Abstract
DesK is a sensor histidine kinase (HK) that allows Bacillus subtilis to respond to cold shock, triggering the adaptation of membrane fluidity via transcriptional control of a fatty acid desaturase. It belongs to the HK family HPK7, which includes the nitrogen metabolism regulators NarX/Q and the antibiotic sensor LiaS among other important sensor kinases. Structural information on different HK families is still scarce and several questions remain, particularly concerning the molecular features that determine HK specificity during its catalytic autophosphorylation and subsequent response-regulator phosphotransfer reactions. To analyze the ATP-binding features of HPK7 HKs and dissect their mechanism of autophosphorylation at the molecular level, we have studied DesK in complex with ATP using high resolution structural approaches in combination with biochemical studies. We report the first crystal structure of an HK in complex with its natural nucleotidic substrate. The general fold of the ATP-binding domain of DesK is conserved, compared with well studied members of other families. Yet, DesK displays a far more compact structure at the ATP-binding pocket: the ATP lid loop is much shorter with no secondary structural organization and becomes ordered upon ATP loading. Sequence conservation mapping onto the molecular surface, semi-flexible protein-protein docking simulations, and structure-based point mutagenesis allow us to propose a specific domain-domain geometry during autophosphorylation catalysis. Supporting our hypotheses, we have been able to trap an autophosphorylating intermediate state, by protein engineering at the predicted domain-domain interaction surface.
Keywords: Bacterial Protein Kinases, Bacterial Signal Transduction, Enzyme Structure, Phosphorylation Enzymes, X-ray Crystallography, Protein Engineering, Protein-Protein Docking Simulation, Structure-based Mutagenesis
Introduction
Histidine kinases3 (HKs)4 are multifunctional dimeric enzymes with highly variable sensing regions and more conserved intracellular cores. In concerted action with a range of specific protein partners called response regulators (RRs), they conform two-component signal transduction systems (TCSs), present in all three major life divisions (1, 2). Although TCSs represent the most abundant signaling system within bacteria, for unknown evolutionary reasons they are absent in higher eukaryotes, including mammals. The only mammalian HK relatives are the nucleus-encoded mitochondrial dehydrogenase kinases, which phosphorylate specific serine residues (3).
Intracellular HK domains catalyze an ATP-dependent protein kinase activity, allowing the HK to autophosphorylate a specific histidine, in a signal-regulated fashion. Once phosphorylated, the cytoplasmic core catalyzes a subsequent phosphotransfer reaction, swapping the histidine-linked phosphate group onto a specific aspartate residue in the receiver domain of the cognate RR. Many HKs catalyze a further third type of reaction, whereby the unphosphorylated HK specifically promotes the dephosphorylation of its cognate phospho-RR through a protein-phosphatase mechanism. Those HKs that are able to catalyze autokinase and RR-phosphatase reactions have been shown to tightly balance these two functions as a key to proper signaling through the HK-controlled pathway (4).
A large array of TCSs is expressed at any given time in a cell. One of the most addressed issues in this regard is the interaction network among HKs and RRs, because most bacterial species express 20–30 HK-RR pairs or even more, and yet any given histidine kinase has only one or two cognate response regulators, implying minimal cross-talk among different pathways (5–7). These specificity issues deserve to be analyzed in greater detail, taking into account the significant global structural conservation among HKs. Atomic level differences cannot be underestimated as the basis of distinctive and specific regulatory and mechanistic determinants, affecting each one of the functional steps in HK-mediated reactions.
Structural analyses reveal a modular architecture in HKs, with variations in domain organization that define HK classes I and II (8). In both groups, the three-dimensional architecture predicts extensive movement of the ATP-binding domain (ABD) to physically interact with the histidine-containing phosphotransfer domain, given that autophosphorylation appears to occur exclusively within the dimer. In class I HKs the phosphotransfer domain immediately precedes the ABD and mediates HK dimerization (hence called the dimerization and histidine phosphotransfer (DHp) domain), whereas in class II enzymes it is not involved in dimerization and several additional domains intervene between the phosphotransfer and ABD domains. Sequence-based classification of the different domains in HKs is also useful in structure/functional analyses. According to the DHp domains, HKs have been grouped into four Pfam families (9): whereas ∼85% can be assigned to the HisKA group (PF00512), ∼10% of all known histidine kinases belong to the HisKA_3 family (PF007730). Concerning the ABDs, HKs have been clustered in 11 families (2), noting that three-dimensional structural information is still lacking for five of them.
In terms of the autophosphorylation mechanism, some HKs like EnvZ and NtrB (10, 11) autophosphorylate following a trans pathway, i.e. one monomer phosphorylating the histidine of the other. Instead, HK853 and PhoR have recently been shown to perform autokinase catalysis in cis (12). Autophosphorylation appears to proceed according to a mutually exclusive behavior, either cis- or trans-acting. Given the fact that the ATP-binding domains of HKs are flexible in the kinase competent state (13), the cis/trans exclusion is likely correlated to specific interdomain configurations at the interaction surface. As clearly demonstrated for the HK-RR interaction, interfaces seem to be crucial to determining specificity in HK signal transduction (7, 12). We wished to explore whether structural constraints play a role in defining specific orientations for interacting surfaces between the ABD and the DHp domains in HKs, eventually relevant during autophosphorylation catalysis.
We focused on the protein DesK from Bacillus subtilis, a class I HK (14) displaying an N-terminal sensor domain (∼150 residues) with almost no extracellular region, other than the loops that connect the four or five transmembrane segments (15). The transmembrane region can sense temperature-modulated fluidity changes of lipid bilayers (13), transmitting the signal toward the C-terminal cytoplasmic catalytic core (∼220 residues), hereafter called DesKC. The cold thermal stimulus is detected by DesK, which then interacts with its cognate RR, DesR, constituting a canonical TCS. In vitro, DesKC has been shown to catalyze three different reactions depending on the phosphorylation states of the partners: its own phosphorylation, phosphotransfer to DesR, and dephosphorylation of phospho-DesR (15). We have recently reported that the switch from a kinase- to a phosphatase-biased activity in full-length DesK is temperature-regulated through the action of the trans-membrane segments (13). In vivo experiments have shown that DesK functions as a kinase at cold temperatures (15, 16), autophosphorylating a conserved residue (His188) in the DHp domain. This phosphoryl group is then transferred to a receiver aspartate in DesR, a DNA-binding RR that then activates the transcription of des (encoding for the acyl lipid desaturase Δ5-Des). Unsaturated fatty acyls in phospholipids, the product of Δ5-Des activity, raise the membrane's fluidity, triggering the switch of DesK from a kinase to a phosphatase state. As a consequence, the concentration of phospho-DesR falls off, leading to shutdown of des transcription (16). DesK is a HisKA_3 HK that belongs to the HPK7 family, together with nitrate/nitrite-linked regulation TCS histidine kinases, namely NarQ and NarX (17), among many others.
We now report the crystal structure of the ABD of DesK at 1.8 Å resolution, disclosing new information about HPK7 histidine kinases. To our knowledge, this is the first experimental structure of a histidine kinase in complex with its physiologic substrate (ATP). Structural analyses, complemented by an extensive in silico study of sequence conservation and phosphotransfer intermediate-state modeling, allow us to extend the current knowledge of HKs. In particular, we pinpoint key residues at the domain-domain interaction surface, enabling us to predict the geometry of the interdomain interface during autophosphorylation. Enzymatic characterization of wild-type DesKC, as well as of selected structure-based point mutants, confirm the functional role of conserved residues in helix α5, far from the catalytic center, strongly suggesting that domain-domain interfaces play a key role in catalysis regulation. Further supporting the structural hypotheses, we engineered cysteine mutants on the basis of the predicted interdomain orientation, allowing us to trap a covalent autophosphorylation intermediate state.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The plasmids used for expression correspond to: the entire cytoplasmic region DesKC (residues 155–367), the point mutant DesKCH188V (as described in Ref. 15), and full-length DesR (see Ref. 18). The engineered Cys mutants (G192C/G334C, Q193C/G334C, and S196C/G334C), as well as DesKCE342A and DesKCE343A were generated using the QuikChange multi site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Mutations were confirmed by DNA sequencing. DesKC and derived mutants were expressed as N-terminal His6 tag fusion proteins, including a TEV protease cleavage site for tag removal, in Escherichia coli strain M15/pREP4. Protein purification was performed with standard Ni2+-affinity chromatography using a HisTrap column (Amersham Biosciences), then dialyzed against an excess volume of 50 mm Tris-HCl, pH 8, 500 mm NaCl, 10% glycerol, and 1 mm DTT at room temperature, in the presence of a 1/40 (w/w) ratio of His-tagged TEV protease (19). A second Ni2+ column was performed, flow-through was concentrated (Vivaspin®, Sartorius), and injected into a HiLoadTM 16/60 Superdex 75 prep grade column (Amersham Biosciences) pre-equilibrated with 50 mm Tris·HCl, pH 8, 300 mm NaCl, and 0.5 mm DTT. After size-exclusion chromatography, the proteins were concentrated to ∼10 mg/ml and stored in aliquots at −80 °C for further use in crystallization trials. His6-tagged DesR was overexpressed in the same E. coli strain and purified following a similar protocol as DesKC, with no proteolytic step.
Serendipitous proteolysis of DesKCH188V resulted in the reproducible crystallization of the ATP-binding domain (DesKABD, spanning residues 243–367). Briefly, DesKCH188V was mixed with DesR (1:1 molar ratio) in 50 mm Tris·HCl, pH 8, 300 mm NaCl, 0.5 mm DTT, 10 mm MgCl2, 5 mm ATP, 5 mm BeF3. Vapor diffusion crystallization in hanging drops was performed by mixing 2 μl of protein solution (10 mg/ml total protein) with 2 μl of reservoir solution (20% polyethylene glycol 3350, 0.2 m NH4Cl). Multicrystals grew very slowly and were further used as microseeds in fresh drops with identical mother liquor, resulting in single crystals of DesKABD, which typically diffracted to ∼1.6–1.8 Å resolution. Single crystals were cryoprotected in mother liquor containing 25% glycerol and 1 m NaI for 60 s, flash frozen in liquid N2, and stored until data collection.
Diffraction Data Collection, Structure Determination, and Refinement
X-ray diffraction data (Table 1) were collected at the Institut Pasteur de Montevideo, at 108 K (Cryostream Series 700, Oxford Cryosystems) with CuKα radiation (1.5418 Å), using a microsource MicroMax007-HF rotating copper anode (Rigaku), Varimax-HF mirrors (Rigaku), and a Mar345 image plate detector (Mar Research). Highly redundant data sets were collected and processed using Mosflm/Scala (20). The structure was solved by single anomalous diffraction using the signal from the iodide atoms. Experimental electron density maps allowed for chain tracing and subsequent phase-restrained model refinement with TLS according to standard procedures. Full details of structure solution and model refinement are given in the supplemental materials.
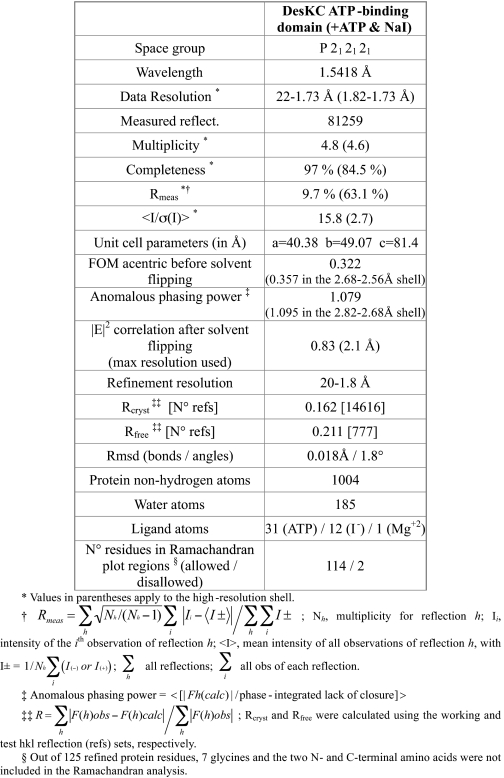
TABLE 1.
Data collection, phasing and refinement statistics
Protein-Protein Docking Calculations
Atomic coordinates of the dimerization DHp domain of DesK (residues 176–242) were obtained from our previous report (13) DesKCH188E PDB 3GIE, except that the Glu188 was replaced in silico by a His (using the most populated rotamers). This procedure was preferred, because 3GIE was solved at higher resolution and is otherwise structurally similar to wt DesKC 3EHF (13). Docking calculations were performed with HADDOCK 2.0 (21). The ATP and Mg2+ were treated as a single residue; corresponding bonds and angles were defined to keep appropriate cation coordination geometry. The DesKCABD model was first subjected to a simulated annealing/molecular dynamics protocol (CNS 1.2 (22)) consisting of heating to 3000 K and a slow cooling to 300 K in 60 steps (3.5 fs). All atomic positions of DesKCABD were restrained, except for the ATP lid (residues 326–334) and the N-terminal residues (residues 243–246). We generated 20 different conformations of these two fragments using different initial velocities. The ATP lid loop ensemble was used to rule out effects associated to ATP lid flexibility, which has been extensively reported, even in the presence of ADP or non-hydrolysable ATP analogues (13, 23, 24). As for the four N-terminal residues in the ABD, allowed flexibility should take into account eventual effects of different initial dispositions on the final interdomain geometry. The used distance restraints (ambiguous interaction restraints (AIR)) were: 1) His188 Nϵ2 of DHp monomer A, linked to ATP Pγ in DesKABD and 2) Lys242 main-chain C of DHp monomer B, with DesKABD Gly243 N, restoring the intrachain peptidic bond. The alternative restraint His188 Nϵ2 of DHp monomer B linked to DesKABD ATP Pγ (modeling a cis-acting autophosphorylation configuration), was also subjected to computations, resulting in lower energy scores and not further analyzed. To test the robustness of the results, independent simulations were run with different initial settings, including alternative rotamers and protonation states for His188, as well as different AIR force constant values. Several reports point to Nϵ2 as the most reactive center engaged in histidine phosphorylation (25, 26). Given that no major differences were recorded comparing Nϵ2 with Nδ1, Nϵ2 was ultimately kept as the atom to define the phospho-His distance restraints in further analyses, although experimental evidence is not available to conclude which imidazole nitrogen is actually reactive during DesK autophosphorylation. A total of 3000 structures was generated during the rigid-body energy-minimization step. The best 300 structures were refined in the semi-flexible annealing step and then subjected to a final refinement with explicit solvent. For the remaining parameters, we used HADDOCK default settings, as well as for the scoring function used to sort structures after the rigid-body and semi-flexible steps. Four replicates were done for this docking protocol to assess whether and how the obtained structures constituted a robust result. The same protocol was used for other DHp states, from the structures of DesKCH188V (3EHH) and DesKC_wt-P (3GIG). Root mean squared deviation (r.m.s.d.) values were computed through tailored (Tool command language) scripting within VMD (27) upon superposition of the Cα atoms of the DHp domain onto the best-scored structure model.
Sequence-based Analyses
A full referenced description is included in the supplemental materials. Briefly, homologous sequences were retrieved from bacterial genomes stored at the IMG system (28) with a standard BLASTp search. Unique sequences (406) were then aligned with the program Muscle. “HisKA_3” histidine kinases were retrieved from Pfam, and 762 unique proteins thereof were aligned with Muscle. The alignments displayed in Fig. 3 are a subset of the original multiple sequence alignment (MSA) of Pfam sequences, obtained by trimming the MSA into a non-redundant set of proteins with a cutoff of 88% pairwise identity. Some long proteins (>700 residues) were also manually removed from this set, and new MSAs were computed with Mafft (29) for the reduced sets. For analyses concentrating in HPK7 “NarQ/DesK-like” proteins (supplemental Fig. S1), sequences sharing the Pfam DHp-ABD architecture (808) were retrieved and formatted into a BLASTp-searchable database. NarQ and DesK were then used as queries against this smaller in-house database. MSAs were mapped onto the molecular surfaces of DesKABD and the DHp domain (using the same DesK models as for the docking calculations).
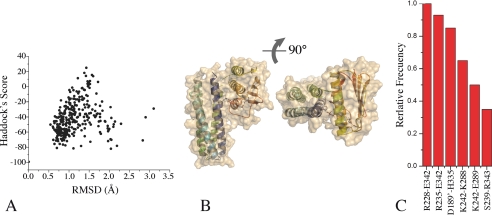
FIGURE 3.
Distance-restrained docking of DesKC ABD and DHp domains. A, the global Haddock score is plotted as a function of r.m.s.d. (calculated with respect to the best scoring model). Note that the best scoring models cluster at <1.5 Å r.m.s.d. B, three-dimensional structure of a representative high-scoring docked model of DesKC in the autophosphorylation state, in two orthogonal views. The solvent-accessible surface is rendered transparent to distinguish the relative organization of the domain-domain configuration (secondary structure elements are highlighted). Residues His188 and ATP are shown as sticks. C, relative frequency of interacting residue pairs in the best 10 docked structures. The histogram is limited to >30% frequencies. Engaged residues are listed, distinguishing with a single prime those belonging to the second protomer within the dimer.
Interdomain Disulfide Covalent Bonding Studies
Oxidation and Cys-disulfide formation of the Cys-engineered mutants (DesKC G192C,G334C, Q193C,G334C, and S196C,G334C) were triggered by incubating 10 μm protein in contact with air. Other procedures were also assayed, seeking for more rapid and higher yield oxidations, although these resulted in more drastic side reactions. Briefly, 10–100 μm hydrogen peroxide, and preformed complexes ∼0.2 mm Cu(II)-(phenanthroline)3 or Cu(II)-(bathocuproine disulfonate)2, were incubated for different times with purified protein. In all cases, thiol oxidation was stopped by incubation with 10 mm N-ethylmaleimide and 1 mm EDTA. Aliquots were analyzed with SDS-PAGE and MALDI-TOF mass spectrometry, with or without previous reduction (100 mm DTT). Thiol air oxidation was the mildest and most specific (data not shown) and selected for detailed analyses. Protein samples from selected SDS-PAGE bands were subjected to tryptic digestion with sequencing grade trypsin (Promega, Madison, WI) at 35 °C, overnight. Peptides were extracted from gels with 60% acetonitrile in 0.2% trifluoroacetic acid. Eluates were concentrated by vacuum centrifugation and desalted using reversed-phase microcolumns (C18 OMIX pipette tips, Varian). Samples were then directly spotted onto the mass spectrometer sample plate with 2 ml of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.2% trifluoroacetic acid). Mass spectra of positive ions were acquired in reflector mode with a MDS SCIEX 4800 MALDI TOF/TOF instrument (Applied Biosystems). Peptide analysis was performed using both Mascot Peptide Mass Fingerprint searching mode (Matrix Science) and GPMAW32 (version 4.02, Lighthouse Data, San Diego, CA). The following modifications were considered: disulfides, N-ethylmaleimide-blocked thiols, reduced thiols, and oxidized methionine.
Enzyme Activity Assays
Autophosphorylation and ATPase activities, resulting in net ATP consumption, were measured with a coupled enzyme assay (30). Briefly, the production of ADP was coupled to the oxidation of NADH by pyruvate kinase and lactate dehydrogenase (Sigma). The coupling system was in large excess to avoid saturation, and the oxidation of NADH was measured at 340 nm in a Cary® UV-visible spectrophotometer (Varian) at 24 °C. The reaction mixtures contained 50 mm HEPES, pH 7.5, 150 mm NaCl, 145 mm KCH3CO2, 8 mm MgCl2, 3 mm phosphoenolpyruvate, 240 mm NADH, 5 units of pyruvate kinase, 23 units of lactate dehydrogenase, and different ATP concentrations, in 200 μl of final volume. In the absence of DesKC the NADH oxidation was stable and linear for several minutes. Before starting the reaction, we preincubated the coupled system with ATP to convert the small initial ADP contamination to ATP. To start the reaction, wild-type DesKC or selected DesKC point mutants were added at 22 μm final concentration. Fitting of raw kinetic data using Michaelis-Menten functions was performed with the program Origin v. 8 (Originlab). In the case of wt DesKC, we could obtain more precise initial rate values by first fitting the biphasic time course to an exponential plus a linear function model (Equation 1),
where the A1 and A2 terms are the amplitudes of the exponential and linear phases, respectively; k2 is the constant rate of the linear phase (k2 < 0), whereas k1 is a substrate concentration-dependent rate coefficient used to calculate the initial velocity V0 for the exponential non-turnover phase in Equation 2.
The parameter f is the stoichiometry factor relating the concentration of phosphorylatable histidine sites with respect to total enzyme concentration [E]. This approach takes advantage of the integral method for determining initial rates with higher precision (31).
For the evaluation of GTP as a potential substrate of DesKC, and given that the coupled enzyme pyruvate kinase is able to use GDP, we followed the same protocol described above, substituting ATP for GTP at identical concentrations. To test if GTP behaves as a competitive inhibitor, eventually indicating DesKC GTP-binding capacity, 5 mm GTP was added in competition assays that contained 0.5 mm ATP.
RESULTS
Crystallization, Experimental Phasing, and Structure Refinement of the ATP-binding Domain of DesK
Orthorhombic crystals of the C-terminal ATP-binding domain of DesK (DesKABD) were initially obtained by serendipitous proteolysis, during DesKC-DesR crystallization assays. Proteolysis was found to be reproducible and may well be linked to the enrichment of a particularly unstable form of DesKC under the conditions used for protein-protein complex formation. The structure was determined at 1.8-Å resolution, phased using single wavelength anomalous diffraction (Table 1), after quick-soaking the crystals with NaI. Direct methods were used to determine the iodide positions. Subsequent phase calculation and refinement resulted in high quality electron density maps (Fig. 1), which allowed for straightforward phase-restrained refinement of the entire protein. Apart from the iodide atoms, the final model includes DesKABD residues 243–367 (following full-length DesK numbering), a bound molecule of ATP-Mg2+, and 185 structural water molecules (Table 1). Co-crystallization with ATP proved to be essential for DesKABD crystal growth. Surprisingly, DesKABD/ADP co-crystallization was unsuccessful, suggesting subtle yet significant conformational differences due to the γ-phosphate. Intrinsic ATPase activity of ABDs in other histidine kinases has precluded the determination of ATP-bound structures. DesK appears to have a particularly lower ATPase activity, a feature enabling direct visualization of the triphospho-nucleoside within its binding pocket. We further looked for nucleotide exchange in the crystals, aiming for detailed information of the putative nucleotide-triggered conformational rearrangements. After incubating in a solution with 5 mm EDTA, the crystals disappeared almost immediately, indeed consistent with a nucleotide-dependent structural reorganization (32). Further attempts to substitute ATP by ADP were performed through soaking competition assays. However, the refined models invariably show a mixture of both nucleotides, with 50% occupancy of ADP in the best cases (data not shown), precluding straightforward analysis of the modifications induced in the protein.
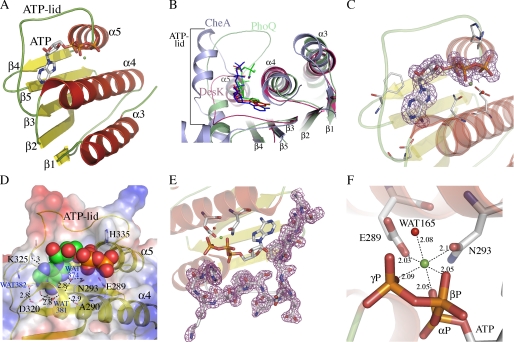
FIGURE 1.
Structure of the ATP-binding domain of DesK and details of its nucleotide-binding pocket. A, schematic representation showing topology and secondary structure elements. α-Helices are colored red, β-strands are yellow, and loops are green. B, superposition of DesKABD (red tones), PhoQ (green tones), and CheA (blue tones) showing the exposure of the corresponding bound nucleotides to the solvent. Note the absence of secondary structure elements in the ATP lid of DesK. A surface representation of these differences is detailed in supplemental Fig. S2. C, 2mFobs − DFcalc electron density map contoured at 1σ, displayed around the ATP molecule, highlighting clear signal for the entire nucleotide. D, solvent-accessible surface representation of DesKABD, with the ATP moiety depicted as van der Waals balls. The surface is colored according to the mapping of electrostatic potential (red = negative, blue = positive). Key protein residues for ATP binding are highlighted in stick representation and explained in the text. The buried water H-bond network is also shown. Numbers correspond to distances in Å. E, 2mFobs − DFcalc map contoured at 1 σ, showing clear electron density for the entire ATP lid loop, ordered in the presence of ATP. F, close-up of the Mg2+ coordination site. Oxygens from the three ATP phosphates, residues Glu289 and Asn293, and water Wat165, are seen closing the octahedral coordination shell of the cation.
Structure Analysis of DesKABD
The ABD of DesK is a globular domain (Fig. 1A), composed of a five stranded β-sheet (β1 to β5) opposed to a layer of three helices (α3–α5, respecting the α-helix numbering, including the DHp domain from the whole cytoplasmic DesKC region (13)). A search with DALI (supplemental Table S1) revealed strong structural similarity (Z scores > 7) to several proteins belonging to the GHL superfamily of slow ATPases (33), including CheA (PDB 1I59), TM0853 (2C2A), PhoQ (1ID0), KinB (3D36), PrrB (1YS3), NtrB (1R62), and SpoIIAB anti-σF factor (1TID and 1TIL), or yet different mitochondrial dehydrogenase kinases (1GKZ and 2BU8). Comparing DesKABD (ABD HPK7 family) with the other families for which three-dimensional structures are currently available (KinB (HPK1), EnvZ (HPK2), PhoQ (HPK3), NtrB (HPK4), or yet, CheA (HPK9)), some differences become immediately obvious: the ABD of DesK is smaller and has only three α-helices, and the ATP lid is correspondingly shorter. The ATP-binding groove is correspondingly shallower; hence, bound ATP is seen significantly more exposed to the bulk solvent (Fig. 1, B and D, and supplemental Fig. S2).
Electron density corresponding to the bound ATP is well defined for the entire molecule (Fig. 1C). Although the adenine ring is deeply buried and stacks against Phe324, the rest of the nucleotide sits on the abovementioned rather open groove. Strictly conserved in HKs, Asp320 is observed hydrogen bonding with the amino N6 of the adenine base as well as with the main chain N of Gly322. These interactions would not allow for GTP binding, which has been otherwise reported in other HKs (34). Further reinforcing this ATP-specificity profile, the nucleotide-binding pocket in DesK displays a conserved water network (35, 36), strongly stabilizing the complex by complete satisfaction of the H-bonding potential of the protein and the adenine base moiety in the site (Fig. 1D). Waters Wat373, Wat374, and Wat382, deeply buried in the pocket, interface the adenine base with a defined array of protein residues, and a fourth water molecule (Wat381) finishes some intra-protein inter-residue H-bonding (Fig. 1D). Amino acids on β3 (the already mentioned Asp320) and β5 (Thr359), define the “floor” of the pocket. Helix α4 is particularly important, contributing several interacting residues (Ala290, Asn293, Lys296, and His297), in this way delimiting one of the pocket's “walls.” Between β3 and α5 (residues 321–334) the loop corresponding to the so-called “ATP lid,” is fully defined in electron density (Fig. 1E), albeit showing higher than average B factors, both on side chains and the main chain. The ATP lid contains some residues that interact with the ATP moiety, such as Lys325 with its main-chain N H-bonding adenine N3. But, the loop is basically loose, prone to interact with other partners. We further analyzed the structure of the DesKABD·ATP complex in comparison with previously reported structures with other nucleotides and analogues (13), to identify regions displaying statistically significant variations in atomic positions (see supplemental materials for details of the comparison methods used). Overall, the domain can be described as globally rigid. In contrast, helix α5, which directly connects the ATP-binding pocket with the other end of the domain (Fig. 1, A and D), is one of the few regions showing largest atomic coordinate variability (supplemental Fig. S3).
The Mg2+ ion, known to be essential for kinase activity in HKs (36), adopts a typical octahedral coordination structure (Fig. 1F), making contacts with the oxygens of the three ATP phosphates, two side chains of residues on α4 (Glu289 and Asn293) and a water molecule (Wat165). This water molecule might play a catalytic role, acting as a proton donor to phosphate γ, in a dissociative “substrate-assisted” mechanism toward phosphotransfer (37, 38). Asn293 is the most conserved residue within the signature motif “N box” in HKs, which together with Glu289 make hydrogen bond interactions with the β-phosphate of ATP.
Other signature motifs on the ABD are also linked to the ATP binding function, such as residues Asp320-Asp/Asn-Gly322 defining box G1, wherein Asp320 interacts with the adenine ring as described above. Box G2 is not well conserved in multiple alignments (Gly336-X-X-Gly339) and is observed in DesKABD just after the ATP lid loop, making van der Waals contacts with the α and β phosphates. Finally, box G3 (Thr355-X-X-Gly-Thr359) at the N terminus of strand β4, defining part of the floor of the adenine binding pocket, also displays poor conservation.
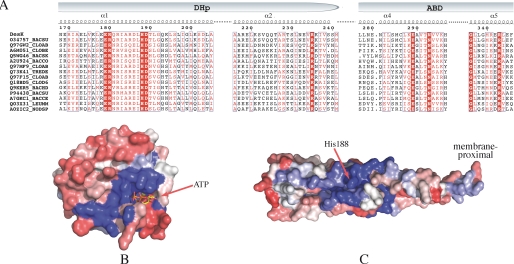
Multiple sequence alignments of DesK with HKs of the HisKA_3 family, allowed us to identify an interesting pattern of conservation (Fig. 2A). Mapping these alignments onto the DesKABD structure (Fig. 2B) revealed the expected conserved residues directly involved in ATP binding and Mg2+ coordination. What caught our attention was a second conserved patch, which is readily identified and distant from the ATP pocket, composed of residues Gly336 to Leu344 on helix α5. A well conserved surface patch is also found on the DHp domain, surrounding the phosphorylatable His (Fig. 2C). According to their respective locations, these amino acid conservation patches on the DHp and on helix α5 of the ABD, are well positioned to participate in complementary surfaces of interaction, upon ABD approximation onto the four-helix bundle during autophosphorylation, ensuring a specific domain-domain geometry.
FIGURE 2.
Sequence alignments and conservation patterns of the HisKA_3 subfamily. A, selected region of a reduced multiple sequence alignment with sequences from the “HisKA_3” Pfam subfamily. Identical residues are shown in bold white on a red background; conserved residues are boxed in red. Pfam IDs are indicated on the left followed by a species code: BACSU, Bacillus subtilis; CLOAB, Clostridium acetobutylicum; CLOBE, Clostridium beijerinckii (strain ncimb 8052); BACSK, Bacillus clausii (ksm-k16); BACCO, Bacillus coagulans (36d1); TREDE, Treponema denticola; CLOD6, Clostridium difficile (630); BACHD, Bacillus halodurans; BACCE, Bacillus cereus cytotoxis (nvh 391–98); LEUMM, Leuconostoc mesenteroides mesenteroides (atcc 8293/ncdo 523); and NODSP, Nodularia spumigena (ccy 9414). B, sequence conservation/variability mapped onto the molecular surface of the ATP-binding domain of DesKC. Color coding is a range from red (variable) to blue (conserved) through an intermediate white. C, same as B, mapped onto the surface of the DHp domain of DesKC.
Protein-Protein Docking: the Autophosphorylation State
We then asked whether the conserved residues could play a role in defining an interaction surface between the two domains of DesKC. Semiflexible domain-domain docking calculations were performed, to identify one or more energetically favorable structures of DesKC with the ABD and the DHp domains in close interaction, providing molecular models that should be compatible with the autophosphorylation intermediate state. We have recently reported structural data corresponding to different functional states along the catalytic cycle of DesK (13), including conformations corresponding to the autokinase-, phosphotransfer-, and phosphatase-competent forms, some of them indeed displaying particular domain-domain interfaces. However, the autophosphorylation state is elusive, because it is a transient conformation, essentially difficult to visualize by experimental methods in its native state. Given that high resolution models are better starting points to undertake docking calculations, we used the ABD coordinates that we are now reporting, to compute protein-protein docking calculations onto the DHp domain (PDB 3GIE, corresponding to the DHp kinase-competent state), using the program Haddock v2.0. It has been proposed that the ATP lid of HKs is important in the autophosphorylation reaction (39). The fact that our DesKABD structure displays no ambiguity in this loop, further highlights its usefulness as a docking probe over previous DesK structures.
Haddock allows for the inclusion of explicit distance restraints. The distance between Nϵ2 of His188 and γP of the ABD-bound ATP was kept restrained within a short range (1–4 Å), in accordance with the expected situation during trans-autokinase catalysis. A second restraint was added between the carbonyl C of Lys242 in the DHp model, and the amino N of Gly243 in the ABD, to include the actual covalent link between both domains in the simulations. The ATP-His188 distance restraints corresponding to a cis-acting autophosphorylation model were also tested. The resulting docking figures displayed less favorable energy scores (data not shown). The distance between the hinge point (residues 242–243) and His188 was significantly larger in the cis models, imposing a severe restriction in the degrees of freedom for the ABD to accommodate to the distance restraints. This resulted in a solvent-exposed His188Nϵ2-ATPγP interaction, with almost no protein environment to assist in catalysis, in sharp contrast with the trans-acting model. Finally, the amino acid conservation patch on the DHp domain (Fig. 2C), matches well with the predicted ABD-binding interface (Fig. 3), only when applying the docking distance restraints in trans.
Further in-depth analyses were pursued with the trans-autophosphorylation models. Analysis of the top 300 structures sorted by score, revealed a clear-cut cluster, with best-scored structures having r.m.s.d. values of <2.5 Å (Fig. 3A). Independent runs of this docking protocol reached basically equal solutions, showing the robustness of the result (supplemental Fig. S4). Considering the output of all runs, two populations could readily be identified, which display only slight differences in the relative orientation of the ATP binding domain (Fig. 3B and supplemental Fig. 4E). The frequency distribution of interacting residue pairs shows a few of them particularly well represented (Fig. 3C). For example, in the best 10 structures from diverse runs, three of these pairs interact in >85% of the models, systematically involving residues spanning α5 and the loop just N-terminal to it. We further analyzed these docking results in view of the previously identified sequence conservation patterns (Fig. 2). Certain residues, poised to interact according to the docking simulations, are indeed highly conserved. The best-scored structures show all ABD α-helices contacting the DHp domain (Fig. 3B). The most represented residue pairs in the docked structures ensembles (Fig. 3C) are as follows: Arg235–Glu342 (93–70%, conservation figures within HiskA_3 Pfam sequences for the two residues, each one, respectively, in the DHp and the ABD domains), Arg228–Glu342 (35–70%), Asp289–His335 (96–15%), Lys242–Glu289 (44–96%), and Asp189–Lys296 (99–52%). These interactions were reproducibly obtained in all top-scoring models from independent runs. In these structures the C-terminal part of the ATP lid interacts with helix α1 of the DHp, through hydrogen bonds between His335 and Asp289 as well as Gly199 with Lys333. Other residues that seem important in making strong interactions are Asp189–Lys296, Arg228–Glu342, Arg235–Glu342, and Lys242–Glu289, again highlighting several of these candidate amino acids located in helix α5 of the ABD. The final configuration of the ATP lid places Gly334 near Gly192, small residues that seem important to reach the necessary close packing between the interacting domains.
The set of interacting residues suggested by these docking calculations is fully consistent with prior knowledge on the interaction surface between HKs and RRs (7, 12), implying that the ABD of DesK would be occluding a portion of the DHp-DesR interaction surface. This is consistent with the fact that DesKC cannot form a complex with DesR, except when operating as a phosphatase or phosphotransferase (13), conformations in which the ABD is immobilized away from the DHp-DesR interface.
Because the DHp domain in DesK is known to adopt different conformations (13), we extended the docking protocols described above to explore possible effects of DHp rearrangements on the predicted interdomain interaction with the ABD. Two different experimental models of the DHp domain were used: DesKCH188V (pdb 3EHH, corresponding to the phosphatase-competent state) and phosphorylated wt DesKC (3GIG, phosphotransferase state). The results suggest that changes in the DHp domain would hamper autophosphorylation, relative to the favorable “kinase-competent” state (supplemental Fig. S5). In the phosphotransferase configuration of the DHp domain, the predicted buried surface area is significantly smaller, predicting a lower interaction energy (supplemental Table S2). Finally, when the DHp corresponds to the phosphatase state, the predicted interaction energy is higher, but the γP of ATP is consistently found farther, distant from His188 (supplemental Table S2 and Fig. S5B), and thus not compatible with autophosphorylation. This holds true even when distance restraints have been kept identical throughout the simulations.
Trapping the Trans-autophosphorylation Intermediate State
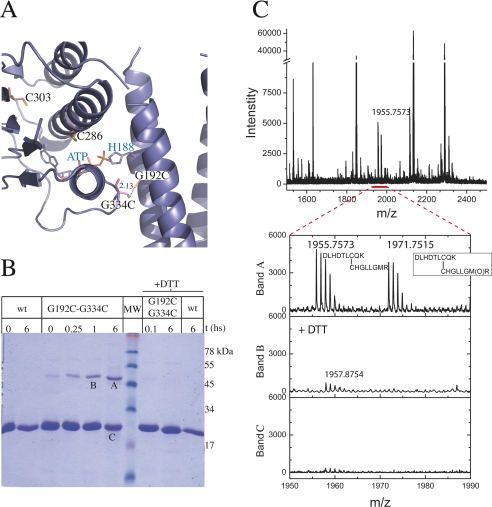
If the docked structures reveal a genuine specific interdomain interface during autophosphorylation, we should be able to trap the intermediate state designing structure-based cysteine mutants (Fig. 4). These engineered proteins should be able to undergo disulfide covalent linking between the ABD and the DHp domains, only if they interact as predicted according to our sequence conservation and docking analyses. With this aim we selected the G192C/G334C double mutant, taking into account the close predicted distance between Gly192 and Gly334 (Fig. 4A). We allowed the purified mutant protein to oxidize spontaneously in contact with air. A species corresponding to the dimer appeared very rapidly (Fig. 4B) and showed a time-dependent accumulation at the expense of the monomeric form in SDS-PAGE. The covalent intermonomeric bond was proved to be sensitive to a reducing agent (Fig. 4B). Longer oxidation times resulted in protein instability, with species of a wide range of apparent molecular weights (data not shown), although the main product was still the dimeric form. To maximize the yield of the covalent dimeric form, we also explored speeding up the oxidation, using H2O2, Cu(II)-(phenanthroline)4, or Cu(II)-(bathocuproine disulfonate)4. However, even with short incubation times, accelerated oxidation conditions resulted in a rather broad range of oligomeric species of difficult analysis (data not shown). This is probably due to the presence of two endogenous cysteines in the ABD, which seem to be required for proper ABD folding (the corresponding Cys to Ser mutants proved to be unstable in solution, data not shown). Given these issues, we could not conclude from SDS-PAGE analyses alone if the putative major disulfide bond was being formed between the two engineered cysteines as planned. To be conclusive, we submitted the air-oxidized dimeric species to MALDI-TOF mass spectrometry, comparing samples that had previously been reduced to those that had not (Fig. 4C). Indeed, the S–S bond was found to be linking the two introduced cysteines, confirming the trapping of an intermediate of the trans-acting autophosphorylation reaction. If this intermediate is specific, in the sense of being dependent on a particular geometric ABD-DHp orientation, the precise position of the engineered cysteines should affect the outcome of the cross-linking experiment. Two variants were thus generated, the double substituted E193C/G334C, as well as S196C/G334C. The former displaces the reactive position on the DHp domain by one residue in the helix, whereas the latter shifts it by one full helical turn. At position 193, the introduced cysteine was not able to induce the formation of any detectable dimeric species (supplemental Fig. S6A). Position 196 allowed for a minor fraction of disulfide bonding (supplemental Fig. S6B), consistent with the fact that it approximates a full helical turn shift, predicting the introduced Cys to point in the same direction as the original targeted position 192. Nonetheless, Cys196 showed a significant reduction in disulfide bond formation kinetics compared with Cys192. Overall, both controls lend strong support to the specificity of the geometric arrangement between the two interacting domains, pointing to the relevance of the trapped intermediate as a bona fide mimetic of the autophosphorylation state.
FIGURE 4.
Disulfide cross-linking assay. A, theoretical model of the mutated G192C,G334C protein in the autophosphorylation conformation, according to domain-domain docking results. Cysteine residues are shown as sticks. Predicted distance between the engineered cysteines, compatible with disulfide bond formation, is marked. B, SDS-PAGE of the cross-linking reaction. wt: wild-type DesKC used as control; G192C,G334C: DesKC double mutant with engineered cysteines (predicted molecular mass of the covalent dimer species: 50 kDa); +DTT: incubation with 0.1 m DTT. MW: molecular weight markers (corresponding masses are depicted on the right in kilodaltons). Air or air + DTT incubations were allowed to proceed for t hours. Note the DTT-induced reversion of the dimeric species to monomers. Bands A, B, and C were subsequently excised from the gel and subjected to mass spectrometry analyses. C, mass spectra of tryptic peptides derived from bands A, B, and C, showing coincidence of experimental and predicted m/z data. The spectrum segment, including peptides containing the expected disulfide bond (detailed in the inset), is enlarged. Met340 is shown to be in the reduced and oxidized forms (m/z 1955.7573 and 1971.7515, respectively).
Residues Involved in Domain-Domain Interaction Modulate Catalysis
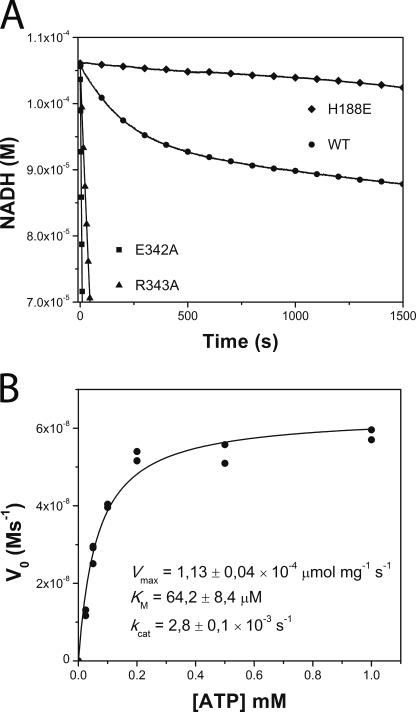
The spatial geometry of the domain-domain interaction surface has thus been predicted by docking calculations and further supported by trapping an intermediate state with structure-based engineered cysteine mutants. The interdomain interface involves a number of residue pairs. To gather experimental evidence on the actual functional role of interface amino acids, we planned a point-mutagenesis approach, focusing on residues not involved in ATP-binding/metal coordination nor in phosphorylatable histidine protonation, because residues involved in these functions should have obvious effects in catalysis, independent from their influence in domain-domain interaction forces. Residues in helix α5 of the ABD, located far from both the ATP-binding cleft and the phosphorylatable histidine, thus become ideal candidates. A particularly well conserved amino acid, present in all the best-scoring ABD-DHp docked complexes, is Glu342. We substituted Glu342 by alanine, and enzymatic ATP consumption activity was measured with a coupled enzyme assay (40). Surprisingly, instead of lowering activity due to the hindrance of the autophosphorylation ABD-DHp interaction, the point mutation resulted in a gain-of-activity effect, increasing >60-fold the rate of ATP consumption with respect to the wild-type enzyme (Fig. 5A). Close examination of our formerly reported DesKC structures (13) shows that Glu342 is involved in interaction surfaces between the ABD and the DHp, both in the phosphatase (3EHH) and the phosphorylated phosphotransferase (3GIG) states. Its role may thus be more complex, engaged in stabilizing other states apart from the autophosphorylation intermediate. To test this hypothesis we included a second point mutant, substituting Arg343 by an alanine. Arg343 has not been predicted to take part in the autophosphorylation interdomain interface, but is otherwise well conserved in the helix α5 patch and is also seen participating in ABD-DHp interactions in states other than the autokinase configuration, much as Glu342 does (13). Indeed, a similar gain-of-activity effect was reproduced, resulting in a >10-fold increase of the ATP consumption rate versus wild-type DesKC (Fig. 5A).
FIGURE 5.
Kinetic characterization of DesKC autophosphorylation. A, temporal course of ATP consumption for wild-type DesKC (WT, ○) and DesKC mutants E342A (■), R343A (▴), and H188E (autophosphorylation inactive, ♦). H188E shows a basal ATPase activity corresponding to the intrinsic activity of the ABD. Wild-type DesKC displays a biphasic behavior, with an initially exponential phase followed by a second linear regime. Note the high ATP consumption rates of E342A and R343A, which showed no biphasic pattern, even in extended incubation times until NADH depletion (not shown in this plot). B, ATP dependence of wt DesKC autophosphorylation initial velocities. Pure DesKC (22 μm) was used. kcat was calculated considering the total concentration of monomer.
This coupled enzyme assay conveys more information (41) than the classically used detection of radioactive phosphate addition onto the histidine during autokinase activity. Rapid kinetics of histidine spontaneous dephosphorylation may have profound effects on the steady-state phospho-histidine, eventually leading to no detection of autophosphorylation, even under high autophosphorylation rate regimes (17). To better understand the increased ATP consumption activity of the α5 point mutants, we decided to undertake a more extensive enzymatic characterization of wt DesKC using this coupled enzyme assay approach.
Enzymatic Characterization of DesKC
Wild-type DesKC showed a robust biphasic ATP consumption curve (Fig. 5A). At this pH phospho-His is stable (42), explaining the relatively early inflection point on the time course: once a first phase of autophosphorylation reaches completion, only a basal ATPase activity remains, which is similar to the ATPase activity of an autophosphorylation inactive mutant (DesKCH188E) (Fig. 5A). The slopes of each phase correspond to ATP consumption rate constants of 2.8 × 10−3 ± 0.1 s−1 and 3.1 × 10−4 ± 0.2 s−1. Attempts to lower the pH to enter into a catalytic regime were unsuccessful due to the intrinsic instability of DesKC. The kinetics observed for wild-type DesKC (Fig. 5B) fully agrees with previous reports using radioactive assays (15), resulting in kinetic parameters that are in the same range as several other HKs (supplemental Table S3). The ratio of ATP equivalents consumed during the exponential phase with respect to total moles of potentially phosphorylatable sites (i.e. DesKC monomeric species) converged reproducibly to ∼0.5, giving strong support to a model of asymmetric phosphorylation. The phosphorylation of one of the monomers within the dimer appears to preclude the second event on the other monomer, in agreement with previous crystallographic evidence (13). Given the structural analysis of the ATP-binding pocket, we also asked whether DesKC is actually able to use GTP as an alternative autophosphorylation substrate. Using the coupled enzyme assay described above, substituting ATP by GTP, no nucleotide consumption was detected (data not shown; detection limit = −1.9 × 10−10 ms−1). To rule out the possibility of GTP binding, competition assays were set, indicating that an excess of GTP did not alter ATP consumption rates to any detectable extent.
DISCUSSION
Recent reports (12) highlight the need to avoid oversimplified generalizations on mechanistic features of histidine kinases, such as cis- or trans-acting autophosphorylation, built on the basis of only a few well known examples. Additional work is thus required to gain direct insight into the wide array of HK proteins and their unpredicted variations of enzymatic and/or regulatory mechanisms. We are now reporting the high resolution x-ray structure of the ATP-binding domain of an HPK7 histidine kinase, in complex with ATP. The molecular features we disclose identify particular elements likely applicable to the full HPK7 family, among which are a rather short connecting segment between the ABD and the DHp domains and a different structural organization on the immediate vicinity of the nucleotide-binding pocket. Five other HK families are still lacking experimental three-dimensional structures.
In a first line of analysis, the structure of DesKABD is, as far as we know, the first HK domain solved in binary complex with its physiologic substrate ATP. The ATP lid loop is much shorter in DesK than in other families (such as the ones including CheA and PhoQ, among others), resulting in a correspondingly higher solvent exposure of the nucleotide (Fig. 1B and supplemental Fig. S2). Two hydrogen bonds engaging the β and γ phosphates contribute to ATP binding in DesK: Gly336[N]–ATP[O3G], Leu337[N]–ATP[O2A]. In addition, ionic interactions are readily identified: Lys296 and His297 on one side, and His335 on the other, display side-chain nitrogens at short interaction distances to the ATP β and γ phosphates. Following proposed mechanisms for nucleotidase and phosphotransfer catalysis, these cationic residues are well positioned to play a functional role in stabilizing the negative charge that builds up on phosphate β upon β–γ bond breakage (37). Although HKs belong to the GHL superfamily of slow ATPases, HK intrinsic ATPase activities have been reported to be much lower than the corresponding ones for DNA gyrase, MutL, or Hsp90. Nevertheless, for poorly documented reasons, there are no available experimental structures of HKs in complex with ATP: the triphospho-nucleosides hydrolyze in the course of the crystallization experiments, seemingly through a residual ATPase activity, even in some cases using “non-hydrolyzable” ATP analogues (24). This has stimulated and justified in silico simulations of ATP·HK complexes (39). The fact that HKs with higher ATPase activity have a key basic residue replacing His335 (e.g. Arg434 in PhoQ or Lys494 in CheA) is consistent with the hypothesized ionic stabilization of the negative charge on phosphate β and could explain a lower ATPase activity in DesK-like HKs. In contrast, previous suggestions (35) ascribed the higher ATPase activity of GHL proteins, such as MutL, to the supposedly conserved substitution of a glutamate residue (Glu29 in MutL) replacing an Asn or His in HKs (Asn385 in PhoQ, and His405 in CheA). Our structure argues against this simple explanation, because the ABD of DesK, displaying still lower intrinsic ATPase activity (allowing us to trap the ATP·DesK complex), shows a glutamate at this position (Glu289), as in true ATPases. This glutamate is actually well conserved in the HPK7 family of HKs (supplemental Fig. S1), as well as in other related enzymes with serine-kinase activity (3). It will be relevant to test whether other HPK7 HKs such as NarX-, NarQ-, ComP-, UhpB-, or LiaS-like (43), display low/null intrinsic ATPase activity, in agreement with our structure-based hypothesis.
The number of DesKC-ATP interactions in the nucleotide-binding pocket denotes a snug fit that allowed us to predict the specific hindrance of GTP binding, in comparison with other HKs able to use the guanosine compound as autophosphorylation substrate (34). This was verified experimentally, observing that DesKC does not use GTP as a substrate, and neither does it bind GTP to outcompete the adenosine species.
In a second line of thought, the results we now report lend experimental support and atomic detail information to the concept that the autophosphorylation reaction in HKs depends on a specific and asymmetric interaction surface between the ATP binding and the dimerization phosphotransfer domains. This hypothesis is supported by sequence conservation, domain-domain interaction predictions, and the consequent trapping of a disulfide-linked covalent dimer.
The conserved surface patch on the DHp domain (Fig. 2C), poised to interact with the ABD in the autophosphorylation reaction, is distinct from the RR-contacting region, according to recent bona fide HK-RR crystal structures (12). This is consistent with reported RR specificity re-wiring results (6), because the authors only mutated seven (or less) residues, all located in the tip of the four-helix bundle, distal from the phosphorylatable histidine. The resultant chimeric HKs maintain autophosphorylation activity while changing RR specificity. The two patches, one restraining autophosphorylation, the other RR-phosphotransfer specificity, appear to overlap only partially in DesKC. Nevertheless, this does predict a competitive exclusion scenario in the HisKA_3 family, whereby a freely moving ABD would represent a higher effective concentration partner than the diffusible RR. Our recent findings of ABD mobility control along the HK catalytic cycle (13) and correlated modulation of RR association are fully consistent with this idea.
A novel function also emerges, which concerns the conserved patch of residues in helix α5 of the ATP-binding domain. Indeed, point mutations E342A and R343A in this region induce significant effects on the enzymatic activity. To our surprise, instead of diminishing autophosphorylation rates due to destabilization of the interdomain complex, ATP consumption was significantly accelerated. This effect is compatible with two possible scenarios: an increased and sustained autophosphorylation activity under a catalytic regime, implying continuous dephosphorylation of the phospho-His, or, instead, an allosteric effect on the ATP-binding pocket, dramatically augmenting the intrinsic ATPase reaction. The latter hypothesis seems less likely, given the solvent-exposed location of the mutated residues and their distance to the ATP-binding pocket. A more direct explanation comes from structural analyses. This particular region of α5 on the ABD domain is directly involved in distinct DHp-ABD interdomain interactions on the other functional states of the enzyme, including both the phosphorylated phosphotransferase-competent state, as well as on the phosphatase configuration (13). Hence, the relative population of each functional state at any given time could well be modulated by these interdomain interactions. By mutating Glu342 and Arg343 we have destabilized the phosphotransferase and/or the phosphatase states to a greater extent than the kinase-competent one. Phospho-histidine might become more labile in the absence of the appropriate switch to an asymmetrically phosphorylated phosphotransferase conformation, resulting in exacerbated ATP consumption. The strong negative cooperativity effect of the phosphorylated monomer onto the unphosphorylated one within the dimer, inhibiting an otherwise symmetric phosphorylation, had been previously suggested by our group (13) and now confirmed by the autophosphorylation reaction stoichiometry. The fact that the phosphorylated state is strongly asymmetric could clearly result in differential modulation of phospho-histidine stability through ABD mobility control and/or local environment modifications. The three-dimensional structure of phosphorylated DesKC (13) shows that the ABD of monomer A (ABD-A) is immobilized, involving residues in helix α5, through interactions with the DHp. The ABD-B, instead, remains freely mobile. The interesting point is that the fixed ABD-A remains in close proximity of the phospho-His of monomer B. Given that DesR is able to interact with the phosphorylated kinase, and that a mobile ABD seems to be able to outcompete DesR binding, the asymmetric phosphotransferase state further supports autophosphorylation happening in trans in DesK. It also shows how α5 in the ABD can directly participate in the stabilization of this state. This hypothesis of differentially populated three states in the sensor kinase awaits further experimental confirmations currently underway, one of the main difficulties being the few residues predicted to stabilize only one of the three states, added to the relative instability of the whole protein prone to aggregation after mutagenesis of key positions.
The prediction of the docked domain-domain configuration guided the design of engineered cysteines, able to promote disulfide bonding only if the predicted conformations are actually sampled in solution. In this way, a putative autophosphorylation intermediate state was indeed trapped, also suggesting that DesK performs the autophosphorylation reaction in trans, one monomer phosphorylating the other within the dimer. Crystallographic and biophysical analyses of this en transfer autophosphorylation state are currently underway. Valuable information will thus become available to understand the autophosphorylation step in histidine kinases and attempt transduction uncoupling at precise steps of the signaling pathways.
Supplementary Material
Acknowledgments
At the Institut Pasteur de Montevideo, we thank Matias Machado for valuable help in using Haddock and Ariel Chaparro for computation scripts, and the staff from the Units of Analytical Biochemistry (mass spectrometry analyses) and Recombinant Proteins (protein production and purification). The plasmids used to express recombinant DesKC and DesR were kindly donated by Diego de Mendoza.
This work was supported in part by Grants FCE2007_219 (to A. B.) and FCE2007_377 (to M. G.) from the Agencia Nacional de Investigación e Innovación, Uruguay (ANII) and Grant ANR-06-PCVI-0009-01 (to A. B.) from the Agence Nationale de la Recherche, France.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs S1–S6 and Tables S1–S3.
The atomic coordinates and structure factors (code 3H6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Recommended trivial name “Protein-Histidine Kinases” (EC 2.7.13.x).
- HK
- histidine kinase
- RR
- response regulator
- TCS
- two-component signal transduction system
- ABD
- ATP-binding domain
- DTT
- dithiothreitol
- MSA
- multiple sequence alignment
- MALDI
- matrix-assisted laser desorption ionization
- TEV
- tobacco etch virus
- TOF
- time-of-flight
- r.m.s.d.
- root mean squared deviation.
REFERENCES
- 1.Plowman G. D., Sudarsanam S., Bingham J., Whyte D., Hunter T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13603–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolanin P. M., Thomason P. A., Stock J. B. (2002) Genome Biol. 3, REVIEWS3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machius M., Chuang J. L., Wynn R. M., Tomchick D. R., Chuang D. T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsing W., Russo F. D., Bernd K. K., Silhavy T. J. (1998) J. Bacteriol. 180, 4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laub M. T., Goulian M. (2007) Annu. Rev. Genet 41, 121–145 [DOI] [PubMed] [Google Scholar]
- 6.Siryaporn A., Goulian M. (2008) Mol. Microbiol. 70, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skerker J. M., Perchuk B. S., Siryaporn A., Lubin E. A., Ashenberg O., Goulian M., Laub M. T. (2008) Cell 133, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta R., Qin L., Inouye M. (1999) Mol. Microbiol. 34, 633–640 [DOI] [PubMed] [Google Scholar]
- 9.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai S. J., Inouye M. (2003) J. Mol. Biol. 329, 495–503 [DOI] [PubMed] [Google Scholar]
- 11.Ninfa E. G., Atkinson M. R., Kamberov E. S., Ninfa A. J. (1993) J. Bacteriol. 175, 7024–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casino P., Rubio V., Marina A. (2009) Cell 139, 325–336 [DOI] [PubMed] [Google Scholar]
- 13.Albanesi D., Martín M., Trajtenberg F., Mansilla M. C., Haouz A., Alzari P. M., de Mendoza D., Buschiazzo A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16185–16190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilwes A. M., Alex L. A., Crane B. R., Simon M. I. (1999) Cell 96, 131–141 [DOI] [PubMed] [Google Scholar]
- 15.Albanesi D., Mansilla M. C., de Mendoza D. (2004) J. Bacteriol. 186, 2655–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilar P. S., Hernandez-Arriaga A. M., Cybulski L. E., Erazo A. C., de Mendoza D. (2001) EMBO J. 20, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noriega C. E., Schmidt R., Gray M. J., Chen L. L., Stewart V. (2008) J. Bacteriol. 190, 3869–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cybulski L. E., del Solar G., Craig P. O., Espinosa M., de Mendoza D. (2004) J. Biol. Chem. 279, 39340–39347 [DOI] [PubMed] [Google Scholar]
- 19.van den Berg S., Löfdahl P. A., Härd T., Berglund H. (2006) J. Biotechnol. 121, 291–298 [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project Number 4. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 21.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 22.Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 23.Bilwes A. M., Quezada C. M., Croal L. R., Crane B. R., Simon M. I. (2001) Nat. Struct. Biol. 8, 353–360 [DOI] [PubMed] [Google Scholar]
- 24.Marina A., Waldburger C. D., Hendrickson W. A. (2005) EMBO J. 24, 4247–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lott J. S., Paget B., Johnston J. M., Delbaere L. T., Sigrell-Simon J. A., Banfield M. J., Baker E. N. (2006) J. Biol. Chem. 281, 22131–22141 [DOI] [PubMed] [Google Scholar]
- 26.Surette M. G., Levit M., Liu Y., Lukat G., Ninfa E. G., Ninfa A., Stock J. B. (1996) J. Biol. Chem. 271, 939–945 [DOI] [PubMed] [Google Scholar]
- 27.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 28.Markowitz V. M., Kyrpides N. C. (2007) Methods Mol. Biol. 395, 35–56 [DOI] [PubMed] [Google Scholar]
- 29.Katoh K., Toh H. (2008) Brief Bioinform. 9, 286–298 [DOI] [PubMed] [Google Scholar]
- 30.Lindsley J. E. (2001) Methods Mol. Biol. 95, 57–64 [DOI] [PubMed] [Google Scholar]
- 31.Espenson J. (1995) Chemical Kinetics and Reaction Mechanisms, 2nd Ed., pp. 15–45, McGraw-Hill Book Co, New York [Google Scholar]
- 32.Dehner A., Furrer J., Richter K., Schuster I., Buchner J., Kessler H. (2003) ChemBioChem 4, 870–877 [DOI] [PubMed] [Google Scholar]
- 33.Dutta R., Inouye M. (2000) Trends Biochem. Sci 25, 24–28 [DOI] [PubMed] [Google Scholar]
- 34.Scaramozzino F., White A., Perego M., Hoch J. A. (2009) J. Bacteriol. 191, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marina A., Mott C., Auyzenberg A., Hendrickson W. A., Waldburger C. D. (2001) J. Biol. Chem. 276, 41182–41190 [DOI] [PubMed] [Google Scholar]
- 36.Stock A. M., Robinson V. L., Goudreau P. N. (2000) Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 37.Kötting C., Blessenohl M., Suveyzdis Y., Goody R. S., Wittinghofer A., Gerwert K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13911–13916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweins T., Geyer M., Scheffzek K., Warshel A., Kalbitzer H. R., Wittinghofer A. (1995) Nat. Struct. Biol. 2, 36–44 [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Xu Y., Shen J., Luo X., Chen J., Chen K., Zhu W., Jiang H. (2005) J. Am. Chem. Soc. 127, 11709–11719 [DOI] [PubMed] [Google Scholar]
- 40.Tawa P., Stewart R. C. (1994) J. Bacteriol. 176, 4210–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tawa P., Stewart R. C. (1994) Biochemistry 33, 7917–7924 [DOI] [PubMed] [Google Scholar]
- 42.Hultquist D. E., Moyer R. W., Boyer P. D. (1966) Biochemistry 5, 322–331 [DOI] [PubMed] [Google Scholar]
- 43.Mascher T., Helmann J. D., Unden G. (2006) Microbiol. Mol. Biol. Rev. 70, 910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.