Abstract
Free methionine-R-sulfoxide reductase (fRMsr) reduces free methionine R-sulfoxide back to methionine, but its catalytic mechanism is poorly understood. Here, we have determined the crystal structures of the reduced, substrate-bound, and oxidized forms of fRMsr from Staphylococcus aureus. Our structural and biochemical analyses suggest the catalytic mechanism of fRMsr in which Cys102 functions as the catalytic residue and Cys68 as the resolving Cys that forms a disulfide bond with Cys102. Cys78, previously thought to be a catalytic Cys, is a non-essential residue for catalytic function. Additionally, our structures provide insights into the enzyme-substrate interaction and the role of active site residues in substrate binding. Structural comparison reveals that conformational changes occur in the active site during catalysis, particularly in the loop of residues 97–106 containing the catalytic Cys102. We have also crystallized a complex between fRMsr and isopropyl alcohol, which acts as a competitive inhibitor for the enzyme. This isopropyl alcohol-bound structure helps us to understand the inhibitory mechanism of fRMsr. Our structural and enzymatic analyses suggest that a branched methyl group in alcohol seems important for competitive inhibition of the fRMsr due to its ability to bind to the active site.
Keywords: Aging, Methionine, Protein Structure, Reductase, X-ray Crystallography, Catalytic Mechanism, Free Methionine-R-sulfoxide Reductase
Introduction
Reactive oxygen species can damage cellular components, including lipids, nucleic acids, and proteins. Damage to proteins by reactive oxygen species is probably due to oxidation of side chains of amino acid residues (1). The sulfur-containing amino acids, methionine and cysteine, are the most sensitive to oxidation. Oxidation of methionine generates a diastereomeric mixture of methionine S-sulfoxide (Met-S-O)3 and methionine R-sulfoxide (Met-R-O) (2). Methionine oxidation is associated with a variety of physiological and pathological processes, such as cellular signaling, aging, and neurodegenerative diseases (3, 4). For example, methionine oxidation activates calcium/calmodulin-dependent protein kinase II in the absence of calcium (5), regulates the life span of yeast, fruit fly, and nematode (6–8), and may advance progression of Alzheimer and Parkinson diseases (9–12).
However, this oxidation can be reversed by the methionine-sulfoxide reductases (Msrs). Two distinct families of Msrs have evolved for the stereospecific reduction of methionine sulfoxides in proteins (13, 14). MsrA catalyzes the reduction of Met-S-O, whereas MsrB reduces Met-R-O. Most organisms from bacteria to humans possess a methionine sulfoxide reduction system that confers upon them the ability to repair oxidative damage and consequently impacts their longevity in oxidative environments (2, 4). In addition, Msrs are involved in the virulence mechanism of some bacterial pathogens, including Mycoplasma genitalium and Neisseria gonorrhoeae (15–17). Recently, an enzyme specific for the reduction of free Met-R-O has been identified from Escherichia coli and named fRMsr (18). This protein is found in unicellular organisms, including Saccharomyces cerevisiae, but absent in multicellular organisms (19). Interestingly, fRMsr contains a GAF domain, which is a ubiquitous motif present in cyclic GMP phosphodiesterases (20). Two variants of fRMsr proteins were detected with different conserved Cys residues (19); type I fRMsrs contain three conserved Cys residues, whereas type II fRMsrs have two.
The structures and catalytic mechanisms of MsrA and MsrB are well characterized (21–24). Although MsrA and MsrB are completely different in sequence and structure, they share a common catalytic mechanism involving formation of a sulfenic acid intermediate on the catalytic Cys, followed by regeneration of the oxidized catalytic Cys. Briefly, a catalytic Cys attacks the sulfur of methionine sulfoxide and forms a sulfenic acid intermediate, with concomitant release of the product, methionine. The catalytic Cys sulfenic acid then forms an intramolecular disulfide bond by interacting with a resolving Cys. The disulfide bond is reduced by reductants, and consequently the enzyme becomes active again. Thioredoxin (Trx) is generally considered the in vivo reductant, whereas dithiothreitol (DTT) can be used in vitro. In contrast, the catalytic mechanism of fRMsr is poorly understood, although previous studies suggested that its catalytic mechanism is similar to those of MsrA and MsrB, involving the common sulfenic acid chemistry.
It has been found that Staphylococcus aureus, a leading cause of hospital- and community-acquired infections, contains a type I fRMsr, three MsrAs, and an MsrB (19, 25). S. aureus fRMsr contains three conserved Cys residues (Cys68, Cys78, and Cys102). Two crystal structures of fRMsrs from E. coli and S. cerevisae are available (Protein Data Bank codes 1VHM (18, 26) and 1F5M (27), respectively). Both structures contain a disulfide bond between Cys68 and Cys102 (numbering is based on the S. aureus fRMsr) in the active sites, suggesting that fRMsrs use Cys residues for catalysis. The active site is enclosed in a small cavity (18, 19, 26, 27). This enclosed cavity supports the apparent substrate specificity for free Met-R-O but not for protein-based forms. Previous studies suggested that Cys78 functions as a catalytic residue, Cys102 as a primary resolving Cys, and Cys68 as a secondary resolving residue (18, 19). However, the roles of these three Cys residues are unclear in the catalysis of fRMsr. Thus, the catalytic mechanism of this enzyme has yet to be elucidated.
In this study, we resolved four structural forms of the S. aureus fRMsr by x-ray crystallography: reduced form (fRMsrred), complexed form with the substrate (fRMsrsub), oxidized form (fRMsrox), and another complexed form with isopropyl alcohol (fRMsrisopro). The first three structures represent different catalytic states of fRMsr. The last structure, fRMsrisopro, helps us to understand the inhibitory mechanism of fRMsr. We also performed biochemical analyses using the wild type S. aureus fRMsr and single and double mutants, in which the three conserved Cys are replaced with Ser. We studied the inhibitory effect of various alcohols on fRMsr. Our structural and enzymatic studies provide insights into the catalytic mechanism of fRMsr with conformational changes that occur during catalysis and into the inhibitory mechanism involving a branched methyl group of alcohols.
EXPERIMENTAL PROCEDURES
Purification, Crystallization, and X-ray Analysis
Gene cloning, protein expression, purification, and crystallization of S. aureus fRMsr have been described elsewhere for the oxidized and isopropyl alcohol-complexed forms of S. aureus fRMsr (fRMsrox and fRMsrisopro) (29). The crystal complexed with isopropyl alcohol was obtained from a crystallization solution consisting of 2 m ammonium sulfate and 10% (v/v) 2-propanol. For the reduced form of fRMsr (fRMsrred), cell pellets were resuspended in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole, 10 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride). The purification procedures were similar to those of oxidized form, but 10 mm DTT was used in the final purification procedure of gel filtration. The crystallization condition comprised 24% polyethylene glycol 3350 and 0.35 m potassium fluoride. The substrate complex form of fRMsr (fRMsrsub) was obtained by soaking 9 mm free Met-R-O in native crystals of mutant C68S fRMsr in which the crystallization condition comprised 26% polyethylene glycol 400 and 0.1 m MES, pH 6.4.
The crystals were soaked in a solution containing 25% (v/v) ethylene glycol used as cryoprotectant and frozen in liquid nitrogen. X-ray diffraction data were collected with an ADSC Quantum CCD 210 detector at beamlines 6C and 4A at Pohang Light Source (Pohang, South Korea). A total range of 360° was covered with 1.0° oscillation and 30-s exposure per frame. The crystal-to-detector distance was set to 150 mm. The data sets were processed and scaled using HKL 2000 (30). The fRMsrred, fRMsrsub, fRMsrox, and fRMsrisopro crystals diffracted to 1.9, 2.3, 1.5, and 1.7 Å, respectively. The detailed statistics are summarized in Table 1.
TABLE 1.
Data collection statistics and refinement statistics
| fRMsrred | fRMsrsub | fRMsrox | fRMsrisopro | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.23986 | 1.23986 | 1.23986 | 1.23986 |
| Space group | P21 | P21 | P6122 | P6122 |
| Unit cell | a = 69.0 | a = 41.6 | a = b = 90.0 | a = b = 89.8 |
| b = 119.6 | b = 87.5 | c = 88.5 | c = 88.8 | |
| c = 80.3 | c = 42.9 | |||
| Resolution range (Å) | 50.0–1.9 (1.93–1.9)a | 50.0–2.3 (2.34–2.3) | 50.0–1.5 (1.55–1.5) | 50.0–1.7 (1.76–1.7) |
| Observed reflections | 454,984 | 64,222 | 980,295 | 479,682 |
| Unique reflections | 98,600 | 13,036 | 33,994 | 23,645 |
| Redundancy | 4.6 (3.2) | 4.9 (3.2) | 28.9 (6.2) | 20.3 (7.6) |
| Completeness (%) | 98.1 (94.5) | 97.5 (92.7) | 98.6 (87.1) | 99.1 (94.8) |
| Rsym (%)b | 7.5 (33.6) | 7.0 (23.6) | 5.9 (33.0) | 6.6 (28.7) |
| I/sigma (I) | 19.2 (2.7) | 13.4 (4.1) | 14.5 (3.2) | 16.0 (5.5) |
| Refinement statistics | ||||
| Rfactor/Rfree (%) | 21.6/25.6 | 22.2/25.2 | 22.1/23.4 | 22.0/24.2 |
| r.m.s. deviation bond (Å) | 0.005 | 0.006 | 0.004 | 0.005 |
| r.m.s. deviation angles (degrees) | 1.2 | 1.2 | 1.1 | 1.2 |
| Mean B factor | 26.4 | 49.7 | 22.8 | 23.9 |
| Ramachandran plot | ||||
| Most allowed region (%) | 88.9 | 85.0 | 90.4 | 91.2 |
| Additional allowed region (%) | 10.9 | 12.0 | 9.6 | 8.8 |
| Generously allowed region (%) | 0.3 | 2.9 | 0 | 0 |
| Disallowed region (%) | 0 | 0 | 0 | 0 |
a Values in parentheses represent the highest resolution shell.
b Rsym = Σhkli|Ihkli − 〈Ihkl〉|/Σhkli Ihkli, where I is the observed intensity, 〈I〉 is the average intensity, and i is counts through all symmetry-related reflections. The crystallographic Rfactor is based on 95% of the data used in refinement, and Rfree is based on 5% of the data withheld for the cross-validation test.
Model Building and Structure Refinement
The crystal structures of fRMsr were solved by molecular replacement methods using CNS (28) and Molrep (31) programs. The coordinates of E. coli fRMsr (Protein Data Bank code 1VHM) (18, 26) were used as the search model. Refinements were performed with several cycles of torsion-angle-simulated annealing, energy minimization, individual B factor refinement, and manual model rebuilding. The models were completed by iterative cycles of model building with Coot (32) and subsequently by refinement with CNS (28). The final models for fRMsrred, fRMsrsub, fRMsrox, and fRMsrisopro yielded Rfactor and Rfree values of 21.6 and 25.6% for fRMsrred, 22.2 and 25.2% for fRMsrsub, 22.1 and 23.4% for fRMsrox, and 22.0 and 24.2% for fRMsrisopro, respectively. Refinement data were validated by the PROCHECK program (33) and are provided in Table 1. All figures were created using CCP4mg (34).
Measurements of Msr Activities
For free Msr activity, NADPH oxidation was monitored as a decrease of A340 at room temperature for 10 min in the reaction mixture. The reaction mixture (200 μl) contained 50 mm sodium phosphate (pH 7.5), 50 mm NaCl, 0.2 mm NADPH, 10 μg of E. coli Trx (Sigma), 14 μg of human Trx reductase 1, 0.1 mm EDTA, 1 mm free Met-R-O or free Met-S-O, and 2 or 10 μg of fRMsr enzyme. Enzyme activity was defined as nmol of oxidized NADPH/min using a molar extinction coefficient of 6220 m−1 cm−1. Km and Vmax values were determined by non-linear regression using GraphPad Prism 5 software.
For peptide Msr activity, dabsylated methionine sulfoxide was used as the substrate in a DTT-dependent reaction. The reaction mixture (100 μl), containing 50 mm sodium phosphate (pH 7.5), 50 mm NaCl, 20 mm DTT, 200 μm dabsyl-Met-R-O or dabsyl-Met-S-O, and 1 μg of fRMsr enzyme, was incubated at 37 °C for 30 min. The reaction product, dabsyl-Met, was analyzed by high pressure liquid chromatography.
For inhibition assays of various alcohols on fRMsr activity, the reaction mixture (200 μl) contained 50 mm sodium phosphate (pH 7.5), 50 mm NaCl, 0.2 mm NADPH, 10 μg of E. coli Trx, 14 μg of human Trx reductase 1, 0.1 mm EDTA, 1 mm free Met-R-O, 1% various alcohols, and 2 μg of fRMsr enzyme. The reaction mixture was incubated at room temperature for 10 min, and the decrease of A340 was monitored.
Preparation of Single or Double Mutant Forms of S. aureus fRMsr
C68S, C78S, C102S, C68S/C78S, and C68S/C102S mutants were generated by site-directed mutagenesis using a pET28a-based wild type construct (29). All constructs were verified by DNA sequencing.
RESULTS AND DISCUSSION
Catalytic Activities of Wild Type and Mutant Forms of S. aureus fRMsr
First, we tested the substrate specificity of S.aureus fRMsr toward free Met-R-O, free Met-S-O, dabsyl-Met-R-O (mimic to peptide Met-R-O), dabsyl-Met-S-O, and dimethyl sulfoxide. The enzyme assay was performed by analyzing NADPH oxidation in the reaction mixture. As expected, S. aureus fRMsr reduced free Met-R-O but could not reduce free Met-S-O, dabsyl-Met-R-O, dabsyl-Met-S-O, or dimethyl sulfoxide, showing the same substrate specificity as E. coli and S. cerevisiae fRMsrs characterized previously (18, 19).
To determine the roles of the three conserved Cys residues (Cys68, Cys78, and Cys102; supplemental Fig. S1) in catalysis, we mutated these residues to Ser, making single or double mutants (C68S, C78S, C102S, C68S/C78S, and C68S/C102S). We assayed the Trx-dependent activities of these mutant fRMsrs and compared them with the wild type. As shown in Table 2, the activity of C68S was 32% of wild type. Unexpectedly, the activity of C78S was 75% of wild type. This Cys residue was previously suggested to be the catalytic residue in E. coli and S. cerevisae fRMsrs (18, 19). Interestingly, C102S had no catalytic activity. Consistent with this result, C68S/C102S had no catalytic activity either, whereas C68S/C78S retained 22% of enzyme activity.
TABLE 2.
Specific activities and kinetic parameters of wild type and mutant forms of S. aureus fRMsr
Enzyme assays were performed using 10 μg of purified proteins as described under “Experimental Procedures.” Values in parenthesis represent the activity relative to wild type. Km and Vmax values were determined by fitting the data to the Michaelis-Menten equation. NA, not assayed.
| Proteins | Specific activity | Km | Vmax |
|---|---|---|---|
| nmol/min/mg protein | μm | nmol/min/mg protein | |
| Wild type | 85 ± 5 (100) | 50 ± 10 | 360 ± 10 |
| C68S | 27 ± 2 (32) | 210 ± 20 | 280 ± 10 |
| C78S | 64 ± 20 (75) | 110 ± 50 | 440 ± 50 |
| C102S | 0 ± 0.3 (0) | NA | NA |
| C68S/C78S | 19 ± 3 (22) | 830 ± 240 | 130 ± 20 |
| C68S/C102S | 0 ± 0.3 (0) | NA | NA |
We then analyzed kinetic parameters of C78S, C68S, and C68S/C78S as well as wild type (Table 2). The Vmax value of C78S was slightly higher than that of wild type; the Km value was 2-fold higher than that of wild type. These data indicate that Cys78 is non-essential for catalysis by fRMsr. The Vmax value was significantly reduced in C68S mutant, whereas the Km value was 4-fold higher, compared with those of wild type. The double C68S/C78S mutant exhibited more decreased Vmax (3-fold lower than wild type) and more increased Km (16-fold higher than wild type).
Thus, in contrast to the previously suggested model, Cys102 is proposed to be the catalytic Cys, Cys68 may serve as the resolving Cys that forms a disulfide bond with Cys102, and Cys78 is a non-essential residue for catalytic function. The above enzymatic data are consistent with our recent findings from S. cerevisae fRMsr that Cys125 (corresponding to Cys102 in S. aureus fRMsr) functions as the catalytic residue, as determined by enzyme and in vivo growth complementation assays (35).
Crystal Structure of the Reduced Form of fRMsr
Previously known structures from both E. coli and S. cerevisae fRMsrs are oxidized forms with a disulfide bond between Cys68 and Cys102 (26, 27). In addition, the E. coli fRMsr structure contains a complex with MES in the active site. Here, we resolved the structure of a reduced form of S. aureus fRMsr (fRMsrred) (supplemental Fig. S2A). The crystal of fRMsrred comprises four dimers in the asymmetric unit. There are several hydrogen bond interactions in the interface region of the dimer structure (Fig. 1A). To assess on quantitative grounds the possibility that these hydrogen bond interactions may stabilize an fRMsrred dimer, the dimer interface was evaluated by using the program PISA (36). This widely used program estimates a dimeric state for fRMsrred (complexation significance score = 1). In particular, this analysis shows that the buried area upon formation of the dimeric assembly is 932.7 Å2, which accounts for 11.8% of the total surface area for each molecule. It should be noted that S. cerevisae fRMsr is also a dimer in solution (27).
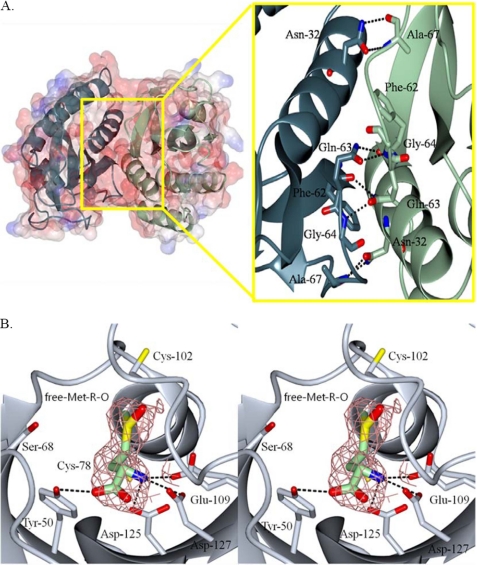
FIGURE 1.
The overall structure of a reduced form of S. aureus fRMsr (A) and structure of substrate-bound active site of C68S fRMsr in stereo (B). In A, a dimer is shown by electrostatic surface and ribbon models (blue, C subunit; light green, D subunit). A close-up view represents the dimer interface region. In the interface region, Asn32, Gln63, Phe62, and Ala67 of the C subunit interact by hydrogen-bonding, respectively, with Ala67, Gly64/Phe62, Gln63, and Asn32 of the other subunit. In B, the ligand, free Met-R-O, is depicted as a light green stick model. The omit electron density of substrate is shown at 1.5 σ. Hydrogen bond interactions between the substrate and the active site residues are indicated by black dotted lines (for details, see “Results and Discussion”).
The carboxamide groups of Asn32 and Gln63 from one subunit form hydrogen bonds with the backbones of Ala67 and Phe62 from the other subunit, respectively. In addition, the side chain of Gln63 of one subunit interacts with the backbone of Gly64 of the other subunit. The overall one-subunit structure of fRMsrred is composed of six antiparallel β-strands (β1–β6) and four α-helices (α1–α4) (supplemental Fig. S2A). The active site contained Trp46, Tyr50, Leu59, Cys68, Cys78, Cys102, Asp103, Ala104, Ser106, Glu109, Asp125, and Asp127 in five antiparallel β-strands, two loops, and one α-helix, where Cys78 is located (Fig. 2C and supplemental Fig. S2A). The side chains of Cys102 and Cys68 are located in the active site. The distances between the sulfur atoms of Cys68 and Cys102, Cys68 and Cys78, and Cys78 and Cys102 are 5.7, 10.1, and 10.2 Å, respectively. On one side of the cavity of the active site, Trp46 and Ala104 form a hydrophobic region, whereas the opposite side displays a hydrophilic region consisting of Tyr50, Glu109, Asp125, and Asp127. The structure of fRMsrred contains several water molecules (Wat) in the active site. Particularly, Wat4 interacts with the side chains of Tyr50 (3.4 Å) and Asp125 (2.9 Å). Also, Wat104 interacts with the side chains of Glu109, Asp125, and Asp127; Wat491 interacts with the side chain of Cys78, Glu109, and Asp125, respectively (Fig. 2C). These interactions involving water molecules in the active site may stabilize the conformation of reduced fRMsr.
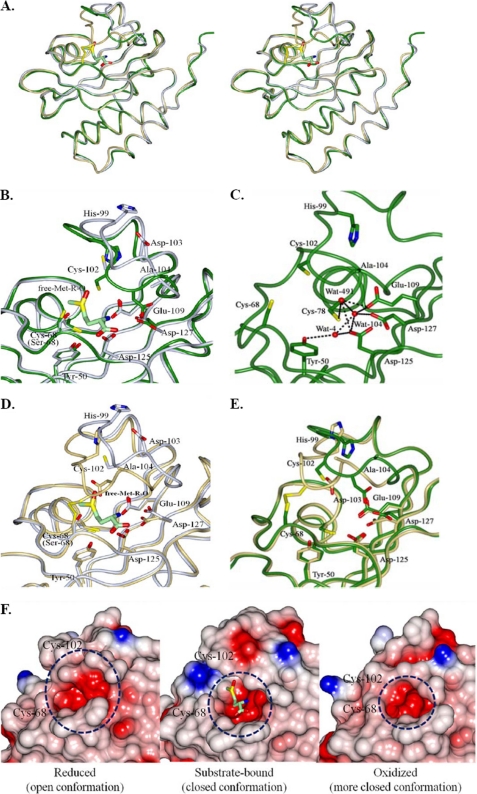
FIGURE 2.
Structural comparison of reduced, substrate-bound, and oxidized S. aureus fRMsrs. A, stereoscopic views showing comparison of overall structures of reduced, substrate-bound, and oxidized S. aureus fRMsrs. The backbone models for reduced (fRMsrred), substrate-bound (fRMsrsub), and oxidized (fRMsrox) forms are shown in green, light blue, and light yellow, respectively. B and C, comparison of active sites between fRMsrred (green) and fRMsrsub (light blue). The active site residues of fRMsrred and fRMsrsub are superimposed (B), and those of fRMsrred are independently shown (C). In C, hydrogen bond interactions among water molecules and active site residues are indicated by dotted lines. D, comparison of active sites between fRMsrsub (light blue) and fRMsrox (light yellow). The active site residues of fRMsrsub and fRMsrox are superimposed. E, comparison of active sites between fRMsrred (green) and fRMsrox (light yellow). The active site residues of fRMsrred and fRMsrox are superimposed. The disulfide bond between Cys68 and Cys102 in fRMsrox is represented by a yellow stick, and substrate Met-R-O in fRMsrsub is shown by a light green stick. F, conformational changes of fRMsrred, fRMsrsub, and fRMsrox. The active site is shown with electrostatic surface models. The surfaces are colored according to the electrostatic potentials from −21 kiloteslas/e (red) to +21 kiloteslas/e (blue). The electrostatic surface potentials were calculated by using APBS (37).
Structure of fRMsr in Complex with the Substrate
Here, we have resolved the first structure of S. aureus fRMsr complexed with the substrate free Met-R-O (fRMsrsub) using C68S fRMsr, which shows a Michaelis-like complex (supplemental Fig. S2B). The sulfoxide moiety of the substrate was clearly shown in the omit electron density map of the active site (Fig. 1B). This structure could lead us to understand the catalytic mechanism of fRMsr, the mode of binding to the substrate, and the roles of the active site residues during catalysis. The structure of fRMsrsub comprises a dimer with the substrate in each subunit of the asymmetric unit. The overall conformation of fRMsrsub, in which Ser replaces Cys68 in the loop of the active site, is conserved with the reduced form of wild type fRMsr (supplemental Fig. S2). However, there are significant conformational changes around the active site, as discussed below.
The substrate Met-R-O is positioned by several hydrogen bonds and stacking interactions. The acidic side chains of Glu109, Asp125, and Asp127 in the hydrophilic region form hydrogen bonds with nitrogen of the substrate (Fig. 1B). In addition, the residue Tyr50 forms a hydrogen bond with the carboxylate group of Met-R-O. The sulfoxide of the substrate is located close to Cys102, pointing toward the sulfur atom of Cys102 (6.7 Å). The thiol of Cys78 points toward the carboxylate of the substrate although located closer to the sulfoxide of the substrate (4.3 Å). Our structural analysis, along with the above enzymatic data, suggests that Cys102 is the catalytic residue of fRMsr. The hydrophobic region involving Ala104 in the active site could accommodate the ϵ-methyl group of the substrate via van der Waals interactions, whereas the hydrophilic region could orient the substrate in the active site via hydrogen bonds with the nitrogen of the substrate. Also, this hydrophilic region may play a role in stabilizing the protonated oxygen atom of the sulfoxide moiety during catalysis. Thus, the hydrophobic pocket of the active site is shown to be essential for binding affinity to the substrate, whereas the hydrophilic region seems important for binding specificity.
Structure of the Oxidized Form of S. aureus fRMsr and a Comparison with Known fRMsr Structures
We also determined the structure of an oxidized form of S. aureus fRMsr (fRMsrox) containing a disulfide bond between Cys68 and Cys102 (supplemental Fig. S2C). The structure of fRMsrox comprises one subunit in the asymmetric unit. However, fRMsrox is found to form a dimer with a crystallographic 2-fold symmetry-related molecule in the unit cell. In fact, the fRMsrred and fRMsrsub structures were grown in the p21 monoclinic space group, whereas the fRMsrox crystal grew in the p6122 hexagonal space group.
We compared the S. aureus fRMsrox with the structures of E. coli and S. cerevisiae fRMsrs previously reported, which are also oxidized forms with a disulfide bond between the above Cys residues. The oxidized E. coli fRMsr contains MES in the active site. S. aureus fRMsr shows 53% amino acid sequence identity with E. coli and S. cerevisiae fRMsrs, respectively (supplemental Fig. S1). The backbone structure of S. aureus fRMsrox could be superimposed on the E. coli and S. cerevisiae fRMsrs with r.m.s. deviations of 1.6 and 5.4 Å, respectively, as determined by CNS (Crystallography and NMR System) (28) for 154 Cα atoms of the overall structures (supplemental Fig. S3A). Interestingly, there were significant differences in a loop region including the catalytic Cys102 (residues 97–106) between S. aureus fRMsrox and E. coli fRMsr (supplemental Fig. S3B). Particularly, positions of His99, Ala101, and Asp103 move away from the corresponding residues of E. coli fRMsr to a distance of 3.3, 4.8, and 3.6 Å, respectively. Also, this loop region was significantly different from that of S. cerevisiae fRMsr (supplemental Fig. S3C). The positions of Ala101 and Asp103 move away from the corresponding residues of S. cerevisiae fRMsr to a distance of 4.2 and 6.1 Å, respectively. Thus, the structural comparison revealed that the catalytic Cys-containing loop region is quite flexible in fRMsr proteins.
Comparison and Conformational Changes of Reduced, Substrate-bound, and Oxidized Forms of fRMsr
We compared the reduced (fRMsrred), substrate-bound (fRMsrsub), and oxidized (fRMsrox) structures of S. aureus fRMsr (Fig. 2), which are representative of the catalytic steps of the fRMsr reaction.
The backbone structure of fRMsrsub could be superimposed on the fRMsrred, with an r.m.s. deviation of 1.4 Å (Fig. 2A). There were significant conformational changes particularly in the loop consisting of residues 97–106 (Fig. 2B). Cys102 and Asp103 of fRMsrsub are the most displaced residues in the loop, shifted by 4.7 and 10.9 Å, respectively. The positions of Ile100, His99, and Lys97 lie at 2.4, 5.1, and 2.2 Å, respectively, from the corresponding residues of fRMsrred. However, the position of Cys78 in fRMsrsub and fRMsrred remains relatively unchanged. The Glu109 residue in fRMsrsub resides at a distance of 3.6 Å from Asp127, whereas in fRMsrred it resides at a distance of 4.7 Å. Cys68 and Cys102 residues in fRMsrsub reside at a distance of 9.7 and 11.7 Å, respectively, from Cys78, whereas in fRMsrred they reside at a distance of 10.1 and 10.2 Å, respectively. Water molecules in the active site of fRMsrred interact with Glu109, Asp125, Asp127, and Tyr50 residues that form hydrogen bonds with nitrogen and the carboxylate group of the substrate (Fig. 2C). When comparing the structure of fRMsrsub with fRMsrred, the substrate Met-R-O in the active site replaces the water molecules occupied in the fRMsrred (Fig. 2, B and C).
We next compared the structure of fRMsrsub with fRMsrox. The backbone structure of fRMsrsub could be superimposed on the fRMsrox, with an r.m.s. deviation of 1.4 Å (Fig. 2A). Significant conformational changes are observed in the loop of residues 97–106 between these two structures (Fig. 2D). Cys102 and Asp103 residues are shifted by 6.7 and 7.8 Å, respectively, between fRMsrsub and fRMsrox. Specifically, in the substrate complex form, the loop moves into the active site compared with the oxidized form, resulting in positioning the thiol group of the catalytic Cys102 toward the entrance of the active site. The positions of Ile100, His99, and Gly98 in fRMsrox lie at 6.6, 7.7, and 3.4 Å, respectively, from the corresponding residues of fRMsrsub. However, the position of Cys78 in fRMsrsub and fRMsrred remains relatively unchanged.
We finally compared fRMsrred structure with fRMsrox structure. The backbone structure of fRMsrred could be superimposed on the fRMsrox with an r.m.s. deviation of 1.5 Å (Fig. 2A). Although the overall structures were well superimposed, there were significant differences in the loop of residues 94–106 (Fig. 2E). The loop in fRMsrred moves into the active site, which results in the positioning of the His99, Cys102, Asp103, and Ala104 residues toward the entrance of active site. Large movements occur in the catalytic residue Cys102 and its neighboring residues Asp103 and Ala104. The Cys102 residues in the oxidized and reduced forms of fRMsr reside at a distance of 4.7 Å from each other. Asp103 and Ala104 in the oxidized and reduced forms reside at a distance of 8.7 and 6.5 Å, respectively, from each other. The side chains of Glu109 and Asp127 in the active site reside farther from each other in fRMsrred (4.9 Å) than in fRMsrox (3.7 Å). Moreover, the distance between Cys78 and Asp127 is changed from 5.8 Å in fRMsrred to 8.3 Å in fRMsrox. In addition, the fRMsrred structure shows movement of the side chain of Cys68 toward the entrance of the active site. Together, the movements of the active site residues (particularly Cys102) determine the conformations of fRMsrred and fRMsrox, leading to an open conformation in fRMsrred and a closed conformation in fRMsrox (Fig. 2F).
Catalytic Mechanism
Our crystal structures of fRMsrred, fRMsrsub, and fRMsrox help in understanding the mode of binding of the substrate Met-R-O to the active site of fRMsr, the roles of the active site residues, and the conformational changes of fRMsr during catalysis. Significant conformational changes of the active site, particularly in the loop including the catalytic Cys, occur in each catalytic step. The reduced form has an open conformation to allow access to the substrate, the substrate-bound form takes a closed conformation after accommodation of Met-R-O, and the oxidized form is turned to a more closed conformation after catalysis (Fig. 2F).
Among the three conserved Cys residues, Cys102 was the most mobile, whereas Cys78, the previously suggested catalytic residue from E. coli and S. cerevisiae fRMsrs (18, 19), was the most immobile. Our enzymatic studies concluded that Cys102 is the catalytic residue. Cys68 is suggested to be the resolving Cys by structural and kinetic analyses. Cys78 had no catalytic function, but this residue may play a role in substrate binding, as judged by the kinetic data (i.e. an increase in Km value in the C78S mutant). Here, we propose that the catalytic mechanism of fRMsr consists of three steps. 1) Cys102 attacks the sulfoxide moiety of Met-R-O and is then oxidized to Cys sulfenic acid. 2) Cys68 interacts with the sulfenic acid intermediate to form a disulfide bond. 3) Finally, the Cys102–Cys68 disulfide bond is reduced by a reductant (typically by Trx), and the fRMsr enzyme activity is regenerated (Fig. 3).
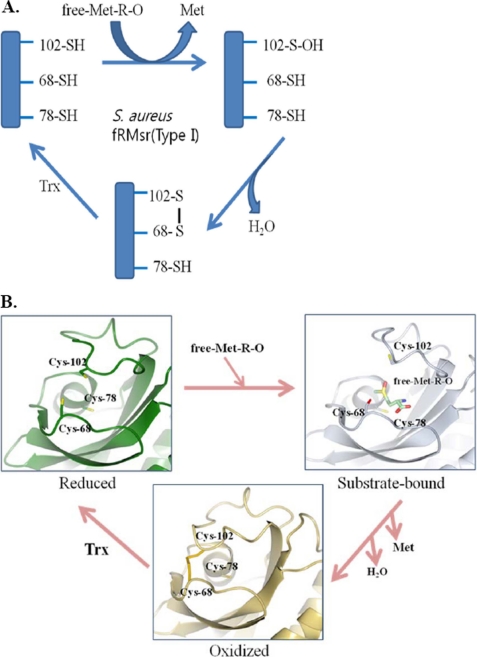
FIGURE 3.
Proposed catalytic mechanism of fRMsr. A, a schematic representation. Catalytic Cys102 attacks free Met-R-O (Met-R-O) and is then oxidized to sulfenic acid. Cys68 acts as a resolving Cys and thus interacts with the Cys sulfenic acid to form a disulfide bond. The resulting Cys102–Cys68 disulfide bond is reduced by a reductant (typically by Trx in vivo or by DTT in vitro), and finally the enzyme becomes active. Cys78 has no catalytic function. B, a structural representation. Reduced fRMsr initially displays an open conformation in the active site, and after binding of the substrate, the enzyme is converted to a closed conformation. Formation of a disulfide bond between Cys102 and Cys68 makes the enzyme more closed.
It should be noted that, in contrast to type I fRMsrs, type II enzymes contain only the conserved Cys78 and Cys102. They lack Cys68. Because our studies revealed no direct function for Cys78 in the catalysis of type I fRMsr, it is questionable whether this residue plays any role in the catalysis of type II fRMsr. It is possible that this Cys78 would function as a resolving Cys, replacing the role of Cys68 in type I enzymes. Thus, biochemical and structural studies of type II fRMsr would be interesting.
Implications of the Mechanism of Action of fRMsr Using a Competitive Inhibitor
We determined another complex structure (fRMsrisopro) that contains isopropyl alcohol in the active site pocket of fRMsr. This crystal was obtained from a crystallization solution consisting of 2 m ammonium sulfate and 10% (v/v) isopropyl alcohol and had one subunit of protein in the asymmetric unit like fRMsrox. The fRMsrisopro structure also contains a disulfide bond formed by Cys68 and Cys102. Interestingly, the binding pattern of isopropyl alcohol is expected to define the location of the substrate binding site (Fig. 4). The hydroxyl group of isopropyl alcohol interacts by hydrogen-bonding with Glu109, Asp125, and Asp127, respectively. This is similar to the hydrogen bond interactions of these residues with the nitrogen of the substrate in fRMsrsub. This structural analysis suggests that isopropyl alcohol could act as a competitive inhibitor of fRMsr enzyme. To test this hypothesis, we assayed the enzyme activities in the presence of 1% ethanol, n-propyl alcohol, or isopropyl alcohol (Table 3). Relative activities of fRMsr in the presence of ethanol and n-propyl alcohol were 91%. However, in the presence of isopropyl alcohol, the enzyme activity significantly decreased to 69%. We further determined kinetic parameters in the presence of 1% isopropyl alcohol. The Km value was 100 ± 30 μm, which is 2-fold higher than that in the absence of isopropyl alcohol. The Vmax value was 420 ± 30 nmol/min/mg of protein, similar to that without isopropyl alcohol. These results indicated that isopropyl alcohol can competitively inhibit fRMsr activity. Moreover, a branched methyl group in isopropyl alcohol may play a role in this inhibitory effect.
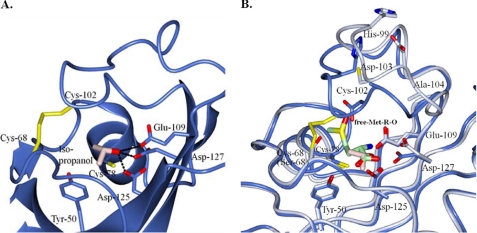
FIGURE 4.
Structure of isopropyl alcohol-bound S. aureus fRMsr and its comparison with the substrate-bound form. A, the active site of fRMsrisopro. Interactions between isopropyl alcohol (Iso-propanol) and active site residues are represented by dotted lines. B, comparison of active sites between fRMsrisopro (light blue) and fRMsrsub (light gray). The active site residues of fRMsrisopro and fRMsrsub are superimposed. Disulfide bonds between Cys68 and Cys102 and isopropyl alcohol in fRMsrisopro are shown by yellow and light pink sticks, respectively, and substrate Met-R-O (Met-R-O) in fRMsrsub is shown by a light green stick.
TABLE 3.
Relative activity of fRMsr enzyme with various alcohols
Enzyme assays were performed in the presence of 1% various alcohols as described under “Experimental Procedures.”
| Alcohol | Relative activity |
|---|---|
| % | |
| None | 100 |
| Ethanol | 91 |
| n-Propyl alcohol | 91 |
| Isopropyl alcohol | 69 |
| n-Butyl alcohol | 87 |
| Isobutyl alcohol | 52 |
We further tested the inhibitory effect with 1% n-butyl alcohol and isobutyl alcohol (Table 3). In the presence of n-butyl alcohol, the relative activity was 87%, similar to that with ethanol or n-propyl alcohol. However, the enzyme activity was significantly inhibited by 50% in the presence of isobutyl alcohol. Together, our results indicate that a branched methyl group in alcohols seems important for competitive inhibition of fRMsr enzyme activity. The branched methyl group of alcohols may be crucial for binding to the active site in order to competitively inhibit the fRMsr activity, suggesting that the methyl group of the substrate may be important for binding affinity to the enzyme shown in the structure of fRMsrsub.
In summary, we have determined the crystal structures of reduced, substrate-bound, oxidized, and inhibitor-bound fRMsrs at atomic resolution levels. Our structural and biochemical studies suggest the catalytic mechanism of fRMsr, where Cys102 acts as the catalytic residue and Cys68 acts as the resolving Cys. Our structures show the mode of binding of the substrate free Met-R-O, the roles of active site residues in catalysis, and the conformational changes of the active site during catalysis, particularly by the loop containing the catalytic Cys102. In addition, our studies with a competitive inhibitor, isopropyl alcohol, predict the mechanism of action of fRMsr, where the methyl group in the substrate or a branched methyl group in alcohols seems important for interaction with the enzyme.
Supplementary Material
Acknowledgments
We thank the staff for assistance during the data collection at beamlines 6C and 4A of Pohang Light Source, South Korea.
This work was supported by a Korea University Grant 2009 (to Y. M. C.) and by Korea Healthcare Technology R&D Project (Ministry for Health, Welfare, and Family Affairs) Grant A090181 (to H. -Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3KSF, 3KSG, 3KSH, and 3KSI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Met-S-O
- methionine S-sulfoxide
- Met-R-O
- methionine R-sulfoxide
- DTT
- dithiothreitol
- Msr
- methionine-sulfoxide reductase
- fRMsr
- free methionine-R-sulfoxide reductase
- Trx
- thioredoxin
- MES
- 2-(N-morpholino)ethanesulfonic acid
- DTT
- dithiothreitol
- r.m.s.
- root mean square
- dabsyl
- 4-(4-dimethylaminophenylazo)-benzolsulfonyl.
REFERENCES
- 1.Stadtman E. R. (2006) Free Radic. Res. 40, 1250–1258 [DOI] [PubMed] [Google Scholar]
- 2.Weissbach H., Resnick L., Brot N. (2005) Biochim. Biophys. Acta 1703, 203–212 [DOI] [PubMed] [Google Scholar]
- 3.Kim H. Y., Gladyshev V. N. (2007) Biochem. J. 407, 321–329 [DOI] [PubMed] [Google Scholar]
- 4.Moskovitz J. (2005) Biochim. Biophys. Acta 1703, 213–219 [DOI] [PubMed] [Google Scholar]
- 5.Erickson J. R., Joiner M. L., Guan X., Kutschke W., Yang J., Oddis C. V., Bartlett R. K., Lowe J. S., O'Donnell S. E., Aykin-Burns N., Zimmerman M. C., Zimmerman K., Ham A. J., Weiss R. M., Spitz D. R., Shea M. A., Colbran R. J., Mohler P. J., Anderson M. E. (2008) Cell 133, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koc A., Gasch A. P., Rutherford J. C., Kim H. Y., Gladyshev V. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minniti A. N., Cataldo R., Trigo C., Vasquez L., Mujica P., Leighton F., Inestrosa N. C., Aldunate R. (2009) Aging Cell 8, 690–705 [DOI] [PubMed] [Google Scholar]
- 8.Ruan H., Tang X. D., Chen M. L., Joiner M. L., Sun G., Brot N., Weissbach H., Heinemann S. H., Iverson L., Wu C. F., Hoshi T., Chen M. L., Joiner M. A., Heinemann S. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnham K. J., Ciccotosto G. D., Tickler A. K., Ali F. E., Smith D. G., Williamson N. A., Lam Y. H., Carrington D., Tew D., Kocak G., Volitakis I., Separovic F., Barrow C. J., Wade J. D., Masters C. L., Cherny R. A., Curtain C. C., Bush A. I., Cappai R. (2003) J. Biol. Chem. 278, 42959–42965 [DOI] [PubMed] [Google Scholar]
- 10.Hou L., Kang I., Marchant R. E., Zagorski M. G. (2002) J. Biol. Chem. 277, 40173–40176 [DOI] [PubMed] [Google Scholar]
- 11.Schöneich C. (2005) Biochim. Biophys. Acta 1703, 111–119 [DOI] [PubMed] [Google Scholar]
- 12.Wassef R., Haenold R., Hansel A., Brot N., Heinemann S. H., Hoshi T. (2007) J. Neurosci. 27, 12808–12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B. C., Dikiy A., Kim H. Y., Gladyshev V. N. (2009) Biochim. Biophys. Acta 1790, 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissbach H., Etienne F., Hoshi T., Heinemann S. H., Lowther W. T., Matthews B., John G., Nathan C., Brot N. (2002) Arch. Biochem. Biophys. 397, 172–178 [DOI] [PubMed] [Google Scholar]
- 15.Dhandayuthapani S., Blaylock M. W., Bebear C. M., Rasmussen W. G., Baseman J. B. (2001) J. Bacteriol. 183, 5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olry A., Boschi-Muller S., Marraud M., Sanglier-Cianferani S., Van Dorsselear A., Branlant G. (2002) J. Biol. Chem. 277, 12016–12022 [DOI] [PubMed] [Google Scholar]
- 17.Sasindran S. J., Saikolappan S., Dhandayuthapani S. (2007) Future Microbiol. 2, 619–630 [DOI] [PubMed] [Google Scholar]
- 18.Lin Z., Johnson L. C., Weissbach H., Brot N., Lively M. O., Lowther W. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9597–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le D. T., Lee B. C., Marino S. M., Zhang Y., Fomenko D. E., Kaya A., Hacioglu E., Kwak G. H., Koc A., Kim H. Y., Gladyshev V. N. (2009) J. Biol. Chem. 284, 4354–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoraghi R., Corbin J. D., Francis S. H. (2004) Mol. Pharmacol. 65, 267–278 [DOI] [PubMed] [Google Scholar]
- 21.Boschi-Muller S., Gand A., Branlant G. (2008) Arch. Biochem. Biophys. 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 22.Kim Y. K., Shin Y. J., Lee W. H., Kim H. Y., Hwang K. Y. (2009) Mol. Microbiol. 72, 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowther W. T., Weissbach H., Etienne F., Brot N., Matthews B. W. (2002) Nat. Struct. Biol. 9, 348–352 [DOI] [PubMed] [Google Scholar]
- 24.Tête-Favier F., Cobessi D., Boschi-Muller S., Azza S., Branlant G., Aubry A. (2000) Structure 8, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 25.Singh V. K., Moskovitz J. (2003) Microbiology 149, 2739–2747 [DOI] [PubMed] [Google Scholar]
- 26.Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., Buchanan S. G., Buchanan M. D., Batiyenko Y., Christopher J. A., Emtage S., Eroshkina A., Feil I., Furlong E. B., Gajiwala K. S., Gao X., He D., Hendle J., Huber A., Hoda K., Kearins P., Kissinger C., Laubert B., Lewis H. A., Lin J., Loomis K., Lorimer D., Louie G., Maletic M., Marsh C. D., Miller I., Molinari J., Muller-Dieckmann H. J., Newman J. M., Noland B. W., Pagarigan B., Park F., Peat T. S., Post K. W., Radojicic S., Ramos A., Romero R., Rutter M. E., Sanderson W. E., Schwinn K. D., Tresser J., Winhoven J., Wright T. A., Wu L., Xu J., Harris T. J. (2005) Proteins 60, 787–796 [DOI] [PubMed] [Google Scholar]
- 27.Ho Y. S., Burden L. M., Hurley J. H. (2000) EMBO J. 19, 5288–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 29.Bong S. M., Moon J. H., Kim H. Y., Kim H. S., Kim A. Y., Chi Y. M. (2009) Acta Crystallogr. F 65, 1120–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1996) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 34.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 35.Kwak G. H., Kim M. J., Kim H. Y. (2010) Biochem. Biophys. Res. Commun. 395, 412–415 [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.