Abstract
The pannexins (Panx1, -2, and -3) are a mammalian family of putative single membrane channels discovered through homology to invertebrate gap junction-forming proteins, the innexins. Because connexin gap junction proteins are known regulators of neural stem and progenitor cell proliferation, migration, and specification, we asked whether pannexins, specifically Panx2, play a similar role in the postnatal hippocampus. We show that Panx2 protein is differentially expressed by multipotential progenitor cells and mature neurons. Both in vivo and in vitro, Type I and IIa stem-like neural progenitor cells express an S-palmitoylated Panx2 species localizing to Golgi and endoplasmic reticulum membranes. Protein expression is down-regulated during neurogenesis in neuronally committed Type IIb and III progenitor cells and immature neurons. Panx2 is re-expressed by neurons following maturation. Protein expressed by mature neurons is not palmitoylated and localizes to the plasma membrane. To assess the impact of Panx2 on neuronal differentiation, we used short hairpin RNA to suppress Panx2 expression in Neuro2a cells. Knockdown significantly accelerated the rate of neuronal differentiation. Neuritic extension and the expression of antigenic markers of mature neurons occurred earlier in stable lines expressing Panx2 short hairpin RNA than in controls. Together, these findings describe an endogenous post-translational regulation of Panx2, specific to early neural progenitor cells, and demonstrate that this expression plays a role in modulating the timing of their commitment to a neuronal lineage.
Keywords: Connexin, Neural Stem Cell, Neurodevelopment, Neurodifferentiation, Protein Palmitoylation, Pannexin
Introduction
Neural stem and progenitor cells (NPCs)3 in the hippocampal formation generate new granule neurons throughout adult life. Multipotential Type I and Type IIa NPCs located in the subgranular zone of the dentate gyrus give rise to intermediate progenitor populations (Type IIb and Type III) before terminal neuronal differentiation (reviewed in Ref. 1; see also the schematic in Fig. 4B). The physiological production (and deletion) of these new neurons is implicated in the consolidation of spatial learning and memory (reviewed in Ref. 2), whereas aberrant or impaired neurogenesis is associated with multiple neurological and cognitive disorders, including epilepsy, mood disorders, neurodegenerative diseases, and ischemia (reviewed in Refs. 3 and 4).
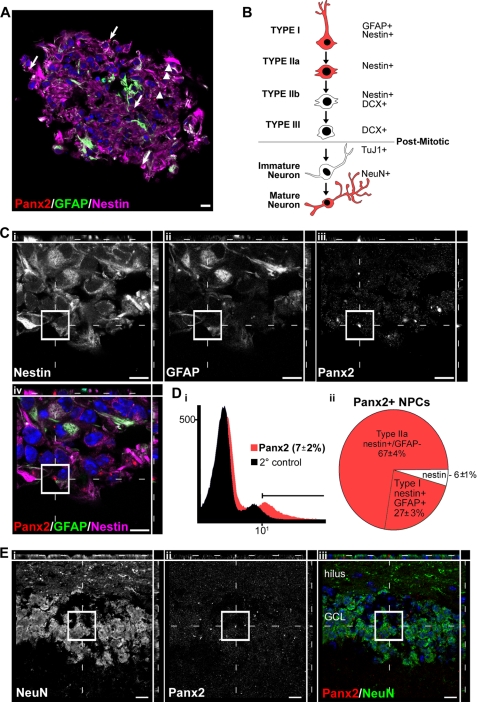
FIGURE 4.
Panx2 expression is restricted to Type I and IIa early stem-like hippocampal neural progenitors and mature neurons and is not expressed in intermediate progenitor populations or immature neurons. A, Panx2 localizes to both Type I NPCs (GFAP+/nestin+; arrows) and Type IIa NPCs (GFAP-negative/nestin+; arrowheads) in neurosphere cultures as assessed by confocal microscopy. B, schematic depicting the lineage progression of hippocampal NPCs over the course of postnatal specification to neurons. The antigenic lineage markers used to identify different cell populations by flow cytometry and immunocytochemistry are indicated. C, higher magnification of a Type I cell expressing nestin (i), GFAP (ii), and a Panx2+ rod-like structure (iii). The merged, pseudocolored image is depicted in iv. A Panx2+ Type IIa cell can be seen in the same field (arrowhead). Panx2 was not detected in DCX+ Type IIb or III NPCs or Tuj1+ immature neurons (supplemental Fig. S2). D, the percentage of cells at DIV 14 expressing Panx2 was quantified (i) and characterized (ii) by flow cytometry analysis. E, because the immature neurons generated in our neurosphere cultures do not mature to NeuN+ cells by DIV 14, expression in adult granule neurons was confirmed in P90 hippocampal tissue. Mature NeuN+ hippocampal granule neurons of the dentate gyrus (i) express Panx2 rod-like structures (ii). The pseudocolored merged image is shown in iii. In all micrographs, Hoechst 33258 was used as a nuclear counterstain (blue). Scale bars, 10 μm. Polyclonal anti-Panx2 (Aviva Systems Biology) was used. GCL, granule cell layer.
The signaling systems that maintain multipotential NPC populations in their “stem-like” state within the adult hippocampal network have only begun to be elucidated (2). One mechanism, the expression of ion-permeable single membrane (ion channels and gap junction hemichannels) as well as double-membrane (gap junction intercellular) channels, is implicated in the control of embryonic and postnatal NPC migration, proliferation, and specification (5–11). Channel proteins, in part, regulate metabolic coupling, adhesion, and calcium wave propagation between NPCs and adjacent terminally differentiated cells (5–11). Pannexins are a family of membrane channel proteins discovered through their homology to the invertebrate family of gap junction proteins, the innexins. Panx1 is expressed in multiple tissues. Panx2 is enriched in the central nervous system. Panx3 is found in skin and cartilage (12–14). Given their sequence homology with innexins and their structural homology with connexins, pannexins were originally proposed to form intercellular channels; however, recent studies suggest that Panx1 and Panx3 more likely form single membrane channels, with Panx1 channels permeable to ions, ATP, and arachidonic acid-related lipids (15–23) (reviewed in Ref. 24).
Little is known about the biology of central nervous system-expressed Panx2. Intriguingly, Panx2 mRNA is detected only in postnatal tissue (25). In the hippocampus, Panx2 mRNA levels increase during the peak period of granule cell neurogenesis (25). Although Panx2 is primarily detected in terminally differentiated neurons, transient expression is observed in glial fibrillary acidic protein (GFAP)-positive cells following brain injury (25–28). No physiological function has yet been ascribed to Panx2 beyond reports that, upon heterologous expression in Xenopus oocytes, Panx2 physically interacts with Panx1 to reduce current amplitude (12, 29). Upon ectopic expression in HEK293 cells, only a small increase in dye uptake is observed in comparison with Panx1 and Panx3 transfection (30).
In this study, we asked whether Panx2 is expressed by postnatal NPCs and whether its expression plays a role in regulating neurogenesis. Using both primary NPC cultures and Neuro2a (N2a) cells to provide functional insight, we show that Panx2 protein expression and post-translational modification modulates the timing of NPC commitment to a neuronal lineage.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary NPC cultures were isolated from postnatal day 3 (P3) C57BL/6 mouse hippocampus and expanded as neurospheres for 14 days in vitro (DIV) as described (10). Hippocampal neurons were cultured as previously described (31) for 7 DIV. N2a cells were cultured in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells (1.8 × 105 cells/60-mm diameter dish) were induced to differentiate to a neuronal phenotype in low serum medium (2% fetal bovine serum) supplemented with 10 μm retinoic acid (32). To label cell membrane, cells were treated with 5 μg/ml Alexa-488-conjugated wheat germ agglutinin (Invitrogen/Molecular Probes) for 5 min at 37 °C in Hanks' buffered salt solution (137 mm NaCl, 5.4 mm KCl, 0.25 mm Na2HPO4, 0.44 mm KH2PO4, 4.2 mm NaHCO3), washed in Hanks' buffered salt solution, and fixed and processed for immunostaining as described below. In loss-of-function experiments, a Panx2-specific (accession number NM_001002005.2) siRNA target sequence (AAGAGCAACTTCATCTTCGAC) was identified using the Ambion siRNA Target Finder (available on-line). The pSilencerTM expression vectors insert design tool was used to design the hairpin siRNA-encoding DNA oligonucleotide inserts (Panx2B-top, 5′-GATCCGAGCAACTTCATCTTCGACTTCAAGAGATCGAAGATGAAGTTGCTCTTA-3′; Panx2B-bottom, 5′-AGCTTAAGAGCAACTTCATCTTCGACCTCTTGAAGTCGAAGATGAAGTTGCTCG-3′) cloned into pSilencerTM 4.1-CMV hygro. Stable clonal lines of empty vector (control) and shRNA-expressing N2a cells were generated, plated on poly-d-lysine-coated coverslips at a density of 1.8 × 105 cells/60-mm diameter dish, and induced to differentiate to a neuronal phenotype as described above. Morphological differentiation was assessed at 0, 8, 16, and 48 h after neuronal induction. Cells were fixed, and brightfield images were captured with a Leica DMXRA2 microscope using a Hamamatsu Orca ER digital camera and Openlab software version 5.05 (Improvision) by an experimenter blinded to the identity of the sample. Quantitation of neurite length and number of neurites/cell was performed on 10 fields/condition at ×20 magnification. At least 400 cells were measured in each condition using the Advanced Measurement Module of Openlab software version 5.05 (Improvision). Percentage differentiation was calculated as the number of cells with neurites of a length >1 times the diameter of the cell soma expressed as a percentage of the total number of cells/field. Counts were performed by two independent investigators blinded as to the identity of samples analyzed. Data from both experimenters were averaged to yield a single data point per field, time point, and condition.
Reverse Transcription-PCR
Total RNA was isolated, and first-strand synthesis and PCR were carried out as described (10) using the following cycling parameters: 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 57 °C for 50 s, and 72 °C for 2 min, and a final step at 72 °C for 7 min. Primers were as follows: 5′-GAGAAAAAGCATACCCGCCAC-3′ and 5′-GGTGAGCAGACATGGAATGA-3′ defining a 269-bp PANX2 amplicon (accession number NM_001002005.2) and 5′-TGGTGCTGAGTATGTCGTGGAGT-3′ and 5′-AGTCTTCTGAGTGGCAGTGATGG-3′ defining a 292-bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplicon (accession number NM_008084.2).
Protein Analyses
Murine hippocampus and cortex were isolated from P0–P90 C57BL/6 mice, P3 NPCs cultured as neurospheres for 14 DIV, N2a cells cultured in complete medium, or N2a cells differentiated for 48 h to a neuronal phenotype. Western analysis was performed as described (10). All procedures were carried out in agreement with the guidelines of the Canadian Council for Animal Care and the University of Ottawa Animal Care Committee. Samples were homogenized in RIPA buffer (10 mm PBS, 150 mm NaCl, 9.1 mm dibasic sodium phosphate, 1.7 mm monobasic sodium phosphate), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 30 μl/ml aprotinin, 10 mm sodium orthovanadate, 100 μg/ml phenylmethylsulfonyl fluoride, and 1 μm NaF) for 30 min and centrifuged for 20 min at 12,000 rpm to remove debris. To assess N-linked glycosylation, lysates were further incubated in 0.5% SDS, 40 mm dithiothreitol and denatured by boiling for 10 min. This protocol also partially depalmitoylates target proteins without significant impact on other post-translational modifications (33, 34). Aliquots of these same lysates were incubated with or without 40 units of N-glycosidase F (New England Biolabs, Pickering, Canada) in a buffer containing 1% Nonidet P-40 and 50 mm sodium phosphate, pH 7.5, for 1 h at 37 °C. A similar protocol was used for Endo H treatment. For dephosphorylation experiments, cells were lysed in a Tris-based RIPA buffer (50 mm Tris-HCl (pH 9.0 at 37 °C) and 10 mm MgCl2 supplemented with 1% Nonidet-P40, 0.1% SDS, 30 μl/ml aprotinin, 100 μg/ml phenylmethylsulfonyl fluoride, and 1 μm NaF) and treated without or with 20 units of shrimp alkaline phosphatase (Promega, Madison, WI) for 1 h at 37 °C. For all Western blots, SDS-PAGE was performed under reducing conditions (dithiothreitol and β-mercaptoethanol) without heating. Palmitoylation was directly assessed by treatment of RIPA lysates with buffer (10 mm PBS) or 1 m hydroxylamine, pH 7.4, a chemical depalmitoylating agent that cleaves thioester linkages, for 20 min at room temperature, followed by Western blot analysis as described (35). Palmitoylation was further quantified by metabolic labeling, immunoprecipitation, and depalmitoylation essentially as described (36) with the following modifications. Neurospheres were labeled with 10 μm BODIPY FL hexadecanoic acid (37) (Invitrogen/Molecular Probes) for 1 h at 37 °C in 95% O2, 5% CO2, followed by lysis in RIPA buffer and immunoprecipitation with polyclonal anti-Panx2 C-term (Zymed Laboratories Inc./Invitrogen). Immunoprecipitation was performed as described (38). After washing with RIPA buffer followed by PBS or 1 m hydroxylamine, pH 7.4, lipids were extracted using a modified Bligh and Dyer protocol (39). Chloroform extracts were evaporated under a constant stream of N2 gas, lipids were resuspended in ethanol, and the concentration of BODIPY FL-C16 released from immunoprecipitates was measured using a BioTek fluorescence microplate reader referenced to a standard curve (485-nm excitation/528-nm emission; BioTek, Winooski, VT). Data were expressed as pm of BODIPY FL-C16/immunoprecipitate.
Primary antibodies used for Western blots were polyclonal anti-Panx2 (1:800; Aviva Systems Biology, San Diego, CA) previously characterized (26) (supplemental Fig. S1), monoclonal anti-GAPDH (1:2000; Imgenex, San Diego, CA), mouse monoclonal anti-Tuj1 (1:10000; Fitzgerald Industries, Concord, MA), mouse monoclonal anti-Sumo-1 (1:1000; Zymed Laboratories Inc./Invitrogen), mouse monoclonal anti-NeuN (1:500; Millipore/Chemicon, Temecula, CA), polyclonal anti-phospho-GSK3β (glycogen synthase kinase 3β, phospho-Ser9) (1:1500; Cell Signaling Technology, Danvers, MA), and polyclonal anti-transferrin (1:2000; Assay Designs/Stressgen, Ann Arbor, MI). Horseradish peroxidase-conjugated secondary antibodies (anti-rabbit and mouse IgGs) diluted 1:2000 were from Jackson Immunoresearch Laboratories (West Grove, PA).
Confocal Immunofluorescence Microscopy
Neurosphere, P7 and P90 mouse brain cryopreservation, serial 10-μm cryosectioning, and immunofluorescence were performed as described (10, 11). Antibodies were diluted in 10 mm PBS supplemented with 0.3% Triton X-100 and 3% bovine serum albumin. Primary antibodies were polyclonal anti-Panx2 (1:250; Aviva Systems Biology), polyclonal anti-Panx2 C-term (1:50; supplemental Fig. S1), rat monoclonal anti-GFAP (1:20; Zymed Laboratories Inc./Invitrogen), mouse monoclonal anti-NeuN (1:100; Millipore/Chemicon), monoclonal anti-nestin (1:50; Chemicon), guinea pig polyclonal anti-doublecortin (DCX; 1:800; Chemicon), monoclonal anti-Tuj1 class III β-tubulin (1:250; RDI), polyclonal anti-NG2 (1:200; Chemicon), rat monoclonal anti-platelet-derived growth factor α receptor (PDGFαR; 1:300; BD Biosciences), monoclonal anti-RIP (1:1000; Chemicon), monoclonal anti-giantin (1:250; Lifespan Biosciences, Seattle, WA), monoclonal anti-calnexin (1:100; BD Biosciences), monoclonal anti-syntaxin (1:100; Sigma), and monoclonal anti-Ki67 (1:250; BD Biosciences). Secondary antibodies were Cy3-conjugated anti-rabbit IgG (1:600), fluorescein isothiocyanate-conjugated anti-rabbit IgG (1:100), Cy3-conjugated anti-guinea pig IgG (1:800), fluorescein isothiocyanate-conjugated anti-mouse IgG (1:100), and Cy5-conjugated anti-mouse IgG (1:400; Jackson ImmunoResearch). Hoechst 33258 (1 μg/ml) was used as a nuclear counterstain. Confocal immunofluorescence imaging was performed on a Zeiss 510 META laser-scanning confocal microscope, and images were captured with Zeiss LSM 5 software version 4.0 SP2 using EC plan-neofluor ×40/1.30 oil DIC M27 or plan-apochromat ×63/1.4 oil DIC M27 objectives with a pinhole size of 1 Airy unit (or equal optical slices for multiple fluorophores) through z-stacks in 0.5-μm optical sections (Zeiss Canada, Toronto, Canada). Three-dimensional digital reconstruction and AVI movies were rendered using Zeiss LSM 5 software version 4.0 SP2 and Axiovision software version 4.7.
Flow Cytometry
NPCs in neurosphere cultures were disaggregated into single cells in versene (Invitrogen) before fixation in 2% formaldehyde. Cell suspensions were permeabilized in 2% fetal bovine serum (PBS/fetal bovine serum) supplemented with 0.18% saponin. Primary antibodies were added directly to aliquots of 105 cells at the following concentrations: Panx2, 1:50 (Aviva Systems Biology); nestin, 1:20; GFAP, 1:20. Following several washes, cells were incubated in secondary antibodies: fluorescein isothiocyanate-conjugated anti-rabbit IgG (1:25), Cy5-conjugated anti-mouse IgG (1:25), and R-phycoerythrin-conjugated anti-rat IgG (1:25; AbD Serotec, Raleigh, NC). Analysis was performed on a Beckman Coulter FC500 flow cytometer with CXP software (Beckman Coulter Canada, Mississauga, Canada).
RESULTS
Panx2 Protein Is Expressed by Type I NPCs in the Neonatal Hippocampus
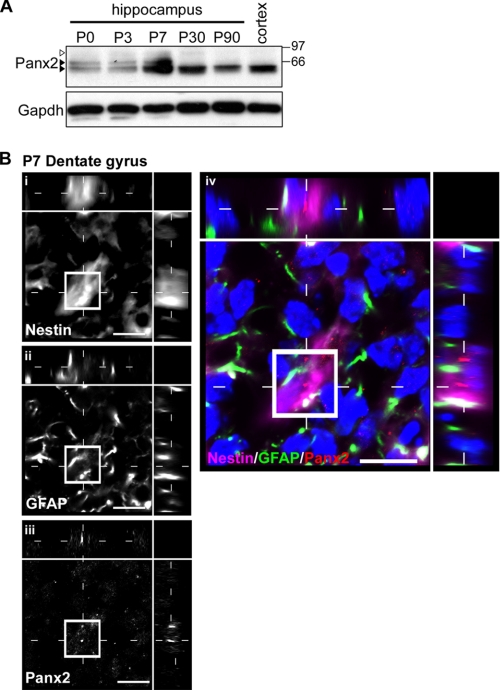
Hippocampal Panx2 protein levels were assessed by Western blotting between postnatal day 0 (P0) and P90 (Fig. 1). In P0–P30 hippocampus, multiple Panx2 species were observed ranging from 60 to 85 kDa (Fig. 1A, black and white arrowheads). An increase in protein expression was evident at P7. In adult brain (P90), the predominant Panx2 species migrated as a single ∼60 kDa band (Fig. 1, lower black arrowhead).
FIGURE 1.
Different Panx2 species are expressed over the course of postnatal hippocampal development. A, three Panx2 species ranging from ∼60 to 85 kDa (arrowheads) are predominant in hippocampal tissue between P0 and P7; two species are detected at P30 (white arrowhead and lower black arrowhead), and a single 60-kDa species predominates at P90, detected also in adult cortex (lower black arrowhead). Membranes were stripped and reprobed for GAPDH as a loading control (lower panel). A representative immunoblot of triplicate experiments is shown. B, Panx2 localizes to neurons (data not shown) and Type I (stem-like) GFAP+/nestin+ NPCs by confocal immunofluorescence of P7 hippocampus. The developing granule cell layer of the dentate gyrus is depicted. Confocal grayscale images with orthogonal views of a triple-labeled Type I NPC are shown in i–iii for each of the primary antibodies used; the pseudocolored merged image is shown in panel iv. Hoechst 33258 was used as a nuclear counterstain (blue). Scale bar, 10 μm. Polyclonal anti-Panx2 (Aviva Systems Biology) was used.
Pyramidal neurons of CA fields of the hippocampus are born embryonically, whereas the majority of granule neurons in the dentate gyrus are born between P3 and P10 (41, 42). Neuronal maturation and synaptogenesis is complete by P30 (43–46). To identify Panx2-expressing cells during the peak period of neurogenesis, double immunolabeling was performed on P7 hippocampal tissue for Panx2 and antigenic markers expressed by stem-like multipotential Type I NPCs as well as mature neuronal markers. In addition to the expected expression in neurons, we found discrete Panx2+ punctae localized to nestin+/GFAP+ Type I NPCs in the developing dentate gyrus (Fig. 1B).
Postnatal NPCs Intrinsically Express Panx2 in Vitro
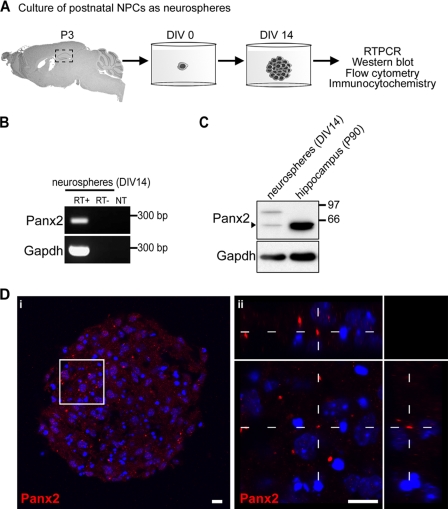
To assess whether Type I hippocampal NPCs intrinsically express Panx2 protein or whether expression depends upon the extrinsic cues provided by the postnatal hippocampal network (47), Type I NPCs were removed from P3 hippocampi and cultured for 14 DIV in suspension (Fig. 2A). Both Panx2 mRNA (Fig. 2B) and protein (Fig. 2C) were readily detected in neurosphere cultures on DIV 14. Panx2 migrated as a ∼60-kDa species in both NPC cultures and adult tissue (Fig. 2B, black arrowhead). Protein expression was elevated in adult tissue compared with NPC cultures (Fig. 2B). As observed in early postnatal hippocampal tissue lysates (Fig. 1A), a higher molecular mass Panx2 protein species (∼85 kDa) was also observed in NPC cultures that was not present in adult tissue (Fig. 2C, white arrowhead). Alternative splicing was ruled out as a potential source of the NPC-specific Panx2 species by Northern blotting (data not shown).
FIGURE 2.
Panx2 is expressed by primary postnatal NPCs cultured as neurospheres. A, schematic of methodology. B, NPCs express Panx2 mRNA. Transcript was detected by reverse transcription-PCR using random-primed RNA extracted from >150 pooled neurosphere cultures. Controls included reactions processed in the absence of reverse transcriptase (RT−) or without template (NT). The same random-primed templates were amplified for GAPDH to confirm template integrity (lower panel). C, Two Panx2 protein species (∼60 kDa (black arrowhead) and ∼85 kDa (white arrowhead)) are detected in neurosphere cultures. The ∼60-kDa species predominates in adult hippocampus. Membranes were stripped and reprobed for GAPDH protein as a loading control (lower panel). A representative immunoblot of triplicate experiments is shown. D, Panx2 expressed in NPC cultures in vitro (i) exhibits a morphology similar to that seen in Type I stem-like NPCs in vivo. The white box indicates the 3× optical zoom inset in ii with orthogonal side views created from a serial confocal z-stack revealing the rod-like Panx2+ structure in greater detail. Three-dimensional reconstructions are provided in supplemental Movies M1 and M2. Hoechst 33258 was used as a nuclear counterstain (blue). Scale bars, 10 μm. Polyclonal anti-Panx2 (Aviva Systems Biology) was used.
Confocal immunofluorescence revealed a similar punctate expression profile in cultured NPCs (Fig. 2D) as observed in vivo (Fig. 1B), confirmed using two Panx2 antibodies directed against two different epitopes, in the first extracellular loop and in the C-terminal intracellular tail, respectively (Fig. 2D and supplemental Fig. S1A). Panx2 invariably appeared as prominent intracellular punctae (Fig. 2D and supplemental Fig. S1B). At higher magnification, these punctae were found to form perinuclear oblong structures ∼1.5 μm in length with a flattened rod-like morphology (supplemental Movies M1 and M2).
Panx2 Is S-Palmitoylated in NPCs
To characterize Panx2 protein expressed in NPCs, we performed biochemical tests for multiple putative post-translational modifications. Online prediction tools identified potential Panx2 phosphorylation, sumoylation, N-glycosylation, and palmitoylation sites that could account for the delayed migration of the NPC-specific species (available on the ExPASy web site).
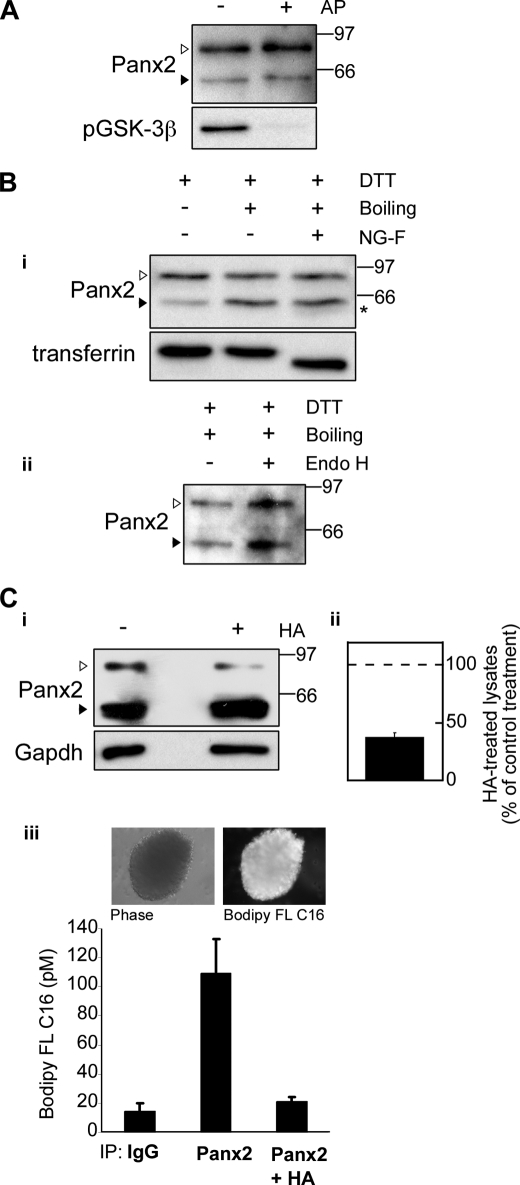
Lysates, prepared from postnatal neurosphere cultures, were dephosphorylated with alkaline phosphatase (AP) (Fig. 3A). Dephosphorylation had no visible effect on the mobility of the NPC-specific species (Fig. 3A, upper panel, white arrowhead). To confirm the efficiency of alkaline phosphatase treatment, blots were stripped and reprobed for phospho-GSK3β (Ser9) (Fig. 3A, lower panel).
FIGURE 3.
NPCs express an S-palmitoylated Panx2 species. Protein lysates were prepared from P3 neurosphere cultures on DIV 14. A, the mobilities of the ∼85-kDa (white arrowhead) and ∼60-kDa species (black arrowhead) were not markedly altered by treatment with alkaline phosphatase (AP). Blots were stripped and reprobed with anti-phospho-GSK3β (Ser9) (pGSK-3β) polyclonal antibody to confirm efficient dephosphorylation (bottom panel). B, the ∼85-kDa species is not N-glycosylated. i, for deglycosylation experiments, lysates were either incubated at 4 °C (first lane) or boiled in the presence of SDS and dithiothreitol (DTT) and incubated at 37 °C with control buffer (second lane) or N-glycosidase F (third lane). Conditions that promote partial depalmitoylation (second and third lanes) induced a shift in the mobility of the ∼85-kDa Panx2 species (white arrowhead) to the ∼60-kDa species (black arrowhead, compare first and second lanes). A small shift to below ∼60 kDa was detected after treatment with N-glycosidase F (asterisk). Blots were stripped and reprobed with anti-transferrin antibody to verify effective deglycosylation (bottom panel). ii, Endo H treatment did not alter Panx2 migration. C, the NPC-specific ∼85-kDa species is a palmitoylated form of Panx2. i, the intensity of the 85-kDa species was markedly reduced following hydroxylamine (HA) treatment. ii, quantitation of this reduction in three independent experiments. Data represent densitometric analysis of the upper band (white arrowhead) standardized to GAPDH and expressed as a percentage of the buffer-treated control. Polyclonal anti-Panx2 (Aviva Systems Biology) was used. iii, to provide direct evidence of palmitoylation, NPCs were metabolically labeled with BODIPY FL hexadecanoic acid (upper panel). Panx2 was immunoprecipitated using the Panx2 C-term polyclonal antibody (Zymed Laboratories Inc./Invitrogen) and treated with buffer or hydroxylamine. Fluorescent palmitate-associated samples after these incubations were extracted and quantified. Data represent pm BODIPY FL hexadecanoic acid extracted from each sample. Panx2 is clearly palmitoylated, and this modification is reversed by treatment with hydroxylamine. All details are as described under “Experimental Procedures.”
To assess sumoylation, Panx2 was immunoprecipitated and blotted with antibodies raised against Sumo1. Although sumoylation was readily detected in input lysates, no evidence of specific sumoylation was observed between the Panx2 and control IgG immunoprecipitates (supplemental Fig. 3).
Previous studies have shown that ectopically expressed Panx1, -2, and -3 in human embryonic kidney cells are glycosylated (16, 30). A minor, potentially N-glycosylated component, of the Panx2 NPC profile was detected following treatment with N-glycosidase F (Fig. 3B, panel i, asterisk). A faint band, migrating just below the mature ∼60-kDa Panx2 species, was consistently observed (Fig. 3B, asterisk, panel i, lane 3), which may indicate endogenous N-glycosylation of the mature species, consistent with previous reports (30). However, deglycosylation did not markedly alter the migration of the NPC-specific upper doublet species (Fig. 3B, upper panel i, white arrowhead, compare lanes 2 and 3). Immunoblots were stripped and reprobed for transferrin to confirm effective deglycosylation (Fig. 3B, lower panel i). Likewise, treatment with Endo H had no effect on Panx2 mobility (Fig. 3C, panel ii).
During the preparation of samples for both N-glycosidase and Endo H treatment, we consistently found that the process of boiling, under reducing conditions, reduced the intensity of the upper Panx2 species and increased the intensity of lower molecular weight species in NPC lysates (Fig. 3B, compare lanes 1 and 2, panel i). This procedure is one means of depalmitoylating proteins (33, 34). To test this hypothesis, we examined the Panx2 sequence for potential palmitoylation sites. Although there is no single palmitoylation consensus sequence, probably due to the large size of the newly discovered DHHC (Asp-His-His-Cys) palmitoylating protein family, conserved patterns can still be predicted (48). Our in silico analysis (CCS-Palm version 2.04) (49) identified at least one potential palmitoylation consensus site (SYLCTYY) at amino acid 246, and several additional sites were detected when analyses were performed at a lower stringency. To directly assess S-palmitoylation, lysates prepared from NPC cultures were treated with 1 m hydroxylamine. Hydroxylamine releases palmitic acid from palmitoylated proteins by chemically cleaving the thioester linkages of fatty acyl groups to cystine (Fig. 3C, panels i–iii). Panx2 lysates were clearly sensitive to hydroxylamine treatment as demonstrated by the reduction in the intensity of the upper Panx2 species to 37.9 ± 3.6% of control (i.e. reactions carried out in the absence of hydroxylamine) quantified in triplicate experiments (Fig. 3C, panels i and ii).
These data were further confirmed by in vivo metabolic labeling of neurosphere cultures using fluorescently labeled palmitate (BODIPY FL hexadecanoic acid; Fig. 3C, panel iii). Protein lysates prepared from labeled cultures were immunoprecipitated with either control IgG or anti-Panx2 C-term antibody. The lipids bound to each sample were extracted (50), and the amount of fluorescent palmitic acid was quantified. We found that 109 ± 34 pm of BODIPY FL palmitate was bound to protein in the Panx2 immunoprecipitates as compared with 15 ± 4 pm associated with the IgG control samples. Furthermore, hydroxylamine completely liberated the S-linked palmitic acid from the Panx2 immunoprecipitates, reducing the Panx2-linked BODIPY FL palmitate concentrations to that of control immunoprecipitates (21 ± 3 pm) (Fig. 3C, panel iii).
Panx2 Protein Expression Is Restricted to Type I and IIa Early Stem-like Hippocampal Neural Progenitors and Terminally Differentiated Mature Granule Neurons in Vitro and in Vivo
Although each neurosphere is derived from a single Type I or, more rarely, a single Type IIa NPC, daughter cells spontaneously adopt distinct lineages over several DIV (supplemental Fig. S2), recapitulating the multistep neurogenic process observed in vivo (Fig. 4B; reviewed in Ref. 1). Confocal immunofluorescence analysis, using well characterized antigenic markers for NPC lineages (Fig. 4B), indicated that Panx2 expression was restricted to the early Type I and IIa stem-like NPCs expressing the filament protein markers nestin and GFAP (Fig. 4, A and C). These results were confirmed by flow cytometry of dissociated cultures. Panx2+ cells made up 7 ± 2% of the total number of cells present in DIV 14 neurosphere culture (Fig. 4D, panel i). Ninety-four percent of the Panx2+ cells expressed nestin (Type IIa NPCs) or co-expressed nestin and GFAP (Type I NPCs) (Fig. 4D, panel ii, and supplemental Fig. S2B). Double labeling with Panx2 and Ki67 confirmed expression in both proliferating and quiescent NPCs (supplemental Fig. S2C). We did not detect Panx2 in DCX+ neuronal progenitors (Type IIb and III) or Tuj1+ immature neurons (supplemental Fig. S2, D and E). It is important to note that our culture conditions were optimized to promote Type I and IIa NPC proliferation and that the TuJ1+ immature neurons that spontaneously differentiate within the neurosphere core do not further develop into mature NeuN+ neurons (10). Although we did not detect Panx2 protein in immature TuJ1+/NeuN-negative immature neurons in vitro or in vivo, Panx2 protein expression was readily detected in mature NeuN+ cells in vivo in the dentate gyrus (Fig. 4E), consistent with previous reports of neuronal expression (28). At no point was Panx2 protein observed in GFAP+/nestin− astrocytes, NG2+/PDGFαR+ oligodendrocyte progenitors, or RIP+ oligodendrocytes (data not shown). Taken together, these data demonstrate that palmitoylated Panx2 is expressed early in the hippocampal neurogenic program by multipotential NPCs but not by their intermediate progeny and is re-expressed (but not palmitoylated) following terminal neuronal differentiation and maturation.
Panx2 Localizes to Different Subcellular Compartments in Multipotential NPCs and Terminally Differentiated Neurons
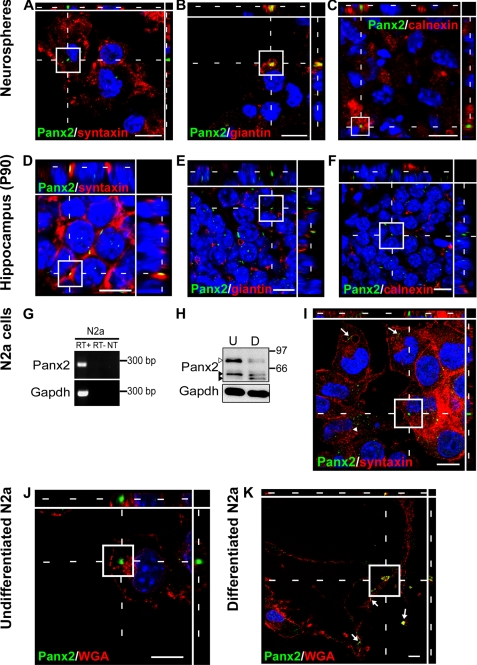
Palmitoylation of neuronal transmembrane proteins, including G-protein-coupled receptors, neuronal cell adhesion molecules, and glutamate receptors, regulates their shuttling between intracellular compartments (reviewed in Ref. 48). To test whether the change in palmitoylation is associated with a shift subcellular localization in Type I NPCs and neurons, we localized Panx2+ rod-like structures to discrete organelles by confocal analysis in combination with immunolabeling for plasma, Golgi apparatus, and ER membranes (Fig. 5). In hippocampal NPCs, Panx2 was not detected at the plasma membrane (Fig. 5A). Instead, Panx2+ rod-like structures colocalized with the Golgi apparatus marker, giantin (Fig. 5B), and also within ER membranes labeled with calnexin (Fig. 5C). In mature hippocampal granule neurons, Panx2 primarily formed rod-like structures in plasma membrane (Fig. 5D) and did not co-localize with giantin (Fig. 5E) or calnexin (Fig. 5F).
FIGURE 5.
Panx2 localizes to different subcellular compartments in Type I and IIa NPCs and mature neurons. Subcellular localization was assessed by confocal microcopy using antigenic markers of or carbohydrate-binding proteins labeling plasma membrane (syntaxin, wheat germ agglutinin), Golgi apparatus (giantin), and endoplasmic reticulum (calnexin). A–C, in NPCs, Panx2 does not localize to the plasma membrane (A) but is found at the Golgi apparatus (B) and associated with ER membranes (C). D–F, in mature granule neurons, Panx2 is found at the plasma membrane (D) but not at Golgi (E) or ER (F) membranes. G–K, this shift in localization is associated with neuronal differentiation. Panx2 mRNA (G, reverse transcription-PCR) and protein (H, Western analysis; I–K, confocal immunofluorescence) are expressed by undifferentiated and differentiated N2a cells (U, undifferentiated; D, differentiated). Both the 85-kDa palmitoylated (white arrowhead) and 60-kDa unpalmitoylated (upper black arrowhead) species are present in undifferentiated N2a cells with intensity of the 85-kDa form reduced following differentiation. A novel Panx2 species is also detected (lower black arrowhead) (H). Abbreviations and controls are as described in the legend to Fig. 2. J and K, further subcellular localization comparisons were made using fluorescently labeled wheat germ agglutinin as a plasma membrane marker. In undifferentiated N2a cells, Panx2 is not detected at the plasma membrane (J). Following 48 h of differentiation with 10 μm retinoic acid, Panx2 colocalizes with wheat germ agglutinin at the plasma membrane in neuritic extensions (K, arrows). This shift in localization is associated with a reduction in Panx2 protein expression, in particular the ∼85-kDa species (white arrowhead, H). In all micrographs, Hoechst 33258 was used as a nuclear counterstain (blue). Scale bars, 10 μm. Polyclonal anti-Panx2 (Aviva Systems Biology) was used.
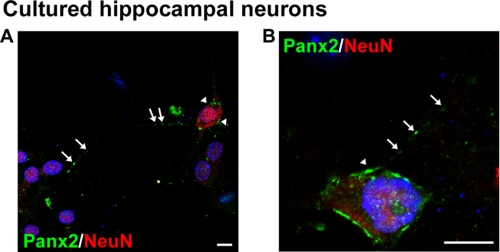
To provide further evidence of a shift in subcellular localization following neuronal differentiation, we employed the mouse neuroblastoma N2a cell model. N2a cells differentiated with retinoic acid in low serum medium express the mature neuronal marker NeuN yet can be easily transfected in their undifferentiated state (32). We found that N2a cells endogenously expressed Panx2 mRNA by reverse transcription-PCR (Fig. 5G) and protein by immunofluorescence and Western analysis (Fig. 5, H and I). In undifferentiated N2a cells, Panx2 formed intracellular rod-like punctae similar to those observed in hippocampal NPCs (Fig. 5, I and J, boxed z-stacks and arrows) but also displayed a dispersed intracellular punctate vesicular pattern not detected in NPC cultures (Fig. 5I, arrowheads). Following a 48-h differentiation to a neuronal phenotype (retinoic acid/low serum), Panx2 relocalized to the plasma membrane of both cell soma and neurites (Fig. 5K). This subcellular redistribution was associated with a reduction in the intensity of the upper species and shift in protein mobility consistent with depalmitoylation (Fig. 5H, upper white arrowhead) as well as the appearance of a de novo ∼60-kDa lower molecular mass species (Fig. 5I, black arrowheads) that may reflect deglycosylation of the mature Panx2 or alterations in other putative post-translational modifications (30). An overall reduction in protein expression was also observed in differentiated N2a cultures. To confirm these data, Panx2 expression was examined in cultured hippocampal neurons by confocal microscopy (Fig. 6, A and B). We found Panx2 immunostaining restricted to NeuN+ neurons at 7 DIV and not the glial cells in our cultures, and it was observed at the plasma membrane of the cell soma as well as in varicosities on neuritic processes.
FIGURE 6.
Panx2 localizes to the plasma membrane of primary hippocampal neurons. A and B, confocal images of hippocampal neuronal cultures immunolabeled for the primarily nuclear marker NeuN and Panx2. The arrows indicate Panx2 present at the plasma membrane of cell soma and along process varicosities. Hoechst 33258 was used as a nuclear counterstain (blue). Scale bars, 10 μm. Polyclonal anti-Panx2 (Aviva Systems Biology) was used.
Panx2 Knockdown Accelerates Neuronal Differentiation
Stable expression of Panx2-targeted shRNA in N2a cells resulted in a ∼50% reduction in protein expression of both the palmitoylated and unpalmitoylated Panx2 species in undifferentiated cultures (Fig. 7, A and C). Remarkably, knockdown accelerated morphological differentiation to a neuronal phenotype following exposure to neurogenic stimuli (Fig. 7B). Eight hours after exposure to retinoic acid, significantly more Panx2 shRNA-expressing cells exhibited a neuronal morphology than empty vector control transfectants (empty vector N2a transfectants, 17.4 ± 3.4%; shRNA-mediated Panx2 knockdown N2a transfectants, 40.4 ± 5.2%; Fig. 7B). Although Panx2 knockdown did not further enhance morphological differentiation at later time points (48 h), stable Panx2 shRNA transfectants expressed higher levels of the mature neuronal marker, NeuN, and lower levels of the immature neuronal marker, TuJ1, than empty vector controls (Fig. 7C). Together, these data demonstrate that the loss of Panx2 from N2a cells accelerates their progression to a neuronal phenotype.
FIGURE 7.
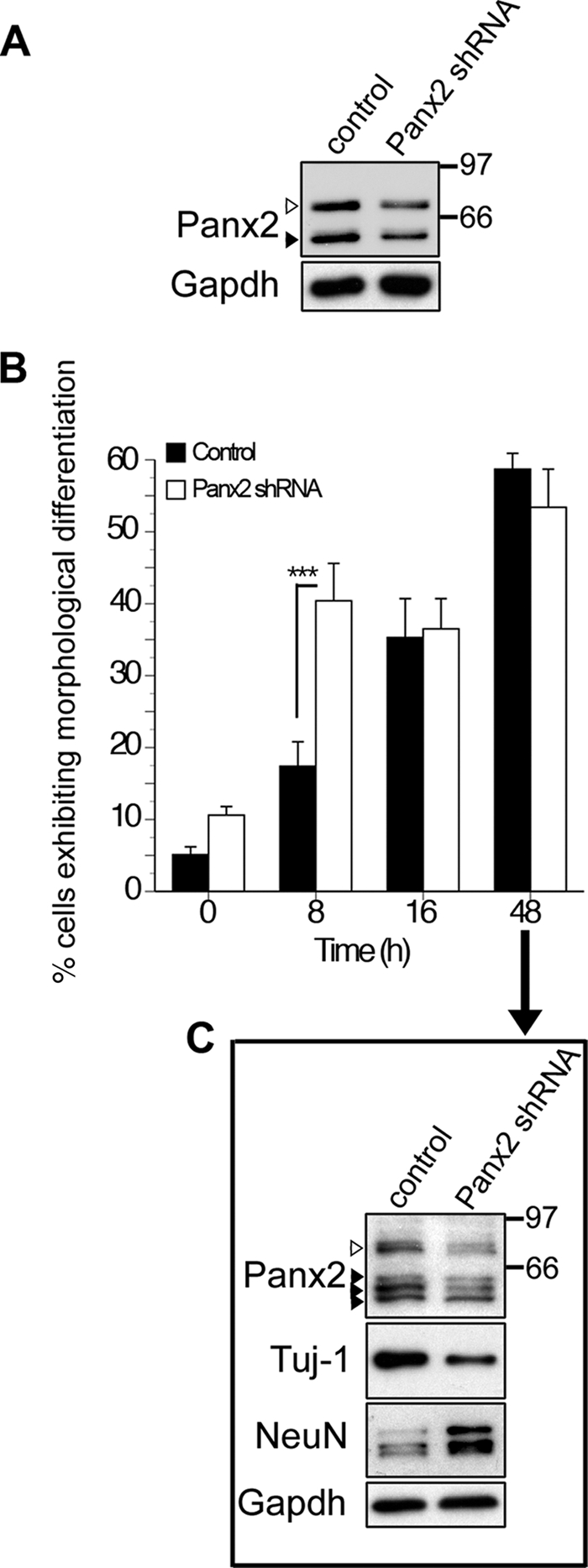
shRNA-mediated knockdown of Panx2 accelerates morphological and biochemical indices of differentiation. A, Western blot of lysates from N2a cells stably transfected with empty vector (first lane) or plasmid encoding Panx2 shRNA (second lane). A knockdown of about 50% is observed in both the ∼85-kDa (white arrowhead) and ∼60-kDa (black arrowhead) Panx2 species. B and C, empty vector controls and Panx2 shRNA-expressing N2a cells were differentiated for up to 48 h with retinoic acid (10 μm) as described under “Experimental Procedures.” Cells were defined as morphologically differentiated if they exhibited one or more neurites of a length greater than the diameter of the cell soma. B, morphological indices of neuronal differentiation appeared earlier when Panx2 expression was suppressed (p < 0.001, two-way analysis of variance, post hoc Bonferroni). C, after 48 h of differentiation, N2a cells expressed multiple Panx2 species reduced in shRNA expressing cells (arrowheads). The white arrowhead indicates the ∼85-kDa species. Although 60% of cells were morphologically differentiated in both groups by this time point (C), N2a cells expressing Panx2 shRNA exhibited higher levels of the mature neuronal antigen, NeuN, and lower levels of Tuj1-reactive βIII tubulin expressed by immature neurons than control cells, indicative of a greater degree of biochemical maturation. Polyclonal anti-Panx2 (Aviva Systems Biology) was used. Error bars, S.E.
DISCUSSION
In this study, we show that Panx2 is dynamically expressed over the course of postnatal hippocampal neurogenesis in vivo and in vitro. At least two Panx2 species are present in stem-like Type I and IIa NPCs that are not detected in their neuronally committed Type IIb and III progeny or in immature neurons. These represent S-palmitoylated and unpalmitoylated species, with S-palmitoylated Panx2 uniquely detected in multipotential NPCs. Only the unpalmitoylated Panx2 species is re-expressed in adult hippocampus following granule neuron maturation. In keeping with evidence that S-palmitoylation regulates intracellular sorting and protein-membrane interactions (48), we found that Panx2 forms rod-like structures in the ER and Golgi, whereas, in adult hippocampal granule cells, Panx2 is expressed at the plasma membrane. This is the first evidence of a cell type-specific post-translational modification of a pannexin associated with a shift in subcellular localization.
In addition to changes in post-translational modification and subcellular localization, we found that shRNA-mediated reduction of Panx2 expression accelerated both morphological and biochemical indices of differentiation in N2a cells. This, along with the absence of Panx2 expression in DCX+ neuronally committed hippocampal NPCs and Tuj-1+ immature neurons, provides evidence that intracellular, palmitoylated Panx2 itself probably plays a modulatory role in neurogenesis, participating either in maintaining “stemness” of multipotential NPCs or, alternatively, in the regulation the timing of their transition to differentiation once exposed to neurogenic stimuli. This potential function is novel for pannexins but has precedent in the innexin literature. Recent work by Yeh et al. (51) has demonstrated that the Caenorhabditis elegans-expressed innexin UNC-7 (and UNC-9) may act as a “corral” sequestering multiple protein components required for presynaptic active zone differentiation to discrete plasma membrane (and perisynaptic) regions during synaptogenesis. It is tempting to speculate that an analogous function for S-palmitoylated Panx2 may be to sequester proteins that promote synaptogenesis at the ER and Golgi in multipotential NPCs, with the loss of Panx2 enabling their redistribution in intermediate progeny, and the re-expression of unpalmitoylated Panx2 at the plasma membrane of mature neurons facilitating the localization of the required presynaptic neuronal machinery to developing active zones over the course of synaptogenesis. Clearly, this hypothesis will require a comprehensive proteomic analysis of Panx2-interacting proteins and their subcellular localization in Type I NPCs and mature neurons.
The formation of rod-like structures at the ER and Golgi apparatus is unusual for a gap junction-associated protein and highlights an important spatial and temporal regulation of protein expression associated with neurogenesis. A growing body of evidence indicates that purine nucleotides act as short range autocrine factors enhancing NPC proliferation via purinergic receptor-mediated mobilization of intracellular calcium stores (52–54). In this context, it is also possible that the regulation of pannexin-mediated hemichannel activity underlies the modulatory role of Panx2 on NPC fate. Not only has Panx1 been shown to form functional hemichannels in GFAP+ glia that release ATP in response to purinergic receptor stimulation (55, 56); ectopically expressed protein has also been shown to form functional calcium-permeable channels in the ER (40). A potential interaction between S-palmitoylated Panx2 and Panx1 in NPCs could target Panx1 to the ER to act as a Ca2+ leak channel (29, 40). Alternatively, expression of unpalmitoylated Panx2 at the plasma membrane could facilitate Panx2-mediated reduction of Panx1 permeability and prime NPCs for neuronal specification (29). It is interesting to note that pharmacological inhibition or siRNA-mediated knockdown of P2X7 ionotropic purinergic receptors also accelerates morphological neuronal differentiation of N2a cells (32), similar to what we observed in the present study. These putative physical and functional interactions may represent redundant mechanisms of regulating Panx1 channel-forming activity and thus modulate the timing of NPC specification. In summary, these data provide the first evidence that Panx2 is uniquely palmitoylated in the Type I and IIa NPCs resident in postnatal mammalian hippocampus and that Panx2 protein expression plays a role in regulating the timing of neuronal differentiation.
Supplementary Material
Acknowledgments
We thank L. Mitchell for assistance in Northern blotting, A. Strom and S. Imbeault for advice in confocal microscopy, R. DesRoches for assistance with lineage assessment, J. Bennett for editorial assistance, Dr. C. Thompson and the Centre for Stroke Recovery for confocal access, Dr. L. Filion for advice in quantitative flow cytometry analysis, and Dr. J. Woulfe for helpful discussion.
This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-62826, the Heart and Stroke Foundation Centre for Stroke Recovery, and Genome Canada operating support (to S. A. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Movies S1 and S2.
- NPC
- neural stem and progenitor cell
- DCX
- doublecortin
- DIV
- day(s) in vitro
- ER
- endoplasmic reticulum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFAP
- glial fibrillary acidic protein
- N2a
- Neuro2a
- Pn
- postnatal day n
- PBS
- phosphate-buffered saline
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- RIPA buffer
- radioimmunoprecipitation assay buffer
- Endo H
- endo-β-N-acetylglucosaminidase H.
REFERENCES
- 1.Kempermann G., Jessberger S., Steiner B., Kronenberg G. (2004) Trends Neurosci. 27, 447–452 [DOI] [PubMed] [Google Scholar]
- 2.Li G., Pleasure S. J. (2010) Curr. Opin. Neurobiol. 20, 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma D. K., Bonaguidi M. A., Ming G. L., Song H. (2009) Cell Res. 19, 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson A., Boekhoorn K., Van Dam A. M., Lucassen P. J. (2008) Genes Brain Behav. 7, Suppl. 1, 28–42 [DOI] [PubMed] [Google Scholar]
- 5.D'Ascenzo M., Piacentini R., Casalbore P., Budoni M., Pallini R., Azzena G. B., Grassi C. (2006) Eur. J. Neurosci. 23, 935–944 [DOI] [PubMed] [Google Scholar]
- 6.Liebau S., Pröpper C., Böckers T., Lehmann-Horn F., Storch A., Grissmer S., Wittekindt O. H. (2006) J. Neurochem. 99, 426–437 [DOI] [PubMed] [Google Scholar]
- 7.Wegner F., Kraft R., Busse K., Härtig W., Schaarschmidt G., Schwarz S. C., Schwarz J., Hevers W. (2008) J. Neurochem. 107, 1056–1069 [DOI] [PubMed] [Google Scholar]
- 8.Walker T. L., White A., Black D. M., Wallace R. H., Sah P., Bartlett P. F. (2008) J. Neurosci. 28, 5240–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias L. A., Kriegstein A. R. (2008) Trends Neurosci. 31, 243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imbeault S., Gauvin L. G., Toeg H. D., Pettit A., Sorbara C. D., Migahed L., DesRoches R., Menzies A. S., Nishii K., Paul D. L., Simon A. M., Bennett S. A. (2009) BMC Neurosci. 10, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melanson-Drapeau L., Beyko S., Davé S., Hebb A. L., Franks D. J., Sellitto C., Paul D. L., Bennett S. A. (2003) J. Neurosci. 23, 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13644–13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. (2000) Curr. Biol. 10, R473–R474 [DOI] [PubMed] [Google Scholar]
- 14.Baranova A., Ivanov D., Petrash N., Pestova A., Skoblov M., Kelmanson I., Shagin D., Nazarenko S., Geraymovych E., Litvin O., Tiunova A., Born T. L., Usman N., Staroverov D., Lukyanov S., Panchin Y. (2004) Genomics 83, 706–716 [DOI] [PubMed] [Google Scholar]
- 15.Bao L., Locovei S., Dahl G. (2004) FEBS Lett. 572, 65–68 [DOI] [PubMed] [Google Scholar]
- 16.Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. (2007) J. Cell Sci. 120, 3772–3783 [DOI] [PubMed] [Google Scholar]
- 17.Penuela S., Celetti S. J., Bhalla R., Shao Q., Laird D. W. (2008) Cell Commun. Adhes. 15, 133–142 [DOI] [PubMed] [Google Scholar]
- 18.Huang Y. J., Maruyama Y., Dvoryanchikov G., Pereira E., Chaudhari N., Roper S. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locovei S., Wang J., Dahl G. (2006) FEBS Lett. 580, 239–244 [DOI] [PubMed] [Google Scholar]
- 20.Thompson R. J., Jackson M. F., Olah M. E., Rungta R. L., Hines D. J., Beazely M. A., MacDonald J. F., MacVicar B. A. (2008) Science 322, 1555–1559 [DOI] [PubMed] [Google Scholar]
- 21.Jiang H., Zhu A. G., Mamczur M., Falck J. R., Lerea K. M., McGiff J. C. (2007) Br. J. Pharmacol. 151, 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locovei S., Bao L., Dahl G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7655–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamura M., Jr., Ruskin D. N., Masino S. A. (2010) J Neurosci. 30, 3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacVicar B. A., Thompson R. J. (2010) Trends Neurosci. 33, 93–102 [DOI] [PubMed] [Google Scholar]
- 25.Vogt A., Hormuzdi S. G., Monyer H. (2005) Brain Res. Mol. Brain Res. 141, 113–120 [DOI] [PubMed] [Google Scholar]
- 26.Wang X. H., Streeter M., Liu Y. P., Zhao H. B. (2009) J. Comp. Neurol. 512, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray A., Zoidl G., Wahle P., Dermietzel R. (2006) Cerebellum 5, 189–192 [DOI] [PubMed] [Google Scholar]
- 28.Zappalà A., Li Volti G., Serapide M. F., Pellitteri R., Falchi M., La Delia F., Cicirata V., Cicirata F. (2007) Neuroscience 148, 653–667 [DOI] [PubMed] [Google Scholar]
- 29.Bruzzone R., Barbe M. T., Jakob N. J., Monyer H. (2005) J. Neurochem. 92, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 30.Penuela S., Bhalla R., Nag K., Laird D. W. (2009) Mol. Biol. Cell 20, 4313–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swayne L. A., Chen L., Hameed S., Barr W., Charlesworth E., Colicos M. A., Zamponi G. W., Braun J. E. (2005) Mol. Cell Neurosci. 30, 339–351 [DOI] [PubMed] [Google Scholar]
- 32.Wu P. Y., Lin Y. C., Chang C. L., Lu H. T., Chin C. H., Hsu T. T., Chu D., Sun S. H. (2009) Cell. Signal. 21, 881–891 [DOI] [PubMed] [Google Scholar]
- 33.Camp L. A., Hofmann S. L. (1993) J. Biol. Chem. 268, 22566–22574 [PubMed] [Google Scholar]
- 34.Schroeder C., Ford C. M., Wharton S. A., Hay A. J. (1994) J. Gen. Virol. 75, 3477–3484 [DOI] [PubMed] [Google Scholar]
- 35.Chamberlain L. H., Burgoyne R. D. (1998) Biochem. J. 335, 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedram A., Razandi M., Sainson R. C., Kim J. K., Hughes C. C., Levin E. R. (2007) J. Biol. Chem. 282, 22278–22288 [DOI] [PubMed] [Google Scholar]
- 37.Thumser A. E., Storch J. (2007) Mol. Cell Biochem. 299, 67–73 [DOI] [PubMed] [Google Scholar]
- 38.Fowler S. L., McLean A. C., Bennett S. A. (2009) Cell Commun. Adhes. 16, 117–130 [DOI] [PubMed] [Google Scholar]
- 39.Ryan S. D., Whitehead S. N., Swayne L. A., Moffat T. C., Hou W., Ethier M., Bourgeois A. J., Rashidian J., Blanchard A. P., Fraser P. E., Park D. S., Figeys D., Bennett S. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20936–20941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanden Abeele F., Bidaux G., Gordienko D., Beck B., Panchin Y. V., Baranova A. V., Ivanov D. V., Skryma R., Prevarskaya N. (2006) J. Cell Biol. 174, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altman J., Bayer S. A. (1990) J. Comp. Neurol. 301, 365–381 [DOI] [PubMed] [Google Scholar]
- 42.Tole S., Christian C., Grove E. A. (1997) Development 124, 4959–4970 [DOI] [PubMed] [Google Scholar]
- 43.Crain B., Cotman C., Taylor D., Lynch G. (1973) Brain Res. 63, 195–204 [DOI] [PubMed] [Google Scholar]
- 44.Steward O., Falk P. M. (1986) J. Neurosci. 6, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toni N., Laplagne D. A., Zhao C., Lombardi G., Ribak C. E., Gage F. H., Schinder A. F. (2008) Nat. Neurosci. 11, 901–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toni N., Teng E. M., Bushong E. A., Aimone J. B., Zhao C., Consiglio A., van Praag H., Martone M. E., Ellisman M. H., Gage F. H. (2007) Nat. Neurosci. 10, 727–734 [DOI] [PubMed] [Google Scholar]
- 47.Alvarez-Buylla A., Lim D. A. (2004) Neuron 41, 683–686 [DOI] [PubMed] [Google Scholar]
- 48.Fukata Y., Fukata M. (2010) Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 49.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wislet-Gendebien S., Visanji N. P., Whitehead S. N., Marsilio D., Hou W., Figeys D., Fraser P. E., Bennett S. A., Tandon A. (2008) BMC Neurosci. 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Yeh E., Kawano T., Ng S., Fetter R., Hung W., Wang Y., Zhen M. (2009) J. Neurosci. 29, 5207–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hogg R. C., Chipperfield H., Whyte K. A., Stafford M. R., Hansen M. A., Cool S. M., Nurcombe V., Adams D. J. (2004) Eur. J. Neurosci. 19, 2410–2420 [DOI] [PubMed] [Google Scholar]
- 53.Mishra S. K., Braun N., Shukla V., Füllgrabe M., Schomerus C., Korf H. W., Gachet C., Ikehara Y., Sévigny J., Robson S. C., Zimmermann H. (2006) Development 133, 675–684 [DOI] [PubMed] [Google Scholar]
- 54.Lin J. H., Takano T., Arcuino G., Wang X., Hu F., Darzynkiewicz Z., Nunes M., Goldman S. A., Nedergaard M. (2007) Dev. Biol. 302, 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iglesias R., Dahl G., Qiu F., Spray D. C., Scemes E. (2009) J. Neurosci. 29, 7092–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suadicani S. O., Iglesias R., Spray D. C., Scemes E. (2009) ASN Neuro. 1, e00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.