Abstract
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway plays important roles in regulating cell motility. TSC2, a downstream target of AKT, is a central player in negatively controlling cell proliferation and protein translation through suppressing the activity of mTOR (mammalian target of rapamycin). However, the function of TSC2 in regulating cell migration remains unclear. Here, we show that TSC2 plays a critical role in the control of cell spreading, polarity, and migration. TSC2-deficient fibroblast cells were impaired in their ability to spread and alter actin cytoskeleton upon stimulation with insulin-like growth factor-1. Using scratch-induced polarization assay, we demonstrate that TSC2(−/−) fibroblast cells polarized poorly toward the wound compared with wild-type cells. Similarly, knockdown of TSC2 expression in colon cancer cells resulted in a marked decrease in cell motility. Functionally, the activation of CDC42- and RAC1-GTPase was largely reduced in TSC2 knock-out fibroblast and TSC2 knockdown cancer cells. Furthermore, overexpression of an activating p110α mutant or short term rapamycin treatment rescued the cell polarization defect in TSC2(−/−) fibroblast cells. Concurrently, the activation of CDC42 and RAC1 increased. The defect in cell migration and CDC42 and RAC1 activation was reversed by reintroducing TSC2 back into TSC2(−/−) fibroblast cells. Taken together, we identified a novel role of TSC2 in controlling cell polarity and migration by regulating CDC42 and RAC1 activation.
Keywords: Cdc42, Cell Migration, Cell Motility, MTOR, Phosphatidylinositol 3-Kinase, Tuberous Sclerosis (Tsc)
Introduction
Tuberous sclerosis is an autosomal dominant disorder characterized by formation of hamartomas in multiple organs, including the brain, kidney, heart, lung, and skin. The mutations are found in genes encoding two tumor suppressors, tuberous sclerosis complexes TSC1 and TSC2, also named harmatin and tuberin, respectively (1). TSC1 and TSC2 function as a complex and exert their tumor suppressor function by negatively regulating the mTOR pathway (2). In the TSC1-TSC2 complex, TSC1 functions as a membrane-tethering anchor protein, whereas TSC2 serves as a GTPase activation protein to promote GTP hydrolysis and inactivation of RHEB, a small G-protein activator of mTOR. Loss of function mutations in either the TSC1 or TSC2 gene lead to abnormal up-regulation of mTOR signaling and are associated with the pathogenesis of the disease. Recent studies have shown that the activity of TSC2 can be regulated by multiple signaling inputs in cells (3). For example, TSC2 lies downstream of the PI3K/AKT2 pathway, and phosphorylation by Akt deceases the GTPase activation protein activity of TSC2 and subsequently actives mTOR (4, 5). On the other hand, phosphorylation by the nutrient-sensing kinase, AMPK, enhances its ability to inactivate RHEB, therefore turning off mTOR-dependent cell growth signaling (6). In addition, TSC2 plays a critical role in controlling the negative feedback regulation of PI3K/Akt signaling (7, 8). Specifically, whereas loss of TSC2 function in tuberous sclerosis and other hamartoma syndromes leads to hyperactivation of mTOR and its downstream substrate p70S6 kinase (p70S6K), activation of AKT is suppressed strongly due to p70S6K-directed down-regulation of IRS-1 protein. This feedback inhibition of PI3K/AKT signaling has been indicated as the major underlying mechanism for the low malignant potential of tumors arisen from TSC2 mutations. Indeed, bypassing the feedback inhibition to activate AKT as in the case of PTEN loss dramatically enhances the tumor malignancy in mouse models (9). Moreover, the negative regulation imposed upon PI3K signaling by TSC2 mutations can be released by treating cells with rapamycin, a specific inhibitor of mTOR (8).
Several previous studies have indicated that TSC1 and TSC2 may play a role in regulating cell migration (10–12). It has been shown that TSC1 interacts with the ezrin-radixin-moesin family of actin-binding proteins and promotes the cell adhesion-mediated activation Rho-GTPase (10). In addition, overexpression of TSC2 results in an increase of Rho-GTP but not RAC1-GTP or CDC42-GTP (11). However, a later study shows that overexpression of TSC2 in TSC2-deficient rat leiomyoma cells promotes RAC activation and inhibition of Rho-GTP (12). The regulatory effect of TSC2 on actin cytoskeleton has been attributed to its ability to interact with TSC1 (12). Collectively, it remains unclear how loss of functional TSC2, as in the case of tuberous sclerosis and other tumors, directly affects cell migration.
The driving force for cell migration is provided by actin cytoskeleton reorganization, a dynamic and complex process orchestrated by activation of small GTPases of the Rho family, particularly Rho, RAC, and CDC42. Among them, Rho activation induces stress fiber formation, RAC promotes actin polymerization to produce lamellipodia, and CDC42 is required for cell polarity (13–15). Many growth factor pathways have been implicated in the regulation of cell migration. Specifically, activation of PI3K downstream of growth factor receptors leads to generation of PtdIns-3,4,5-P3, and this lipid product serves as a scaffold at the leading edge of the cells to facilitate the compartmentalization of effector proteins required for directional sensing and cell polarity (16). The establishment and maintenance of cell polarity is essential in directing cell movement, and inhibition of CDC42 activity prevents cell polarization and results in slower migration. Moreover, PI3K and PtdIns-3,4,5-P3 have been linked to the positive feedback activation of RAC at the front edge of migrating neutrophils (17). Dysregulation of coordinated activation of Rho family GTPases often leads to defects in cell migration. However, hyperactivation of Rho family GTPases has frequently been utilized by cancer cells to promote cell invasion and metastasis (18).
In this study, we examined the loss of TSC2 expression on cell migration and polarization. Using TSC2(−/−) fibroblast cells, we showed that loss of TSC2 inhibits cell polarization and migration. Similarly, knockdown of TSC2 in colon cancer cells resulted in decreased migration rate. Because TSC2 is involved in controlling the signaling amplitude of PI3K/AKT pathway through feedback regulation, we addressed whether loss of TSC2-mediated inhibition of PI3K signaling contributes to the migration defect. Furthermore, we determined the activation of RAC1 and CDC42 in TSC2(−/−) fibroblasts and TSC2 knockdown colon cancer cells.
EXPERIMENTAL PROCEDURES
Reagents
The retroviral expression plasmid for p110αH1074R, pBabe-p110αH1074R, and the pBabe-puro control vector were obtained from Addgene. The wild-type and TSC2(−/−) rat embryonic fibroblast (REF) cells were generously provided by Dr. Jin Q. Cheng (H. Lee Moffitt Cancer Center and Research Institute). The retroviral expression plasmid for wild-type human TSC2, pBabe-TSC2, was a gift from Dr. Sourav Ghosh (The University of Arizona College of Medicine). The following rabbit antibodies were from Cell Signaling: anti-TSC2, anti-AKT (detect total AKT protein), and phospho-AKT (the Ser473 site). The anti-RAC1 and anti-CDC42 mouse monoclonal antibodies were purchased from Millipore. The anti-GM-130 monoclonal antibody was obtained from BD Transduction Laboratories. The anti-γ tubulin monoclonal antibody was from Sigma-Aldrich.
Cells
The following cell culture media were used for the cell lines specified: human colon cancer HCT116 cells, McCoy's 5A medium; human colon cancer KM20 cells, minimum Eagle's medium plus sodium pyruvate, nonessential amino acids, and essential vitamin; 293T cells, Dulbecco's modified Eagle's medium; and REF cells, Dulbecco's modified Eagle's medium/F12 (50:50). All media were supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin. Transient transfection of HCT116 cells with small interfering RNA targeting TSC2 (siGENOME SMARTpool siRNA, Dharmacon) was carried out using Lipofectamine 2000 (Invitrogen) by following the manufacturer's protocol. The final concentration of small interfering RNA used was 50 nm.
Generation of TSC2 Knockdown Cells Using Lentivirus-mediated Delivery of shRNA
The shRNAs for the human TSC2 gene used in this study were constructed in a pLKO.1-puro vector and obtained from Addgene. The targeting sequence is 5′-CACTGGCCTTGGACGGTATTG-3′. A plasmid carrying a nontargeting sequence was used to create the control cells. For virus packaging, the control or TSC2-specific shRNA constructs were co-transfected with Mission lentiviral packing mix (Sigma-Aldrich) into 293T cells using FuGENE 6. The colon cancer cells including HCT116 and KM20 were infected with virus containing media, and the stable knockdown cells were obtained through selection with puromycin.
Generation of p110αH1074R- or TSC2-overexpressing Stable Cell Lines
293T cells were co-transfected with pBabe-puro vector, pBabe-p110αH1074R, or pBabe-TSC2, and a packaging plasmid pCL (Addgene) using FuGENE 6. The cell medium was collected at 72 h post-transfection and used directly to infect wild-type and TSC2(−/−) REF cells. The stable REF cells were selected using puromycin.
Cell Spreading Assay
REF cells were starved in Dulbecco's modified Eagle's medium/F12 with 0.1% bovine serum albumin for 2 h and collected by trypsinization. The cells were plated onto fibronectin-coated glass coverslips for the indicated times. The phase images were taken with a 10× objective (Nikon TE2000). The spread cells were defined as cells with extended processes and not phase-bright, whereas the nonspread cells were round and phase-bright. The percentage of spreading cells were calculated accordingly.
Wound Healing and Cell Polarization Assays
A monolayer of confluent REF cells grown in a 6-well culture plate was wounded with a linear scratch by a sterile 200-μl pipette tip. The phase-contrast images were taken at 0 and 15 h after the scratch wounds were made (4× objective, Nikon TE2000). To monitor cell polarization, REF cells were grown on coverslips and allowed to reach confluency. A larger wound was made using a sterile 1-ml pipette tip. The cells were washed and returned to regular growth medium with 10% fetal bovine serum for 15 h. Immunofluorescence staining was performed as described previously (14). Briefly, the cells were fixed in 4% paraformaldehyde and permeabilized using 0.3% Triton X-100. The localization of the Golgi network was detected using the anti-GM-130 antibody and visualized using Alexa488-conjugated secondary antibody (Invitrogen). Actin was stained using Texas Red-conjugated phalloidin. The nuclei of the cells were stained with 4′,6-diamidino-2-phenylindole-containing mounting medium. Images were taken using a Nikon TE2000 inverted microscope with a 20× objective. The cells with Golgi localized within 120° sectors facing the wound were scored positive as polarized cells.
Time-lapse Live Cell Imaging and Analysis
To record the migration of live REF cells in the wound healing assay, a monolayer of confluent cells grown in a glass-bottom culture dish were wounded as described above. The cells at the edge of the scratch wound were monitored at 37 °C using a Nikon BioStation IM equipped with a CO2 incubation chamber. Time-lapse phase images were taken every 30 min for 15 h with a 20× objective. The movement of cells was tracked and analyzed using Nikon Element AR software.
Trans-well Migration Assay
The control and TSC2 knockdown colon cancer cells were cultured overnight in serum-free medium. The next day, cells were trypsinized, resuspended in serum-free medium containing 0.1% bovine serum albumin, and added to the top well of each migration chamber with an 8-μm pore size membrane (Corning). The migration was induced by the presence of collagen (15 μg/ml) and IGF-1 (20 ng/ml) in the bottom chambers. To assess the effect of rapamycin on cell migration, rapamycin (20 nm) or dimethyl sulfoxide were included in the bottom-chamber medium. After 5 or 18 h (for HCT116 and KM20 cells, respectively), the inserts were fixed in methanol and stained with 0.5% crystal violet. The cells that have not migrated were removed from the top chambers using cotton swaps. The numbers of migrated cells were counted using an inverted microscope.
Pulldown Assay for Activated CDC42 and RAC1
To assess the amount of activated CDC42 and RAC1 in REF or colon cancer cells, GST pulldown assays were performed using the GST-tagged p21 binding domain of PAK1 (GST-PBD) as described previously (19). Briefly, GST-PBD fusion proteins were expressed in bacteria and purified using glutathione-Sepharose. The REF and colon cancer cells were grown to ∼70% confluency in regular growth medium containing fetal bovine serum. For rapamycin treatment, the cells were incubated in medium containing 20 nm rapamycin for 5 h. Subsequently, the cells were lysed in lysis buffer (50 mm Tris, pH 7.4, 100 mm NaCl, 1% Nonidet P-40, 10% glycerol, 2 mm MgCl2, and protease inhibitor cocktails), and soluble proteins were incubated with purified GST-PBD to pull down activated CDC42 and RAC. The amount of total and active CDC42 and RAC1 was detected using CDC42 and RAC1 antibodies.
RESULTS
Loss of TSC2 Expression Inhibits Cell Spreading and Disrupts Actin Cytoskeleton Rearrangement
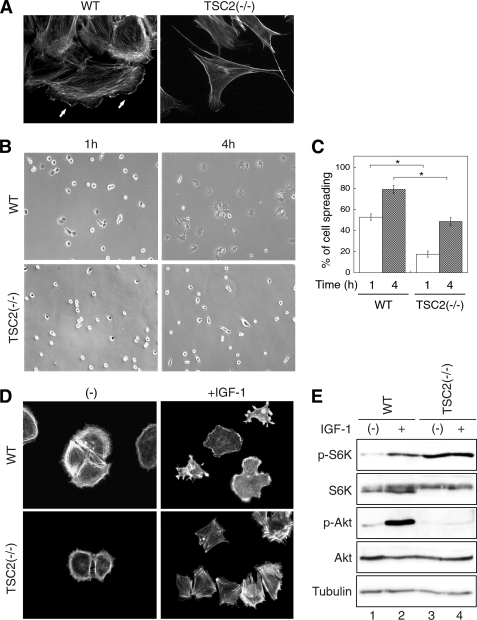
During the experiments to examine the negative feedback regulation mediated by TSC2 using TSC2(−/−) rat fibroblast cells, we were intrigued by the observation that the morphology of WT and TSC2(−/−) cells showed striking differences. When adhered to the culture plates, the formation of lamellipodia and filopodia was readily observed in WT REF cells (Fig. 1A, lamellipodia indicated by solid arrows), whereas TSC2(−/−) cells showed elongated spindle-like morphology, however, apparently completely lacking lamellipodia. The numbers of filopodia were largely reduced and observed only at the end of cell protrusions in TSC2(−/−) cells (Fig. 1A). To determine the effect of TSC2 deletion on cell spreading, WT and TSC2(−/−) cells were trypsinized, reseeded onto fibronectin-coated coverglasses, and monitored for the morphological changes during the time course of cell attachment. As shown in Fig. 1B, a marked difference in the appearance of cell spreading on fibronectin was observed at 1 and 4 h. At the 1-h time point, ∼50% of WT cells showed spread out morphology with clear formation of lamellipodia and filopodia, and >80% of the cells are fully spread out by 4 h. In contrast, <20% of TSC2(−/−) cells at 1 h, and 50% at 4 h appeared to establish attachments with fibronectin, and an elongated morphology was observed as well (Fig. 1C).
FIGURE 1.
Loss of TSC2 expression inhibits cell spreading and actin rearrangement. A, morphology of WT and TSC2(−/−) REF cells on culture plates. WT and TSC2(−/−) REF cells grown on glass coverslips were fixed and stained with Texas Red-conjugated phalloidin. Formation of lamellipodia was observed in WT cells as indicated by solid arrows. B, WT and TSC2(−/−) REF cells were serum-starved for 2 h and replated onto fibronectin-coated coverslips for 1 and 4 h. Representative phase contrast images were taken from the WT and TSC2(−/−) cells after spreading for indicated time. C, quantitative representation of summarized results obtained from three independent experiments. The percentage of cell spreading was calculated as described under “Experimental Procedures.” For statistical analysis, the data obtained from the TSC2(−/−) cells were compared with the WT cells, and the p values were determined by two-sample t tests. Data represent the mean ± S.E. (n = 3; *, p < 0.05). D, after allowing cells to spread on fibronectin-coated coverslips for 1 h, one set of the cells were treated with IGF-1 (20 ng/ml) for 5 min. The cells were subsequently fixed and stained with Texas Red-conjugated phalloidin. Images shown were obtained with a 20× objective. E, activation of PI3K signaling is attenuated in TSC2(−/−) REF cells. WT and TSC2(−/−) REF cells were serum-starved for 2 h and subsequently stimulated with IGF-1 (20 ng/ml) for 10 min. The cells lysates were prepared and analyzed for the phosphorylation status of p70S6K and AKT.
Next, we determined whether loss of TSC2 affects actin cytoskeleton organization. WT and TSC2(−/−) cells were reseeded onto fibronectin-coated coverglasses as in the cell-spreading experiments and treated with IGF-1 after 1 h of spreading. The cells were stained with Texas Red-conjugated phalloidin to reveal the growth factor-induced rearrangement of actin cytoskeleton. Compared with untreated WT cells, TSC2(−/−) cells showed a more contracted actin structure lacking detectable actin staining other than in the cortical region (Fig. 1D, left panels). Upon IGF-1 treatment, a striking difference in actin cytoskeleton rearrangement was observed in TSC2(−/−) cells compared with WT cells. Whereas the actin structure was rearranged to localize to the cell margin upon formation of lamellipodia and filopodia in WT cells, abundant actin stress fiber formation was observed in TSC2(−/−) cells, and no lamellipodia and filopodia were formed (Fig. 1D, right panels). Taken together, these observations indicate that TSC2 plays an important role in the regulation of actin cytoskeleton assembly, including lamellipodia and filopodia formation to affect cell spreading and morphology. To confirm that the activation of PI3K signaling is attenuated largely in TSC2(−/−) cells, the cells were subjected to IGF-1 treatment and analyzed for activation of p70S6K and AKT. Consistent with previous findings (7, 8), loss of TSC2 expression resulted in hyperactivation of p70S6K and impaired activation of AKT indicating that TSC2(−/−) cells are controlled by the p70S6K-dependent negative feedback regulation.
Loss of TSC2 Expression Inhibits Directional Cell Migration and Polarization
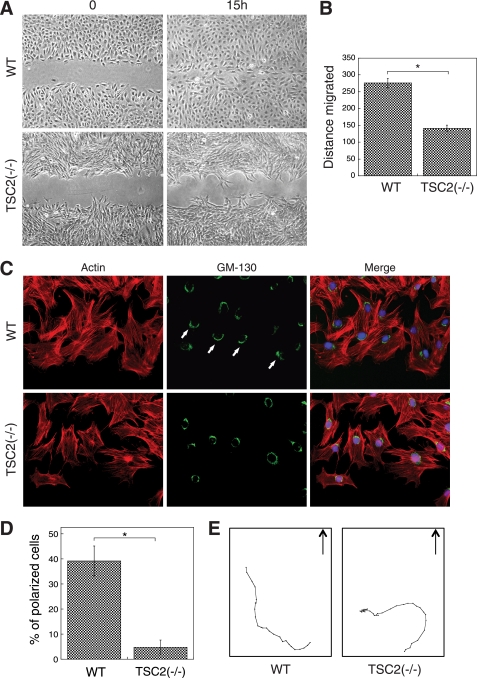
To examine the role of TSC2 in regulating cell migration, the rate of REF cell migration was determined using in vitro wound-healing assays. A confluent monolayer of WT or TSC2(−/−) REF cells were scratched and allowed to migrate into the wound area for 15 h. As shown in Fig. 2A, although the wound area was healed completely by WT cells, a significant gap remained for TSC2(−/−) cells. The rate of migration was ∼50% slower for TSC2(−/−) cells compared with WT cells (Fig. 2B).
FIGURE 2.
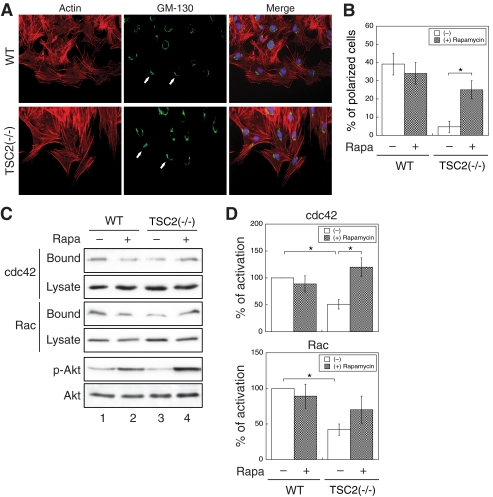
Loss of TSC2 expression inhibits cell migration and prevents cell polarization. The confluent monolayer of WT and TSC2(−/−) REF cells were scratched with a sterile pipette tip. A, shown are phase-contrast images showing the size of the wounds at 0 and 15 h. B, shown is a quantitative representation of summarized results obtained from three independent scratch assay experiments. The distance migrated (μm) was measured and quantified using Nikon Element software. Data represent the mean ± S.E. (n = 3; *, p < 0.05). C, 15 h post-wounding, WT and TSC2(−/−) REF cells were stained with the anti-GM-130 antibody and visualized using the Alexa488-conjugated secondary antibody (GM-130 panels). The actin was visualized using Texas Red-conjugated phalloidin (Actin panels). The nuclei were stained with 4′,6-diamidino-2-phenylindole. The scratch wounds were located parallel to the bottom of each image. The polarized cells with Golgi facing the wound are marked with solid arrows. D, the quantitative results showing the percentage of polarized cells are shown. For each experiment, six to eight images were taken along the scratch, and 100–120 total cells located at the edge of the wound were counted. The cells with Golgi orientated facing the wound are considered polarized. Data represent the mean ± S.E. (n = 3; *, p < 0.05). E, representative trace of a single migrating WT or TSC2(−/−) cell is shown. Time-lapse images of migrating live cells were taken every 30 min for 15 h during cell migration, and the position of a single cell was tracked at each time point using Nikon Element software. The cells start out from the bottom of the graphs, and the arrows indicate the direction of the cell movement if moving toward the wound.
We next examined the role of TSC2 in controlling polarity establishment using the wound-healing assay. In this assay, directional movement of cells is initiated by a scratch through the cell monolayer, and cells at the wound edge polarize toward the wound, allowing cells to migrate in a direction perpendicular to the wound (15). In our study, polarization of the cells is visualized by reorientation of the Golgi forward to face the wound. As shown in Fig. 2, C and D, >40% of WT REF cells clearly polarized along the edge of the wound as the Golgi moved to the front of the cell facing the wound. In marked contrast, almost all of TSC2(−/−) cells failed to polarize with <5% cells showing reorientation of the Golgi. Interestingly, when tracking the cell movement over the entire course of the wound-healing assay, we found that WT cells migrate toward the wound in a more persistent and efficient manner, whereas TSC2(−/−) cells tended to move around in all directions (Fig. 2E and supplemental Movies 1 and 2). Although the total distance traveled by WT and TSC2(−/−) cells was not significantly different, the final distance of a single cell from its starting origin was decreased by 30–50% in TSC2(−/−) cells. Taken together, these results suggest that loss of TSC2 expression blocks the establishment of cell polarization, resulting in a large decrease in the rate of cell migration.
Knockdown of TSC2 in Colon Cancer Cells Inhibits Cell Migration
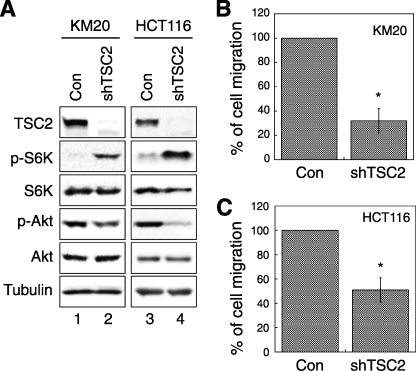
To further confirm the role of TSC2 in regulating cell migration, we generated stable TSC2 knockdown cells using lentivirus-based shRNA. Fig. 3A shows that TSC2 protein expression was decreased in both KM20 (lanes 1 and 2) and HCT116 (lanes 3 and 4) colon cancer knockdown cells. As reported previously, whereas phosphorylation at Thr389 of p70S6K is up-regulated in TSC2 knockdown cells, phosphorylation of Akt is inhibited indicating the activation of the negative feedback loop upon loss of TSC2 expression (Fig. 3A). Because both colon cancer cell lines are from epithelial cell origin, the rate of migration is difficult to measure within a reliable time frame with the wound-healing assay. Thus, trans-well migration assays were performed to determine whether altering TSC2 expression affects cell migration. Consistent with the results obtained from REF cells in the wound-healing assay, knockdown of TSC2 largely reduced the rate of cell migration. The number of cells migrated to the lower chamber decreased by ∼65 and ∼50% for KM20 and HCT116 cells, respectively (Fig. 3, B and C). To confirm the specificity of TSC2-specific shRNA, HCT116 cells were transiently transfected with a pool of small interfering RNAs targeting endogenous TSC2. Consistent with the results obtained from the stable knockdown cells, depletion of TSC2 expression reduced the number of cells migrated by ∼70% as assessed using trans-well migration assays (data not shown). Collectively, our findings indicate that loss of TSC2 negatively affects cell migration.
FIGURE 3.
Knockdown of TSC2 in colon cancer cells impairs cell migration. A, Western blots of cell lysates prepared from TSC2 stable knockdown cells (shTSC2) were analyzed using the following antibodies to show the phosphorylation status or the total protein expression: TSC2, phospho-p70S6K (Thr389 site, p-S6K), p70S6K (S6K), phospho-Akt (Ser473 site, p-Akt), Akt, and γ tubulin antibodies. B, KM20 control and TSC2 knockdown cells were subjected to a two-chamber trans-well migration assay. The numbers of migrated cells were quantified and normalized to the control cells. C, HCT116 control and TSC2 knockdown cells were subjected to a two-chamber migration assay. The numbers of migrated cells were quantified, normalized to the control (Con) cells, and expressed as percentage of migration. Data represent mean ± S.E. (n = 4; *, p < 0.05).
Loss of TSC2 Expression Suppresses RAC1 and CDC42 Activation
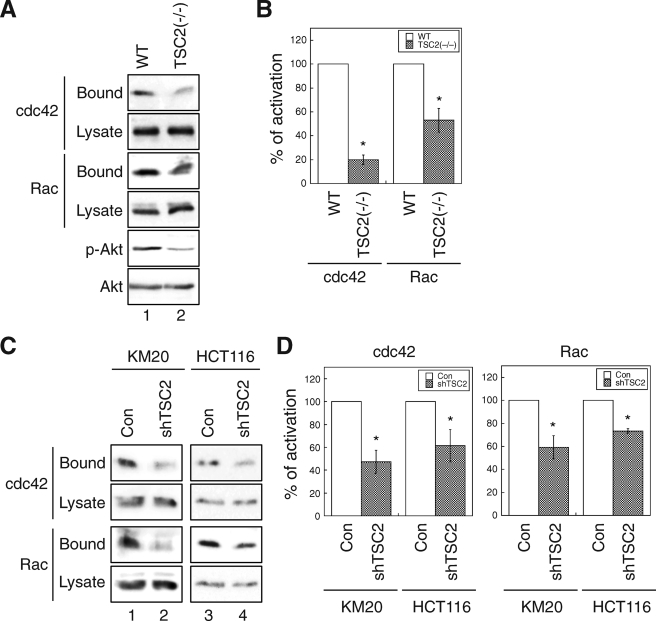
It is accepted generally that alteration of the function of Rho family GTPases leads to defects in cell migration. Particularly, activation of RAC is linked to formation of lamellipodia, and CDC42 is required to establish cell polarity in migration (13, 15). As we found that TSC2(−/−) REF cells lack lamellipodia formation when spread on fibronectin (Fig. 1) and polarize poorly (Fig. 2), GST pulldown activity assays were performed to examine whether loss of TSC2 expression affects the activation of RAC1 and CDC42 in REF and colon cancer cells. As shown in Fig. 4, A and B, the amount of activated CDC42 and RAC1 (GST-PBD bound) was decreased by ∼80 and ∼50%, respectively, in TSC2(−/−) cells compared with WT cells. Similarly, CDC42 and RAC1 activation was reduced to 50–70% of the control level in TSC2 knockdown KM20 (Fig. 4C, lanes 1 and 2) and HCT116 cells (lanes 3 and 4) as well (Fig. 4D). These results suggested that TSC2 expression is required to maintain CDC42 and RAC1 activation in cells, and this decrease in CDC42 and RAC1 activity may account for the cell migration defects observed in TSC2 knock-out and knockdown cells.
FIGURE 4.
Loss of TSC2 expression represses the activation of CDC42 and RAC1. A, WT and TSC2(−/−) REF cells were analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The amount of activated and total CDC42 and RAC1 in the cells were detected using the anti-CDC42 and anti-RAC1 antibodies on Western blots. The GST-PBD bound panels represent the activated fraction of CDC42 and RAC1, and 10% of cell lysates were used to show the total expression of CDC42 and RAC1 (Lysate panels). The p-Akt panel shows the phosphorylation level of AKT at Ser473 in the cell lysates. B, the percentage of activation was quantified by normalizing the level of activated CDC42 and RAC1 (Bound panels) to the total proteins in the lysates (Lysate panels). Data shown in the graph represent the mean ± S.E. (n = 3; *, p < 0.05). C, the control and TSC2 knockdown colon cancer cells, including KM20 and HCT116, were analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The activated and total CDC42 and RAC1 were analyzed as in A. D, the percentage of activation was quantified by normalizing the level of activated CDC42 and RAC1 (Bound panels) to total proteins in the lysates (Lysate panels). Data shown in the graphs represent the mean ± S.E. (n = 4; *, p < 0.05).
Rapamycin Treatment Partially Rescues the Polarization Defect in TSC2(−/−) REF Cells
Because it has been well documented that PI3K and its lipid product PtdIns-3,4,5-P3 closely regulate cell migration, and loss of TSC2 inhibits PI3K/AKT activity through activation of mTOR and p70S6K (8), we hypothesized that loss of TSC2-induced inhibition of cell motility may be due to the negative feedback regulation of PI3K. As discussed above, treating cells with rapamycin releases TSC2-mediated inhibition of PI3K signaling. Here, we examined whether the defect in cell polarization in TSC2(−/−) REF cells can be rescued by rapamycin. WT and TSC2(−/−) REF cells were scratched and allowed to polarize along the edge of the wound in the presence or absence of rapamycin. In the presence of rapamycin, polarization of WT REF cells was slightly less efficient. However, whereas TSC2(−/−) cells failed to polarize under the control condition, the percentage of cells polarized increased from ∼5% to more than 25% as indicated by the Golgi reorientation in the presence of rapamycin (Fig. 5, A and B).
FIGURE 5.
Rapamycin treatment partially rescues the polarization and CDC42/RAC1 activation defects in TSC2(−/−) REF cells. A, the confluent monolayer of WT and TSC2(−/−) REF cells were scratched with a sterile pipette tip. Rapamycin (Rapa; 20 nm) was added to cell medium. 15 h after wounding, the WT and TSC2(−/−) REF cells were stained with the anti-GM-130 antibody and visualized using the Alexa488-conjugated secondary antibody (GM-130 panels). The actin was visualized with Texas Red-conjugated phalloidin (Actin panels). The nuclei were stained with 4′,6-diamidino-2-phenylindole. The scratch wounds were located parallel to the bottom of each image. The polarized cells with Golgi facing the wound are marked with solid arrows. B, the quantitative results showing the percentage of polarized cells are shown. For each experiment, six to eight images were taken along the scratch, and 100–120 total cells located at the edge of the wound were counted. The cells with Golgi orientated facing the wound are considered polarized. Data represent the mean ± S.E. (n = 3; *, p < 0.05). C, WT and TSC2(−/−) REF cells were treated with dimethyl sulfoxide or rapamycin (20 nm) for 5 h and analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The amount of activated and total CDC42 and RAC1 in the cells were detected using the anti-CDC42 and anti-RAC1 antibodies on Western blots. The GST-PBD bound panels represent the activated fraction of CDC42 and RAC1, and 10% of cell lysates were used to show the total expression of CDC42 and RAC (Lysate panels). The p-Akt and Akt panels show the phosphorylation level of AKT at Ser473 and total AKT expression in the cell lysates, respectively. D, the percentage of activation was quantified by normalizing the level of activated CDC42 and RAC1 (Bound panels) to total proteins in the lysates (Lysate panels). Data shown in the graphs represent the mean ± S.E. (n = 3; *, p < 0.05).
We next determined the activation of CDC42 and RAC1 in the presence of rapamycin. Consistent with previous studies, treating cells with rapamycin released the negative regulation of PI3K/AKT signaling as phosphorylation of AKT increased in the presence of rapamycin (Fig. 5C). Under the same condition, the amount of activated CDC42 was significantly increased in TSC2(−/−) cells (Fig. 5C, lanes 3 and 4, and D). Activation of RAC1 was increased with rapamycin treatment in TSC2(−/−) cells as well, although the difference compared with untreated cells was not significant (Fig. 5, C and D). In addition, the morphology of TSC2(−/−) cells was not altered upon rapamycin treatment. Taken together, our results showed that the negative feedback loop activated by loss of TSC2 contributes to the impaired cell polarization observed in TSC2(−/−) cells, and treating the cells with rapamycin allows the re-establishment of polarity during cell migration.
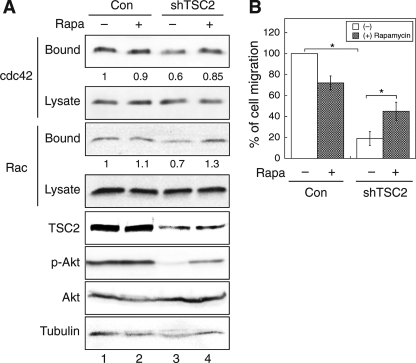
Rapamycin Treatment Increases the Rate of Migration in TSC2 Knockdown Colon Cancer Cells
To confirm the effects of rapamycin on TSC2(−/−) cells, we examined whether rapamycin treatment improves the rate of migration in TSC2 knockdown colon cancer cells. In addition, the activation of CDC42 and RAC1 was determined in the control and TSC2 knockdown KM20 cells upon rapamycin treatment. As shown in Fig. 6A, treating cells with rapamycin had no effect on the amount of activated CDC42 and RAC1 in control cells (lanes 1 and 2), whereas activation of CDC42 and RAC1 was increased largely in the TSC2 knockdown cells to a similar level as in the control cells (lanes 3 and 4). In addition, the control and TSC2 knockdown KM20 cells were subjected to trans-well migration assay, and the rate of migration was determined by counting the cells migrated to the lower chamber. As reported in previous studies (21), treating cells with rapamycin reduced the rate of cell migration in the control cells. However, the number of TSC2 knockdown cells migrated increased by 2-fold when treated with rapamycin (Fig. 6B). Similar results were obtained with HCT116 control and TSC2 knockdown cells (data not shown). Collectively, these data suggest that rapamycin may have a positive effect on cell migration if the cells are under the control of negative feedback regulation.
FIGURE 6.
Rapamycin treatment partially rescues the migration and CDC42/RAC1 activation defects in TSC2 knockdown colon cancer cells. A, KM20 control (Con) and TSC2 knockdown cells were treated with dimethyl sulfoxide or rapamycin (20 nm) for 5 h and analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The amount of activated and total CDC42 and RAC1 in the cells were detected using the anti-CDC42 and anti-RAC1 antibodies on Western blots. The GST-PBD bound panels represent the activated fraction of CDC42 and RAC1, and 10% of cell lysates were used to show the total expression of CDC42 and RAC1 (Lysate panels). The expression of TSC2, p-AKT (phospho-Ser473), total AKT, and tubulin are detected in the cell lysates. The percentage of activation was quantified by normalizing the level of activated CDC42 and RAC1 (Bound panels) to total proteins in the lysates (Lysate panels), and indicated by the numbers below the representative panels. B, KM20 control and TSC2 knockdown cells were subjected to trans-well migration analysis in the presence or absence of rapamycin in the lower chamber. The numbers of migrated cells were quantified, normalized to the control cells, and expressed as percentage of migration. Data represent mean ± S.E. (n = 3; *, p < 0.05). Rapa, rapamycin.
Overexpression of an Active Mutant of p110α Partially Rescues the Polarization Defect in TSC2(−/−) REF Cells
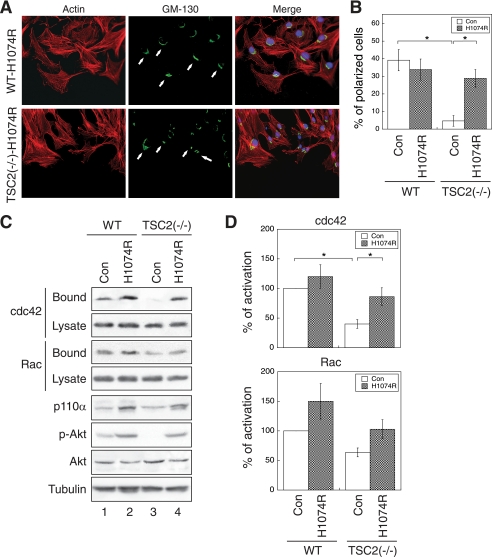
The above data suggest that TSC2-mediated feedback inhibition of PI3K may contribute to the cell polarization and migration defect seen in TSC2-deficient cells. We next tested whether overexpression of an active mutant of the catalytic subunit of PI3K (PIK3CA), p110α-H1074R, can improve cell polarization. The H1074R mutation in the PIK3CA gene was first identified in colorectal cancer patients (22). Cancer cells carrying this mutant PIK3CA have reduced dependence on growth factors, and apoptosis is inhibited (22). The p110α-H1074R mutant was stably expressed in WT and TSC2(−/−) REF cells using retrovirus-mediated infection. The control cells were generated by infecting cells with a retrovirus encoding the blank vector. The ability of the stable cells to establish polarity was examined using the wound-healing assay as described above. Fig. 7A shows that the WT control and H1074R-overexpressing cells polarized similarly as the parental cells (Fig. 2), and ∼40% of the cells established polarity toward the wound (Fig. 7B). On the other hand, whereas polarization of the control TSC2(−/−) cells was largely abolished (Fig. 2B, and similar as the parental cells shown in Fig. 2), overexpression of H1074R mutant markedly increased the percentage of polarized cells from ∼5% to ∼30% (Fig. 7B).
FIGURE 7.
Overexpression of an active p110α mutant of PI3K rescues the polarization and CDC42/RAC1 activation defects in TSC2(−/−) REF cells. A, WT and TSC2(−/−) REF cells infected with control (Con) retrovirus or a retrovirus encoding an oncogenic activating mutant of p110α, H1047R, were subjected to polarization assays as described in Fig. 2. The cells were stained with the anti-GM-130 antibody and visualized using the Alexa488-conjugated secondary antibody (GM-130 panels). The actin was visualized with Texas Red-conjugated phalloidin (Actin panels). The nuclei were stained with 4′,6-diamidino-2-phenylindole. The scratch wounds were located parallel to the bottom of each image. The polarized cells with Golgi facing the wound are marked with solid arrows. B, shown are the quantitative results showing the percentage of polarized cells. For each experiment, six to eight images were taken along the scratch, and 100–120 total cells located at the edge of the wound were counted. The cells with Golgi orientated facing the wound are considered polarized. Data represent the mean ± S.E. (n = 3; *, p < 0.05). C, the control and p110α-H1047R-infected WT and TSC2(−/−) REF cells were analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The amount of activated and total CDC42 and RAC1 in the cells were detected using the anti-CDC42 and anti-RAC1 antibodies on Western blots. The GST-PBD bound panels represent the activated fraction of CDC42 and RAC1, and 10% of cell lysates were used to show the total expression of CDC42 and RAC1 (Lysate panels). The expression of p110α in the cell lysates was detected using the anti-p110α antibody. The p-AKT and AKT panels show the phosphorylation level of AKT at Ser473 and total AKT expression, respectively. D, the percentage of activation was quantified by normalizing the level of CDC42 and RAC1 activation (Bound panels) to total proteins in the lysates (Lysate panels). Data shown in the graphs represent the mean ± S.E. (n = 3; *, p < 0.05).
To investigate whether the overexpression of H1074R mutant restores CDC42 and RAC1 activation in TSC2(−/−) cells, we performed a GST-PBD pulldown assay to determine the amount of activated CDC42 and RAC1 in the control and H1074R-overexpressing cells. Moderate overexpression of p110α-H1074R was detected with the anti-p110α antibody using Western blot analysis (Fig. 7C, p110α panel). In addition, phosphorylation of AKT was increased in H1074R-overexpressing cells (Fig. 7C, p-Akt panel) suggesting that the H1074R mutant is able to bypass loss of TSC2-mediated negative inhibition of Akt. Furthermore, CDC42 activation is significantly increased in H1074R overexpressing TSC2(−/−) cells to a similar level as seen in the control WT cells (Fig. 7D, Cdc42 graph). The amount of activated RAC1 is increased upon overexpression of H1074R as well, although the difference was not significant compared with the control TSC2(−/−) cells (Fig. 7D, Rac graph). Taken together, these results suggest that the active mutant of p110α rescues the polarization defect of TSC2(−/−) cells likely through activation of CDC42.
Re-expression of TSC2 Rescues the Migration Defect in TSC2(−/−) REF Cells
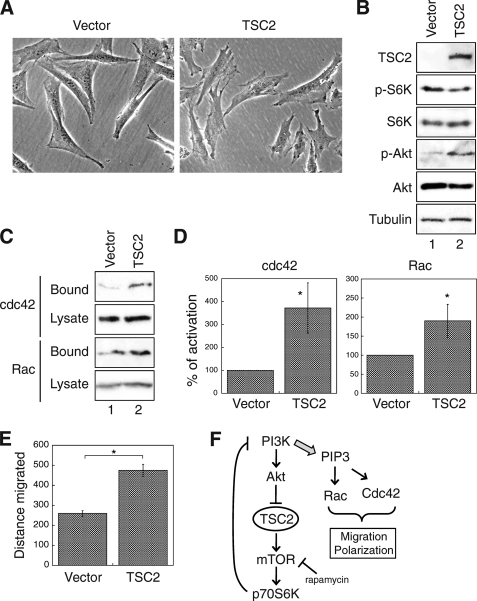
To confirm the specificity of TSC2-dependent regulation on cell migration, we reintroduced the wild-type human TSC2 gene into TSC2(−/−) REF cells via retrovirus-mediated infection. First, re-expression of TSC2 changed the morphology of TSC2(−/−) REF cells. Whereas the vector infected cells maintained the triangular and elongated shape as observed in parental cells, more lamellipodia and filopodia formation was detected in cells re-expressing TSC2 (Fig. 8A). The negative feedback loop was effectively blocked as shown by the expression of TSC2 and the alteration of p70S6K and AKT phosphorylation (Fig. 8B). In addition, the GST-PBD pulldown assays were performed to determine the amount of activated CDC42 and RAC1 in the vector- and TSC2-expressing cells. The results showed that re-expression of TSC2 resulted in a 2–4-fold increase of active CDC42 and RAC1 (Fig. 8, C and D) in cells suggesting that the activation of CDC42 and RAC1 was rescued by TSC2. Furthermore, the rate of cell migration was significantly improved in cells re-expressing TSC2 (Fig. 8E).
FIGURE 8.
Re-expression of TSC2 rescues the migration defection in TSC2(−/−) REF cells. A, phase-contrast images were taken from stable TSC2(−/−) REF cells infected with control (Con) vector or wild-type TSC2. Images shown were obtained with a 20× objective. B, expression of TSC2 and phosphorylation status of p70S6K and Akt in stable TSC2(−/−) REF cells infected with control vector or TSC2 (lanes 1 and 2, respectively). C, the control vector or TSC2-infected TSC2(−/−) REF cells were analyzed for CDC42 and RAC1 activation using GST-PBD pulldown assays. The amount of activated and total CDC42 and RAC1 in the cells were detected using the anti-CDC42 and anti-RAC1 antibodies on Western blots. The GST-PBD bound panels represent the activated fraction of CDC42 and RAC1, and 10% of cell lysates were used to show the total expression of CDC42 and RAC1 (Lysate panels). D, the percentage of activation was quantified by normalizing the level of CDC42 and RAC1 activation (Bound panels) to total proteins in the lysates (Lysate panels). Data shown in the graphs represent the mean ± S.E. (n = 4; *, p < 0.05). E, wound healing assays were performed to determine the rate of migration in the vector or TSC2-infected TSC2(−/−) REF cells. The quantitative results are summarized and shown in the graph. The distance migrated (μm) was measured and quantified using Nikon Elements software. Data represent the mean ± S.E. (n = 3; *, p < 0.05). F, shown is a diagram showing signaling pathways involved in TSC2-dependent regulation of cell polarity and migration. Our study here demonstrates that loss of TSC2 triggers the negative feedback regulation in which the activation of PI3K is largely diminished. This leads to decreased activation of RAC1 and CDC42 and migration defects. Treating cells with rapamycin, overexpression of a constitutively active p110α or re-expression of TSC2 rescue the migration defect by releasing or bypassing the negative regulation.
In summary, we demonstrate that TSC2 plays an important role in regulating cell polarity and migration by controlling the negative feedback loop. Loss of TSC2 expression results in a decrease of PI3K activation and subsequently reduced activation of CDC42 and RAC1. Thus, TSC2 positively regulates the rate of cell migration and polarization via maintaining the activation of CDC42 and RAC1 (Fig. 8F).
DISCUSSION
Loss of function mutations in the TSC2 gene are responsible for the pathogenesis of tuberous sclerosis and other hamartoma syndromes (1). TSC2 functions as a tumor suppressor by negatively controlling the mTOR-signaling pathway. In addition, loss of TSC2 is known to activate the negative feedback regulation of PI3K/AKT signaling (23). While the antigrowth function of TSC2 has been well characterized, it is less clear how TSC2 regulates cell migration and polarization. In the current study, we focused on elucidating the effect of TSC2 knockdown on cell motility. Fibroblasts lacking TSC2 display significantly reduced ability to spread and migrate. Similarly, knockdown of TSC2 expression in colon cancer cells results in a decrease in cell migration. Moreover, loss of TSC2 expression largely inhibits the establishment of cell polarity. These migration defects correlate with decreased CDC42 and RAC1 activation in TSC2 deficient cells. Furthermore, treating cells with rapamycin or overexpression of an active mutant p110α allows TSC2(−/−) cells to polarize, suggesting that loss of TSC2-induced negative feedback regulation of PI3K plays a major role in inhibiting cell polarization. Finally, re-expression of a human wild-type TSC2 gene in TSC2(−/−) RFE cells rescues the migration defect, confirming the role of TSC2 in regulating cell migration.
It has been shown previously that TSC1 inhibits RAC1 activation and TSC2 blocks this inhibition upon binding to TSC1 in Eker rat leiomyoma cells (12). Consistent with this report, we demonstrate that RAC1 activation is inhibited in both TSC2(−/−) REF and TSC2 knockdown colon cancer cells. More importantly, we show for the first time that CDC42 activation is greatly reduced upon loss of functional TSC2 expression. It has been shown that integrin-mediated activation of CDC42 controls cell polarity in migrating astrocytes (15). In this study, we find that change in CDC42 activity tightly correlates with polarization of rat fibroblast cells during migration, and loss of TSC2 largely inhibits CDC42 activation. It has been shown that activation of PI3K and production of its lipid product PtdIns-3,4,5-P3 are required for activation of CDC42 (17). Because lack of cell polarization in TSC2(−/−) cells is rescued by rapamycin treatment or overexpression of an active p110α mutant in our experiments, we conclude that loss of TSC2-induced decrease of PI3K activity, and the subsequent decrease of PtdIns-3,4,5-P3 is likely responsible for down-regulation of CDC42 and the polarity defect.
In the case of RAC1 activation, many studies have placed RAC1 downstream of PI3K as well (13, 16). Thus, inhibition of RAC1 activation in TSC2-deficient cells is likely the result of TSC2-mediated feedback inhibition of PI3K. This may account for lack of lamellipodia formation in TSC2(−/−) REF cells. Interestingly, similar alteration in cell morphology have been reported in RAC1-null mouse embryonic fibroblasts. Both lack of lamellipodia formation and elongated morphology are observed in RAC1-null mouse embryonic fibroblast cells (24, 25). Consistent with previous studies that genetic deletion of RAC1 results in delayed cell spreading and slower migration rate (24, 25), we find that TSC2(−/−) REF cells spread and migrate much slower compared with WT cells. However, the rate of migration is not significantly accelerated in TSC2(−/−) REF cells either treated with rapamycin or overexpressing the p110α-H1074R mutant (data not shown). This may due to the fact that only a moderate increase of RAC1 activation is achieved (Figs. 5D and 7D). Indeed, either rapamycin or p110α-H1074R are not sufficient to alter the morphology of TSC2(−/−) REF cells. In addition, recent studies show that TSC2 can directly suppress mTORC2 activity (23, 26), and mTORC2 activity is known to regulate actin cytoskeleton organization (27). Further studies are needed to determine whether the loss of the TSC2-induced migration defect is associated with the ability of TSC2 to regulate mTORC2.
Previous studies indicate that activation of Rac1 and RhoA is mutually inhibitory in cells (28–30). Indeed, we find that lysophosphatidic acid-induced RhoA activity is elevated in TSC2(−/−) REF and TSC2 knockdown colon cancer cells (data not shown), whereas CDC42 and RAC1 activities are decreased. Most recently, using real-time imaging of biosensors for small GTPases, Machacek et al. (31) elegantly demonstrate that assembly of cell protrusion initially requires RhoA activity, followed by increased activation of CDC42 and RAC1 to ensure coordinated cell movement. Thus, the overall defect of cell migration observed in TSC2-deficient cells is likely a combinational effect of activity imbalance among CDC42, RAC1, and RhoA.
Loss of TSC2 is known to play a major role in up-regulating the negative feedback inhibition of the PI3K/AKT pathway. Because AKT signaling is largely diminished by this feedback inhibition, tumors lacking TSC2 are usually benign and rarely metastasize to other organs (1). Here, we show that cell migration is also impaired in TSC2-deficient cells, thus, providing an underlying molecular mechanism for lack of aggressiveness in mutant TSC2-related tumors. However, if the limitation set by TSC2 mutations can be overcome, as in the case of PTEN deletion, the severity of the tumor enhances dramatically (9). Consistent with this notion, we also find that knockdown of TSC2 has no effect on cell migration in PTEN-deleted U87 glioblastoma cells (data not shown). Furthermore, we show that rapamycin treatment reduces cell migration in control colon cancer cells. During this short term rapamycin treatment (5 h), no significant change in CDC42 and RAC1 activity is observed (Figs. 5D and 6A). Thus, this rapamycin-induced decrease of migration is likely due to suppression of p70S6K activity as reported previously in rhabdomyosarcoma cells (21). Further studies are needed to elucidate the potential role of mTOR in regulating CDC42 and RAC1 activation. More importantly, rapamycin treatment releases the inhibition of migration in TSC2 knockdown cells (Fig. 6B). As rapamycin and its derivatives are being tested clinically for their anti-cancer activity (20), caution should be taken, as our results suggest that rapamycin treatment may increase the metastatic potential of tumors lacking TSC2. In summary, our study has identified a novel role of TSC2 in regulating cell polarization and migration. In addition to increased cell growth mediated by loss of TSC2 expression, TSC2 plays a positive role in maintaining the balance of small GTPases to ensure coordinated cell migration.
Supplementary Material
Acknowledgment
We thank Dr. Kathleen O'Connor (University of Kentucky) for providing the GST-PBD expression construct and helping with BioStation-related experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA133429 (to T. G.). This work was also supported by American Cancer Society Research Scholar Grant RSG0822001TBE (to T. G.) and NIH Grant R01CA104748 (to B. M. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies 1 and 2.
- PI3K
- phosphatidylinositol 3-kinase
- IGF
- insulin-like growth factor
- p70S6K
- p70S6 kinase
- PtdIns-3,4,5-P3
- phosphatidylinositol 3,4,5-phosphate
- REF
- rat embryonic fibroblast
- shRNA
- short hairpin RNA
- GST
- glutathione S-transferase
- GST-PBD
- GST-tagged p21 binding domain of PAK1
- WT
- wild-type.
REFERENCES
- 1.Curatolo P., Bombardieri R., Jozwiak S. (2008) Lancet 372, 657–668 [DOI] [PubMed] [Google Scholar]
- 2.Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoki K., Guan K. L. (2009) Hum. Mol. Genet. 18, R94–R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 5.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 6.Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 7.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning B. D. (2004) J. Cell Biol. 167, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning B. D., Logsdon M. N., Lipovsky A. I., Abbott D., Kwiatkowski D. J., Cantley L. C. (2005) Genes Dev. 19, 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb R. F., Roy C., Diefenbach T. J., Vinters H. V., Johnson M. W., Jay D. G., Hall A. (2000) Nat. Cell Biol. 2, 281–287 [DOI] [PubMed] [Google Scholar]
- 11.Astrinidis A., Cash T. P., Hunter D. S., Walker C. L., Chernoff J., Henske E. P. (2002) Oncogene 21, 8470–8476 [DOI] [PubMed] [Google Scholar]
- 12.Goncharova E., Goncharov D., Noonan D., Krymskaya V. P. (2004) J. Cell Biol. 167, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobes C. D., Hall A. (1995) Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 14.Nobes C. D., Hall A. (1999) J. Cell Biol. 144, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne-Manneville S., Hall A. (2001) Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]
- 16.Cain R. J., Ridley A. J. (2009) Biol. Cell 101, 13–29 [DOI] [PubMed] [Google Scholar]
- 17.Van Keymeulen A., Wong K., Knight Z. A., Govaerts C., Hahn K. M., Shokat K. M., Bourne H. R. (2006) J. Cell Biol. 174, 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson R., Pedersen E. D., Wang Z., Brakebusch C. (2009) Biochim. Biophys. Acta 1796, 91–98 [DOI] [PubMed] [Google Scholar]
- 19.Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 20.Huang S., Houghton P. J. (2003) Curr. Opin Pharmacol. 3, 371–377 [DOI] [PubMed] [Google Scholar]
- 21.Liu L., Li F., Cardelli J. A., Martin K. A., Blenis J., Huang S. (2006) Oncogene 25, 7029–7040 [DOI] [PubMed] [Google Scholar]
- 22.Samuels Y., Diaz L. A., Jr., Schmidt-Kittler O., Cummins J. M., Delong L., Cheong I., Rago C., Huso D. L., Lengauer C., Kinzler K. W., Vogelstein B., Velculescu V. E. (2005) Cancer Cell 7, 561–573 [DOI] [PubMed] [Google Scholar]
- 23.Huang J., Manning B. D. (2009) Biochem. Soc. Trans. 37, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo F., Debidda M., Yang L., Williams D. A., Zheng Y. (2006) J. Biol. Chem. 281, 18652–18659 [DOI] [PubMed] [Google Scholar]
- 25.Vidali L., Chen F., Cicchetti G., Ohta Y., Kwiatkowski D. J. (2006) Mol. Biol. Cell 17, 2377–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J., Dibble C. C., Matsuzaki M., Manning B. D. (2008) Mol. Cell. Biol. 28, 4104–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 28.Rottner K., Hall A., Small J. V. (1999) Curr. Biol. 9, 640–648 [DOI] [PubMed] [Google Scholar]
- 29.Arthur W. T., Burridge K. (2001) Mol. Biol. Cell 12, 2711–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta Y., Hartwig J. H., Stossel T. P. (2006) Nat. Cell Biol. 8, 803–814 [DOI] [PubMed] [Google Scholar]
- 31.Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009) Nature 461, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.