Abstract
Mammalian myosin IXb (Myo9b) has been shown to exhibit unique motor properties in that it is a single-headed processive motor and the rate-limiting step in its chemical cycle is ATP hydrolysis. Furthermore, it has been reported to move toward the minus- and the plus-end of actin filaments. To analyze the contribution of the light chain-binding domain to the movement, processivity, and directionality of a single-headed processive myosin, we expressed constructs of Caenorhabditis elegans myosin IX (Myo9) containing either the head (Myo9-head) or the head and the light chain-binding domain (Myo9-head-4IQ). Both constructs supported actin filament gliding and moved toward the plus-end of actin filaments. We identified in the head of class IX myosins a calmodulin-binding site at the N terminus of loop 2 that is unique among the myosin superfamily members. Ca2+/calmodulin negatively regulated ATPase and motility of the Myo9-head. The Myo9-head demonstrated characteristics of a processive motor in that it supported actin filament gliding and pivoting at low motor densities. Quantum dot-labeled Myo9-head moved along actin filaments with a considerable run length and frequently paused without dissociating even in the presence of obstacles. We conclude that class IX myosins are plus-end-directed motors and that even a single head exhibits characteristics of a processive motor.
Keywords: Actin, Calmodulin, Enzymes, Molecular Motors, Multifunctional Protein, Myosin, Rho
Introduction
Myosins form a large superfamily of actin-based molecular motors that is composed of at least 35 classes (1). Class IX myosins arose in metazoa after the separation of the fungi (1). Invertebrates contain a single myosin class IX gene with the exception of the Drosophila species that have lost their class IX myosin. Bony fishes contain four myosin IX genes and other vertebrates, including mammalia two genes. The two class IX myosins in mammals, Myo9a2 and Myo9b, exist in multiple splice variants (2). Myo9a has been shown to play a role in epithelial differentiation and morphology whereas Myo9b regulates the migration of macrophages and possibly other immune cells (3, 4). Class IX myosins share a similar structure with the myosins of the other classes, containing a head region, a calmodulin/light chain-binding domain, and a tail region. Additionally, class IX myosins carry some unique features, including a large N-terminal extension preceding the head domain and a long insertion within the head domain in loop 2. The tail region comprises a C1 zinc-binding domain and a RhoGAP domain. Because of this RhoGAP domain, class IX myosins are involved in signal transduction regulating the dynamics of the actin cytoskeleton (2, 5).
Mammalian Myo9b, the only class IX myosin studied so far in vitro, exhibits unique mechano-chemical properties. It has been reported to take multiple successive steps along actin filaments without dissociating, indicating that it is a processive motor (6–8). This is remarkable because Myo9b is a single-headed myosin. Other myosins that move processively on actin filaments, such as myosin V, dimeric myosin VI, and myosin VII, are two-headed myosins, and their processivity can be explained by a hand-over-hand mechanism in which the two heads bind coordinately to actin filaments (9–18). However, this mechanism cannot account for the processivity of a single-headed Myo9. It has been proposed that the large insertion in loop 2 of the Myo9b motor domain acts as an actin tether that prevents dissociation from the actin filament in the weak binding states during the processive movement (6–8, 19–21). Indeed, Myo9b binds to F-actin with a relatively high affinity in the ATP-bound state that for other myosins represents a weak actin-binding state (20, 22, 23). The critical importance of the unique insertion for the binding of Myo9b to F-actin has been further demonstrated by the findings that deletion of the insertion lowers F-actin affinity considerably and that the isolated loop 2 insertion of Myo9b binds to F-actin with high affinity (21, 22). In the two-headed processive motors the release of ADP is rate-limiting so that these myosins spend most of their cycling time in the high F-actin affinity ADP-bound state. The rate-limiting step of the Myo9b ATPase cycle is unlike in any other characterized myosin ATP hydrolysis (20, 22). Myosins in the ATP-bound state typically exhibit a weak affinity for F-actin, and this would not be compatible with processive movement.
Currently, the direction of movement of Myo9 along the polar actin filaments is controversial. Native Myo9b immunoadsorbed from human leukocyte extracts moved toward the plus-end of the actin filament (24). However, a truncated recombinant Myo9b was reported to move toward the minus-end of the actin filament (6). The reason(s) for this difference in Myo9b directionality is not known.
To get a better understanding of the motor properties of class IX myosins, one has to rely on recombinant proteins. However, the production of active recombinant mammalian class IX myosin motors has proven to be difficult. Therefore, we set out to study Myo9 (HUM-7) from Caenorhabditis elegans. Invertebrates contain a single class IX myosin that is more closely related to mammalian Myo9b than Myo9a (1). Currently, it is not known how well motor properties are conserved within class IX. For other myosins it has been noted that even within a given class different members can have quite distinct motor properties.
In the other classes of myosins the light chain-binding domain serves as a lever arm determining step size and hence velocity of movement and processivity. Furthermore, its orientation determines directionality of movement. To analyze the function of the light chain-binding region in the potentially single-headed processive class IX myosins, we studied the mechano-chemical properties of constructs with and without the light chain-binding domain. Here, we report that Myo9 is a plus-end-directed motor and shows characteristics of a processive motor irrespective of the presence or absence of the light chain-binding domain. Furthermore, we identified a calmodulin-binding site in the extended loop 2 of the Myo9-head that is unique to class IX myosins.
EXPERIMENTAL PROCEDURES
Construction of Plasmids and Generation of Recombinant Baculoviruses
Total RNA was isolated from C. elegans, and a cDNA fragment encoding amino acids 1–1043 of C. elegans Myo9 was obtained using reverse transcription-PCR and the oligonucleotides 5′-CGGGATCCATGTCATTTGACTCAATATCAGCTGG-3′ and 5′-CGGGGCCCAATCTCGCCGACTCGCTTGCGAACC-3′. The C terminus was modified by the addition of a nucleotide sequence encoding an Avi tag peptide (GLNDIFEAQKIEWHE) for site-specific biotination and a FLAG tag peptide (DYKDDDDK) for purification. The construct was subcloned into the transfer vector pFastBacTM1 (Invitrogen). A similar strategy was applied to obtain the Myo9-head construct encompassing amino acids 1–959 of C. elegans Myo9 followed by three alanine residues, the 15 residues Avi tag, three glycine residues, and the FLAG tag. Recombinant baculovirus DNA was generated by the Bac-to-Bac® method as described previously and transfected into Spodoptera frugiperda (Sf9) cells (21). Individual viruses were isolated by end point dilution and then amplified three times. Final virus titers were determined before infection of Sf9 cells for protein expression. The generation of recombinant baculovirus encoding rat calmodulin was described previously (22).
Protein Expression, Purification, and Biotination
400 ml of Sf9 cells (1 × 106 cells ml−1) cultured in Grace's medium with 10% fetal calf serum were co-infected with the Myo9-head or Myo9-head-4IQ and rat calmodulin recombinant baculoviruses at a multiplicity of infection of 4 for the Myo9 viruses and 8 for the calmodulin virus. Infected Sf9 cells were collected after 48–60 h and washed once with phosphate-buffered saline. The consecutive steps were performed at 4 °C. Cells were resuspended in 40 ml of lysis buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 2 mm MgCl2, 1 mm EGTA, 10% glycerol, 1 mm dithiothreitol, 2 mm ATP, 0.1 mg ml−1 Pefabloc, 0.01 mg ml−1 leupeptin, 0.02 unit ml−1 aprotinin) and lysed by sonication (three times 30 pulses at 100% and a frequency of 0.6). The homogenate was clarified by centrifugation at 45,000 × g for 15 min and recentrifugation at 170,000 × g for 40 min. Occasionally, 5 μg ml−1 calmodulin was added to the cleared lysate before it was loaded onto a column of 0.4 ml of preequilibrated anti-FLAG M2 affinity-agarose (Sigma-Aldrich). After passing it over the column four times, the column was washed twice with 5 ml of lysis buffer and twice with 5 ml of assay buffer (20 mm Hepes, pH 7.4, 50 mm KCl, 2 mm MgCl2, 1 mm EGTA, 10% glycerol, 1 mm dithiothreitol). The myosin was eluted with 0.05 mg ml−1 soluble FLAG peptide (Sigma-Aldrich) in assay buffer.
BirA ligase was prepared as described by Howarth and Ting (25). Biotination of Myo9 constructs was performed while they were immobilized on the FLAG-agarose. After loading the lysate onto the column, the column was washed twice with 5 ml of lysis buffer and once with 6 ml of 100 mm potassium phosphate buffer, pH 7.4. The Myo9 constructs were biotinated with 10 μm BirA ligase, 20 μm biotin, 2 mm ATP, and 100 mm potassium phosphate, pH 7.4, at 4 °C for 2 h or at room temperature for 1 h. Free BirA ligase, biotin, and ATP were removed by washing the column twice with 5 ml of lysis buffer and twice with 5 ml of assay buffer. The biotinated myosin was eluted as described above. Alternatively, biotination of Myo9 constructs was achieved in vivo by coexpressing Myo9 constructs, calmodulin, and BirA ligase in Sf9 cells with 0.2 mg ml−1 biotin in the Grace's medium. The purification of biotinated Myo9 constructs was performed as described above.
Protein concentrations were determined by the Bradford assay using BSA as a standard. Purified proteins were stored at 4 °C and used within 2 days.
Myosin II HMM was prepared according to Margossian and Lowey (26). Rat calmodulin was expressed in Escherichia coli and purified as described before (22). Rabbit skeletal muscle actin was prepared according to Pardee and Spudich (27). G-actin concentration was determined by absorption at 290 nm (ϵ = 26,600 m−1 cm−1) (28). G-actin was polymerized in 10 mm Hepes, pH 7.4, 50 mm KCl, 2 mm MgCl2, 2 mm NaN3, and 1 mm β-mercaptoethanol. Inactivated N-ethylmaleimide-HMM was prepared according to Meeusen and Cande (29).
Mass Spectrometry
Purified Myo9-head was separated by SDS-PAGE and stained with Coomassie Blue. The 17-kDa protein band was excised and subjected to tryptic digestion as described previously (30). Liquid chromatography-tandem mass spectrometry analysis was carried out essentially as described in Naumann et al. (31) with the following modifications. Nano-liquid chromatography was performed on an Ultimate 3000 system (Dionex, Sunnyvale, CA) using the same solvent gradients as described (31). Mass spectrometry was done on an LTQ Orbitrap XL (Thermo, Bremen, Germany) mass spectrometer. For peptide analysis the “big5” method was used performing a full scan first and then five consecutive tandem mass spectrometry fragmentations on the five most abundant peptide ions determined from the full scan. Protein identification was performed using the SEQUEST software (Thermo). Identifications were considered significant if they passed the following criteria: XCorr charge 1, ≥ 1.75; XCorr charge 2, ≥ 2.5; XCorr charge 3, ≥ 3.5; and additionally, two distinct peptides per protein have to be identified.
Calmodulin Overlay
GST fusion proteins encompassing residues 1–168, 708–828, 737–778, and 774–828 of C. elegans Myo9 were amplified by PCR and cloned into pGEX4T-1. In the fusion proteins encoding amino acids 708–828, arginine residues 716 and 721 were mutated to glutamine residues, tryptophan residue 717 and phenylalanine residue 730 to serine residues, respectively, using QuikChange mutagenesis. Peptides 712–731, 712–731Q716/Q721, and 719–739 were generated by primer annealing and insertion into pGEX4T-1. The GST fusion proteins were expressed in E. coli, and cell homogenates were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were either incubated with a rabbit anti-GST antibody (Sigma-Aldrich) followed by a secondary antibody coupled to horseradish peroxidase or with biotinated bovine brain calmodulin (Calbiochem) followed by streptavidin-horseradish peroxidase in the presence of 0.1 mm CaCl2. The membranes were incubated for 45 min with TBST (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 0.05% Tween 20, and 0.1 mm CaCl2) and 5% nonfat dry milk and then overnight at 4 °C in TBST and 5% nonfat dry milk supplemented with biotinated calmodulin (400 ng ml−1). After three 10-min washes with TBST, the membranes were further incubated for 45 min with streptavidin-horseradish peroxidase in TBST before being washed again with TBST (3 × 10 min). The calmodulin-binding peptides were detected with Supersignal West Pico chemiluminescence substrate solution (Thermo) and recorded using the chemiluminescence reader Fujifilm LAS-1000 Intelligent Dark Box (Raytest/Fujifilm).
ATPase Assays
Purified Myo9 constructs were cleared by ultracentrifugation (150,000 × g for 10 min) immediately before use. Steady-state Mg2+-ATPase activities were determined by the NADH-coupled assay. The assays were performed at 20 °C either in the absence or presence of 50 μm free Ca2+ in assay buffer including 10 μm exogenous calmodulin to saturate free light chain-binding sites (also for the Myo9-head construct) and 0.2 mm NADH, 2 mm phosphoenolpyruvate, 3.3 units ml−1 lactate dehydrogenase, 2.3 units ml−1 pyruvate kinase, various actin concentrations (0–20 μm), and 0.1–0.2 μm Myo9-head or 0.4–0.5 μm Myo9-head-4IQ. Assays were started by the addition of 2 mm ATP, and Pi-dependent reduction of NADH concentration was followed photometrically at 340 nm for 10 min. The Vmax and Kactin values were determined by fitting the data to the Michaelis-Menten equation.
Preparation of Dual-fluorescence-labeled Actin Filaments
Dual-fluorescence-labeled actin filaments were prepared according to Herm-Götz et al. (32) with small modifications. Briefly, G-actin labeled with Alexa Fluor 488-maleimide (Invitrogen) was mixed with gelsolin (a generous gift from Setsuko Fujita-Becker, Heidelberg) at a ratio of 165:1 (mol/mol) and incubated on ice for 20 min. Actin polymerization was initiated by adding 100 mm KCl at room temperature. After a 10-min incubation, the resulting F-actin seeds were stabilized by the addition of equimolar amounts of unlabeled phalloidin. Because the plus-end was blocked by gelsolin, F-actin elongation only occurred at the minus-end. For elongation, a 10-fold excess of unlabeled G-actin was added to gelsolin-capped seeds and first incubated on ice for 5 min before elongation was induced for 10 min at room temperature by adding 100 mm KCl. Actin filaments were stabilized by the addition of equimolar amounts of tetramethyl rhodamine isothiocyanate-phalloidin to added G-actin and stored overnight at 4 °C. The dual-fluorescence-labeled actin filaments were diluted to a final actin concentration of 5–10 nm immediately before use.
In Vitro Motility Assay
Gliding filament assays were preformed at room temperature, basically as described by Toyoshima et al. (33) but with some modifications. A flow cell was made from a glass slide and a coverslip, separated by two strips of double-sided tape. The glass slide was precoated with 0.5 mg ml−1 BSA and the coverslip with nitrocellulose. All reagents were prepared in assay buffer (AB) containing 25 mm imidazole, pH 7.4, 25 mm KCl, 4 mm MgCl2, 10 mm dithiothreitol, and 1 mm EGTA or an EGTA-Ca2+ buffer system to give 0.1 mm free Ca2+ concentration. Reagents were introduced in 30-μl volumes into the flow cell except for the solution containing myosin that was added in a volume of 10 μl. For Myo9 constructs, reagents were introduced in the following order: 0.5 mg ml−1 biotinated BSA (Sigma-Aldrich), 0.1% pluronic, 0.5 mg ml−1 streptavidin, the indicated amounts of myosin, 6.5 nm fluorescently labeled F-actin, AB buffer, and AB buffer supplemented with 2 mm ATP and 0.5% methylcellulose. Except for biotinated BSA, pluronic, and streptavidin, the AB buffer contained additionally 10 μm calmodulin and an oxygen-scavenging system (2.5 mg ml−1 glucose, 100 μg ml−1 glucose oxidase, 20 μg ml−1 catalase). Surface myosin densities given assume that every molecule introduced into the flow cell was adsorbed to the surface of the coverslip and that none got denatured. For gliding assays with myosin II HMM, reagents were introduced into the chamber as follows: 0.03–0.06 μg μl−1 myosin II HMM, 0.5 mg ml−1 BSA, 2 μm unlabeled F-actin, AB buffer with 2 mm ATP, AB buffer, 6.5 nm fluorescence-labeled actin, AB buffer, and finally AB buffer supplemented with 20 μm ATP and 0.5% methylcellulose. All solutions were introduced into the flow cell for 2 min except for the AB washes. Images were recorded every 10 s. Filaments moving continuously for at least three frames were used to calculate the gliding velocities. Velocities of individual filaments were determined with the Retrac program.
Quantum Dot Stepping Assay
Flow cells were prepared as described for the in vitro motility assay. First, 0.9 mg ml−1 N-ethylmaleimide-HMM was introduced and incubated for 2 min. Then, the surface was blocked by the addition of 1% pluronic. Tetramethyl rhodamine isothiocyanate-phalloidin-labeled F-actin (6.5 nm) was introduced, and flow cells were washed immediately with AB buffer. Biotinated Myo9-head was preincubated with 20 nm streptavidin-coated quantum dots (qdots) 525 (Invitrogen) in a molar ratio of 2 or 4 qdots/Myo9-head for 3 min in the presence of 2 mm ATP. At a mixing ratio of 2 qdots/Myo9-head ∼91% of qdots do not carry more than one Myo9-head. At a ratio of 4:1, this number increases to ∼97%. These percentages were calculated using a binomial distribution. After introduction of this premix into the flow cell, images were recorded every 5 s. The characteristic run length λ of qdots moving along actin filaments was determined by fitting of the function P(x) = a e−x/λ with P(x) being the probability of a qdot moving a distance x along the filament.
RESULTS
Expression and Purification of Myo9 Constructs
Sequencing of the C. elegans Myo9 cDNA that was obtained by reverse transcription-PCR revealed that it differed from the HUM-7 amino acid sequence at two positions. Residues 608–615 (VSPISPFW) were missing, and residues 731KKSAG735 were replaced by 731KSESAG736. Our deduced Myo9 amino acid sequence changes matched perfectly with the predicted Myo9 sequences for Caenorhabditis briggsae and Caenorhabditis remanei (1). An alignment of this sequence with those of the previously characterized rat and human Myo9b is shown in supplemental Fig. S1.
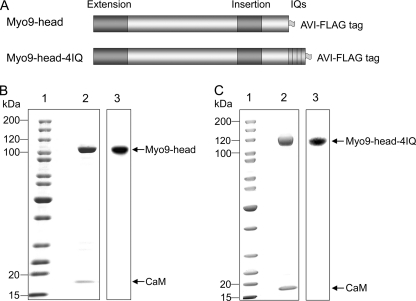
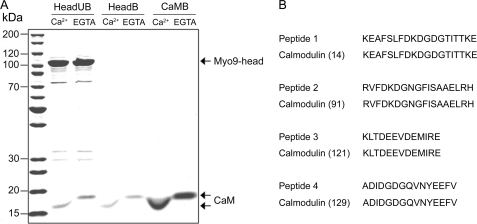
We expressed two different motor constructs of C. elegans Myo9, Myo9-head and Myo9-head-4IQ, together with calmodulin in Sf9 insect cells using a baculovirus expression system (Fig. 1A). The FLAG-tagged proteins were affinity-purified (Fig. 1, B and C). Purifications yielded 100–250 μg of Myo9-head and 100–150 μg of Myo9-head-4IQ from 400 ml of Sf9 cells. Both constructs could be quantitatively biotinated (Fig. 1, B and C). Surprisingly, a protein co-migrating on SDS-PAGE with calmodulin was detected not only in preparations of Myo9-head-4IQ, but also in preparations of the Myo9-head. Similar to authentic calmodulin, the co-purified 17-kDa protein was resistant to boiling and demonstrated a calcium-dependent shift in electrophoretic mobility (Fig. 2). Additional proof that this 17-kDa protein represents calmodulin was obtained by mass spectrometry. Derived peptides matched exactly to peptides of calmodulin (Fig. 2). Taken together, these results demonstrate that calmodulin is co-purified with the Myo9-head. The stoichiometry of calmodulin associated with Myo9-head was determined to be 1.1 ± 0.1 (means ± S.E.). By contrast, the purified Myo9-head-4IQ construct was associated with 3.8 ± 0.3 (means ± S.E.) molecules of calmodulin.
FIGURE 1.
Design and purification of C. elegans Myo9 constructs. A, schematic representation of the expressed C. elegans Myo9 constructs Myo9-head and Myo9-head-4IQ. B and C, purified and biotinated Myo9-head and Myo9-head-4IQ proteins. Purified proteins were separated by SDS-PAGE and either stained with Coomassie Blue (lanes 2) or transferred to a membrane and visualized with streptavidin-horseradish peroxidase (lanes 3). Lane 1, molecular mass markers; the positions of the heavy chains Myo9-head, Myo9-head-4IQ, and the calmodulin light chain (CaM) are indicated.
FIGURE 2.
Calmodulin co-purifies with the Myo9-head. A, purified Myo9-head and authentic calmodulin either boiled for 5 min or not and centrifuged. The supernatants of unboiled Myo9-head (HeadUB), boiled Myo9-head (HeadB), and authentic boiled calmodulin (CaMB) were separated by SDS-PAGE in standard sample buffers containing either 5 mm Ca2+ or 2 mm EGTA. The positions of Myo9-head heavy chain and calmodulin (CaM) are indicated. B, comparison of the peptide masses obtained from the 17-kDa protein that was co-purified with Myo9-head and peptides from rat calmodulin. Numbers in parentheses indicate the positions of the first displayed amino acid in the calmodulin sequence.
Conserved N-terminal Peptide in Extended Loop 2 Binds Calmodulin
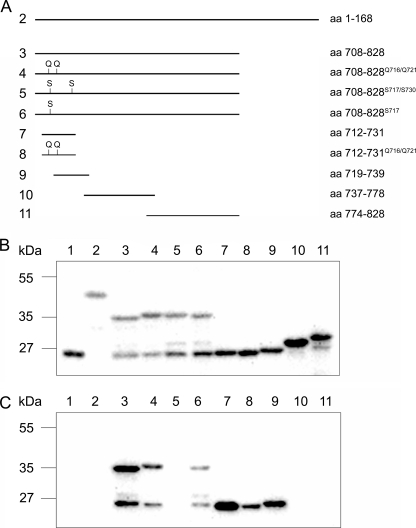
We reasoned that class IX-specific head sequences were the most likely candidates for binding calmodulin. Therefore, we expressed the N-terminal extension and different fragments of the extended loop 2 (insertion) as GST fusion proteins in E. coli and performed calmodulin overlay assays. As shown in Fig. 3, calmodulin bound to the insertion but not to the N-terminal extension. A double mutation of two highly conserved arginine residues in the N-terminal region of the insertion to glutamines (R716Q and R721Q) significantly reduced the amount of bound calmodulin. Point mutations of a highly conserved tryptophan and phenylalanine residue in the N-terminal region of the insertion to serine residues (W717S and F730S) completely abolished the binding of calmodulin. Mutation of the tryptophan residue (W717S) alone significantly decreased the amount of bound calmodulin. Furthermore, a systematic fragmentation of the insertion revealed that calmodulin solely binds to the N terminus of the insertion. The short peptide encompassing amino acids 712–731 at the N terminus bound calmodulin potently, and the interaction was significantly inhibited upon mutation of the arginine residues 716 and 721 to glutamine residues. An overlapping peptide shifted by 7 residues encoding amino acid residues 719–739 also demonstrated a clearly reduced affinity for calmodulin. These results indicate that residues 712–731 comprise the calmodulin-binding site and that residues 716/721, 717, and 730 contribute to the binding of calmodulin. These residues are highly conserved within class IX myosins (supplemental Fig. S2), suggesting that all class IX myosins have this calmodulin-binding site in common.
FIGURE 3.
N-terminal region of Myo9 loop 2 binds calmodulin. A, schematic overview of Myo9-head fragments that were fused to GST and expressed in E. coli. B and C, cell homogenates separated on SDS-PAGE, transferred to polyvinylidene difluoride membrane, and either incubated with anti-GST antibody (B) or biotinated calmodulin in the presence of 0.1 mm CaCl2 (C). Binding was visualized by chemiluminescence using either secondary antibodies or streptavidin that was coupled to horseradish peroxidase. Lane 1, GST; lanes 2–11, fusion proteins indicated in A.
Ca2+ Regulates Steady-state F-actin-activated ATPase Activity
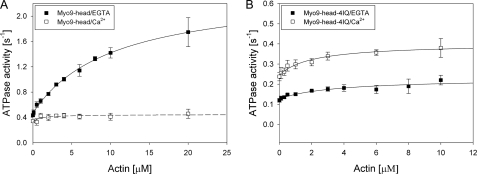
Steady-state F-actin-activated Mg2+-ATPase activity was determined for the Myo9-head and the Myo9-head-4IQ in the absence and presence of 50 μm free Ca2+ at 20 °C using a NADH-coupled assay (Fig. 4 and Table 1). In the absence of F-actin, the Myo9-head had a basal ATPase activity of 0.47 ± 0.01 s−1. The ATPase activity was increased 5-fold by F-actin to a Vmax of 2.40 ± 0.10 s−1. The deduced Kactin was 10.38 ± 1.03 μm, and the second-order rate constant for F-actin binding (kapp) was 0.19 μm−1 s−1. The Myo9-head-4IQ construct displayed a lower basal ATPase activity of 0.13 ± 0.01 s−1 that was just slightly activated by F-actin to a Vmax of 0.23 ± 0.03 s−1. The determined Kactin for the Myo9-head-4IQ construct was 3.55 ± 2.27 μm and the kapp 0.03 μm−1 s−1. These results suggest that both the basal and F-actin-activated ATPase activities of Myo9 are negatively regulated by the light chain-binding domain.
FIGURE 4.
F-actin-activated Mg2+-ATPase activity of Myo9 constructs is sensitive to calcium. The steady-state ATPase activities of the Myo9-head (0.1–0.2 μm) (A) and the Myo9-head-4IQ (0.4–0.5 μm) (B) were measured in the absence (filled squares) or presence (open squares) of 50 μm free Ca2+ in assay buffer containing 20 mm Hepes, pH 7.4, 50 mm KCl, 2 mm MgCl2, 1 mm EGTA, 10% glycerol, 1 mm dithiothreitol, 10 μm exogenous calmodulin, and 0–20 μm F-actin at 20 °C using a NADH-coupled assay. Data are the means from three or four independent preparations, and the error bars indicate S.E. The data points were fitted with a hyperbola. Please note the different scales for the ordinates in A and B.
TABLE 1.
ATPase activity and gliding velocity of C. elegans Myo9 constructs
Mg2+-ATPase activity was determined at 20 °C in the absence and presence of 50 μm free Ca2+. The gliding velocity of actin filaments was determined at room temperature in the absence and presence of 0.1 mm free Ca2+. The errors presented are standard errors calculated from the results of repeated experiments of 3 or 4 independent preparations for the ATPase assay and >7 independent preparations for the actin gliding assay. Dash (–) indicates that no motility was detected.
| Basal | Vmax | Kactin | kapp | Velocity | |
|---|---|---|---|---|---|
| s−1 | s−1 | μm | μm−1s−1 | nm s−1 | |
| Head/EGTA | 0.47 ± 0.01 | 2.40 ± 0.10 | 10.38 ± 1.03 | 0.19 | 23.81 ± 3.02 |
| Head/Ca2+ | 0.33 ± 0.02 | 0.45 ± 0.05 | 1.08 ± 1.03 | 0.10 | – |
| Head-4IQ/EGTA | 0.13 ± 0.01 | 0.23 ± 0.03 | 3.55 ± 2.27 | 0.03 | 24.44 ± 4.17 |
| Head-4IQ/Ca2+ | 0.25 ± 0.01 | 0.40 ± 0.02 | 2.06 ± 0.69 | 0.08 | 60.07 ± 15.75 |
Next, we tested the effect of Ca2+ binding to calmodulin associated with the head and the light chain-binding domain on the ATPase activity. In the presence of free Ca2+, the Myo9-head exhibited a basal ATPase activity (0.33 ± 0.02 s−1) comparable with that in the absence of free Ca2+ (0.47 ± 0.01 s−1), but it demonstrated a 5-fold reduced maximal F-actin-activated ATPase activity (0.45 ± 0.05 s−1). The deduced Kactin was 1.08 ± 1.03 μm, and the kapp 0.10 μm−1 s−1. The basal ATPase activity of the Myo9-head-4IQ construct was slightly increased in the presence of free Ca2+ to 0.25 ± 0.01 s−1 (from 0.13 s−1). The activation of the ATPase activity by F-actin was unchanged, and the Vmax of this construct was somewhat higher because of the increased basal activity (0.40 ± 0.02 s−1). Hence, in the presence of free Ca2+, the Vmax was comparable for the two constructs (Fig. 4 and Table 1). The Kactin for the Myo9-head-4IQ was 2.06 ± 0.69 μm, slightly lower compared with the Kactin determined under EGTA conditions. The calculated kapp with 0.08 μm−1 s−1 was 3-fold higher than under EGTA conditions. These results show that free Ca2+ regulates ATPase activities differentially in the two constructs and that the binding of calcium to the calmodulin associated with the Myo9-head significantly affects the F-actin-activated ATPase activity.
Ca2+ Regulates Mechanochemical Activity of Myo9
We determined the mechanochemical activity of truncated Myo9 constructs in the in vitro actin gliding assay. To observe directional actin filament gliding, it was necessary to attach the motors site-specifically to the surface. This was achieved via the C-terminal Avi tags that could be biotinated either in vitro or in vivo with BirA ligase. The biotination efficiency was ∼90% (data not shown). Both Myo9-head and Myo9-head-4IQ supported robust movement of actin filaments upon specific biotin-streptavidin linkage. More than 80% of the filaments moved on surfaces coated with Myo9-head (supplemental Movie 1), whereas about 40% of the filaments moved on the surfaces coated with Myo9-head-4IQ (supplemental Movie 2). The gliding velocities of the actin filaments varied substantially from preparation to preparation. The mean gliding velocities were determined to be 23.81 ± 3.02 nm s−1 for the Myo9-head and 24.44 ± 4.17 nm s−1 for the Myo9-head-4IQ. However, some preparations yielded clearly faster velocities, and when purified simultaneously, the Myo9-head-4IQ construct supported higher velocities than the Myo9-head construct (Fig. 5). This result suggests that the light chain-binding domain may serve as a lever arm in class IX myosins increasing step size. Myo9-head-4IQ supported equal or faster movement than Myo9-head, although its measured ATPase rate was significantly slower.
FIGURE 5.
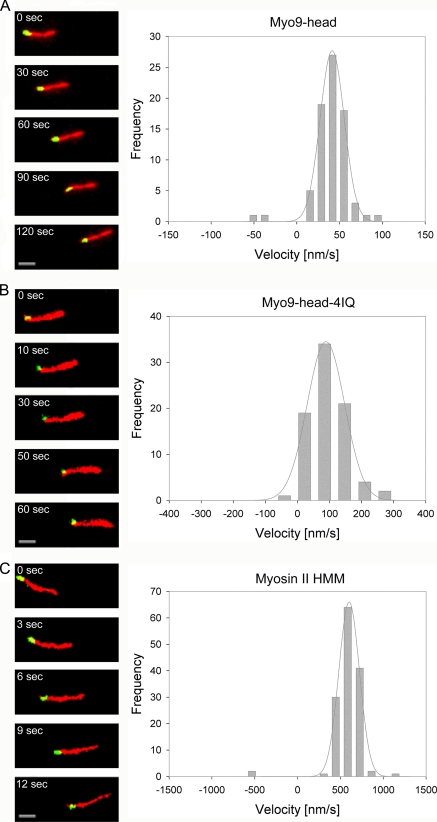
Myo9 moves to the plus-end of actin filaments. Movement of dual-fluorescence-labeled actin filaments was supported by Myo9-head (A), Myo9-head-4IQ (B), and myosin II HMM (C). Individual frames from time-lapse movies are shown on the left. Elapsed time (seconds) is indicated in each panel. Scale bars, 2 μm. The green tip marks the plus-end of an actin filament. Myo9-head, Myo9-head-4IQ, and myosin II HMM move filaments with their green tips trailing, indicating that both Myo9 and myosin II are plus-end-directed motors (see also supplemental movies). Histograms on the right display the number of filaments moving with the plus-end trailing (positive velocities) and leading (negative velocities), respectively. The histograms were fitted with a Gaussian function (gray lines).
To test the effect of Ca2+ on the mechanochemical activity of the two Myo9 constructs, the in vitro actin gliding assay was also performed in the presence of 0.1 mm free Ca2+. Consistent with the ATPase activity, the Myo9-head no longer supported gliding of actin filaments in the presence of free Ca2+. In contrast, the gliding velocity of actin filaments propelled by Myo9-head-4IQ was elevated by Ca2+. The velocity increased from 24.44 ± 4.17 nm s−1 to 60.07 ± 15.75 nm s−1. The Myo9-head-4IQ supported more robust movement in the presence than in the absence of Ca2+. The changes in velocity were observed independently of whether chambers were initially prepared with 0.1 mm Ca2+ or chambers containing EGTA were flushed with Ca2+.
C. elegans Myo9 Motor Constructs Move toward Plus-end of Actin Filaments
The direction of movement along actin filaments was determined for the two Myo9 motor constructs that differ in the presence or absence of the light chain-binding domain. For that purpose, polarity-marked actin filaments were created by dual-fluorescence labeling of the filaments (Fig. 5). Most of the filaments exhibited plus-end-directed movement (positive velocity) in the presence of the truncated Myo9 constructs (>95%). As shown in Fig. 5, actin filaments moved on surfaces coated with either biotinated Myo9-head or Myo9-head-4IQ with the minus-ends leading (also see supplemental Movies 3 and 4). The uniformity of the polarity-marked actin filaments was verified by using myosin II HMM, an established plus-end-directed motor (Fig. 5 and supplemental Movie 5). To reduce the velocity of actin gliding driven by myosin II HMM, the ATP concentration was decreased to 20 μm. This resulted in a gliding velocity of 601.8 ± 3.9 nm s−1 and a mostly plus-end-directed movement proving the quality of the dual labeling of the filaments. The few actin filaments that appeared to support minus-end-directed movements were likely mislabeled because of the lack of a gelsolin cap allowing either polymerization at the plus-end or annealing with the minus-end of another filament.
Myo9 Head Construct Demonstrates Characteristics of Processive Movement
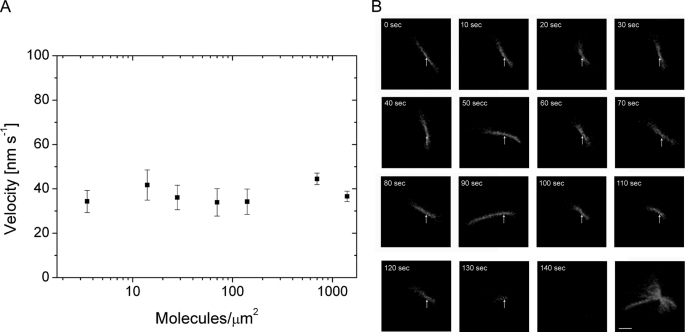
To study whether Myo9 is a processive motor, actin gliding assays were performed over a range of Myo9-head surface densities. Myo9-head supported continuous movement of actin filaments from a high density (>103 molecule μm−2) to an extremely low density (4 molecules μm−2), and the velocity of actin gliding did not change with different surface densities of Myo9-head (Fig. 6A). These are characteristics typical of a processive motor. Another indication for processivity of the Myo9-head was that at low surface densities the moving actin filaments exhibited nodal pivoting (Fig. 6B and supplemental Movie 6). At very low myosin densities, actin filaments attached to the surface with a single contact point. During movement the actin filaments swiveled around the contact point, indicating that they were tethered to the surface by a single Myo9-head molecule. When the end of the actin filament reached the nodal point, the filament diffused away from the surface as shown in Fig. 6B. This result supports the notion that the Myo9-head is a processive motor.
FIGURE 6.
Myo9-head exhibits characteristics of a processive motor. A, gliding velocity of actin filaments does not change as a function of Myo9-head density. The surfaces were coated with different concentrations of biotinated Myo9-head. The gliding velocities of actin filaments (n = 5–40) were quantified, and the average velocities (nm s−1) are represented as means ± S.E. on the y axis. B, nodal pivoting of an actin filament at low Myo9-head densities is shown. An actin filament is pivoting around a single contact point (arrow) while it moves unidirectionally. As the end of the actin filament passes the nodal point, the filament diffuses away from the surface (2nd to last panel). The last panel is the overlay of all the frames shown. Elapsed time (seconds) is indicated in each panel. Scale bar, 2 μm.
Movement of Quantum Dot-labeled Myo9 Head along Actin Filaments
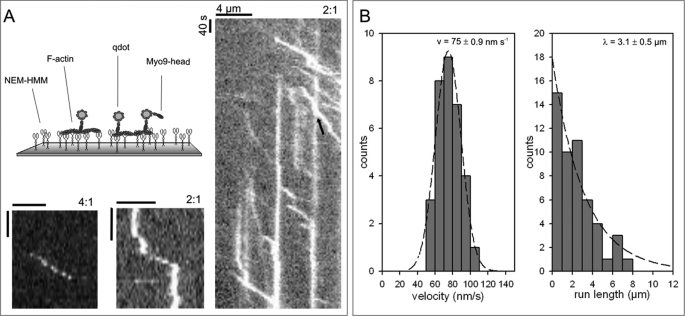
The actin gliding assay visualizes motor activity indirectly by monitoring the movement of the actin filaments rather than the motor molecules themselves. To observe the movement of Myo9 along F-actin directly, we established a myosin stepping assay (Fig. 7). We labeled biotinated Myo9-head with fluorescent streptavidin-coated qdots in a molar ratio of 2 qdots/Myo9-head. When this mixture was introduced into a flow cell with immobilized actin filaments, single qdots could be observed moving along the actin filaments with considerable run lengths of up to 8 μm and a characteristic run length λ of 3.1 ± 0.5 μm (Fig. 7B). Even at a mixing ratio of 4 qdots/Myo9-head we were still able to detect qdot movement for 4.5 μm (Fig. 7A). The average velocity of moving qdots was 75 ± 0.9 nm s−1 (Fig. 7B). This was faster compared with the mean actin filament gliding velocity of 23.81 ± 3.02 nm s−1. We noted several different modes of Myo9 movement (Fig. 7A): qdots (i) associated, moved, and dissociated; (ii) moved, paused, and moved again; (iii) moved and stopped. The observed stopping of qdots on the track is noteworthy. It might be explained by (i) an inactive motor molecule that is additionally bound to the qdot; (ii) the motor molecule entering an immotile, actin-associated state; or (iii) the qdot itself binding unspecifically to the actin filament. Irrespective of the explanation, these immotile qdots served as obstacles and enabled us to analyze how obstacles affect the Myo9-head-driven motility of qdots (Fig. 7A, kymograph representing a “high traffic” situation) (supplemental Movie 7). In most of the cases when a qdot reached an obstacle, it stopped but did not dissociate. In some cases, we were able to detect stop-and-go events with delay times ranging from 20 to 45 s, meaning that Myo9-head qdots successfully passed an obstacle that was attached to the actin filament.
FIGURE 7.
Movement of qdot-labeled Myo9-head along actin filaments. A, biotinated Myo9-head was mixed with streptavidin-coated qdots at a ratio of 2:1 or 4:1 (qdot/Myo9-head) as indicated. Qdot-labeled Myo9-head was added to immobilized actin filaments as shown in the schematic illustration, and images were recorded every 5 s. Exemplary space-time plots (kymographs) of low traffic (left) and high traffic (right) situations are shown. Arrowhead indicates a qdot passing an obstacle. Scale bars represent 4 μm and 40 s, respectively. B, histograms of run length and velocity are plotted. Velocities fit a Gaussian distribution with a mean of 75 ± 0.9 nm s−1, and the average run length was determined to be 3.1 ± 0.5 μm. Data are derived from six experiments from at least three different preparations.
DISCUSSION
By cloning and expressing Myo9 from C. elegans we were able to characterize the motor properties of the head region of a class IX myosin. Although the head construct contained no calmodulin-binding IQ-motifs, calmodulin was co-purified in roughly stoichiometric amounts together with the Myo9-head. The novel calmodulin-binding site was mapped to a peptide sequence in the N-terminal region of loop 2 (50/20-kDa junction) that conforms to the 1-8-14 subclass of calmodulin-binding domains (34). Mutation of the two residues Trp717 and Phe730 abolished the binding of calmodulin. This is in agreement with the notion that these two residues serve as anchors for the C- and N-terminal domains of calmodulin, respectively (35, 36). The identified calmodulin-binding sequence is highly conserved among the class IX myosins and present in class IX myosins only. Therefore, it is predicted that calmodulin binds to the head regions of all class IX myosins. Indeed, the head region of rat Myo9b was also observed to bind calmodulin.3 This calmodulin serves a regulatory role as the binding of Ca2+ to the Myo9-head virtually abolished activation of the ATPase activity by F-actin and prevented the gliding of actin filaments. How apocalmodulin influences the motor properties of the class IX myosins remains to be determined and will be addressed in future work. It could e.g. influence the interaction with F-actin, as the loop 2 in rat Myo9b has been shown to regulate the interaction with actin filaments (21). In class VI myosin, calmodulin was found to be associated with a unique insert between the converter and the IQ-motif that repositions the lever arm and reverses directional movement of myosin VI toward the minus-end of actin filaments (37, 38). A human recombinant Myo9b construct that was truncated in the tail region was reported to move toward the minus-end of actin filaments (6). By contrast, native full-length Myo9b was reported to be a plus-, and not a minus-end directed motor, suggesting that the tail domain of Myo9b regulates motor directionality (24). However, in the present study we found that both the isolated head region and the head region including the light chain-binding domain of C. elegans Myo9 moved toward the plus-end of actin filaments, although they were missing the tail region. Therefore, we propose that class IX myosins are plus-end-directed motors independently of the presence or absence of the tail region. Furthermore, the extended loop 2 and the binding of calmodulin to loop 2 do not cause a reversal of Myo9 directionality.
The purified Myo9-head-4IQ construct contained four calmodulin molecules. Assuming that one calmodulin is associated with loop 2 in the head region, this would imply that one IQ-motif is not saturated with calmodulin. Inspection of the amino acid sequences of the four IQ-motifs revealed that IQ1, IQ2, and IQ4 are predicted to bind both the N- and C-terminal lobes of calmodulin in a compact conformation whereas IQ3, because of the lysine residue at position 7 of the consensus sequence IQXXXRGXXXR, is predicted to bind calmodulin in an extended conformation at the C-terminal lobe only (39). Therefore, IQ3 might have lost its calmodulin molecule during purification. The addition of the light chain-binding region reduced the basal and even more the actin-activated ATPase activity, suggesting that it regulates motor activity. Interestingly, addition of free Ca2+ increased the basal ATPase activity of the Myo9-head-4IQ and accelerated actin filament gliding. A comparison of the steady-state ATPase activity and the velocity in the gliding assay of the C. elegans Myo9-head-4IQ protein with those of analogous or similar mammalian Myo9b constructs revealed a remarkable similarity (6, 20, 21). However, mammalian Myo9b constructs were analyzed in the absence of Ca2+ only.
Making mammalian Myo9b unique is its reported ability to move processively as a single-headed molecule (6–8). It is not known how a single-headed myosin is able to step forward without dissociating. For other myosins it has been demonstrated that the light chain-binding domain serves as a lever arm and that step size is linearly related to lever arm length (40–43). Our present finding demonstrating that the Myo9-head-4IQ construct supports faster actin gliding compared with the Myo9-head construct is in accordance with a lever arm-dependent stepping mechanism for Myo9. Larger steps caused by a longer lever arm will lead to faster gliding of actin filaments. In the double-headed processive myosins Va and myosin VI the lever arm length also influences processivity. A certain length of the lever arm is needed to allow for processive movement (44–46). Interestingly, the Myo9-head exhibited several characteristics typical of a processive motor. In the actin gliding assay the Myo9-head could be diluted to very low concentrations, and the velocity of actin gliding did not change with different concentrations. At high dilutions, actin filaments could be observed pivoting around a single attachment point while they were moving, and they dissociated when the end of the filament passed the attachment point. Quantum dots associated with maximally a few Myo9-head molecules were observed to move along actin filaments over considerable distances with an average run length of 3.1 μm. This run length is comparable with that of the dimeric processive motor myosin Va (47). Continuous movement for such long distances implicates that motility is driven by a coordinated mechanism to prevent detachment and not by independently bound motor molecules. A peculiar behavior of the Myo9-head-coated qdots was that they often attached to actin filaments and did not move until some time later or that they stopped moving without dissociating. Occasionally, we observed that moving qdots were able to maneuver around obstacles in the form of immotile qdots. The ability of passing obstacles on the track has also been reported for kinesin-1 (48). However, in contrast to kinesin-1 we did not observe an increased probability of dissociation from the filament when Myo9-head reached an obstacle. This shows that the mechanism of Myo9 movement offers enough flexibility to deal with the high traffic situation as it is given in a living cell.
Class IX myosins exhibit fascinating motor properties. The molecular mechanism of their movement is still a conundrum, but the results reported here emphasize that a unique mechanism must be operating and represent a starting point for elucidating that mechanism.
Supplementary Material
Acknowledgments
We thank A. Freitag, U. Honnert, M. Müller, P. Nikolaus, and B. Lohmann for skillful technical assistance. We thank A. Karabatsiakis for the isolation of the C. elegans Myo9 cDNA; B. Naumann and M. Hippler of the Institute for Plant Biotechnology and Biochemistry, Westfalian Wilhelms-University, for performing the mass spectrometry analysis; and Dr. U. Pieper for helpful discussions. We thank S. Fujita-Becker (Heidelberg) for the protocol describing the generation of polarity marked actin filaments and for a gift of gelsolin. A. Y. Ting (Boston) provided the cDNA for BirA ligase.
This work was supported by the Deutsche Forschungsgemeinschaft Grant DFG BA 1354/9-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Movies 1–7, and additional references.
G. Kalhammer and M. Bähler, unpublished observations.
- Myo9a and Myo9b
- myosin IXa and IXb, respectively
- AB
- assay buffer
- BSA
- bovine serum albumin
- GST
- glutathione S-transferase
- qdot
- quantum dot
- HMM
- heavy meromyosin.
REFERENCES
- 1.Odronitz F., Kollmar M. (2007) Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bähler M. (2000) Biochim. Biophys. Acta 1496, 52–59 [DOI] [PubMed] [Google Scholar]
- 3.Abouhamed M., Grobe K., San I. V., Thelen S., Honnert U., Balda M. S., Matter K., Bähler M. (2009) Mol. Biol. Cell 20, 5074–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanley P. J., Xu Y., Kronlage M., Grobe K., Schön P., Song J., Sorokin L., Schwab A., Bähler M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12145–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Boom F., Düssmann H., Uhlenbrock K., Abouhamed M., Bähler M. (2007) Mol. Biol. Cell 18, 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue A., Saito J., Ikebe R., Ikebe M. (2002) Nat. Cell Biol. 4, 302–306 [DOI] [PubMed] [Google Scholar]
- 7.Post P. L., Tyska M. J., O'Connell C. B., Johung K., Hayward A., Mooseker M. S. (2002) J. Biol. Chem. 277, 11679–11683 [DOI] [PubMed] [Google Scholar]
- 8.Nishikawa M., Nishikawa S., Inoue A., Iwane A. H., Yanagida T., Ikebe M. (2006) Biochem. Biophys. Res. Commun. 343, 1159–1164 [DOI] [PubMed] [Google Scholar]
- 9.Mehta A. D., Rock R. S., Rief M., Spudich J. A., Mooseker M. S., Cheney R. E. (1999) Nature 400, 590–593 [DOI] [PubMed] [Google Scholar]
- 10.Walker M. L., Burgess S. A., Sellers J. R., Wang F., Hammer J. A., 3rd, Trinick J., Knight P. J. (2000) Nature 405, 804–807 [DOI] [PubMed] [Google Scholar]
- 11.Rock R. S., Rice S. E., Wells A. L., Purcell T. J., Spudich J. A., Sweeney H. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13655–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forkey J. N., Quinlan M. E., Shaw M. A., Corrie J. E., Goldman Y. E. (2003) Nature 422, 399–404 [DOI] [PubMed] [Google Scholar]
- 13.Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003) Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 14.Okten Z., Churchman L. S., Rock R. S., Spudich J. A. (2004) Nat. Struct. Mol. Biol. 11, 884–887 [DOI] [PubMed] [Google Scholar]
- 15.Park H., Li A., Chen L. Q., Houdusse A., Selvin P. R., Sweeney H. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie P., Dou S. X., Wang P. Y. (2006) Biophys. Chem. 122, 90–100 [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Kovács M., Sakamoto T., Zhang F., Kiehart D. P., Sellers J. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5746–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto T., Webb M. R., Forgacs E., White H. D., Sellers J. R. (2008) Nature 455, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinhard J., Scheel A. A., Diekmann D., Hall A., Ruppert C., Bähler M. (1995) EMBO J. 14, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambara T., Ikebe M. (2006) J. Biol. Chem. 281, 4949–4957 [DOI] [PubMed] [Google Scholar]
- 21.Struchholz S., Elfrink K., Pieper U., Kalhammer G., Honnert U., Grützner A., Linke W. A., Liao W., Bähler M. (2009) J. Biol. Chem. 284, 3663–3671 [DOI] [PubMed] [Google Scholar]
- 22.Nalavadi V., Nyitrai M., Bertolini C., Adamek N., Geeves M. A., Bähler M. (2005) J. Biol. Chem. 280, 38957–38968 [DOI] [PubMed] [Google Scholar]
- 23.Post P. L., Bokoch G. M., Mooseker M. S. (1998) J. Cell Sci. 111, 941–950 [DOI] [PubMed] [Google Scholar]
- 24.O'Connell C. B., Mooseker M. S. (2003) Nat. Cell Biol. 5, 171–172 [DOI] [PubMed] [Google Scholar]
- 25.Howarth M., Ting A. Y. (2008) Nat. Protoc. 3, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margossian S. S., Lowey S. (1982) Methods Enzymol. 85, 55–71 [DOI] [PubMed] [Google Scholar]
- 27.Pardee J. D., Spudich J. A. (1982) Methods Cell Biol. 24, 271–289 [DOI] [PubMed] [Google Scholar]
- 28.Houk T. W., Jr., Ue K. (1974) Anal. Biochem. 62, 66–74 [DOI] [PubMed] [Google Scholar]
- 29.Meeusen R. L., Cande W. Z. (1979) J. Cell Biol. 82, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hippler M., Klein J., Fink A., Allinger T., Hoerth P. (2001) Plant J. 28, 595–606 [DOI] [PubMed] [Google Scholar]
- 31.Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 32.Herm-Götz A., Weiss S., Stratmann R., Fujita-Becker S., Ruff C., Meyhöfer E., Soldati T., Manstein D. J., Geeves M. A., Soldati D. (2002) EMBO J. 21, 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyoshima Y. Y., Kron S. J., Spudich J. A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7130–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 35.Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. (1992) Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 36.Simonovic M., Zhang Z., Cianci C. D., Steitz T. A., Morrow J. S. (2006) J. Biol. Chem. 281, 34333–34340 [DOI] [PubMed] [Google Scholar]
- 37.Bahloul A., Chevreux G., Wells A. L., Martin D., Nolt J., Yang Z., Chen L. Q., Potier N., Van Dorsselaer A., Rosenfeld S., Houdusse A., Sweeney H. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4787–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ménétrey J., Bahloul A., Wells A. L., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. (2005) Nature 435, 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terrak M., Rebowski G., Lu R. C., Grabarek Z., Dominguez R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warshaw D. M., Guilford W. H., Freyzon Y., Krementsova E., Palmiter K. A., Tyska M. J., Baker J. E., Trybus K. M. (2000) J. Biol. Chem. 275, 37167–37172 [DOI] [PubMed] [Google Scholar]
- 41.Ruff C., Furch M., Brenner B., Manstein D. J., Meyhöfer E. (2001) Nat. Struct. Biol. 8, 226–229 [DOI] [PubMed] [Google Scholar]
- 42.Purcell T. J., Morris C., Spudich J. A., Sweeney H. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14159–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhler D., Ruff C., Meyhöfer E., Bähler M. (2003) J. Cell Biol. 161, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto T., Wang F., Schmitz S., Xu Y., Xu Q., Molloy J. E., Veigel C., Sellers J. R. (2003) J. Biol. Chem. 278, 29201–29207 [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto T., Yildez A., Selvin P. R., Sellers J. R. (2005) Biochemistry 44, 16203–16210 [DOI] [PubMed] [Google Scholar]
- 46.Liao J. C., Elting M. W., Delp S. L., Spudich J. A., Bryant Z. (2009) J. Mol. Biol. 392, 862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hodges A. R., Krementsova E. B., Trybus K. M. (2007) J. Biol. Chem. 282, 27192–27197 [DOI] [PubMed] [Google Scholar]
- 48.Telley I. A., Bieling P., Surrey T. (2009) Biophys. J. 96, 3341–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.