human SHP gene (NROB2)
Mutation nomenclature was numbered based on GenBankTM cDNA NM_021969.2 and protein NM_068804.1 sequences. In the open reading frame of the coding region, nucleotide +1 corresponds to the A of the ATG start codon. In the intron region, nucleotide +1 corresponds to the 1st nucleotide of the intron. In the 3′-UTR region, nucleotide +1 corresponds to the 1st nucleotide downstream of the stop codon. In the 5′-UTR and promoter region, nucleotide −1 corresponds to the 1st nucleotide upstream of the start codon. The italic underlined number indicates the CADASIL subject without the Notch3 mutation. The asterisks indicate six novel variants identified in this study. The numbers under Controls and CADASIL Patients indicate subject numbers. The frequency numbers represent control (1st) and CADASIL subjects (2nd), respectively.
| Region | Codon | Nucleotide change | Amino acid change | Controls (n = 34) | CADASIL patients (n = 50) | Frequency |
|---|---|---|---|---|---|---|
| % | ||||||
| Exon 1 | 38 | c.113G→A* | p.R38H* | 1 (3) | 0 | 8.8, 0 |
| Exon 1 | 170 | c.510A→T* | p.K170N* | 5 (7, 13, 261, 262, 273) | 1 (210) | 14.7, 2 |
| Exon 1 | 171 | c.512G→C | p.G171A | 3 (1, 6, 8) | 2 (98, 116) | 5.88, 4 |
| Intron | Intron1265T→A* | 1 (8) | 3 (55, 69, 141) | 2.94, 6 | ||
| 3′-UTR | 3′-UTR101C→G* | 1 (2) | 1 (69) | 2.94, 2 | ||
| 3′-UTR | 3′-UTR186T→C* | 2 (2, 8) | 4 (90, 116, 179, 180) | 5.88, 8 | ||
| Promoter | Pro-423C→T* | 1 (1) | 1 (116) | 2.94, 2 |
TABLE 2.
Summary of the mutations already published for the SHP (NROB2) gene
| Exon | Mutation | Effect | References |
|---|---|---|---|
| 1 | H53fsdel10 | Nishigori et al. (16) | |
| 1 | L98fsdel9insAC | ||
| 1 | 100C→T | R34X | |
| 1 | 583G→T | A195S | |
| 1 | 637C→T | R213C | |
| 1 | 647G→A | R216H | |
| 1 | 169C→T | R57W | |
| 1 | 566G→A | G189E | |
| Promoter | −394C→T | Cao and Hegele (14) | |
| Promoter | −195delCTGA | ||
| 1 | 541G→C | G171A | |
| 2 | 903C→T | ||
| Promoter | −195delCTGA | Hung et al. (17) | |
| 1 | R34G | ||
| 1 | R36C | ||
| 1 | 541G→C | G171A | |
| 1 | 541G→C | G171A | Mitchell et al. (15) |
| 1 | 100C→G | R34G | Echwald et al. (18) |
| 1 | 278G→A | G93D | |
| 1 | 415C→A | P139H | |
| 1 | 65C→T | Y22Y | |
| 1 | 339G→A | P133P | |
| 1 | 512G→C | G171A | |
| 1 | 100C→T | R34X | Enya et al. (19) |
| 1 | 112C→T | R38C | |
| 1 | 134G→C | R45P | |
| 1 | 157_166del | His53AlafsX50 | |
| 1 | 160C→T | R54C | |
| 1 | 169C→T | R57W | |
| 1 | 292_300delinsAC | L98TfsX6 | |
| 1 | 314T→G | V105G | |
| 1 | 512G→C | G171A | |
| 1 | 532G→A | D178N | |
| 2 | 566G→A | G189E | |
| 2 | 583G→T | A195S | |
| 2 | 618G→A | W206X | |
| 2 | 637C→T | R213C | |
| 2 | 647G→A | R216H |
Altered Expression and Half-life of SHP Mutants
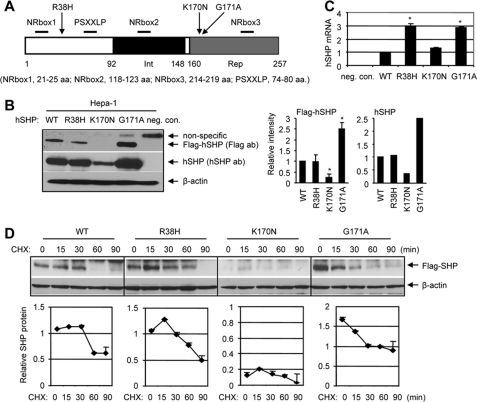
The mutant p.R38H is located in the N terminus, whereas p.K170N and p.G171A are in the repression domain of the SHP protein (Fig. 2A). We constructed FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, and FLAG-SHPG171A expression vectors and examined steady-state SHP protein using anti-FLAG antibodies and Western blots. Mouse hepatoma Hepa-1 cells had barely detectable endogenous mouse SHP (7, 10), thus, provided a good model system to determine the level of exogenously overexpressed hSHP. Unexpectedly, the mutant FLAG-SHPK170N protein at 48 h post-transfection was lost compared with wild-type SHP protein, whereas mutant FLAG-SHPG171A was expressed at about 2–3-fold higher levels than FLAG-SHPWT (Fig. 2B). Those exogenously expressed hSHP proteins were further examined with anti-hSHP antibodies, which detected SHPK170N expression at a constant low level compared with SHPWT, SHPR38H, and SHPG171A. The mRNA levels of SHPR38H and SHPG171A were about 3-fold higher than SHPWT and SHPK170N (Fig. 2C), suggesting enhanced SHP gene transcription. Thus, the increased level of p.G171A protein may in part be associated with its increased transcription, and the decreased p.K170N protein is likely due to its degradation by post-translational modification.
FIGURE 2.
Expression of mutant SHP proteins and their half-life. A, a diagram shows the structure of SHP and location of R38H, K170N, and G171A. The receptor interaction (Int) and repression (Rep) domains are indicated by black and gray boxes, respectively. The proximal locations of NRbox1, NRbox2, NRbox3, and PSXXLP are indicated. aa, amino acids. B, left, shown is a Western blot analysis of the hSHP protein in Hepa-1 cells that overexpressed with FLAG expression plasmids containing each hSHP mutation (SHPR38H, SHPK170N, and SHPG171A) using anti-FLAG antibodies (ab). The experiments were repeated three times with anti-FLAG antibodies and one time with anti-hSHP antibodies. neg. con., cells transfected with equal amounts of empty vector; no bands with correct size were detected by both antibodies. Right, shown is a quantitative analysis of each band intensity on the left (n = 3; *, p < 0.01 relative to WT). The results were expressed as -fold changes relative to SHPWT, which was set as 1. C, shown is a real-time quantitative PCR analysis of hSHP mRNA in mouse Hepa-1 cells overexpressed with SHPWT, SHPR38H, SHPK170Nv and SHPG171A expression vectors. The results were compared with SHPWT, which was set as 1 (*, p < 0.01). neg. con., cells transfected with equal amounts of pcDNA3 empty vector. D, top, Western blots to determine the half-life of mutant SHP proteins. Hepa-1 cells were transfected with each FLAG-SHP vector then treated with cycloheximide (CHX, 50 μm), and SHP was detected with anti-FLAG antibodies. Bottom, quantitative analysis of each band intensity at the top.
A recent report has shown that SHP undergoes rapid degradation with a short half-life in HepG2 cells (∼30 min) (32). We examined the half-life of each SHP mutant in Hepa-1 cells followed by treatment of cells with protein synthesis inhibitor cycloheximide. The half-life of FLAG-SHPWT was about 30 min, which extended to about 60 min by p.R38H and p.G171A (Fig. 2D). FLAG-SHPK170N exhibited the lowest expression. The higher basal level and longer half-life of FLAG-SHPG171A suggest a slow degradation rate and increased protein stability.
Altered Subcellular Localization of SHP Mutants
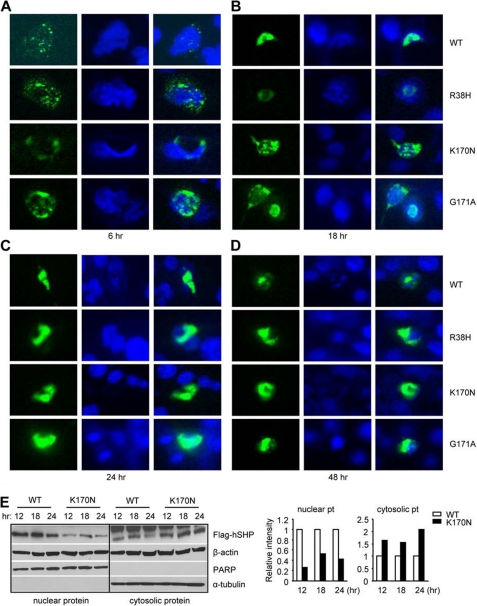
To access the effect of each mutation on SHP subcellular localization, we constructed GFP-SHPWT, GFP-SHPR38H, GFP-SHPK170N, and GFP-SHPG171A expression vectors, overexpressed them into Hepa-1 cells, and monitored the location of SHP proteins by green fluorescence under microscopy. As early as 6 h post-transfection, GFP-SHPWT, GFP-SHPR38H, and GFP-SHPK170N were observed in the cytoplasmic compartment, which did not overlay with 4′,6-diamidino-2-phenylindole stained nucleus, whereas some of the GFP-SHPG171A were found in the nucleus (Fig. 3A). By 18 h after transfection, the nuclear localization of all four SHP proteins was detected (Fig. 3B), although in a large number of cells GFP-SHPR38H and GFP-SHPK170N was still retained in the cytosol. Interestingly, GFP-SHPWT and GFP-SHPG171A were constantly observed in the nucleus at 24 (Fig. 3C) and 48 h (Fig. 3D) post-transfection, whereas most of the GFP-SHPR38H appeared in the cytosol with a few overlaying with the nuclear staining, and the majority of GFP-SHPG171A were found outside the nucleus. Similar results were observed in HeLa cells (not shown).
FIGURE 3.
Subcellular localization of SHP mutants in Hepa-1 cells. A–D, GFP-SHPWT, GFP-SHPR38H, GFP-SHPK170N, and GFP-SHPG171A expression vectors were constructed and overexpressed into mouse Hepa-1 cells. The location of GFP-SHP proteins was monitored by green fluorescence under a microscope at 6 (A), 18 (B), 24 (C), and 48 h (D) post-transfection. E, left, Western blots to determine SHP protein in the nuclear and cytosolic extracts in Hepa-1 cells transfected with FLAG-SHPWT and FLAG-SHPK170N expression vectors using anti-FLAG antibodies are shown. β-Actin, poly(ADP-ribose) polymerase (PARP), and α-tubulin were used as markers and internal controls to detect both nuclear and cytosolic (β-actin), nuclear (PARP), or cytosolic proteins (a-tubulin), respectively. Right, a quantitative analysis of each band intensity is shown on the left and expressed as -fold change relative to WT at each time point, which was set as 1. pt, protein.
These results suggest a possible scenario that p.R38H and p.K170N may alter the ability of SHP to efficiently translocate to the nucleus. We focused on SHPK170N because it showed a significant down-regulation (Fig. 2B). The nuclear FLAG-SHPK170N levels were low at all the time points compared with FLAG-SHPWT, whereas it showed higher levels in the cytosolic fractions (Fig. 3E). On the other hand, the levels of FLAG-SHPK170N were significantly down-regulated 48 h post-transfection (Fig. 2B), suggesting it may have undergone degradation.
Altered Ubiquitination and Acetylation of SHP Mutants
The Lys-170 lysine residue in SHP is well conserved in several species including human and mouse (Fig. 1C). Although Lys-170 was not identified as a major ubiquitination site in a recent study (32), it remains an interesting question if Lys-170 plays a role in SHP protein ubiquitination and if p.K170N mutation alters such process. The antibodies against the FLAG tag produced nonspecific bands in Hepa-1 cells (Fig. 2B) but appeared very specific in HeLa cells (not shown). Thus, we used HeLa cells for the coimmunoprecipitation and Western blotting in Fig. 4.
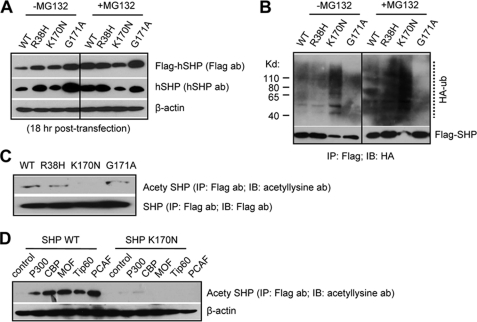
FIGURE 4.
Ubiquitination (A and B) and acetylation (C and D) assays to determine SHP degradation. A, HeLa cells were transfected with FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, or FLAG-SHPG171A expression vectors in the absence (−) or presence of MG132 (+, 5 μm) for 18 h. SHP proteins were determined using anti-FLAG or anti-SHP antibodies (ab) and Western blots. B, HeLa cells were cotransfected with FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, or FLAG-SHPG171A expression vectors together with an HA-ubiquitin (HA-ub) plasmid. FLAG-SHP was immunoprecipitated from cell extracts with anti-FLAG antibodies (IP), and ubiquitinated SHP in the immunoprecipitates was detected by Western blots (IB) with anti-HA antibodies. Positions of ubiquitinated SHP proteins are indicated by a dotted line. C, plasmids expressing the indicated SHP mutants were transfected into HeLa cells. Lysates were immunoprecipitated with anti-FLAG antibodies and blotted with anti-acetyllysine antibodies. The blot was then stripped and blotted with anti-FLAG antibodies. Because of the low basal level of SHPK170N, double amounts of FLAG-SHPK170N plasmids were transfected to keep the FLAG-SHPK170N level similar as other SHP proteins. D, plasmids expressing SHPWT and SHPK170N were co-transfected with acetyltransferases p300, CBP, MOF, Tip60, and PCAF into HeLa cells. Acetylation assays was performed as in C.
Consistent with the results in Hepa-1 cells (Fig. 2B), FLAG-SHPK170N was undetectable in HeLa cells 48 h post-transfection (not shown). We collected proteins 18 h post-transfection, and FLAG-SHPK170N was easily detected (Fig. 4A, left). FLAG-SHPR38H was expressed at a higher level than FLAG-SHPWT. This expression pattern was similar to the mRNA results (Fig. 2C). Because the levels of FLAG-SHPR38H and FLAG-SHPWT proteins appeared similar at 48 h post-transfection, the results suggest that p.R38H may affect FLAG-SHPR38H degradation through an unknown mechanism that is not associated with its half-life (Fig. 2D). Treatment of MG132 in cells elevated FLAG-SHPWT and FLAG-SHPR38H (Fig. 4A, right). In contrast, blocking proteasomal degradation by MG132 did not increase FLAG-SHPK170N. The results suggest that K170N affected SHP protein degradation.
We next performed a cell-based ubiquitination assay. In the absence of MG132, the levels of ubiquitinated FLAG-SHPWT and FLAG-SHPR38H were similar that were enhanced by MG132 treatment (Fig. 4B), consistent with the report in Hepa1c cells (32). On the contrary, the p.K170N mutation robustly increased ubiquitinated FLAG-SHPK170N, as indicated by the ladder of high molecular weight forms of FLAG-SHPK170N, suggesting that the Lys to Asn conversion at 170 position made FLAG-SHPK170N more susceptible to proteasomal degradation, which may contribute to its down-regulation. Interestingly, the p.G171A mutant decreased SHP ubiquitination compared with SHP wild type.
Protein acetylation of lysine residues is one of the most ubiquitous protein modification (33). Because the lysine 170 of SHP did not appear as an ubiquitination site, we analyzed if it may serve as a potential site for acetylation. In the presence of histone deacetylases (HDACs) inhibitor trichostatin A, an acetylated SHP, was observed in cells overexpressed with FLAG-SHPWT, FLAG-SHPR38H, and FLAG-SHPG171A but not with FLAG-SHPK170N (Fig. 4C). Co-expression of acetyltransferases including p300, CBP, MOF, Tip60, and PCAF markedly increased acetylated SHPWT but not SHPK170N (Fig. 4D). The data suggest that SHP is likely an acetylated protein, and the mutation K170N significantly impaired SHP acetylation.
Altered Repressive Function of SHP Mutants
Next, we used three naturally occurring promoters and ERRγ, LRH-1, and HNF4α responsive reporter systems to examine the inhibitory activity of SHP.
We recently showed that expression of wild-type SHP significantly decreased ERRγ transactivation of the YY1 gene promoter (11). As expected, expression of wild-type SHP markedly decreased ERRγ activation of the YY1 promoter (Fig. 5A). The mutants p.R38H and p.G171A exhibited similar activity, whereas p.K170N lost activity relative to SHP WT. Contrary to the observation with ERRγ in Fig. 5A, all three mutants displayed comparable or even stronger ability relative to SHP WT in inhibiting LRH-1 (Fig. 5B), suggesting each mutation maintained its repressive activity in the context of interaction with LRH-1.
FIGURE 5.
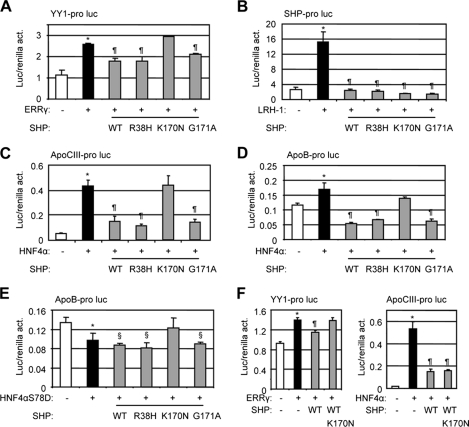
Functional analysis of SHP mutants. Transcriptional regulation of the three mutations p.R38H, p.K170N, and p.G171A was examined using luciferase reporter assays. act, activity. A, YY1 promoter luciferase reporter was co-expressed with ERRγ and each mutant SHP expression construct in Hepa-1 cells. B, SHP promoter luciferase reporter was co-expressed with LRH-1 and each mutant SHP expression construct in Hepa-1 cells. C, apoCIII promoter luciferase reporter was co-expressed with HNF4a and each mutant SHP expression construct in Hepa-1 cells. D and E, apoB promoter luciferase reporter was co-expressed with HNF4a (D) or a point mutation HNF4aS78D (E) and each mutant SHP expression construct in Hepa-1 cells. F, left, YY1 promoter luciferase reporter was co-expressed with ERRγ in the presence of SHP WT and K170N mutant. Right, apoCIII promoter luciferase reporter was co-expressed with HNF4α in the presence of SHP WT and K170N mutant. Luc activities were determined and normalized to Renilla activities. Data are expressed as the means ± S.D. (*, p < 0.01 versus the white bar; ¶, p < 0.01 versus the black bar; §, p < 0.01 versus the white bar). pro, promoter; luc, luciferase reporter.
We further examined the effects of SHP mutants on HNF4α transactivation of the apoCIII promoter (21). Consistent with the previous report (19), p.G171A retained wild-type SHP transcriptional repression of HNF4α activity, as did the p.R38H mutant (Fig. 5C). In contrast, p.K170N exhibited no repressive activity. To confirm this observation in a different promoter context, the apoB promoter luciferase reporter was used (5, 21). Mutants p.R38H and p.G171A behaved similar to SHP WT to inhibit the activity of wild-type HNF4α below the basal level, whereas p.K170N lost its inhibition on HNF4α transactivation of the apoB promoter (Fig. 5D). The point mutation HNF4αS78D could not bind to its response element on the apoB promoter and, thus, did not activate it (Fig. 5E), WT, p.R38H, and p.K170N SHP still showed intrinsic repression function, which was not observed with p.K170N.
Co expression of p.K170N with SHP WT blocked SHP WT inhibition of ERRγ, but not HNF4α activity (Fig. 5F), suggesting it functioned as a dominant negative SHP specifically on ERRγ activation of YY1 promoter. Taken together, the results suggest that we cannot simply conclude whether a SHP mutation results in alteration of SHP repressive function based on one promoter analysis. Differential inhibitory activity may occur depending on its interaction partner in a promoter-specific manner.
Altered in Vitro Interaction of SHP Mutants with ERRγ, LRH-1, HNF4α, and EID1 Using GST Pulldown Assays
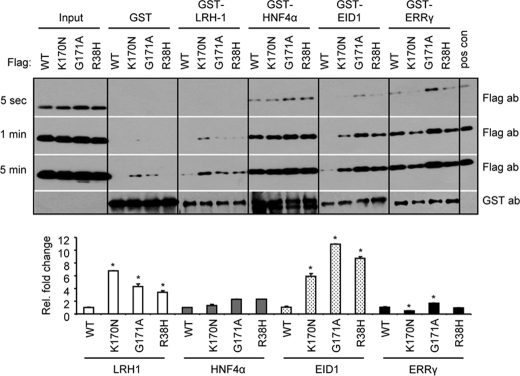
On the basis of the results presented earlier, we propose that the inability of p.K170N to repress ERRγ and HNF4α could not be fully explained by its degradation, because it still showed inhibition of LRH-1, but may likely be in part associated with its altered interaction with the receptors. Concerning Arg-38, a recent study showed that this residue belongs to a set of basic residues at the N-terminal region of SHP that may be involved in the recruitment of acidic co-repressors such as EID1 (23, 27). To this end, a GST pulldown assay was employed with GST-LRH-1, GST-HNF4α, GST-EID1, and GST-ERRγ, and in vitro translated FLAG-SHPWT, FLAG-SHPR38H, FLAG-SHPK170N, and FLAG-SHPG171A to provide a more quantitative measure of the interactions.
We did not detect any interaction between GST alone and the four in vitro translated SHPs (Fig. 6, 5 s and 1 min exposure), although longer exposure produced weak nonspecific bands (5-min exposure). Interestingly, p.G171A and p.R38H mutations enhanced FLAG-SHPG171A and FLAG-SHPR38H interaction with GST-LRH-1, which became even stronger by the p.K170N mutation, as compared with FLAG-SHPWT (5-min exposure). These data are consistent with the somewhat increased repression of LRH-1 by those mutants (Fig. 5B).
FIGURE 6.
GST pulldown assay to determine in vitro interaction of SHP mutants with LRH-1, HNF4a, EID1, and ERRγ. FLAG-SHPWT, FLAG-SHPK170N, FLAG-SHPG171A, and FLAG-SHPR38H were in vitro translated and used to interact with GST fusion proteins GST-LRH-1, GST-HNF4a, GST-EID1, and GST-ERRγ, which were expressed from bacterial E. coli BL21/DE3/RIL. Blots from three different exposure times (5 s, 1 min, and 5 min) are presented. Lower panel, -fold changes relative to WT in each group, which was set as 1 (*, p < 0.01). ab, antibody.
The interaction of FLAG-SHPK170N with GST-HNF4α did not differ from that of SHP WT; however, the interaction between FLAG-SHPG171A or FLAG-SHPR38H and GST-HNF4α was slightly increased (5-s exposure). This result suggests that the lost inhibition of p.K170N on HNF4α activity (Fig. 5, C–D) is unlikely due to the alteration of their interaction.
A weak interaction was noted between FLAG-SHPWT and GST-EID1 as indicated by a faint band in the blot after a longer exposure time (5-min exposure). Similar to LRH-1, all three mutants showed robustly increased association with GST-EID1 compared with FLAG-SHPWT (1-min exposure), with FLAG-SHPG171A showing the highest association.
A reduction in association of FLAG-SHPK170N with GST-ERRγ was revealed by this analysis, whereas an elevated association between FLAG-SHPG171A and GST-ERRγ was identified. The impaired interaction between FLAG-SHPK170N and GST-ERRγ may in part contribute to SHP decreased repressive activity (Fig. 5A).
Altered Recruitment of Co-repressors by SHP Mutants
It has been shown that SHP represses transcriptional activity via recruitment of histone deacetylases (34) and a chromatin remodeling complex involving Sin3A (35, 36). These results prompt us to ask whether the recruitment of those co-repressors by SHPK170N mutant is altered.
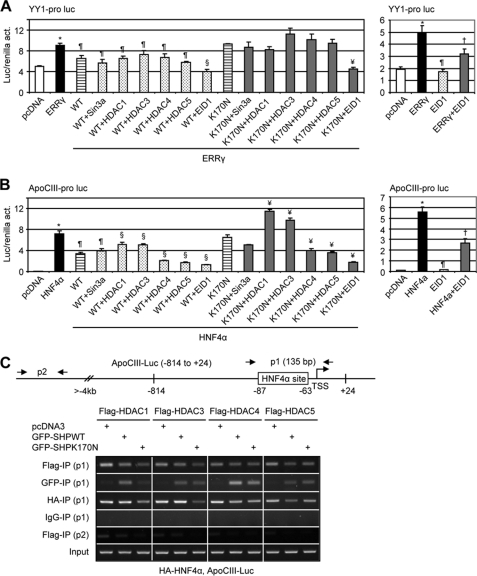
We investigated Sin3α, four different HDACs representative of the main two classes (class I and class II), and EID1. As shown in Fig. 7A, left, when co-transfected with Sin3α, HDAC1, HDAC3, HDAC4, or HDAC5, the activity of SHPWT or SHPK170N on ERRγ was not altered compared with Fig. 5A. Interestingly, both SHPWT and SHPK170N exhibited stronger repressive effects in the presence of EID1 co-expression, and a similar effect was observed with HNF4α in the apoCIII promoter (Fig. 7B, left). Surprisingly, in the absence of SHP, EID1 also inhibited both ERRγ and HNF4a activity (Fig. 7, A and B, right). Thus, EID1 may also function as a repressor of ERRγ and HNF4a.
FIGURE 7.
Co-repressor recruitment by SHP mutants. A and B, shown are transient transfection assays to determine the effects of co-repressors on WT and SHPK170N inhibition of YY1 (A) and apoCIII (B) promoter reporters. Luc activities were determined and normalized to Renilla activities (act.). Data are expressed as the means ± S.D. (*, p < 0.01 versus white bar; ¶, p < 0.01 versus black bar; §, p < 0.01 versus WT striped bar; ¥, p < 0.01 versus K170N striped bar; †p < 0.01 versus black bar). Pro, promoter; Luc, luciferase reporter. C, chromatin immunoprecipitation assays determine the recruitment of co-repressors on the apoCIII promoter by HNF4a and SHP. HeLa cells were co-transfected with FLAG-HDAC1, -3, -4, and -5, GFP-SHPWT and GFP-SHPK170N, HA-HNF4α, and apoCIII promoter (−814 to +24), and anti-FLAG, anti-GFP, and anti-HA antibodies were used to co-immunoprecipitate each corresponding protein, respectively. Two sets of primers were designed for chromatin immunoprecipitation assays. P1 could detect coimmunoprecipitation (IP) of each protein on exogenously overexpressed apoCIII promoter, whereas p2 was located 4 kb upstream from transcriptional start site (TSS) and, thus, served as a negative control.
Another interesting observation was the further decreased SHPWT and SHPK170N repression of HNF4α by co-expression of class II HDAC4 and HDAC5 but released repression by co-expression of class I HDAC1 and HDAC3 (Fig. 7B, left). This is somewhat unexpected, because the recruitment of HDAC1 has been implicated in SHP-mediated repression of androgen receptor (AR) and estrogen receptor α transcriptional activity (34). We then investigated the ability of SHPWT and SHPK170N to recruit those repressors to the apoCIII promoter when they interact with SHP using chromatin immunoprecipitation assays.
The recruitment of HDAC1 and HDAC3, but not HDAC4 and HDAC5, to the apoCIII promoter was reduced by SHPK170N (Fig. 7C, FLAG-IP, p1). The band intensity appeared to be higher in pcDNA3-transfected cells compared with SHPWT in each HDAC group, which may be resulted from a weak nonspecific amplification, as a faint band was also detected by p2 (FLAG-IP) in the pcDNA3 group. The co-recruitment of SHPK170N to the apoCIII promoter in the presence of HDAC1 was significantly decreased as compared with SHPWT (GFP-IP, p1). The co-recruitment of HNF4α with SHPK170N was markedly lower in the presence of HDAC1 and HDAC3 (HA-IP, p1). Overall, the SHPK170N mutant markedly impaired the recruitment of SHP, HNF4α, HDAC1, and HDAC3 to the apoCIII promoter, which may result in de-repression and, thus, the activation of the promoter.
Molecular Dynamics Simulations of SHP Wild Type and Mutants
To shed light on the molecular mechanisms responsible for the impaired activity of K170N mutation, molecular dynamics simulations were carried out on SHPK170N, SHPG171A, and SHP wild type, respectively.
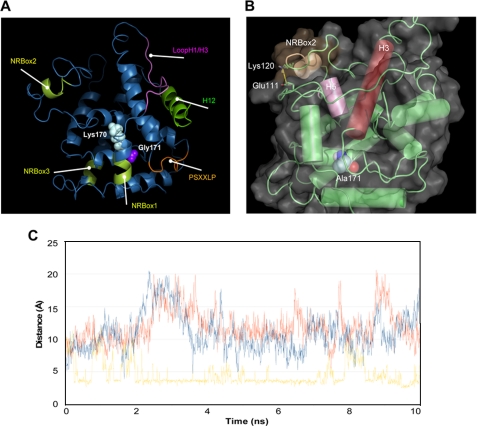
We started, in particular, from the reported homology model of SHP (27, 37) and the observation that some structural elements of the receptor are involved in mediating its biological functions by regulating the interaction with nuclear receptor partners and the recruitment of transcriptional inhibitors as well as chromatin remodeling factors (23, 34, 35, 38–40). Accordingly, we comparatively investigated the effect of K170N and G171A mutations (Fig. 8A) on the stability of the three nuclear receptor boxes, helix H12, and the conserved PSXXLP motif, recently reported to mediate the interaction between LRH-1 and the closest SHP homolog DAX1 (41).
FIGURE 8.
Molecular dynamics simulations of SHP mutants. A, locations of the Lys-170 and Gly-171 in the structure of SHP show the structural elements involved in mediating the biological functions of SHP are also shown. B, a salt bridge interaction in the NR2box motif is shown whose formation was promoted by G171A replacement. C, analysis is shown of the Glu-111—Lys-120 salt bridge interaction in SHP wild type (blue line), SHPK170N (red line), and SHPG171A (yellow line) along the 10 ns of molecular dynamic simulation. The salt bridge is considered to be formed if the distance between the carboxylic oxygen atoms of Glu111 and the side chain nitrogen atom of Lys-120 is below a cut-off value of 3.5 Å.
Table 3 reports the averages and the S.D of the root mean square deviations of backbone atoms of the receptor and, more specifically, of the selected structural motifs. From the inspection of the table, it was found that each mutant induces a different pattern of stabilization (low root mean square deviations) or destabilization (high root mean square deviations) of the structural elements, with SHPG171A stabilizing the nuclear receptor boxes and destabilizing the H12 and PSXXLP motif, whereas SHPK170N promoting the conformational destabilization of all the structural elements of the receptor with respect to SHP wild type. Because both K170N and G171A mutations are remotely localized from the selected structural elements, it is likely that the observed stabilization and destabilization effects may arise from intramolecular interactions that originate from the stabilization of the hydrophobic packing upon the G171A replacement and the loss of the salt bridge between Lys-170 and Asp-103 in SHPK170N, respectively.
Furthermore, Table 3 shows the impressive stabilization of the NRbox2 by SHPG171A. This effect may be ascribed to hydrophobic interactions in the core of SHP that upon G171A replacement propagate through helices H3 and H5 to the NRbox2 where they favor the formation of a salt bridge interaction between residues Glu-111 and Lys-120 (24.52% occupancy, Fig. 8, B and C).
It is worth noting that the NRbox2 has been reported as specifically binding the coactivator recruitment site of some nuclear receptors such as HNF-4α, progesterone receptor, glucocorticoid receptor, and estrogen receptor α (39). In agreement with an emerging paradigm in the field of protein-protein interactions (41), the G171A replacement may selectively influence the specificity of such binding by remotely controlling the conformational stability of the NRbox2.
DISCUSSION
Our studies for the first time identified six novel variants in the promoter, coding region, intron, and 3′-UTR of the SHP gene in human normal and CADASIL-like subjects. When compared with other identified SHP mutations in human metabolic diseases (14–19), these nucleotide changes occurred with a surprisingly high frequency, suggesting a potentially important function of SHP in brain related disease. Unfortunately, no rare variants of the SHP gene were exclusive to patients with CADASIL. Although we cannot link the etiology of CADASIL to mutations of SHP in the present study, we cannot completely rule out this possibility if a larger cohort of subjects would be analyzed. Nonetheless, our studies provide additional evidence that genetic variations in SHP are common among human subjects.
Functional studies of the mutant proteins showed that the mutant p.R38H (c.113G→A) maintained SHP repressive activity of ERRγ, LRH-1, and HNF4α. This is somewhat anticipated, because amino acid histidine exists at position 38 in the mouse and opossum SHP protein, whereas the human and rat SHP proteins have arginine at this position (see Fig. 1C). Thus, substitution of His for Arg in the hinge region of the SHP protein would not be expected to alter the SHP function. Interestingly, this mutation, despite located in the N-terminal region, enhanced SHP interaction with LRH-1, HNF4α, and EID1, as demonstrated using GST pulldown assays, and extended the half-life of SHP. However, its nuclear localization seemed to be impaired. It is possible that this site may affect SHP nuclear translocation, although the underlying mechanism remains to be determined.
The p.G171A mutant was identified in one control subject in a previous report (19). Its activity was determined as normal based on its ability to inhibit HNF4α-mediated transactivation of the HNF1α promoter activity in HepG2 cells. Consistent with the observation in this report, the p.G171A mutant showed comparable ability as SHP WT to inhibit the activity of ERRγ, LRH-1, and HNF4α. The most striking alteration is a drastic increase of its protein level, which may be in part associated with its increased gene transcription, protein half-life, and slow degradation. In addition, p.G171A strengthened the SHP interaction with all four SHP interaction partners that we analyzed, i.e. ERRγ, LRH-1, HNF4α, and EID1. Structurally, this mutation showed an impressive effect in stabilizing NRbox2 located in the interaction domain of SHP, a region that was suggested to play a critical role in coactivator recruitment of several other nuclear receptors (39), as determined by the molecular dynamics simulations of SHP. Therefore, this naturally occurring SHP mutation appears to be a critical determinant of SHP protein stability.
Nuclear receptors are a class of transcription factors modulated by acetylation and deacetylation. For instance, it has been shown that CBP/p300 acetylates the AR, and deacetylation of AR by HDAC1 represses the activation function of AR (42, 43). Estrogen receptor α is also acetylated at certain lysine residues by p300 (44), and increased acetylation of estrogen receptor α by HDAC inhibitors enhances its transcriptional activity (45). We found that p.K170N SHP underwent rapid degradation than other SHP mutants. Mechanistically, we identified Lys-170 as a potential acetylation site and provided the first evidence that SHP Lys-170 acetylation may serve to promote SHP protein stability. Unacetylated lysine 170, due to its conversion to Asn, then makes SHP prone to ubiquitination, which ultimately leads to the destruction of the receptor. Taken together, it is reasonable to posit that deacetylation of the lysine 170 residue in SHP makes the receptor accessible to ubiquitination, leading to SHP elimination by a mechanism similar to that observed for AR and ER.
On the other hand, K170N mutant also severely affected SHP nuclear translocation and its intrinsic repressive function of ERRγ and HNF4α, suggesting the critical role of this lysine residue in maintaining the overall functionality of SHP. A further support of this notion is provided by structural analysis of SHPK170N showing that this mutation promoted the conformational destabilization of all the structural elements of the receptor. Deacetylation of p.K170N also prevented SHP from efficiently recruiting other co-repressors including HDAC1 and HDAC3, contributing to its lost repression function.
In summary, we identified several novel SHP mutants in human subjects and elucidated their functional and structural changes. To the best of our knowledge, this is the first report showing nuclear protein SHP undergoing impaired function caused by natural occurring single mutation.
Supplementary Material
Acknowledgments
We thank the sequencing core facility at the University of Utah School of Medicine for support. We are very grateful to the generous plasmid gifts from Drs. Frances Sladek, Collin Clyne, John Chiang, Chiwai Wong, Eckardt Treuter, Akiyoshi Fukamizu, Uwe Borgmeyer, Timothy Osborne, and Jianyuan Luo. We thank the neurologists from Hospital University Central Asturias for contribution to this work. We thank Dr. Viswanath Gunda for help in reading the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- SNP
- single nucleotide polymorphism
- HDAC
- histone deacetylases
- MD
- molecular dynamic
- AR
- androgen receptor
- hSHP
- human SHP
- CADASIL
- cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- CBP
- cAMP-response element-binding protein (CREB)-binding protein
- GST
- glutathione S-transferase
- UTR
- untranslated region
- WT
- wild type
- ERR
- estrogen-related receptor
- HNF
- hepatocyte nuclear factor
- PCAF
- p300/CBP-associated factor.
REFERENCES
- 1.Markward N. J. (2007) AMIA Annual Symposium Proceedings, Chicago, IL, November 10–14, 2007, p. 1041, American Medical Informatics Association, Bethesda, MD [Google Scholar]
- 2.Lee H. K., Lee Y. K., Park S. H., Kim Y. S., Park S. H., Lee J. W., Kwon H. B., Soh J., Moore D. D., Choi H. S. (1998) J. Biol. Chem. 273, 14398–14402 [DOI] [PubMed] [Google Scholar]
- 3.Seol W., Chung M., Moore D. D. (1997) Mol. Cell. Biol. 17, 7126–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D. D. (2005) Cell Metab. 2, 227–238 [DOI] [PubMed] [Google Scholar]
- 5.Huang J., Iqbal J., Saha P. K., Liu J., Chan L., Hussain M. M., Moore D. D., Wang L. (2007) Hepatology 46, 147–157 [DOI] [PubMed] [Google Scholar]
- 6.He N., Park K., Zhang Y., Huang J., Lu S., Wang L. (2008) Gastroenterology 134, 793–802 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Xu P., Park K., Choi Y., Moore D. D., Wang L. (2008) Hepatology 48, 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Soto J., Park K., Viswanath G., Kuwada S., Abel E. D., Wang L. (2010) Mol. Cell. Biol. 30, 1341–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song G., Wang L. (2008) PloS One 3, e3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G., Wang L. (2008) Nucleic Acids Res. 36, 5727–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song G., Wang L. (2009) PloS One 4, e6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song G., Zhang Y., Wang L. (2009) J. Biol. Chem. 284, 31921–31927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song G., Wang L. (2009) PloS One 4, e7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H., Hegele R. A. (2002) J. Hum. Genet. 47, 445–447 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell S. M., Weedon M. N., Owen K. R., Shields B., Wilkins-Wall B., Walker M., McCarthy M. I., Frayling T. M., Hattersley A. T. (2003) Diabetes 52, 1276–1279 [DOI] [PubMed] [Google Scholar]
- 16.Nishigori H., Tomura H., Tonooka N., Kanamori M., Yamada S., Sho K., Inoue I., Kikuchi N., Onigata K., Kojima I., Kohama T., Yamagata K., Yang Q., Matsuzawa Y., Miki T., Seino S., Kim M. Y., Choi H. S., Lee Y. K., Moore D. D., Takeda J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung C. C., Farooqi I. S., Ong K., Luan J., Keogh J. M., Pembrey M., Yeo G. S., Dunger D., Wareham N. J., O'Rahilly S. (2003) Diabetes 52, 1288–1291 [DOI] [PubMed] [Google Scholar]
- 18.Echwald S. M., Andersen K. L., Sørensen T. I., Larsen L. H., Andersen T., Tonooka N., Tomura H., Takeda J., Pedersen O. (2004) Hum. Mutat. 24, 381–387 [DOI] [PubMed] [Google Scholar]
- 19.Enya M., Horikawa Y., Kuroda E., Yonemaru K., Tonooka N., Tomura H., Oda N., Yokoi N., Yamagata K., Shihara N., Iizuka K., Saibara T., Seino S., Takeda J. (2008) Hum. Mutat. 29, E271–E277 [DOI] [PubMed] [Google Scholar]
- 20.Fouillade C., Chabriat H., Riant F., Mine M., Arnoud M., Magy L., Bousser M. G., Tournier-Lasserve E., Joutel A. (2008) Hum. Mutat. 29, 452. [DOI] [PubMed] [Google Scholar]
- 21.Hwang-Verslues W. W., Sladek F. M. (2008) Mol. Endocrinol. 22, 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li K., Wang R., Lozada E., Fan W., Orren D. K., Luo J. (2010) PloS One 5, e10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Båvner A., Johansson L., Toresson G., Gustafsson J. A., Treuter E. (2002) EMBO Rep. 3, 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentschke M., Süsens U., Borgmeyer U. (2002) Biochem. Biophys. Res. Commun. 299, 872–879 [DOI] [PubMed] [Google Scholar]
- 25.Hirota K., Sakamaki J., Ishida J., Shimamoto Y., Nishihara S., Kodama N., Ohta K., Yamamoto M., Tanimoto K., Fukamizu A. (2008) J. Biol. Chem. 283, 32432–32441 [DOI] [PubMed] [Google Scholar]
- 26.Matsukuma K. E., Wang L., Bennett M. K., Osborne T. F. (2007) J. Biol. Chem. 282, 20164–20171 [DOI] [PubMed] [Google Scholar]
- 27.Macchiarulo A., Rizzo G., Costantino G., Fiorucci S., Pellicciari R. (2006) J. Mol. Graph. Model. 24, 362–372 [DOI] [PubMed] [Google Scholar]
- 28.Schrödinger L. C. C. (2009) Maestro, 2009 Ed., New York [Google Scholar]
- 29.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 30.Deleted in proof
- 31.Shaw D. E. (2009) Desmond Molecular Dynamics System, 2.2 Ed., New York, NY [Google Scholar]
- 32.Miao J., Xiao Z., Kanamaluru D., Min G., Yau P. M., Veenstra T. D., Ellis E., Strom S., Suino-Powell K., Xu H. E., Kemper J. K. (2009) Genes Dev. 23, 986–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X. J. (2004) Nucleic Acids Res. 32, 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gobinet J., Carascossa S., Cavaillès V., Vignon F., Nicolas J. C., Jalaguier S. (2005) Biochemistry 44, 6312–6320 [DOI] [PubMed] [Google Scholar]
- 35.Kemper J. K., Kim H., Miao J., Bhalla S., Bae Y. (2004) Mol. Cell. Biol. 24, 7707–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao J., Fang S., Lee J., Comstock C., Knudsen K. E., Kemper J. K. (2009) Mol. Cell. Biol. 29, 6170–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawson M. I., Xia Z., Jiang T., Ye M., Fontana J. A., Farhana L., Patel B., Xue L. P., Bhuiyan M., Pellicciari R., Macchiarulo A., Nuti R., Zhang X. K., Han Y. H., Tautz L., Hobbs P. D., Jong L., Waleh N., Chao W. R., Feng G. S., Pang Y., Su Y. (2008) J. Med. Chem. 51, 5650–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortlund E. A., Lee Y., Solomon I. H., Hager J. M., Safi R., Choi Y., Guan Z., Tripathy A., Raetz C. R., McDonnell D. P., Moore D. D., Redinbo M. R. (2005) Nat. Struct. Mol. Biol. 12, 357–363 [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Choi M., Suino K., Kovach A., Daugherty J., Kliewer S. A., Xu H. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9505–9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boulias K., Talianidis I. (2004) Nucleic Acids Res. 32, 6096–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sablin E. P., Woods A., Krylova I. N., Hwang P., Ingraham H. A., Fletterick R. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18390–18395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaughan L., Logan I. R., Neal D. E., Robson C. N. (2005) Nucleic Acids Res. 33, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu M., Wang C., Reutens A. T., Wang J., Angeletti R. H., Siconolfi-Baez L., Ogryzko V., Avantaggiati M. L., Pestell R. G. (2000) J. Biol. Chem. 275, 20853–20860 [DOI] [PubMed] [Google Scholar]
- 44.Cui Y., Zhang M., Pestell R., Curran E. M., Welshons W. V., Fuqua S. A. (2004) Cancer Res. 64, 9199–9208 [DOI] [PubMed] [Google Scholar]
- 45.Wang C., Fu M., Angeletti R. H., Siconolfi-Baez L., Reutens A. T., Albanese C., Lisanti M. P., Katzenellenbogen B. S., Kato S., Hopp T., Fuqua S. A., Lopez G. N., Kushner P. J., Pestell R. G. (2001) J. Biol. Chem. 276, 18375–18383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.