Abstract
In humans, mutations in fibrillin-1 result in a variety of genetic disorders with distinct clinical phenotypes. While most of the known mutations in fibrillin-1 cause Marfan syndrome, a number of other mutations lead to clinical features unrelated to Marfan syndrome. Pathogenesis of Marfan syndrome is currently thought to be driven by mechanisms due to haploinsufficiency of wild-type fibrillin-1. However, haploinsufficiency-driven mechanisms cannot explain the distinct phenotypes found in other fibrillinopathies. To test the hypothesis that mutations in fibrillin-1 cause disorders through primary effects on microfibril structure, two different mutations were generated in Fbn1 in mice. One mutation leads to a truncated fibrillin-1 molecule that is tagged with green fluorescent protein, allowing visualization of mutant fibrillin-1 incorporated into microfibrils. In heterozygosity, these mutant mice demonstrate progressive fragmentation of the aortic elastic lamellae and also display fragmentation of microfibrils in other tissues. Fibrillin-2 epitopes are also progressively revealed in these mice, suggesting that fibrillin-2 immunoreactivity can serve as a marker for microfibril degradation. In contrast, a second mutation (in-frame deletion of the first hybrid domain) in fibrillin-1 results in stable microfibrils, demonstrating that fibrillin-1 molecules are not required to be in perfect register for microfibril structure and function and that the first hybrid domain is dispensable for microfibril assembly. Taken together, these results suggest that perturbation of microfibril structure may underlie one of the major features of the Marfan syndrome: fragmentation of aortic elastic lamellae.
Keywords: Connective Tissue, Electron Microscopy (EM), Extracellular Matrix, Fibrillin, Genetic Diseases, Mouse Genetics, Marfan Syndrome
Introduction
Fibrillins are ubiquitous extracellular matrix molecules that assemble into microfibrils (1–3) and target growth factors to the extracellular matrix (4–6). More than 1000 mutations have been identified in the gene for fibrillin-1 (FBN1) (7). Most of these mutations cause the Marfan syndrome (OMIM #154700), an autosomal dominant disorder of connective tissue, with major features in the musculoskeletal, cardiovascular, and ocular systems and minor features in the lung and skin.
Mutant fibrillin-1 initiates disease-causing changes in the extracellular matrix by decreasing the level of functional microfibrils (8, 9) and activating transforming growth factor-β signaling (10, 11). Mutant fibrillin-1 may result in a substandard threshold of microfibrils by interfering with the assembly of microfibrils (12, 13). Alternatively, mutant fibrillin-1 may assemble, but incorporation of mutant fibrillin-1 may destabilize microfibrils (14, 15). Cell culture studies, using human Marfan fibroblasts, have shown many different effects of mutations on total fibrillin-1 synthesis and deposition into the extracellular matrix (16). Because fibrillin-1 is such a large molecule and most mutations cause single amino acid changes or small deletions, it has been impossible to distinguish mutant from wild-type fibrillin-1 protein. Therefore, dominant-negative mechanisms by which mutant fibrillin-1 may decrease the level of functional microfibrils have been difficult to determine, using human fibroblast cultures.
Five Fbn1 mutant mouse lines have been described. The mutations in these mouse lines, relationship to human mutations, and phenotypes in heterozygosity and homozygosity are listed in Table 1. These Fbn1 mutant mouse lines have established the threshold requirements for fibrillin-1 in postnatal tissue homeostasis. They include: mgΔ, a hypomorphic deleted allele (17); mgR, a hypomorphic wild-type allele (8); mgN, a null allele (18), and C1039G, a mutated missense allele (9). In homozygosity, mgΔ and mgN mice represent a severe early postnatal form of Marfan syndrome that is due to extreme deficiency of fibrillin-1. Homozygous mgR mice replicate severe disease that manifests during adult life. Heterozygous C1039G mice demonstrate histological features of aortic disease, but live a normal life span and do not manifest severe clinical disease; however, homozygous C1039G mice die during the perinatal period (9). This series of Fbn1 mutant mice indicated that haploinsufficiency for wild-type fibrillin-1 is the critical threshold requirement for tissue homeostasis (9).
TABLE 1.
Comparison of Fbn1 mutant mouse models
| Model (and reference) | Mutation | Human phenotype | Heterozygous mouse phenotype | Homozygous mouse phenotype |
|---|---|---|---|---|
| mgΔ (17) | Deletion of exons 19–24; extreme hypomorph | Not applicable | Grossly normal | Early postnatal death |
| mgR (8) | Low expression of wild-type Fbn1 | Not applicable | Grossly normal | Death from aortic dissection during adulthood |
| mgN (18) | Null allele | Not applicable | Grossly normal | Early postnatal death |
| C1039G (9) | Missense mutation substituting glycine for cysteine | Classic Marfan syndrome | Aortic dilatation and fragmentation of elastic lamellae; skeletal muscle and mitral valve abnormalities | Early postnatal death |
| Tsk (33) | Large in-frame duplication | Not applicable | Tight skin; skeletal abnormalities | Early embryonic death |
| GT-8 (this report) | Truncation, tagged with eGFP | Not applicable | Fragmentation of aortic elastic lamellae | Early postnatal death |
| H1Δ (this report) | Deletion of exon 7 | Not applicable | Grossly normal | Normal life span |
The prediction that mutant fibrillin-1 interferes with the assembly of microfibrils (12, 13) was tested once in mice. Overexpression of mutant human FBN1 showed incorporation of the human fibrillin-1 into microfibril structures, using human specific fibrillin-1 antibodies, but no apparent effects of the mutant human fibrillin-1 were found on microfibril structure or function in these transgenic mice (9). Therefore, it was concluded that haploinsufficiency for wild-type fibrillin-1, rather than production of mutant protein, is the primary determinant of failed microfibrillar assembly (9). However, this experiment may have been unsuccessful, because a much higher level of expression from the mutant human transgene was required to negatively affect microfibril assembly or structure in the presence of two endogenous wild-type Fbn1 alleles.
Available Fbn1 mouse lines targeted by homologous recombination have not fully addressed the fate of the mutant fibrillin-1 protein. Mice in which Fbn1 exons 19–24 were deleted in-frame (mgΔ) were generated to replicate the dominant-negative pathogenic model of Marfan syndrome (17). Mutant fibrillin-1 in homozygous mgΔ mice was shown to assemble microfibrils, indicating that domains encoded by exons 19–24 are unnecessary for microfibril assembly (17). However, because the neomycin resistance gene substantially lowered expression of the mutant allele, it was impossible to conclude whether or not mutant fibrillin-1 exerts dominant-negative effects in heterozygous mgΔ mice, which were phenotypically indistinguishable from wild type.
To gain further insight into the effects of mutant fibrillin-1 on the assembly and stability of microfibrils, we generated two novel lines of knock-in mice. In one line, fibrillin-1 is truncated and tagged with enhanced green fluorescent protein (eGFP),2 allowing us to conclusively demonstrate that mutant fibrillin-1 is incorporated into microfibrils. In heterozygosity, these mice develop features of Marfan syndrome and, in homozygosity, do not survive past the early postnatal period. In a second line of mutant mice, exon 7 is deleted, resulting in the in-frame deletion of the first hybrid domain in fibrillin-1. To our surprise, these mice survived normally, even in homozygosity, and manifested no apparent defects in microfibril structure or function. To test whether mutant fibrillin-1 can interfere with assembly and function of microfibrils, we performed in vitro assays, and we also bred the truncated Fbn1 mice onto an Fbn2 null background. Results clearly demonstrate a major role for dominant-negative effects of Fbn1 mutations on microfibril structure in the pathogenesis of the Marfan syndrome.
EXPERIMENTAL PROCEDURES
Mice
All materials used for the generation of these mice originated from C57BL/6 mice. Targeting vectors (shown in Fig. 1, A and B) were designed to create two lines of Fbn1 mutant mice. Targeting vectors were generated using C57BL/6 genomic DNA and were electroporated into Bruce 4 embryonal stem (ES) cells. ES cells were screened by Southern blotting for successful homologous recombination. Selected ES cells were injected into blastocysts to generate chimeras. Chimeras were bred to obtain germ line transmission. Fbn1-targeted mice were genotyped by Southern blotting. The Neo gene used for selection was flanked by FRT sites and was removed by breeding targeted mice to C57BL/6 mice containing a transgene for FLPe recombinase. C57Bl/6 mice in which Cre recombinase was knocked into the Rosa 26 locus were bred to heterozygous Fbn1 floxed mice to yield mice in which the Fbn1 mutation was expressed in all cells. Floxed as well as mutant mice were generated by Ozgene Pty. Ltd. (Bentley, Australia).
FIGURE 1.
Targeting vectors and resulting effects on fibrillin-1 in germ line mutant mice. A, in the GT-8 targeting vector (top), a poly(A) tail, eGFP, and a neomycin selection cassette (PGK-Neo, flanked by FRT sites) were placed in the intron between exons 34 and 35. A polyglycine linker (GGGG) and exon 33 splice acceptor site were engineered 5′ to the inverted coding region for eGFP. Cre-mediated recombination of special lox sites (lox66 and lox77) resulted in the inversion of eGFP behind exon 32, separated from exon 32 by the polyglycine linker. The neomycin selection cassette was removed after breeding to Flpe transgenic mice. In the H1Δ targeting vector (bottom), exon 7 is flanked by loxP sites. Cre-mediated recombination of the loxP sites results in deletion of exon 7 and the neomycin selection cassette. B, domain modules contained within the truncated GT-8 fibrillin-1 and within H1Δ fibrillin-1 are depicted, with wild-type fibrillin-1 shown for comparison.
To generate the first line of mice in which fibrillin-1 is truncated and tagged with eGFP, special lox sites were incorporated into the targeting vector (Fig. 1A). Cre-mediated recombination resulted in the inversion of the lox 66 and lox 77 sites, inverting at the same time the eGFP coding sequence into frame after exon 32, and the truncation of fibrillin-1 molecules. A polyglycine linker was engineered to invert between exon 32 and eGFP to facilitate proper folding of the last fibrillin domain and of eGFP. A line of mice called GT-8 (“green truncated” from founder mouse 8) was established and maintained on a C57BL/6 background. For this study, heterozygous GT-8 mice were bred to yield wild-type, heterozygous, and homozygous littermates for analyses. Genotyping was by PCR. To distinguish the wild-type Fbn1 allele, a primer pair (CCGTGGAATCTAAAACCTTGGAG and TGGGAATGATGTGGTGAGAGCC) was used to amplify intronic sequence between exons 34 and 35, yielding a 370-bp band from wild-type Fbn1 and no band from homozygous GT-8 Fbn1. Another primer pair (GTGGGTTCCATTAGAGCATTCATC and GGTGAGAGCCTGTATTGTTTCCTC) was used to amplify sequences flanking the inverted exons 34 and 33, yielding a 591-bp fragment from GT-8 Fbn1 and no band from wild-type Fbn1. Heterozygous mice were identified by the presence of both the 370-bp wild-type band and the 591-bp inverted band.
A second line of mice was generated by Cre-mediated removal of Fbn1 exon 7, flanked by loxP sites, in all cells. This line of mice is called H1Δ (for hybrid 1 domain deletion), and it was maintained on a C57Bl/6 background. For this study, heterozygous H1Δ mice were bred to yield wild-type, heterozygous, and homozygous littermates for analyses. Genotyping was by PCR. An intronic primer pair (TTGGAATGACAGGCTGTGGCAC and TTCGCTGTGTTTCTACAAGGCAG) flanking exon 7 was used to amplify a 655-bp band from the deleted allele and a 1116-bp band from the wild-type allele. Mice heterozygous for the mutant allele displayed both bands, whereas wild-type or homozygous mice showed only a single band.
Fbn2+/− mice (19) were bred to GT-8/+ to yield GT-8/+;Fbn2+/− mice. Doubly heterozygous mutant mice were bred to Fbn2−/− or Fbn2+/− female mice, and all progeny were genotyped by PCR. Fbn2 mutant mice were maintained on a 129/sv background. Fbn2 alleles were amplified by PCR using the following primer pair for exon 1: F2ex1F3 (TGTCTCCAGCCCTACTTCGT) and F2ex1R3 (CCTCGGAGTATTTCCTGCTG). This primer pair yielded a 231-bp Fbn2 exon1 band from the wild-type allele. Primers to amplify the neomycin gene were Neo-F (GGAGAGGCTATTCGGCTATGACTG) and Neo-R (CTCTTCGTCCAGATCATCCTGATC). This primer pair yielded a 436-bp band from the mutant allele, which retained a neo cassette. For genotyping progeny, both exon 1 and neomycin primer pairs were utilized to distinguish between Fbn2 wild-type, heterozygous, and homozygous pups. In addition to these two different PCR approaches used to distinguish Fbn2 genotypes, Fbn2 null mice consistently displayed syndactyly (19). All experiments were conducted according to protocols approved by the Oregon Health & Science Institutional Animal Care and Use Committee.
Real-time Quantitative PCR
75 mg of dissected mouse skin was dropped into 1 ml of TRIzol reagent (Invitrogen) and ground with a micro pestle (Kimble-Chase, Vineland, NJ) into smaller pieces. RNA extraction was performed according to the manufacturer's protocol. A subsequent sample purification step was included using the RNeasyTM kit (Qiagen), and residual cDNA contamination was removed from each sample by using the Turbo DNA-freeTM kit (Ambion, Austin, TX). RNA samples were quantified by photospectrometry, and 1.0 μg of RNA per sample was reverse transcribed using the Bio-Rad iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA). Samples were amplified in triplicates using the iTaqTM SYBR Green Supermix (Bio-Rad) in an iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad). Analysis of data was performed using the 2−ΔΔCt method (20) and quantitated relative to the ARBP0 gene. Gene expression was normalized to littermate wild-type control mice, which provided an arbitrary constant for comparative fold expression.
Western Blotting
Neonatal dermal fibroblast cultures were established for all individual pups in a litter, and equal numbers of cells were passaged and plated into wells. Medium was replaced with serum-free medium for 24 h. Proteins present in the serum-free medium from the individual cultures were trichloroacetic acid-precipitated, applied to 5% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-fibrillin-1 (pAb 9543) or with anti-GFP (RDI Division of Fitzgerald Industries, Concord, MA), using previously described methods (21).
Cell Cultures
Fibroblasts were derived from explant cultures of neonatal skin obtained from all pups in various GT-8 and H1Δ litters. Explant cultures represented all genotypes. Cells were cultured in Dulbecco's modified Eagle's medium (MediaTech, Hendon, VA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin/streptomycin (MediaTech).
Quantitation of Fibrillin-1 and Fibrillin-2 by Sandwich ELISA
P0 dermal fibroblasts were cultured in Dulbecco's modified Eagle's medium, 10% FBS. Eight-well chamber slides were seeded at a density of 100,000 cells/well in 500 μl of Dulbecco's modified Eagle's medium, 10% FBS and incubated for 72 h. Medium was collected and stored at −20 °C. Samples were analyzed by a modified sandwich ELISA. Antibody pairs for fibrillin-1 consisted of Sepharose-coupled recombinant peptide rF20 (22), affinity-purified polyclonal antibody (pAb) 9543, and rF18 (22) affinity-purified pAb 9543. Antibody pairs for fibrillin-2 were generated by affinity-purifying pAb 0868 on Sepharose-coupled recombinant peptide rF47 (21) and by affinity-purifying pAb 1747 on Sepharose-coupled rF37 (23). The antibodies were then biotinylated or AP-conjugated using the Lightning-Link system (Innova Biosciences, Cambridge, UK) according to the manufacturer's protocol. 96-well plates were coated with 5 μg/ml streptavidin (Thermo Scientific, Rockford, IL), followed by incubation with the biotinylated antibodies (0.25 μg/ml in TBS-T, 5% FBS). Medium samples were 2-fold serially diluted in TBS-T (range: undiluted to 1/128 dilution). Standard curves were generated by serially diluting recombinant peptides (rF11 for fibrillin-1 and rF37 for fibrillin-2) in TBS-T, 10% FBS (0–125 ng/ml). Incubation with AP-conjugated antibodies (0.05 μg/ml in TBS-T, 5% FBS) completed the sandwich, and detection of bound materials was by color generation using the ELISA Amplification System (Invitrogen). Absorbance was measured using a Molecular Devices Emax plate spectrophotometer and was then converted to micrograms/ml, according to standard curve values. Calculations to generate standard curves and linear regression equations were performed with Excel software (Microsoft). Statistical analysis and graphs were generated with GraphPad Prism 5.0 for Windows (GraphPad, San Diego, CA). The ten cell lines were grouped by genotype, and the mean concentrations of fibrillin-1 were compared using the Kruskal-Wallis test for a nonparametric analysis of variance. Because no significant difference was found between the groups, post-testing was not performed.
Light Microscopy
Thick sections (25 μm) of 2-month-old GT-8/+ tendon were examined for eGFP fluorescence using a Leica confocal microscope. Thin sections of skin were examined using a Zeiss Axiovert 200M microscope, and micrographs were recorded digitally using AxioVision software (version 4.5). eGFP fluorescence was captured from untreated sections. For evaluation of aortic root tissues, 0.5-μm sections were stained with a combination of toluidine blue and Basic Fuchsin.
Immunofluorescence microscopy of tissue sections, using affinity-purified pAb 9543 (anti-fibrillin-1), affinity-purified pAb 0868 (anti-fibrillin-2), and anti-GFP (RDI Division of Fitzgerald Industries, Concord, MA), was performed as previously described (24). Secondary antibodies were Alexa 488 anti-rabbit, Alexa 488 anti-mouse, and Alexa 568 anti-rabbit (Invitrogen Molecular Probes, Eugene, OR). For cell culture immunofluorescence, 1 × 105 cells in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin were plated onto 4-well Permanox Lab-Tek chamber slides (Nalge Nunc, Rochester, NY) and were grown for 4–13 days. Slides were fixed in cold methanol for 10 min and stained as previously described (24). Slides were coverslipped with ProLong Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen Molecular Probes).
Electron Microscopy
Immunoelectron microscopy of early postnatal and 2-month-old GT-8/+ skin, labeled with anti-GFP (RDI), anti-fibrillin-1 (pAb 9543), or anti-fibrillin-2 (pAb 0868), followed by 5 nm secondary gold conjugate (Amersham Biosciences), was performed as previously described (25). Aligned tilt series were acquired from sections 500 nm thick at a magnification of 29,000× using an FEI Tecnai G2 transmission electron microscope operated at 200 KV. The micrographs were recorded automatically using a bottom-mounted FEI Eagle 2K charge-coupled device camera. The micrographs were taken in a tilt range with a 2° increment from 40° to +40° and a 1° increment from −40° to −60° and from +40 to +65°. Images were aligned using FEI Inspect-3D software.
RESULTS
Expression and Secretion of Fibrillin-1 in GT-8 Mutant Mice
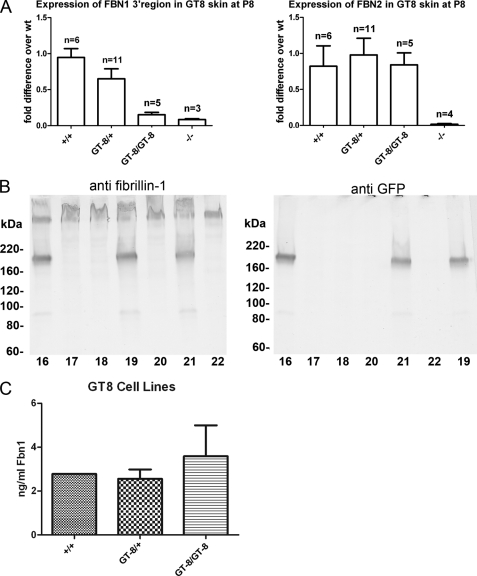
Mice in which fibrillin-1 is truncated and tagged with eGFP were generated using homologous recombination. A diagram of the targeted Fbn1 locus is shown in Fig. 1A. After breeding floxed mice to mice that express Cre recombinase, recombination inverts the region in between the lox66 and lox77 sites, bringing eGFP into frame after exon 32. This inversion truncates fibrillin-1 after the 17th calcium-binding epidermal growth factor-like domain (cbEGF17), adds a polyglycine linker region to the end of cbEGF17, and tags the truncated molecule with eGFP (Fig. 1A).
In this study, we characterized a line of mice called GT-8 (“green truncated” from founder mouse 8) in which the mutation was passed through the germ line. These mice were generated by breeding floxed mice to mice in which Cre recombinase was knocked into the Rosa 26 locus, presumably resulting in recombination in all cells. To test the extent of recombination in the GT-8 line of mice, quantitative PCR was performed using primers directed toward the 3′-end of Fbn1. The 3′-end of Fbn1 should be missing in the mutant allele. At the same time, levels of Fbn2 were also quantitated. Controls included RNA from Fbn1 null and Fbn2 null mice. Results from these experiments showed that there are only insignificant amounts, if any, of residual wild-type Fbn1 expressed in the homozygous GT-8 mice (Fig. 2A). In addition, no compensatory up-regulation of Fbn2 was detected, similar to results previously obtained from Fbn1 null mice (18) (Fig. 2A).
FIGURE 2.
Expression and secretion of GT-8 fibrillin-1. A, left panel: using 3′ Fbn1 primers, quantitative PCR analyses were used to quantitate expression of Fbn1. The -fold differences were calculated compared with average levels in wild-type mice. mRNA from skin of Fbn1−/− mice was used as a control. Levels of 3′ Fbn1 in homozygous GT-8 mice were not significantly different from −/− mice, indicating that recombination of mutant alleles occurred in most, if not all, cells. Right panel: quantitative PCR of Fbn2 mRNA levels showed no significant differences between wild-type, heterozygous, and homozygous GT-8 mice. mRNA from Fbn2−/− mice was included as a control. B, Western blotting of proteins secreted into the medium by skin fibroblast cultures was performed using pAb 9543 (anti-fibrillin-1) (left panel) and anti-GFP (right panel). Blotting with anti-GFP established the identity of the truncated fibrillin-1. These fibroblast cultures were established from a single litter. Fibroblasts from pups 17, 18, 20, and 22 secreted only wild-type fibrillin-1, whereas fibroblasts from pups 16, 19, and 21 secreted both wild-type and eGFP-tagged and -truncated fibrillin-1. Blotting with pAb 9543 demonstrated that the wild-type and truncated fibrillin-1 were present in approximately equal amounts in the medium of heterozygous cultures. C, quantitation of fibrillin-1 was determined by sandwich ELISA. Antibody pairs were directed toward the N-terminal half of fibrillin-1, so that both wild-type and truncated fibrillin-1 would be captured. Skin fibroblast cultures from a litter of GT8 mice were plated (100,000 cells/well) and incubated for 72 h. The mean concentration (and standard deviation) of all cell lines for each genotype is shown (n = 1 for +/+; n = 4 for GT-8/+; n = 5 for GT-8/GT-8). Fibrillin-1 was detected in all cell lines and did not vary significantly by genotype (p = 0.56). Fibrillin-2 was not detectable in the cell culture media (data not shown).
Adding the eGFP tag to fibrillin-1 might have perturbed the folding of the molecule and caused intracellular retention and degradation. To check levels of fibrillin-1 protein, fibroblasts were established from offspring of +/+ X GT-8/+ and GT-8/+ X GT-8/+ breeding pairs. Immunoblotting with antibodies specific for the N-terminal half of fibrillin-1 (pAb 9543) showed that similar levels of wild-type and truncated fibrillin-1 were secreted and accumulated into the medium (Fig. 2B). To more precisely quantitate the amount of fibrillin-1 protein secreted into the medium, sandwich ELISAs were performed. Antibody pairs were directed toward the N-terminal half of fibrillin-1, so that both wild-type and truncated fibrillin-1 would be captured. The levels of total fibrillin-1 secreted into the medium were similar in wild-type, heterozygous, and homozygous GT-8 fibroblast cultures (Fig. 2C), indicating that the GT-8 mutant allele is not poorly expressed and that substantial amounts of GT-8 fibrillin-1 are not abnormally retained inside the cell or degraded. Fibrillin-2 was not detectable in any of the cell lines (data not shown).
Survival of GT-8 Mutant Mice
From GT-8/+ × GT-8/+ breeding, the numbers of wild-type, heterozygous, and homozygous mice that were born were close to the expected Mendelian frequencies. Of 458 pups, 115 were wild type (25%), 251 were heterozygous (55%), and 92 were homozygous (20%). Homozygous GT-8 pups died primarily between P9 and P18; with most deaths occurring around P14. 28 heterozygous mice survived longer than 1 year. Of these 28 mice >1 year old, 7 died of natural causes, 14 were still alive, and 7 were sacrificed for analyses. In addition, 4 of the 251 heterozygous mice died naturally between 1 and 8 months of age. The remaining mice were sacrificed for experiments.
Matrix Incorporation of eGFP-tagged and Truncated Fibrillin-1 in GT-8 Mice
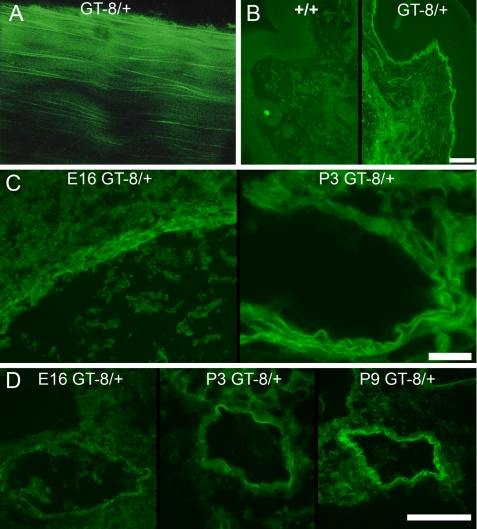
Because the eGFP tag is ∼27 kDa arranged in a cylindrical “beta-can,” it is possible that addition of the eGFP tag might itself interfere with assembly or function of fibrillin-1. However, our studies have not revealed any evidence for specific eGFP functional interference. Instead, we found that the incorporation of the truncated fibrillin-1 into typical fibrillin-1 microfibril patterns was easily visualized, because of the eGFP tag. eGFP fluorescence was observed by whole mount confocal microscopy of tendon (Fig. 3A) and by epifluorescence microscopy of skin sections (Fig. 3B). To our surprise, eGFP fluorescence was undetectable in most fetal and early postnatal tissues, even though fibrillin-1 immunostaining suggests that fibrillin-1 is an abundant and ubiquitous component of connective tissues during development. Anti-GFP immunostaining was equivalent to fibrillin-1 immunostaining during development and into the early postnatal period (data not shown and Fig. 4), demonstrating the higher sensitivity of immunofluorescence, but eGFP fluorescence was mostly too weak to be detected using epifluorescence microscopy. eGFP fluorescence in heterozygous GT-8 mice showed that the highest concentrations of fibrillin-1 are first accumulated in blood vessels, compared with all other tissues examined. eGFP fluorescence was visible in the aorta by E16 and became progressively more intense through early postnatal life (Fig. 3C). In peripheral blood vessels, we noted increasing green fluorescence from E16 through P9 (Fig. 3D). Accumulation of eGFP fluorescence in other tissues was not seen until after 1 month of age (data not shown). During the early postnatal period, homozygous tissues emitted more eGFP fluorescence than heterozygous tissues, as expected (data not shown). Because fibrillin microfibrils are notoriously difficult to solubilize, these results are the first data to show relative quantitative differences in fibrillin-1 in tissues.
FIGURE 3.
Emitted eGFP fluorescence from various GT-8/+ tissues. A, unstained whole mount confocal microscopy of 2-month GT-8/+ tendon shows incorporation of mutant eGFP-tagged fibrillin-1 into long microfibrils in the longitudinal axis of the tendon. B, unstained 2-month wild-type (+/+) and GT-8/+ skin thin sections show specific incorporation of mutant eGFP-tagged fibrillin-1 into typical microfibril patterns in the dermis and at the dermal-epidermal junction. Scale bar = 50 μm. C, unstained sections of aorta from E16 and P3 heterozygous GT-8 mice revealed green fluorescence, visible at E16, which intensified from E16 to the early postnatal period. Scale bar = 20 μm. D, unstained peripheral blood vessels also accumulated large amounts of fibrillin-1 after birth and demonstrated increasing amounts of green fluorescence in heterozygous mice during the early postnatal period. Scale bar = 50 μm.
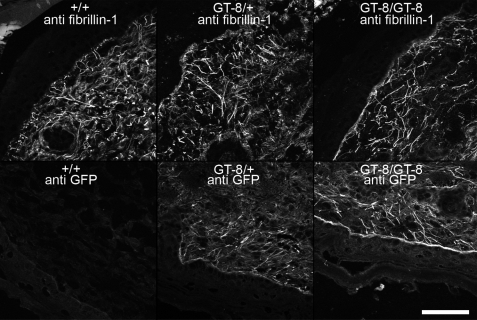
FIGURE 4.
Microfibril staining patterns were similar in P1 skin from wild-type, heterozygous, and homozygous GT-8 mice. Anti-fibrillin-1 (pAb 9543) staining patterns revealed abundant long microfibrils in the skin of all genotypes (top panels). Anti-GFP staining patterns were similar to the anti-fibrillin-1 staining in heterozygous (GT-8/+) and homozygous (GT-8/GT-8) skin sections, demonstrating the incorporation of truncated fibrillin-1 into microfibril patterns. Wild-type (+/+) skin sections were unstained by anti-GFP antibodies. Scale bar = 50 μm.
Microfibril Assembly in GT-8 Mice
To check for any visible differences in microfibril structure, multiple tissues from mice of all three genotypes were examined at different time points (P0–P11). In Fig. 4, we show results from skin, because microfibril staining patterns are particularly distinctive in this tissue. Anti-fibrillin-1 (pAb 9543) immunostaining of newborn and early postnatal skin showed no differences between littermate wild-type, heterozygous, and homozygous GT-8 microfibrils (Fig. 4). In addition, anti-GFP immunostaining in heterozygous and homozygous newborn skin was equivalent to anti-fibrillin-1 staining in wild-type littermate skin (Fig. 4). No abnormalities in microfibril abundance or staining pattern were detected during the early postnatal period (P0–P3).
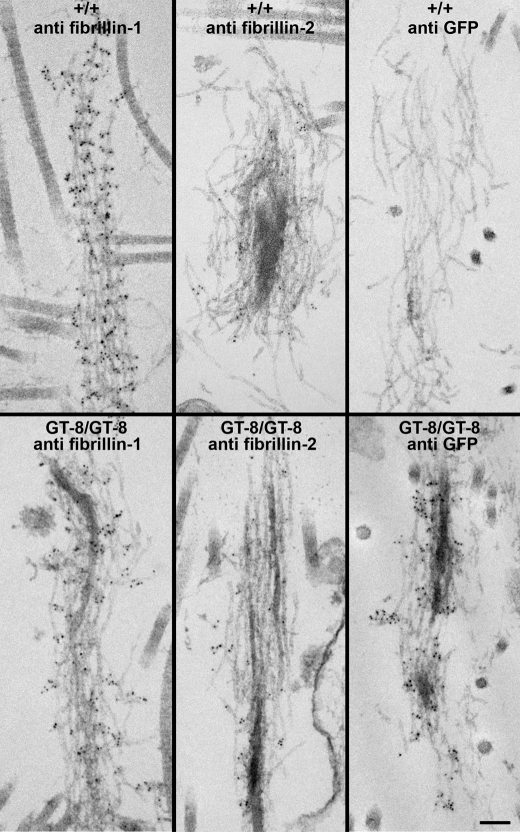
Confirmation that the eGFP fluorescence patterns and immunostaining patterns were due to incorporation of the truncated fibrillin-1 into microfibrils was obtained from electron microscopic immunolocalization. Antibodies specific for GFP decorated fibrillin microfibrils in elastic fibers (Fig. 5) as well as in bundles of microfibrils without amorphous elastin (data not shown). In addition, the ultrastructural appearance of microfibrils in heterozygous and homozygous GT-8 skin was identical to wild-type microfibrils at P0 (Fig. 5). Based on these data, we conclude that truncated fibrillin-1 contains sufficient information to be incorporated into microfibrils in vivo and does not interfere with the assembly of microfibrils. However, compared with wild-type microfibrils, mutant microfibrils did not demonstrate clear periodicity of anti-fibrillin-1 immunogold labeling (Fig. 5).
FIGURE 5.
Ultrastructure of microfibrils in P0 skin from wild-type and homozygous GT-8 mice. Immunogold labeling of wild-type (+/+) and homozygous (GT-8/GT-8) skin demonstrated the incorporation of truncated fibrillin-1 into microfibrils. Microfibrils in the homozygote were well labeled both with anti-fibrillin-1 (pAb 9543) and with anti-GFP. At P0, anti-fibrillin-2 (pAb 0868) showed only minimal immunogold labeling in both the wild-type and homozygous skin. Although the ultrastructural appearance of the mutant microfibrils was similar to wild-type, the periodicity of pAb 9543 immunogold labeling was not obvious along the lengths of mutant microfibrils. Scale bar = 100 nm.
To test matrix assembly in vitro, fibroblast cultures were established from littermate neonatal skins. Wild-type, heterozygous, and homozygous GT-8 cultures were stained with antibodies specific for fibrillin-1 (pAb 9543), fibrillin-2 (pAb 0868), and for GFP. Although wild-type fibroblasts assembled abundant fibrillin-1 fibrils (Fig. 6A), heterozygous GT-8 fibroblasts assembled fewer fibrillin-1 fibrils (Fig. 6B), and homozygous GT-8 fibroblasts failed to assemble a fibrillin-1 fibril matrix (Fig. 6C). eGFP-tagged and -truncated fibrillin-1 was assembled into fibrils by heterozygous GT-8 fibroblasts (Fig. 6E) but not by homozygous GT-8 fibroblasts (Fig. 6F). None of these cultures accumulated fibrillin-2 fibrils (data not shown). Therefore, in the absence of fibrillin-2, truncated fibrillin-1 assembled into fibrils in heterozygous fibroblast cultures and fewer total fibrillin-1 fibrils were detectable. However, in the absence of fibrillin-2 and also wild-type fibrillin-1, truncated fibrillin-1 could not assemble fibrils. We conclude that either truncated fibrillin-1 can interfere with normal fibrillin-1 fibril assembly in the heterozygous cultures or heterozygous cultures have concentrations of wild-type fibrillin-1 that are below those required for fibril assembly.
FIGURE 6.
In vitro assembly of mutant fibrillin-1. Fibroblasts were established from early postnatal littermate skin. GT-8 wild-type (A and D), heterozygous (B and E), and homozygous (C and F) cultures were stained with anti-fibrillin-1 (pAb 9543) (A–C) or with anti-GFP (D–F). Results showed that, in contrast to wild-type fibroblasts, heterozygous cells assembled fewer fibrillin-1 positive fibrils and that truncated fibrillin-1 was assembled into fibrils. Homozygous fibroblasts failed to assemble fibrillin-1 fibrils in vitro. After immunostaining with anti-fibrillin-1, H1Δ wild-type (G), heterozygous (H), and homozygous (I) demonstrated abundant fibrillin-1 fibrils. Scale bar = 50 μm.
Dominant-negative Influence of GT-8 Fibrillin-1
To test in vivo for dominant-negative effects of truncated fibrillin-1, GT-8/+ mice were bred to Fbn2+/− mice to yield doubly heterozygous mice. The resulting mice were viable and fertile. Doubly heterozygous mice were bred to Fbn2+/− and Fbn2−/− females. Table 2 shows the numbers of pups born from these crosses, their genotypes, and the expected and actual percentages of genotypes represented at birth (P0). Results indicated some embryonic lethality for the GT-8/+;Fbn2−/− genotype, especially when the mother was Fbn2−/−. Moreover, GT-8/+;Fbn2−/− pups that were born were not viable. They were found dead or were dying. When Fbn1+/− mice were crossed with Fbn2+/− mice, 50% of the expected Fbn1+/−;Fbn2−/− mice were born and were viable (18), indicating that half the amount of normal fibrillin-1, with no fibrillin-2, marks the threshold amount of fibrillin required for development. Similar numbers of GT-8/+;Fbn2−/− mice may survive through fetal development. However, in the absence of fibrillin-2, truncated fibrillin-1 exerts a dominant-negative effect on wild-type fibrillin-1 that is incompatible with survival after birth.
TABLE 2.
Expected and actual births of GT-8/+;Fbn2−/− pups
| GT-8/+;Fbn2+/− × Fbn2−/− |
||||
|---|---|---|---|---|
| Fbn1+/+;Fbn2+/− | Fbn1+/+;Fbn2−/− | GT-8/+;Fbn2+/− | GT-8+/−;Fbn2−/−a | |
| Total (68) | 21 | 15 | 30 | 2 |
| Expected % | 25% | 25% | 25% | 25% |
| Actual % | 31% | 22% | 44% | 2.9% |
| GT-8/+;Fbn2+/− × Fbn2+/− |
||||||
|---|---|---|---|---|---|---|
| Fbn1+/+;Fbn2+/+ | Fbn1+/+;Fbn2+/− | Fbn1+/+;Fbn2−/− | GT-8/+;Fbn2+/+ | GT-8/+;Fbn2+/− | GT-8/+;Fbn2−/−a | |
| Total (98) | 13 | 38 | 8 | 9 | 22 | 8 |
| Expected % | 12.5% | 25% | 12.5% | 12.5% | 25% | 12.5% |
| Actual % | 13% | 39% | 8.2% | 9.2% | 22% | 8.2% |
a GT-8/+;Fbn2−/− pups died right after birth.
Microfibril Stability and Elastic Fiber Fragmentation in GT-8 Mice
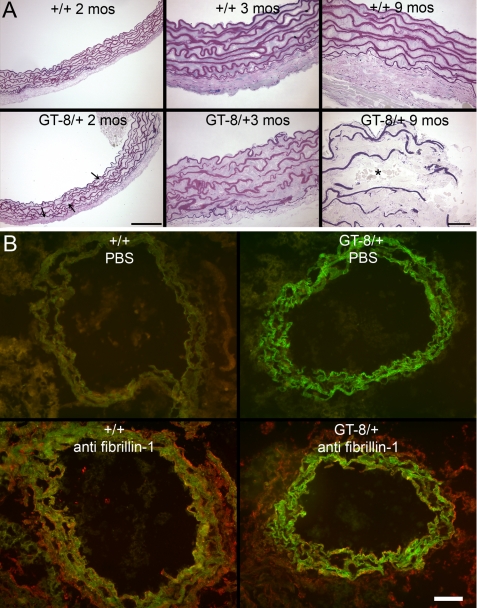
In GT-8/+ mice, compared with their wild-type littermates, fragmentation of the aortic elastic lamellae was observed from 2 months of age, becoming progressively more severe with increasing age (Fig. 7A). In contrast to typical dermal or perichondrial elastic fibers, microfibrils are not easily visualized by electron microscopy on the surfaces of aortic elastic lamellae (26). To determine the extent to which fibrillin microfibrils contribute to aortic elastic lamellae, we examined sections of P3 GT-8/+ aortas. In contrast to thin red immunostaining with anti-fibrillin-1, thick bands of eGFP fluorescence were emitted from the elastic lamellae in GT-8/+ mice, revealing that fibrillin-1 is within the amorphous elastin as well as on the surfaces of the lamellae (Fig. 7B).
FIGURE 7.
Fragmentation and eGFP fluorescence of aortic root elastic lamellae in heterozygous GT-8 mice. A, aortic root morphology was evaluated in wild-type and heterozygous GT-8 littermates at various time points. At 2 months of age, breaks in the elastic lamellae could easily be found in heterozygous mice (arrows). These breaks in the elastic lamellae became more severe by 3 months of age in heterozygous mice. By 9 months of age, breaks in the heterozygous lamellae could be found across whole planes of the aortic wall, resulting in infiltration of blood cells (*) into the smooth muscle media. Scale bars = 100 μm. B, unstained P3 wild-type and heterozygous GT-8 aortic root demonstrated eGFP fluorescence throughout the width of the heterozygous elastic lamellae. Wild-type and heterozygous aortic root sections immunostained with anti-fibrillin-1 (pAb 9543) showed red immunofluorescence in the adventitia and red or yellow (merged green and red) immunofluorescence in the internal elastic lamina and on the surfaces of the elastic lamellae. However, anti-fibrillin-1 staining did not appear to fully penetrate into the elastic lamellae. Scale bar = 50 μm.
In previous studies (27), we found that fibrillin-2 epitopes were revealed in postnatal microfibrils after tissues were digested with crude collagenase. In addition, we showed that fibrillin-2 epitopes are available in postnatal Fbn1 null tissues, in contrast to tissues from wild-type littermates (27). Because crude collagenase cleaves fibrillin-1 at multiple specific sites (28), we proposed that molecules of fibrillin-1 and fibrillin-1-associated microfibrillar proteins normally mask fibrillin-2 epitopes in postnatal microfibrils (27). In addition, these results led to the hypothesis that fibrillin-2 immunoavailability can serve as a marker for microfibril fragmentation.
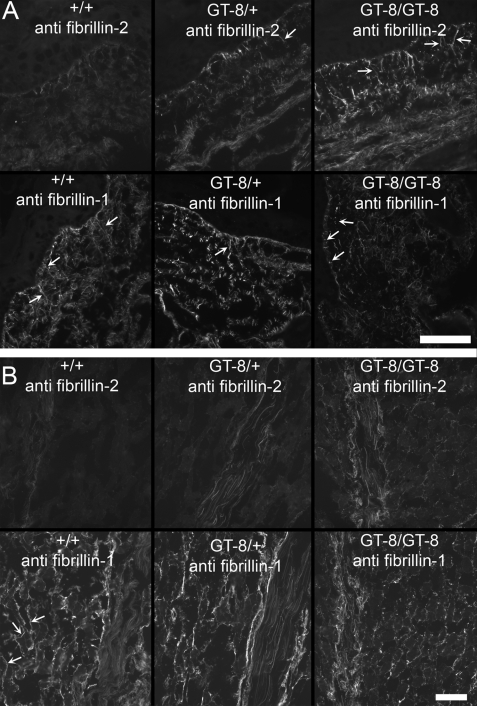
To determine whether incorporation of the mutant fibrillin-1 targets microfibrils for proteolytic degradation (14, 15), we tested whether fibrillin-2 immunostaining is revealed in GT-8/+ mouse tissues. Fig. 8 shows that fibrillin-2 immunostaining was revealed in P8 GT-8 heterozygous and homozygous skin (A) and in P8 GT-8 heterozygous and homozygous skeletal muscle and tendon (B). In contrast, wild-type littermate tissue sections were mostly negative for fibrillin-2 immunostaining. In addition, comparison of fibrillin-1 staining patterns in wild-type, heterozygous, and homozygous mice suggested increased fragmentation of fibrillin-1 staining patterns according to genotype in heterozygous and homozygous tissues (Fig. 8).
FIGURE 8.
Fibrillin-2 immunostaining is revealed in P8 GT-8 mutant mice. A, P8 wild-type, heterozygous, and homozygous GT-8 skin sections were stained with anti-fibrillin-2 (pAb 0868) and with anti-fibrillin-1 (pAb 9543). Fibrillin-2 immunostaining was barely visible in wild-type sections, but long fibrillin-2-positive microfibril fiber bundles (arrows), inserting perpendicularly to the dermal-epidermal junction, were revealed in heterozygous and homozygous GT-8 sections. Fibrillin-2 immunostaining was more abundant in homozygous skin sections than in heterozygous skin sections. In contrast, fibrillin-1 immunostaining showed abundant long microfibril fiber bundles (arrows) in the wild-type skin sections, but in heterozygous and homozygous sections, fibrillin-1 immunostaining appeared fragmented, with very few long fibrils at the dermal-epidermal junction. Because early postnatal heterozygous skin sections showed abundant long fibrillin-1 microfibril fiber bundles (Fig. 4), microfibril assembly was apparently uncompromised. Therefore, P8 sections likely indicate that fragmentation of fibrillin-1 has occurred. B, P8 wild-type, heterozygous, and homozygous GT-8 sections of skeletal muscle and tendon were also examined, and similar results were obtained. Fibrillin-2 immunostaining was concealed in wild-type sections and revealed in heterozygous and homozygous skeletal muscle and tendon. In contrast, fibrillin-1 immunostaining appeared to be more fragmented in heterozygous and homozygous tissues than in the wild-type tissues. This was particularly noticeable in the skeletal muscle where fibrillin-1 immunostaining typically envelopes individual skeletal muscle fibers (arrows). Scale bars = 50 μm.
Light microscopic results were also confirmed at the ultrastructural level. Fig. 9 shows P8 skin microfibrils labeled with anti-fibrillin-1 and anti-fibrillin-2. In contrast to minimal labeling of wild-type microfibrils, there was abundant labeling of microfibrils with fibrillin-2 antibodies in P8 homozygous skin. Immunogold labeling of microfibrils with fibrillin-1 antibodies detected a loss of periodicity in P8 homozygous microfibrils compared with wild-type microfibrils, indicating a disturbance of microfibril structure (Fig. 9). To further evaluate microfibril structure, an aligned tilt series was performed. Three-dimensional images of wild-type dermal microfibrils, labeled with anti-fibrillin-1 antibodies, clearly showed elastic fibers suspended in a basket-like network of microfibrils (supplemental movie). In contrast, homozygous GT-8 skin lacked the basket-like network of fibrillin microfibrils (supplemental movie). Therefore, we conclude that, after microfibril assembly of truncated fibrillin-1 in GT-8 mutant mice, microfibril structure is perturbed, and fragmentation of fibrillin-1 results in the unmasking of fibrillin-2 and the degradation of the microfibril network.
FIGURE 9.
Ultrastructure of microfibrils in P8 wild-type and homozygous GT-8 mice. Immunogold labeling of P8 wild-type and homozygous GT-8 littermate skin demonstrated differences in microfibril ultrastructure. Wild-type microfibrils with fibrillin-1 antibodies showed periodic gold labeling, which was not detectable on fibrillin-1-labeled microfibrils in homozygous skin. In contrast to wild-type microfibrils, which displayed minimal immunogold labeling, homozygous microfibrils were well labeled with fibrillin-2 antibodies. Revealing of fibrillin-2 epitopes indicated a perturbation of microfibril structure. Scale bar = 100 nm.
Microfibril Assembly and Stability in H1Δ Mice
The first hybrid domain in fibrillin-1 contains a free cysteine residue (29) and mediates binding interactions between fibrillin-1 and latent transforming growth factor-β-binding proteins (LTBPs) (24). We tested whether H1Δ fibrillin-1, which is shorter than wild-type fibrillin-1 (Fig. 1B), would perturb microfibril assembly in vivo. In heterozygosity, H1Δ mice tested the hypothesis that fibrillin-1 molecules are required to be in perfect register for microfibril assembly and stability. Homozygous H1Δ mice tested whether the free cysteine present in the first hybrid domain is required for microfibril assembly. The numbers of heterozygous and homozygous H1Δ mice that were born from heterozygous breeding were close to expected Mendelian frequencies. Of 228 pups, 53 (23%) were wild-type, 105 (46%) were heterozygous, and 70 (31%) were homozygous. Both mutant genotypes survived well past 1 year of age. All other homozygous Fbn1 mutant mice die in the early postnatal period, including GT-8 homozygotes (reported in the present study), with the exception of homozygous tight skin or tsk mice, which die during early embryogenesis. Therefore, the compatibility of a homozygous Fbn1 mutation with a normal life span was surprising.
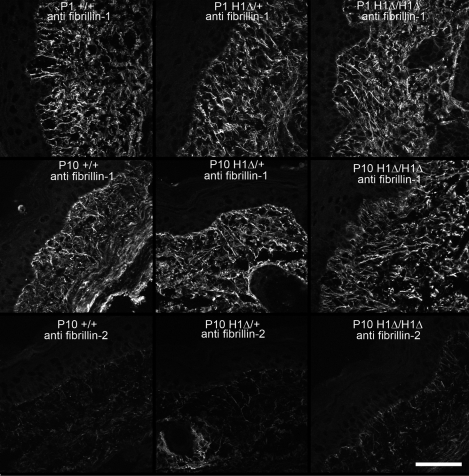
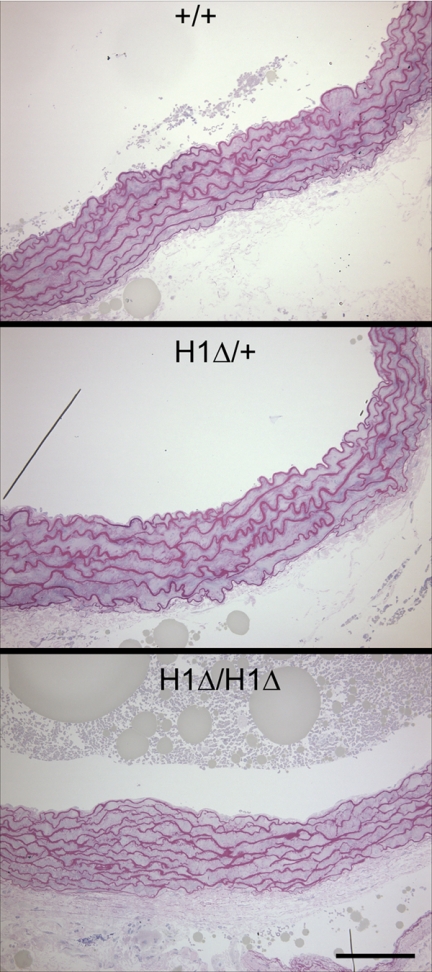
Examinations of microfibril assembly in vitro showed that H1Δ fibrillin-1 did not interfere with fibrillin-1 fibril assembly in heterozygosity (Fig. 6H, compared with wild-type littermate fibroblasts in Fig. 6G) and that homozygous H1Δ fibrillin-1 could form fibrillin-1 fibrils (Fig. 6I). Comparison of fibrillin-1 microfibril immunostaining in various tissues at different early postnatal time points confirmed these in vitro observations. Fig. 10 shows apparently normal fibrillin-1 microfibril staining patterns in skin at P1 and at P10, indicating normal assembly of fibrillin-1 in vivo. In addition, P10 mutant skin failed to show fibrillin-2 immunostaining, indicating that microfibril structure is stable during the early postnatal period, in contrast to GT-8 mutant mouse tissues (Fig. 8). In addition, examination of aortic elastic lamellae did not show significant fragmentation, even at time points greater than 1 year of age (Fig. 11), and ultrastructural analyses showed no visible differences in microfibril structure (data not shown).
FIGURE 10.
Microfibril structure in H1Δ mutant mice. Fibrillin-1 immunostaining of P1 and P10 wild-type, heterozygous, and homozygous H1Δ skin demonstrated no differences in abundance or pattern of microfibril fiber bundles. Fibrillin-2 immunostaining of P10 wild-type, heterozygous, and homozygous H1Δ skin did not reveal fibrillin-2 in H1Δ mutant mice, indicating that microfibril structure is stable in the mutants during the early postnatal period. Scale bar = 50 μm.
FIGURE 11.
Aortic root morphology in H1Δ mutant mice. Aortic root morphology in 15-month wild-type, heterozygous, and homozygous mice showed minimal fragmentation of the elastic lamellae. Scale bar = 100 μm.
DISCUSSION
We used an in vivo approach to answer certain controversial questions related to the fate of mutant fibrillin-1. Early in vitro work showed that fibroblasts established from individuals with the Marfan syndrome failed to properly assemble fibrillin-1 microfibrils, suggesting a mechanism to explain the fragmented appearance of fibrillin-1 microfibrils observed in Marfan skin (30). This hypothesis was supported by transfecting mutant fibrillin-1 into cell cultures and perturbing fibrillin-1 fibril assembly (13). However, in later studies using transgenic mice, overexpression of mutant fibrillin-1 did not interfere with microfibril assembly and did not cause Marfan syndrome (9). Hence, instead of a dominant-negative model in which mutant fibrillin-1 interferes with the assembly of wild-type fibrillin-1, haploinsufficiency of wild-type fibrillin-1 became the preferred mechanism initiating pathogenesis in the Marfan syndrome (9). In addition to these two models (dominant-negative interference versus haploinsufficiency driven mechanisms), a third model was proposed. In this alternative model, mutant fibrillin-1 is assembled into microfibrils, but the mutation destabilizes the structure of fibrillin-1 to the extent that microfibrils can be slowly degraded over time, leading to the clinical features of the Marfan syndrome (14, 15).
We chose two strategies to approach these issues. First, we designed a mutation, a truncation of fibrillin-1 in the “neonatal” region of the molecule (32), that we predicted would result in severe Marfan phenotypes, to test whether mutant fibrillin-1 can interfere with microfibril assembly. To follow the fate of the mutant fibrillin-1, we added an eGFP tag at the end of the truncated molecule. Second, we chose to delete a single fibrillin-1 domain, the first hybrid domain, that has been proposed to mediate intermolecular disulfide bond formation, an initial step in assembly of microfibrils (29). In addition, the first hybrid domain was shown to be responsible for interactions between LTBPs and fibrillin-1 (24). Neither of these two mutations is found in humans with the Marfan syndrome. In Table 1, these mutations are listed along with previously published Fbn1 mutant mice.
Our results using GT-8 mice demonstrated that truncated fibrillin-1 is secreted at the same level as wild-type fibrillin-1 and is clearly assembled into microfibrils. We showed evidence that assembly of truncated fibrillin-1 results in fragmentation of elastic lamellae in the aorta of heterozygous GT-8 mice and fragmentation of fibrillin-1 immunostaining in skin, skeletal muscle, and tendon. In addition, we showed the concomitant unmasking of fibrillin-2 epitopes in the skin, skeletal muscle, and tendon, and we confirmed these light microscopic results at the ultrastructural level. Electron microscopy of an aligned tilt series was used to reveal an intricate network of dermal microfibrils, in which elastic fibers are suspended, and this method allowed us to show that this novel network of dermal microfibrils was lost in the GT-8 homozygous mutant mice. Based on these results, we conclude that truncated fibrillin-1 does not interfere with assembly of microfibrils, but instead exerts a dominant-negative influence on the stability of microfibrils.
One potential mechanism by which the cell might sense a mutant microfibril environment was suggested by immunolocalization experiments, which showed the loss of periodic labeling with anti-fibrillin-1 antibodies (Figs. 5 and 9). In our initial description of fibrillin-1, we noted that the fibrillin-1 periodic immunolabeling of microfibrils matched the collagen banding period (1). We hypothesize that loss of periodic labeling in GT-8 homozygous mice at P0 reflects abnormal integration of microfibrils into the surrounding collagenous environment. This loss of appropriate integration could be mediated by proteins that connect the collagenous environment to microfibrils through interactions mediated by domains missing from the truncated fibrillin-1 molecules. Cellular responses to this abnormal matrix environment could include secretion of proteases in an attempt to remodel and repair the matrix, such that by P8, the loss of periodicity was accompanied by loss of fibrillin-1 immunolabeling, revealing of fibrillin-2 epitopes, and the loss of microfibril bundles.
H1Δ fibrillin-1 was also clearly assembled into microfibrils, again refuting the model whereby mutant fibrillin-1 interferes with microfibril assembly. However, in this case, mutant fibrillin-1 did not result in fragmentation of the aortic elastic lamellae or fragmentation of fibrillin-1 immunostaining in other tissues. In addition, fibrillin-2 epitopes remained masked in H1Δ mutant tissues. These results clearly demonstrate that neither the free cysteine residue present in the first hybrid domain (29) nor perfect registration of fibrillin molecules is required for microfibril assembly. Moreover, H1Δ mutant mice survived in homozygosity, reinforcing the conclusion that this mutation does not adversely perturb microfibril structure or function.
H1Δ is the first report of a mutation in mouse Fbn1 that does not cause early death in homozygosity. We have also generated a second targeted mutation in mice in Fbn1, an in-frame deletion of three domains, which survives in homozygosity.3 In comparison with previous Fbn1 mutant mice (Table 2), our results clearly show that mutations can result in different consequences for microfibril structure with different types of ensuing pathologies. Support for this conclusion is now forthcoming in humans. Mutations in FBN1 can cause Stiff Skin Syndrome, which does not share the clinical features of Marfan syndrome (31), and Weill-Marchesani syndrome, which presents with many clinical features that are the opposite those of Marfan syndrome.3 Neither of these fibrillinopathies demonstrates fragmented microfibrils, which may be required to initiate pathogenesis of the Marfan syndrome.
Fibrillin-1 haploinsufficiency-driven mechanisms include the abnormal activation of transforming growth factor-β signaling due to fibrillin-1 deficiency (10, 11). It was reasoned that, in the absence of critical threshold amounts of fibrillin-1, appropriate sequestration of the large latent transforming growth factor-β complex is lost, because there are insufficient amounts of fibrillin-1 to mediate required interactions with LTBPs (10). This hypothesis will be rigorously tested using H1Δ mice, in which the major residues contributing to the binding sites for LTBPs (24) are deleted.
The addition of novel Fbn1 mutant mice helps to define a spectrum in which disease-causing mechanisms due to haploinsufficiency or due to specific effects on microfibril structure or function can be determined. Results presented in this report now suggest that mutations in fibrillin-1 that destabilize microfibril structure underlie the pathological features of the Marfan syndrome, whereas mutations that do not destabilize microfibril structure do not result in Marfan phenotypes. Because haploinsufficiency of fibrillin-1 may lead to deleterious alterations in the connective tissue in which microfibrils serve critical integrative functions, it is clearly possible that certain cellular responses to a fibrillin-1 haploinsufficient environment are both similar to and different from cellular responses to unstable microfibrils. The available series of Fbn1 mutant mice can be used to determine these similarities and differences and to more precisely identify the threshold requirements for microfibril function.
Supplementary Material
Acknowledgment
We thank Dr. Frank Koentgen and his excellent scientific staff at Ozgene for generating the founder mice used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AR46811 and RC1HL100608-01 (to L. Y. S.), P01 AR049698 (to F. R. and L. Y. S.), and RO1 AR42044 (to F. R.). This work was also supported by the Shriners Hospitals for Children (to D. R. K. and to L. Y. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental movies.
G. Sengle and L. Y. Sakai, manuscript in preparation.
- eGFP
- enhanced green fluorescent protein
- ELISA
- enzyme-linked immunosorbent assay
- Fbn
- fibrillin gene
- FBS
- fetal bovine serum
- H1Δ
- hybrid 1 deletion
- GT-8
- green truncated Fbn1 from found 8
- LTBP
- latent transforming growth factor-β-binding protein
- pAb
- polyclonal antibody
- rF
- recombinant fibrillin
- TBS
- tris-buffered saline
- TBS-T
- tris-buffered saline with Tween 20
- qPCR
- quantitative PCR
- E
- embryonic day
- P
- postnatal day.
REFERENCES
- 1.Sakai L. Y., Keene D. R., Engvall E. (1986) J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., Apfelroth S. D., Hu W., Davis E. C., Sanguineti C., Bonadio J., Mecham R. P., Ramirez F. (1994) J. Cell Biol. 124, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corson G. M., Charbonneau N. L., Keene D. R., Sakai L. Y. (2004) Genomics 83, 461–472 [DOI] [PubMed] [Google Scholar]
- 4.Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. (2003) J. Biol. Chem. 278, 2750–2757 [DOI] [PubMed] [Google Scholar]
- 5.Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. (2005) J. Biol. Chem. 280, 27970–27980 [DOI] [PubMed] [Google Scholar]
- 6.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faivre L., Collod-Beroud G., Loeys B. L., Child A., Binquet C., Gautier E., Callewaert B., Arbustini E., Mayer K., Arslan-Kirchner M., Kiotsekoglou A., Comeglio P., Marziliano N., Dietz H. C., Halliday D., Beroud C., Bonithon-Kopp C., Claustres M., Muti C., Plauchu H., Robinson P. N., Adès L. C., Biggin A., Benetts B., Brett M., Holman K. J., De Backer J., Coucke P., Francke U., De Paepe A., Jondeau G., Boileau C. (2007) Am. J. Hum. Genet. 81, 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira L., Lee S. Y., Gayraud B., Andrikopoulos K., Shapiro S. D., Bunton T., Biery N. J., Dietz H. C., Sakai L. Y., Ramirez F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3819–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Judge D. P., Biery N. J., Keene D. R., Geubtner J., Myers L., Huso D. L., Sakai L. Y., Dietz H. C. (2004) J. Clin. Invest. 114, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neptune E. R., Frischmeyer P. A., Arking D. E., Myers L., Bunton T. E., Gayraud B., Ramirez F., Sakai L. Y., Dietz H. C. (2003) Nat. Genet. 33, 407–411 [DOI] [PubMed] [Google Scholar]
- 11.Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. (2006) Science 312, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietz H. C., McIntosh I., Sakai L. Y., Corson G. M., Chalberg S. C., Pyeritz R. E., Francomano C. A. (1993) Genomics 17, 468–475 [DOI] [PubMed] [Google Scholar]
- 13.Eldadah Z. A., Brenn T., Furthmayr H., Dietz H. C. (1995) J. Clin. Invest. 95, 874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhardt D. P., Mechling D. E., Boswell B. A., Keene D. R., Sakai L. Y., Bächinger H. P. (1997) J. Biol. Chem. 272, 7368–7373 [DOI] [PubMed] [Google Scholar]
- 15.Reinhardt D. P., Ono R. N., Notbohm H., Müller P. K., Bächinger H. P., Sakai L. Y. (2000) J. Biol. Chem. 275, 12339–12345 [DOI] [PubMed] [Google Scholar]
- 16.Aoyama T., Francke U., Dietz H. C., Furthmayr H. (1994) J. Clin. Invest. 94, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira L., Andrikopoulos K., Tian J., Lee S. Y., Keene D. R., Ono R., Reinhardt D. P., Sakai L. Y., Biery N. J., Bunton T., Dietz H. C., Ramirez F. (1997) Nat. Genet. 17, 218–222 [DOI] [PubMed] [Google Scholar]
- 18.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. (2006) J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arteaga-Solis E., Gayraud B., Lee S. Y., Shum L., Sakai L., Ramirez F. (2001) J. Cell Biol. 154, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21.Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., Sakai L. Y. (2003) J. Biol. Chem. 278, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. (1996) J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 23.Keene D. R., Maddox B. K., Kuo H. J., Sakai L. Y., Glanville R. W. (1991) J. Histochem. Cytochem. 39, 441–449 [DOI] [PubMed] [Google Scholar]
- 24.Ono R. N., Sengle G., Charbonneau N. L., Carlberg V., Bächinger H. P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D. B., Ramirez F., Chu M. L., Sakai L. Y. (2009) J. Biol. Chem. 284, 16872–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai L. Y., Keene D. R. (1994) Methods Enzymol. 245, 29–52 [DOI] [PubMed] [Google Scholar]
- 26.Dingemans K. P., Teeling P., Lagendijk J. H., Becker A. E. (2000) Anat. Rec. 258, 1–14 [DOI] [PubMed] [Google Scholar]
- 27.Charbonneau N. L., Jordan C. D., Keene D. R., Lee-Arteaga S., Dietz H. C., Rifkin D. B., Ramirez F., Sakai L. Y. (2010) J. Biol. Chem. 285, 20242–20251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo C. L., Isogai Z., Keene D. R., Hazeki N., Ono R. N., Sengle G., Bächinger H. P., Sakai L. Y. (2007) J. Biol. Chem. 282, 4007–4020 [DOI] [PubMed] [Google Scholar]
- 29.Reinhardt D. P., Gambee J. E., Ono R. N., Bächinger H. P., Sakai L. Y. (2000) J. Biol. Chem. 275, 2205–2210 [DOI] [PubMed] [Google Scholar]
- 30.Hollister D. W., Godfrey M., Sakai L. Y., Pyeritz R. E. (1990) N. Engl. J. Med. 323, 152–159 [DOI] [PubMed] [Google Scholar]
- 31.Loeys B. L., Gerber E. E., Riegert-Johnson D., Iqbal S., Whiteman P., McConnell V., Chillakuri C. R., Macaya D., Coucke P. J., De Paepe A., Judge D. P., Wigley F., Davis E. C., Mardon H. J., Handford P., Keene D. R., Sakai L. Y., Dietz H. C. (2010) Science Transl. Med. 2, 23ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putnam E. A., Cho M., Zinn A. B., Towbin J. A., Byers P. H., Milewicz D. M. (1996) Am. J. Med. Genet. 62, 233–242 [DOI] [PubMed] [Google Scholar]
- 33.Siracusa L. D., McGrath R., Ma Q., Moskow J. J., Manne J., Christner P. J., Buchberg A. M., Jimenez S. A. (1996) Genome Res. 6, 300–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.