Abstract
Lymphoid-specific protein tyrosine phosphatase (Lyp), a member of the protein tyrosine phosphatase (PTP) superfamily of enzymes, is an important mediator of human-leukocyte signaling. Lyp has also emerged as a potential anti-autoimmune therapeutic target, owing to the association of a Lyp-activating mutation with an array of autoimmune disorders. Toward the goal of generating a selective inhibitor of Lyp activity that could be used for investigating Lyp’s roles in cell signaling and autoimmune-disease progression, here we report that Lyp’s PTP domain can be readily sensitized to target-specific inhibition by a cell-permeable small molecule. Insertion of a tetracysteine-motif-containing peptide at a conserved position in Lyp’s catalytic domain generated a mutant enzyme (Lyp-CCPGCC) that retains activity comparable to that of wild-type Lyp in the absence of added ligand. Upon addition of a tetracysteine-targeting biarsenical compound (FlAsH), however, the activity of the Lyp-CCPGCC drops dramatically, as assayed with either small-molecule or phosphorylated-peptide PTP substrates. We show that FlAsH-induced Lyp-CCPGCC inhibition is potent, specific, rapid, and independent of the nature of the PTP substrate used in the inhibition assay. Moreover, we show that FlAsH can be used to specifically target overexpressed Lyp-CCPGCC in a complex proteomic mixture. Since the mammalian-cell permeability of FlAsH is well established, it is likely that FlAsH-mediated inhibition of Lyp-CCPGCC will be useful for specifically targeting Lyp activity in engineered leukocytes and autoimmune-disease models.
Keywords: Biarsenicals, FlAsH, Protein tyrosine phosphatases, Lyp, Protein engineering, Inhibitor sensitization
1. Introduction
Protein tyrosine phosphatases (PTPs) catalyze the dephosphorylation of phosphotyrosine, a central signal-transduction control element in metazoan biology.1 Improperly regulated PTP activity has been implicated as a causative agent in a range of human diseases, including leukemia, solid-tumor cancers, diabetes, and autoimmune disorders.2–6 While many disease-associated mutations in PTP-encoding genes are loss-of-function mutations, gain-of-function PTP mutations are of particular interest from a therapeutic perspective; it is this relatively small subset of PTP disease-associated mutants that one could envision as therapeutic targets for small-molecule PTP inhibitors. Among the best characterized of PTPs that have been linked to activating disease-associated mutations is the lymphoid-specific protein tyrosine phosphatase (Lyp, also called PTPN22), which is expressed predominantly in leukocytes and is a negative regulator of T-cell activation.7 Recently, a flurry of studies has uncovered associations between a single-nucleotide polymorphism (SNP) in the PTPN22 gene and a range of autoimmune disorders, including type-I diabetes,8, 9 rheumatoid arthritis,10 Graves disease,11 myasthenia gravis,12 and systemic lupus erythematosus (SLE).13 The SNP common to all of these associations encodes an arginine-to-tryptophan mutation (R620W) that increases Lyp’s PTP activity.14 This putative connection between high Lyp activity and autoimmune disease has become even more compelling with the recent discovery of a PTP-activity-lowering polymorphism (R263Q) that confers protection against SLE.15 Taken together, these studies present the exciting possibility that Lyp inhibitors could represent an important class of anti-autoimmune therapeutics. Moreover, these genetic data highlight the need for chemical tools that can be used to study the poorly understood connection between Lyp activity and autoimmune-disease progression.
PTP-inhibitor discovery is inherently difficult due to two recurring problems observed with many active-site-directed inhibitors: lack of target specificity (classical PTP catalytic domains share a significant degree of sequence and structural homology with one another) and poor bioavailability (many PTP-binding pharmacophores contain negatively charged phosphotyrosine mimetics that lower an inhibitor’s cellular permeability).6, 16–18 Nevertheless, several groups have recognized the potential therapeutic impact of target-specific Lyp inhibitors, and significant efforts toward a Lyp-specific inhibitor have recently been undertaken.19–23 While the compounds that have been identified from these studies provide important templates for further optimization and discovery, Lyp-inhibitor discovery is a newly emerging field, and chemical tools that can control Lyp activity in cells with high potency and target selectivity are still needed.
Our lab has recently described a systematic strategy for engineering novel inhibitor sensitivity in PTPs.24–28 In our approach, a mutation (point mutation and/or peptide insertion) in a target PTP sensitizes the enzyme to inhibition by a small molecule that does not inhibit wild-type PTPs. When a non-deleterious, inhibitor-sensitizing mutation is discovered, the mutant and inhibitor constitute a specific ligand/receptor pair that can be used to study the cellular roles of the engineered—but functionally “wild-type-like”—PTP targets. Toward this end, we have previously reported that PTP domains can be sensitized to noncompetitive inhibition by a compound that has no significant affinity for wild-type PTPs26—namely, FlAsH, a cell-permeable biarsenical compound that binds to cysteine-rich peptides.29, 30 Specifically, insertion of a FlAsH-binding hexapeptide (CCPGCC) at position 187 of a model PTP (TCPTP) was shown to confer strong FlAsH sensitivity on the enzyme.26 Although the 187 insertion position is distal from TCPTP’s active site, the insertion mutant’s novel inhibitor sensitivity could be structurally rationalized,31 as Glu187 lies at the end of a conserved PTP loop (the WPD loop) that closes upon substrate binding, properly positioning a mechanistically important aspartate residue (the “D” of WPD) for the PTP reaction. Binding of FlAsH to the TCPTP insertion mutant may impede proper closure of the WPD loop, in a manner that is consistent with other noncompetitive inhibitors that target natural allosteric sites in PTPs.32, 33
The fundamental nature of the WPD loop in the structure and mechanism of PTP domains augurs well for the prospect of using FlAsH as an inhibitor of engineered PTPs beyond TCPTP: the WPD loop is one of the most conserved regions in PTP catalytic domains and it can be readily identified from primary sequence alignments.34, 35 Indeed, we have recently shown that PTPs from six distinct PTP sub-families can be sensitized to FlAsH inhibition.27 In all cases, the engineered PTPs respond to FlAsH with high selectivity and potency via a conserved mechanism that is independent of the particular PTP that has been sensitized to inhibition.27
A key advantage of inhibitor-design strategies that exploit similarities in protein family members (in this case, the conserved WPD loop of the PTP domain) is that successful engineering can potentially be applied to any protein-family member,36 and new targets can be selected as complementary biological data reveal their importance. The autoimmune-association studies noted above have brought the need for Lyp inhibitors to the fore, and primary sequence analysis suggests that Lyp may be amenable to FlAsH-sensitization.34 Here we describe the generation and characterization of a sensitized mutant of Lyp, which will provide a novel tool for the elucidation of Lyp’s functions in engineered cells and, potentially, for the pharmacological validation of Lyp as an anti-autoimmune drug target.
2. Materials and methods
2.1. General
FlAsH was synthesized as described previously.26, 29, 37 Absorbance measurements were made on a Molecular Devices Versamax 96-well plate reader. Fluorescence measurements were made on either an ISS K2 multi-frequency phase fluorometer or a Molecular Devices Spectra Max M5 96-well fluorescence plate reader. Gel imaging and analyses were performed on a Syngene InGenius gel-documentation system. Curve fitting was carried out with SigmaPlot 11.0. Errors bars and ± values in all figures and tables represent the standard deviations of at least three independent trials.
2.2. Cloning and mutagenesis
A PCR product encoding the Lyp catalytic domain (residues 1–294) was amplified from full-length PTPN22 cDNA (Open Biosystems, Cat#: IHS1382-8404627) with PfuTurbo DNA polymerase (Stratagene) using the following primers (5′ to 3′): ATCCTGAATTCCATGGACCAAAGAGAAATTCTGCAGAAG and ATCCTAAGCTTCATCTGTCTCTTAAATAGTTCTAATACAGC. The PCR product and pET-21b were doubly digested with EcoRI and HindIII, gel purified, and ligated using T4 DNA ligase, yielding the Lyp-expressing plasmid pZEW001. Insertional mutagenesis was carried out by QuikChange™ essentially as described for TCPTP26 with pZEW001 as template and the following mutagenic primers (5′to 3′): GACCATGATGTACCTTGCTGCCCGGGCTGCTGCTCATCTATAGACCCT and AGGGTCTATAGATGAGCAGCAGCCCGGGCAGCAAGGTACATCATGGTC, yielding the Lyp-CCPGCC-encoding plasmid pZEW014.
2.3. Lyp expression and purification
BL21(DE3)-codonPLUS-RIL E. coli (Stratagene) containing either pZEW001 or pZEW014 were grown overnight at 37°C in LB. Cultures were diluted, grown to mid-log phase (OD600 = 0.5), induced with 0.2 mM isopropyl-1-thio-β-D-galactopyranoside, and shaken at 23°C for 20 h. The cells were harvested by centrifugation, resuspended in binding buffer (50 mM Tris pH 7.9, 500 mM NaCl, 5 mM Imidazole), and lysed by French Press at ~2000 psi. Lysates were clarified by centrifugation, and enzyme purifications were carried out using SwellGel Nickel Chelated Discs (Pierce) according to the manufacturer’s instructions. The protein solutions obtained were exchanged into storage buffer (50 mM 3,3-dimethylglutarate pH 7.0, 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol), concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C. SDS-PAGE was used to estimate enzyme concentrations, by comparison of pixel counts of the major 37 kD band in the enzyme preparation to those of a reference protein (BSA) run on the same gel. Inspection of SDS-PAGE gels yielded estimates of >90% and >50% purity for Lyp and Lyp-CCPGCC, respectively. Nickel-affinity-chromatography-purified yields of both enzymes were approximately 2 mg/L of culture.
2.4. PTP activity and inhibition assays (pNPP substrate)
2.4.1. Michaelis-Menten kinetic assays
Activity assays on para-nitrophenyl phosphate (pNPP) were carried out at 25 °C in low protein-binding tubes. Solutions of wild-type Lyp or Lyp-CCPGCC (2.5 µM) Lyp were incubated in 1×PTP buffer (50 mM 3,3-dimethylgluterate pH 7.0, 1 mM EDTA, 50 mM NaCl) for 2.5 h in the presence of 10 µM FlAsH or DMSO vehicle. Solutions were then diluted in 1×PTP buffer, and PTP reactions were initiated by the addition of 20 µL pNPP (varying concentrations) to 180 µL of the enzyme solution (enzyme concentration in assay: 500 nM). Reactions were quenched by the addition of 40 µL 5 M NaOH. 200 µL of reaction mixture were then loaded onto a 96-well plate and the absorbance at 405 nm was measured. Kinetic constants were determined by fitting the data to the Michaelis-Menten equation.
2.4.2. FlAsH-concentration-dependence inhibition assays
FlAsH-concentration-dependence assays were carried out at 9 mM pNPP, 250 nM Lyp or Lyp-CCPGCC, and varying concentrations of FlAsH in 1×PTP buffer. After a 2 h incubation of enzyme and FlAsH (or vehicle control), pNPP was added. The reactions were quenched and quantified as described above.
2.4.3. Kinetics of FlAsH inhibition
Reactions were carried out in a total volume of 200 µL at 25°C. 30 µL of premixed solutions of FlAsH and pNPP were aliquoted onto a 96-well plate. 170 µL of an enzyme and buffer solution were added such that the final concentrations were 250 nM enzyme, 10 mM pNPP, and 0.5–5 µM FlAsH in 1×PTP buffer. The increase in absorbance at 405 nm was monitored with readings every 10 seconds for 45 minutes. Reaction velocity (At) for each dosage of FlAsH was then determined at each time point, tx, by calculating the slope of the linear regression over the one-minute interval tx−30 ≤ tx ≤tx+30. Relative activity (At/A0) was obtained by dividing the reaction velocity in the presence of each FlAsH dosage by the no-FlAsH control. The pseudo-first-order rate constant (kobs) was determined for each FlAsH concentration by fitting the data to Equation 1,38 in which A0 and A∞ represent the maximum and minimum reaction velocities, respectively:
| (1) |
Each kobs value was plotted against FlAsH concentration and the data were fit to Equation 2,38 in which KI is the apparent inhibition constant, ki is the inactivation rate constant, and [F] is the concentration of FlAsH:
| (2) |
2.5. PTP activity and inhibition assays (DADEpYLIPQQG substrate)
The PTP activities of Lyp and Lyp-CCPGCC on the phosphopeptide DADEpYLIPQQG were measured by monitoring the time-dependent increase of the peptide’s fluorescence at 305 nm, essentially as described.39 Briefly, solutions of 59 nM enzyme and 294 nM FlAsH (or DMSO vehicle only) were incubated for 1.5 h in peptide buffer (50 mM 3,3-dimethylglutarate pH 7, 125 mM NaCl, 1 mM EDTA). PTP reactions were initiated by the addition of DADEpYLIPQQG (EMD) to the enzyme-FlAsH solution such that the final reaction concentrations were 250 nM FlAsH (or control), 50 nM enzyme, and 15 µM peptide. The increase in fluorescence at 305 nm was then measured (λexc= 280 nm, λem= 305 nm, 0.5 nm slit widths) for 20 minutes or until substrate was consumed. To extract kinetic parameters, the intensity of fluorescence was converted to product concentration ([p]) for each time point (t) and fit to Equation 3,39 in which [Lyp] is the total concentration of enzyme in the assay (50 nM) and [p]∞ is the final concentration of product (15 µM):
| (3) |
2.6. Fluorescence
2.6.1. Kinetics of Lyp-CCPGCC-induced FlAsH fluorescence
Wild-type Lyp or Lyp-CCPGCC (250 nM in 1×PTP buffer) was mixed with FlAsH (500 nM) in 1×PTP buffer, and FlAsH fluorescence values (excitation: 510 nm, emission: 529 nm) of the resulting solutions were measured every 10 seconds. The displayed data sets were normalized by the subtraction of a FlAsH-only (no protein) control monitored over the same time range.
2.6.2. Measurement of the Lyp-CCPGCC/FlAsH apparent dissociation constant (KDapp)
Solutions of 25 nM FlAsH and Lyp-CCPGCC ranging in concentration from 39–2500 nM were incubated in 1×PTP buffer for 2.5 h at room temperature. The FlAsH fluorescence values (excitation: 510 nm, emission: 540 nm) of the solutions were measured in 96-well plates, corrected by subtraction of the fluorescence from a FlAsH-only (no-protein) control, and normalized to estimate the percentage of ligand complexed to protein at each Lyp-CCPGCC concentration (ϕ). To estimate the apparent dissociation constant of the Lyp-CCPGCC/FlAsH interaction, the data were fit to Equation 4, which is derived from first principles for a reversible, tight-binding inhibitor:40, 41
| (4) |
[F] and [Lyp] represent the total concentrations of FlAsH and enzyme in the assay, respectively.
2.6.3. Fluorescence of Lyp-expressing cells
Pellets of E. coli cells expressing either Lyp or Lyp-CCPGCC were prepared as described above (section 2.3.) and frozen at −80 °C. Pellets from 15 mL of culture were resuspended and diluted to OD600 = 6.9 (1 cm path length) in 1×PTP buffer containing 10 µM FlAsH. After 2.5 h at room temperature, the FlAsH fluorescence values (excitation: 510 nm, emission: 540 nm) of the suspensions were measured in 96-well plates and corrected by subtraction of the fluorescence from a FlAsH-only (no-cell) control.
2.6.4. In-gel detection of Lyp-CCPGCC/FlAsH fluorescence
Pellets of E. coli cells expressing either Lyp or Lyp-CCPGCC were freeze-thawed and FlAsH-treated (10 µM) as described above (section 2.6.3.). After 2.5 h at room temperature, the cell suspensions were mixed 3:1 with 4×loading buffer (4×LDS, Invitrogen), boiled for 10 minutes, and loaded on a 4–12% Bis-Tris SDS-PAGE gel (Invitrogen). Following electrophoresis, fluorescent bands were visualized on a gel-documentation system using a UV transilluminator for excitation and a 500–600 nm emission filter.
3. Results and Discussion
3.1. Design of and characterization of a FlAsH-sensitized Lyp mutant
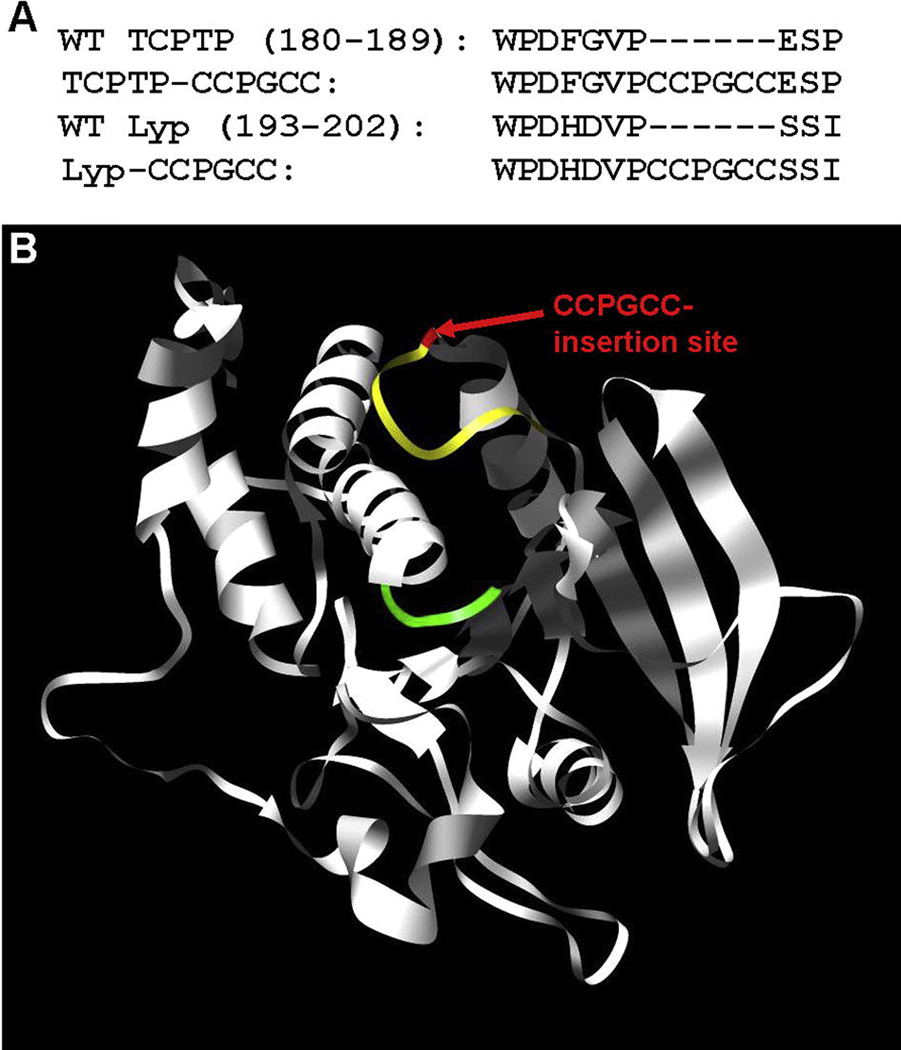
Lyp is a class-I cysteine-based, classical cytoplasmic PTP.34 In addition to a canonical WPD loop, Lyp possesses significant catalytic-domain homology to both TCPTP and the other PTPs to which FlAsH sensitization has been applied.19, 35 To design a potentially FlAsH-sensitized version of Lyp we aligned its primary sequence and catalytic-domain structure with TCPTP, the model PTP on which our PTP-sensitization was developed (Figure 1A). Both sequence comparison and inspection of Lyp’s three-dimensional structure (Figure 1B) suggest that a CCPGCC insertion analogous to the one that has been successful for sensitizing TCPTP may work for the Lyp target as well. To test this idea, we generated the putatively FlAsH-sensitized insertion mutant Lyp-CCPGCC (Figure 1A) by insertional mutagenesis and expressed it from E. coli as a six-histidine tagged protein.
Figure 1.
Design of a FlAsH-sensitized Lyp mutant. (A) Amino-acid sequence alignment of the WPD-loop region of a previously FlAsH-sensitized PTP, TCPTP (wild-type: WT, sensitized: TCPTP-CCPGCC), with that of wild-type Lyp and Lyp-CCPGCC. (B) Ribbon diagram of the Lyp catalytic domain (PDB code: 2QCJ19). Lyp’s canonical WPD loop is shown in yellow with the position of the Lyp-CCPGCC insertion shown in red. For perspective, the phosphotyrosine-binding loop of the PTP active site is shown in green.
For a mutant enzyme/inhibitor pair to be useful in chemical biology it is imperative that the mutated protein retains activity in the absence of the drug.25, 42 To investigate the possible effect of the CCPGCC mutation on Lyp’s PTP activity, we determined the Michaelis-Menten kinetic parameters for wild-type and Lyp-CCPGCC with the artificial PTP substrate para-nitrophenyl phosphate (pNPP). We found that the insertion mutant retains almost full catalytic activity in the absence of FlAsH, with a catalytic rate-constant value (kcat) that is within a factor of two of the corresponding wild-type construct (Table 1). (The wild-type Lyp catalytic domain has been enzymatically characterized by others with pNPP previously,19 and our construct yielded kinetic constants that were in good agreement with literature data.) The tolerance of Lyp to the six-amino-acid insertion, indicated by its relatively low 1.7-fold reduction in kcat, is interesting in light of the fact that Lyp’s closest homolog, PEST, was substantially more affected by the analogous CCPGCC insertion (wild type PEST: kcat = 0.59 s−1; PEST-CCPGCC: kcat = 0.081 s−1).27 These findings suggest that primary-sequence alignments alone will not constitute a suitable guide for prediction of which PTPs are most amenable to FlAsH sensitization via CCPGCC insertion.
Table 1.
Inherent PTP activities of wild-type Lyp and Lyp-CCPGCC catalytic domains
| Enzyme | kcat (s−1)a | KM (mM)a | kcat (s−1)b | KM (µM)b |
|---|---|---|---|---|
| Wild-type Lyp | 0.49 ± 0.04 | 4.86 ± 0.39 | 4.16 ± 0.56 | 8.12 ± 2.63 |
| Lyp-CCPGCC | 0.27 ± 0.02 | 5.05 ± 0.35 | 2.25 ± 0.56 | 17.8 ± 1.8 |
With pNPP used as the PTP substrate.
With DADEpYLIPQQG used as the PTP substrate.
To further test the suitability of Lyp-CCPGCC as an inhibitor-sensitized PTP, we measured its activity with a more physiologically relevant substrate, the phosphopeptide DADEpYLIPQQG, which corresponds to the auto-phosphorylation site of the epidermal growth factor receptor (EGFR988–998). With the phosphopeptide substrate we again found that, in the absence of FlAsH, Lyp-CCPGCC has only a slightly lower activity than wild-type Lyp (virtually identical 1.8- and 1.7-fold drops in kcat relative to wild-type Lyp on peptide and pNPP, respectively; see Table 1). These data show that the catalytic activity of the CCPGCC mutant is “wild-type-like” in activity independent of substrate and suggest that, absent FlAsH, Lyp-CCPGCC will be capable of dephosphorylating the tyrosine-phosphorylated substrates that Lyp encounters in a mammalian cell.
3.2. Target-specific inhibition of Lyp-CCPGCC by FlAsH
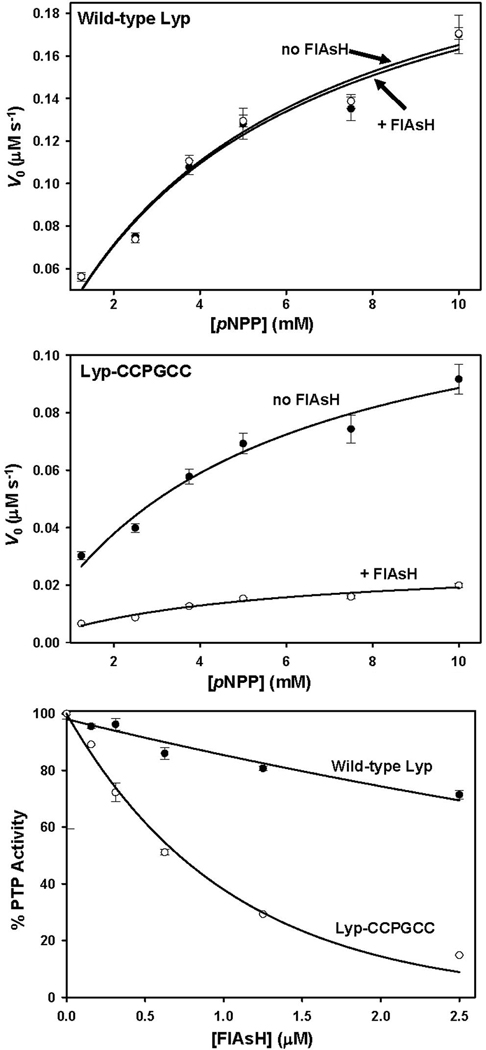
We next investigated the effects of FlAsH treatment on wild-type Lyp and Lyp-CCPGCC. In previous studies from our lab, FlAsH was found to have no discernible effect upon a panel of wild-type PTPs.27 Consistent with this precedent, the kinetic parameters of wild-type Lyp activity with pNPP were indistinguishable in the presence and absence of FlAsH (Table 2, Figure 2A). By contrast, Lyp-CCPGCC is strongly inhibited by FlAsH: upon incubation with a 4-fold molar excess of FlAsH, the specificity constant of Lyp-CCPGCC activity (kcat/KM) dropped 4.5-fold (Table 2, Figure 2B). As previously observed with other FlAsH-sensitized PTPs, Lyp-CCPGCC inhibition appears to be noncompetitive in nature, as the loss of catalytic efficiency is due completely to a decrease in kcat (Table 2).
Table 2.
PTP activities of wild-type Lyp and Lyp-CCPGCC catalytic domains after pre-incubation with FlAsH
| Enzyme | kcat (s−1)a | KM (mM)a | kcat (s−1)b | KM (µM)b |
|---|---|---|---|---|
| Wild-type Lyp | 0.49 ± 0.02 | 4.92 ± 0.35 | 3.84 ± 1.11 | 7.55 ± 4.48 |
| Lyp-CCPGCC | 0.057 ± 0.004 | 4.83 ± 0.44 | NDc | NDc |
With pNPP used as the PTP substrate.
With DADEpYLIPQQG used as the PTP substrate.
ND: Unable to determine kinetic parameters due to low activity; see Figure 3B.
Figure 2.
Target-specific inhibition of Lyp-CCPGCC. (A) WT Lyp (2.5 µM) or (B) Lyp-CCPGCC (2.5 µM) was incubated in the absence (closed circles) or presence of FlAsH (10 µM, open circles), diluted, and assayed for activity with the PTP substrate pNPP at the indicated concentrations. C) Concentration-dependence of FlAsH-induced inhibition: Wild-type Lyp (250 nM, closed circles) or Lyp-CCPGCC (250 nM, open circles) was incubated in the absence or presence (indicated concentrations) of FlAsH. The resulting solutions were assayed for activity with pNPP (9 mM). “% PTP Activity” is defined as the initial velocity in the presence of FlAsH divided by the initial velocity of the vehicle-only (100%) control.
To test whether the degree of Lyp-CCPGCC inhibition was dependent on the FlAsH dosage, we performed a concentration-dependence inhibition assay (Figure 2C). We observed that the degree of inhibition by FlAsH is highly concentration-dependent, with fifty-percent inhibition (IC50) achieved at approximately 625 nM FlAsH. It should be noted that FlAsH so potently inhibits Lyp-CCPGCC that the IC50 value approaches the concentration of enzyme in the assay (250 nM Lyp-CCPGCC, the lowest concentration that can be reasonably used in the pNPP-based assay). Under conditions of such potent, almost stoichiometric, inhibition, measured IC50 values are highly dependent on the enzyme concentration used, and, therefore, of little fundamental significance. For example, under the conditions of the inhibition experiment shown in Figure 2C (250 nM Lyp-CCPGCC), it is theoretically impossible that an inhibitor which binds enzyme in a 1:1 manner could demonstrate a 50% inhibition value of less than 125 nM, half the enzyme concentration. As described below (section 3.4), more sensitive fluorescence-based assays reveal a substantially tighter Lyp-CCPGCC/FlAsH interaction than would be inferred from the inhibition data.
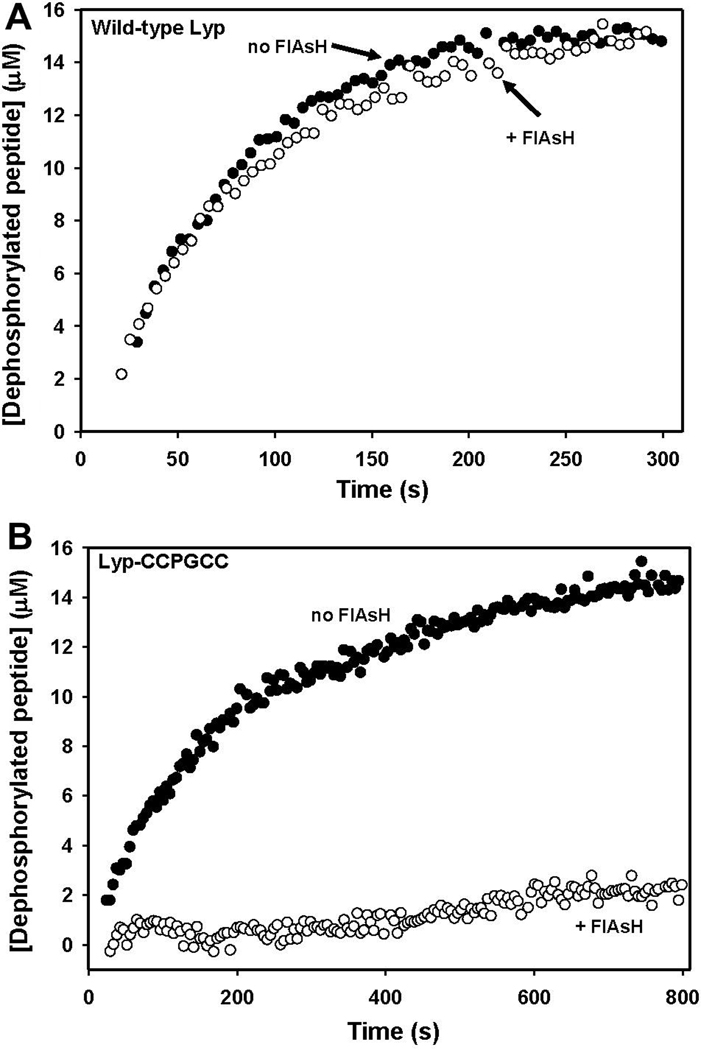
If Lyp-CCPGCC inhibition is to be useful in a cellular context, it is important that FlAsH, the target-specific inhibitor of Lyp-CCPGCC, acts in a substrate-independent manner. To ensure that the FlAsH sensitivity observed with pNPP was not substrate-dependent we investigated Lyp-CCPGCC’s FlAsH sensitivity with DADEpYLIPQQG. Indeed, the engineered FlAsH sensitivity of Lyp-CCPGCC proved to be substrate-independent (Table 2 and Figure 3). When incubated with FlAsH, the activity of the mutant dropped dramatically and specifically (as with pNPP, FlAsH had no significant effect upon wild-type). The FlAsH-induced inhibition of DADEpYLIPQQG dephosphorylation is essentially complete: following incubation with a five-fold molar excess of FlAsH, the magnitude of Lyp-CCPGCC inhibition is so great that the data could not be fit to derive kinetic constants (Figure 3B). Thus, FlAsH-induced inhibition of peptide dephosphorylation by Lyp-CCPGCC is consistent with the potency observed with pNPP and suggests that FlAsH-inhibition could be effective in cellulo, where PTP substrates are more chemically similar to DADEpYLIPQQG than to pNPP. And, taken together, these data showing the potency, specificity, and substrate independence of Lyp-CCPGCC inhibition imply that FlAsH could be used in a cellular context to target Lyp-CCPGCC without substantial off-target PTP inhibition.
Figure 3.
Target-specific inhibition of DADEpYLIPQQG dephosphorylation. (A) Wild-type Lyp (59 nM) or (B) Lyp-CCPGCC (59 nM) was incubated in the absence (closed circles) or presence of FlAsH (294 nM, open circles). After 90 minutes, DADEpYLIPQQG (15 µM) was added to the enzyme solutions and peptide dephosphorylation was monitored by fluorescence.
3.3. Kinetics of Lyp-CCPGCC inhibition by FlAsH
In all of the inhibition experiments described above, Lyp-CCPGCC was allowed to pre-incubate with FlAsH prior to the measurement of PTP activity. Because FlAsH is not a rapid, reversible protein binder—biarsenicals make covalent sulfur-arsenic bonds with their peptide targets29, 43—inhibition of Lyp-CCPGCC is presumably time-dependent, and the timescale of FlAsH action on Lyp-CCPGCC is a potentially complicating issue when using the compound for controlling protein function.
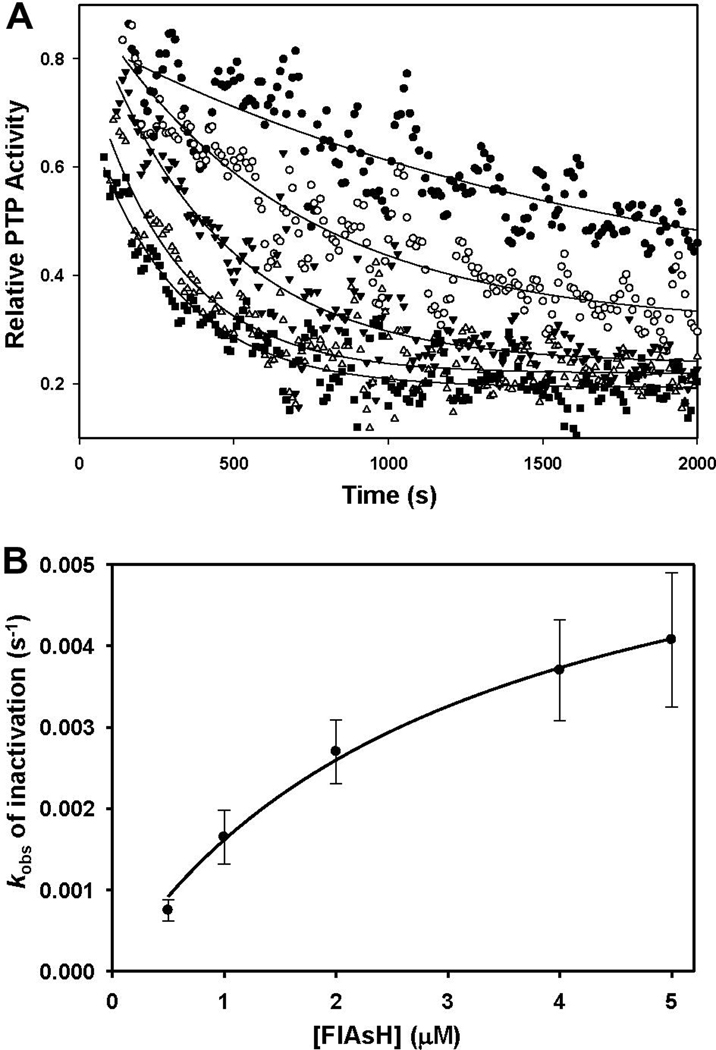
To assess the rate of Lyp-CCPGCC inhibition by FlAsH, we used a continuous assay of pNPP hydrolysis to probe enzyme activity and time-dependent inhibition concurrently.44 We continuously monitored the PTP reaction at various concentrations of FlAsH and determined that the slopes of the resulting progress curves decreased over time, indicating a time-dependent drop in the concentration of catalytically active Lyp-CCPGCC. The rate at which these progress curves fell was dependent on the concentration of FlAsH, and the exponential decay of relative activity suggested a first-order dependence on time (Figure 4A). Analysis of the resulting pseudo-first-order rate constants (kobs) as a function of FlAsH concentration showed that Lyp-CCPGCC inhibition displays saturation kinetics (Figure 4B). The nonlinear relationship between kobs and inhibitor concentration suggests a two-step model of inhibition in which FlAsH and Lyp-CCPGCC first associate in a loose, reversible manner; then, in a second step, undergo a slow conversion to an irreversibly inhibited form. Fitting the inhibition data to this model (see section 2.4.3.) yielded values for the apparent inhibition constant (KIapp) of 3.04 ± 1.05 µM and an inactivation rate constant (ki) of 0.0067 ± 0.0022 s−1. These derived measures of inactivation kinetics compare favorably with other “slow-binding” PTP inhibitors; for example, a recently reported class of aryl vinyl sulfones and sulfonates inactivate PTP domains with KI and ki values that are roughly 100-fold higher and 5-fold lower, respectively.38
Figure 4.
Kinetics of FlAsH-induced Lyp-CCPGCC inhibition. (A) Solutions of Lyp-CCPGCC (250 nM) and pNPP (10 mM) were combined with varying concentrations of FlAsH (closed circles: 0.5 µM, open circles: 1.0 µM, closed triangles: 2.0 µM, open triangles: 4.0 µM, squares: 5.0 µM) and assayed for PTP activity by continuous measurement of absorbance at 405 nm. Relative activity at each time point was computed as the slope of the reaction curve in the presence of FlAsH divided by the slope of the reaction curve for a no-FlAsH control. The curves shown derive from the fitting of the individual time points to a model of exponential activity decay with a first-order dependence on time. (B) Pseudo-first order rate constants were derived from the curve fitting described above and plotted against the relevant indicated concentrations of FlAsH.
3.4. Fluorescence characterization of Lyp-CCPGCC/FlAsH binding
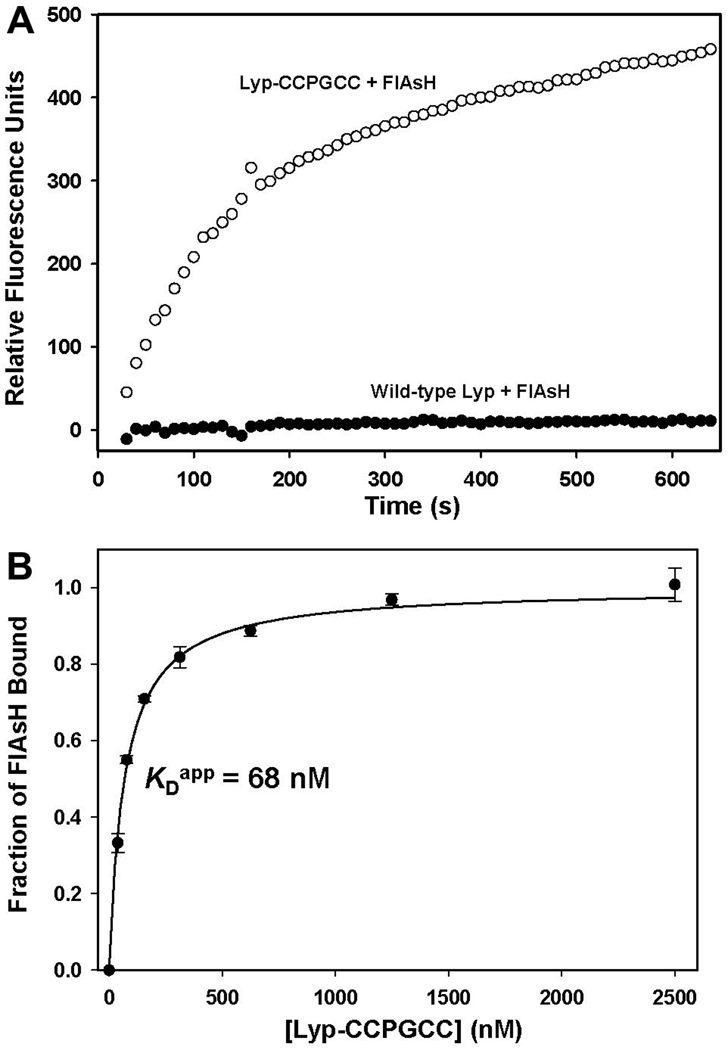
FlAsH was developed by Tsien and co-workers as a fluorescence-based protein-visualization tool, and binding of FlAsH to its tetracysteine target sequence (either in the context of a short peptide or a protein) gives rise to a dramatic increase in the compound’s fluorescence.29, 30 While our primary interest lies in control of Lyp activity, not in Lyp visualization, the putative increase in FlAsH fluorescence upon Lyp-CCPGCC binding potentially provides a useful tool for characterizing the kinetics and thermodynamics of FlAsH’s interactions with the target enzyme. Indeed, we found that incubation of Lyp-CCPGCC with FlAsH leads to a rapid increase in fluorescence over the course of about 10 minutes (Figure 5A). By contrast, no substantial fluorescence increase was observed upon incubation of wild-type Lyp with FlAsH (Figure 5A). These data suggest that FlAsH fluorescence can be a useful tool for specifically detecting the Lyp-CCPGCC/FlAsH interaction, either for in vitro biophysical studies, or in complex proteomic or cellular mixtures.
Figure 5.
Lyp-CCPGCC-induced FlAsH fluorescence. (A) Wild-type Lyp (250 nM, closed circles) or Lyp-CCPGCC (250 nM, open circles) was mixed with FlAsH (500 nM), and FlAsH fluorescence was monitored over time. The displayed data sets were normalized by the subtraction of a FlAsH-only (no protein) control. (B) Determination of the Lyp-CCPGCC/FlAsH apparent dissociation constant: FlAsH (25 nM) was incubated with the indicated concentrations of Lyp-CCPGCC for 2.5 hours and the FlAsH fluorescence of the resulting solutions was measured.
We used the Lyp-CCPGCC-induced increase in FlAsH fluorescence to estimate the apparent binding constant of the Lyp-CCPGCC/FlAsH interaction with greater sensitivity than could be achieved in pNPP-based inhibition assays. (As noted earlier, the high potency of FlAsH complicates the extraction of true inhibition-constant values from percent-inhibition values.) To estimate the apparent dissociation constant in an assay that was not constrained by detection of Lyp-CCPGCC activity, we incubated a constant amount of FlAsH (25 nM) with varying amounts of Lyp-CCPGCC and measured the endpoint FlAsH-fluorescence values of the resulting solutions. As shown in Figure 5B, even at very low Lyp-CCPGCC concentrations FlAsH is largely protein-bound, and a binding-model fit of the fluorescence data yields an apparent dissociation (KDapp) of 68 nM, a remarkably tight interaction for an inhibitor of Lyp activity. (The few known selective inhibitors of wild-type Lyp have inhibition constants in the range of 1–5 µM.19, 20) In a cell, the concentration of FlAsH required for potent Lyp-CCPGCC inhibition would depend on Lyp-CCPGCC’s cellular concentration. Nevertheless, our in vitro data predict that FlAsH-induced Lyp-CCPGCC inhibition in a mammalian cell would require administration of FlAsH at low concentrations, compared to analogous experiments with known Lyp inhibitors, potentially reducing the off-target effects inherent to applications of small-molecule drugs to complex systems.
3.5. Targeting of Lyp-CCPGCC in a complex proteomic mixture
Having established that FlAsH inhibits purified Lyp-CCPGCC, we next sought to determine whether FlAsH could specifically target Lyp-CCPGCC in the context of a complex proteome, such as a cell preparation. Since a direct in-cell reporter of Lyp activity is not available, we took advantage of Lyp-CCPGCC-induced FlAsH fluorescence as a means to potentially detect Lyp-CCPGCC/FlAsH interactions in a complex milieu. Although Lyp visualization is not the primary goal of the current work (FlAsH-mediated visualization of Lyp could be accomplished simply by placing a CCPGCC tag at Lyp’s N- or C-terminus37), the sensitivity of fluorescence detection could provide a powerful test of FlAsH’s ability to specifically target Lyp-CCPGCC in the presence of thousands of potentially competing biomolecules.
To investigate the ability of FlAsH to bind Lyp-CCPGCC in a complex mixture, we incubated freeze-thawed wild-type Lyp-expressing and Lyp-CCPGCC-expressing E. coli cells in the presence of FlAsH and measured the fluorescence of the resulting cell suspensions. (Although the mammalian cell permeability of FlAsH is well established,37 we found that a freeze-thaw cycle was necessary for the timely penetration of FlAsH into E. coli cells.) We found that wild-type-Lyp-expressing cells give rise to a significant FlAsH fluorescence when compared to a FlAsH/buffer control solution (Figure 6A). Although wild-type Lyp does not itself cause FlAsH to fluoresce (Figure 5A), our observed FlAsH fluorescence in the presence of Lyp-expressing bacteria is consistent with the previous finding that E. Coli cells endogenously express at least one protein (SlyD) that binds FlAsH and induces its fluorescence.40, 45 We hypothesize, therefore, that the FlAsH fluorescence from Lyp-expressing bacteria results from the SlyD/FlAsH interaction, with possible low level contributions from other E. Coli gene products.
Figure 6.
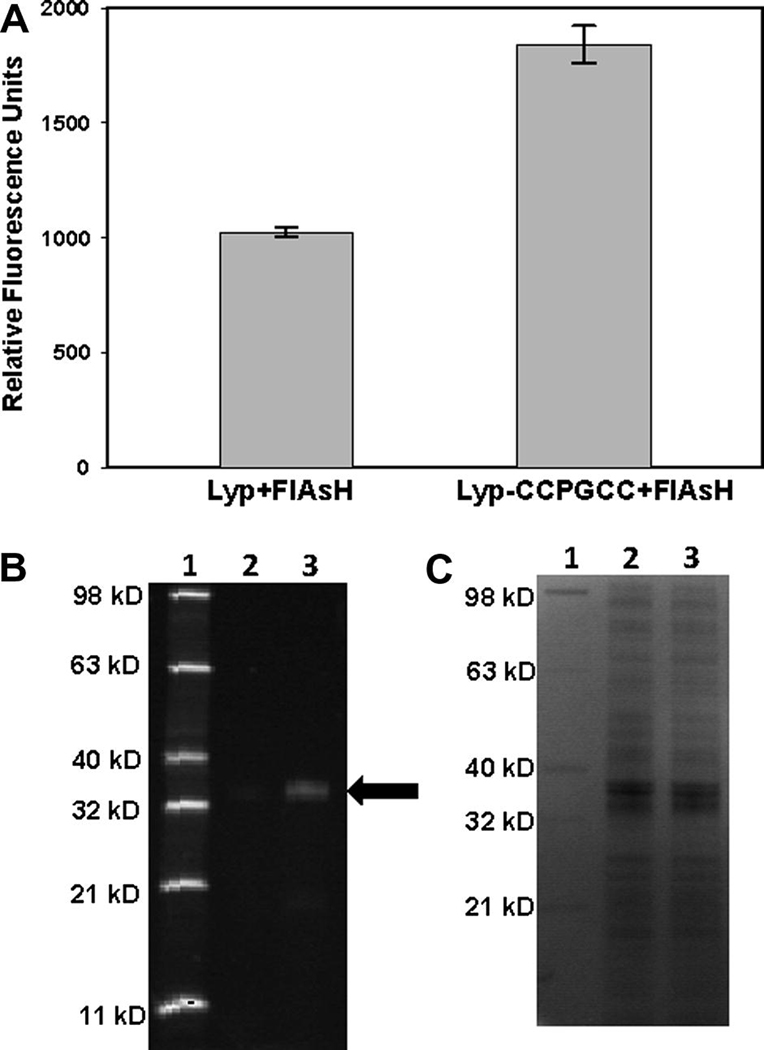
Targeting Lyp-CCPGCC in E. coli cell preparations. (A) Freeze-thawed E. coli cells expressing either wild-type Lyp or Lyp-CCPGCC were incubated in the presence of FlAsH (10 µM). After 2.5 hours, the FlAsH-fluorescence values of the cell suspensions were measured. (B, C) E. coli cells expressing either Lyp or Lyp-CCPGCC were prepared and FlAsH-treated as in (A) and subsequently lysed. Cellular proteins were separated by SDS-PAGE. FlAsH-labeled proteins were detected by fluorescence (B), followed by visualization of all proteins in the same gel by Coomassie staining (C). Lane 1: Fluorescent protein standard (Invitrogen), Lane 2: Lysate from Lyp-expressing cells, Lane 3: Lysate from Lyp-CCPGCC-expressing cells. The black arrow indicates the prominent fluorescent 37-kD band that is enriched in Lyp-CCPGCC-expressing lysates.
Importantly, measured FlAsH fluorescence in the presence of Lyp-CCPGCC-expressing E. coli was reproducibly greater (approximately 80%) than that of wild-type-Lyp expressing cells (Figure 6A). To further investigate the origins of the greater bulk fluorescence in the Lyp-CCPGCC-containing proteome, we separated proteins from the FlAsH-treated cell preparations by SDS-PAGE. By in-gel detection of FlAsH fluorescence, we observed a single band at 37 kD—the molecular weight of Lyp’s catalytic domain—that is enriched in lysates from the Lyp-CCPGCC expressing cells (Figure 6B). (Overexposure of the image allows for detection of other low-fluorescence bands in both lanes, including one whose observed molecular weight of 27 kD is consistent with that of SlyD, data not shown.) Since the two cell populations used in these experiments differ only by the presence of six amino acids in a single protein, the observed differences in bulk and 37-kD FlAsH fluorescence unambiguously derive from FlAsH binding to its target sequence in the Lyp-CCPGCC. These results represent the first demonstration that FlAsH can be used to target PTPs for inhibition in complex proteomic mixtures and lay the groundwork for future PTP targeting in eukaryotic cells.
4. Conclusions
Lymphoid-specific protein tyrosine phosphatase (Lyp) is an important leukocyte-signaling molecule and a putative anti-autoimmune therapeutic target. Small molecules that can specifically control cellular Lyp activity are thus important chemical-biology tools, both in basic signaling studies and in the potential drug-target validation of Lyp. We have shown that insertional mutagenesis can be used to generate a Lyp mutant (Lyp-CCPGCC) that is sensitive to inhibition by a small molecule (FlAsH) which does not inhibit Lyp or any other wild-type PTP tested to date. Lyp-CCPGCC was rationally engineered to display a FlAsH-binding peptide that does not disrupt its inherent PTP activity in the absence of added ligand. Upon addition of FlAsH, the activity of Lyp mutant is strongly inhibited, even under experimental conditions in which FlAsH and Lyp-CCPGCC are present at almost equal concentrations. We have also shown that Lyp-CCPGCC can be targeted in cell preparations of E. coli that express the target protein. Lyp-CCPGCC/FlAsH thus represents an “orthogonal” PTP/inhibitor pair that can be used to control Lyp activity, potentially providing a tool for elucidating Lyp’s functions in mammalian cells and engineered organisms.
Acknowledgments
This research was supported by the National Institutes of Health (2 R15 GM071388-02) and Amherst College. The authors thank Prof. Patricia O’Hara for helpful conversations and for the use of her lab’s K2 fluorometer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tonks NK. Nat. Rev. Mol. Cell Biol. 2006;7:833. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 2.Jiang ZX, Zhang ZY. Cancer Metastasis Rev. 2008;27:263. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Zhang ZY. Drug Discov. Today. 2007;12:373. doi: 10.1016/j.drudis.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Zhang ZY, Lee SY. Expert Opin. Investig. Drugs. 2003;12:222. doi: 10.1517/13543784.12.2.223. [DOI] [PubMed] [Google Scholar]
- 5.Easty D, Gallagher W, Bennett DC. Curr. Cancer Drug Targets. 2006;6:519. doi: 10.2174/156800906778194603. [DOI] [PubMed] [Google Scholar]
- 6.Bialy L, Waldmann H. Angew. Chem. Int. Ed. 2005;44:3814. doi: 10.1002/anie.200461517. [DOI] [PubMed] [Google Scholar]
- 7.Vang T, Miletic AV, Bottini N, Mustelin T. Autoimmunity. 2007;40:453. doi: 10.1080/08916930701464897. [DOI] [PubMed] [Google Scholar]
- 8.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. Nat. Genet. 2004;36:337. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 9.Bottini N, Vang T, Cucca F, Mustelin T. Semin. Immunol. 2006;18:207. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Begovich AB, Carlton VEH, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang QQ, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SYP, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. Am. J. Hum. Genet. 2004;75:330. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, Ball SG, James RA, Quinton R, Perros P, Pearce SHS. J. Clin. Endocrinol. Metab. 2004;89:5862. doi: 10.1210/jc.2004-1108. [DOI] [PubMed] [Google Scholar]
- 12.Vandiedonck C, Capdevielle C, Giraud M, Krumeich S, Jais JP, Eymard B, Tranchant C, Gajdos P, Garchon HJ. Ann. Neurol. 2006;59:404. doi: 10.1002/ana.20751. [DOI] [PubMed] [Google Scholar]
- 13.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VEH, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW. Am. J. Hum. Genet. 2004;75:504. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, Mustelin T, Bottini N. Nat. Genet. 2005;37:1317. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 15.Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, Dasgupta J, Hartiala J, Zhao L, Ortego-Centeno N, D'Alfonso S, Arnett FC, Wu H, Gonzalez-Gay MA, Tsao BP, Pons-Estel B, Alarcon-Riquelme ME, He YT, Zhang ZY, Allayee H, Chen XJS, Martin J, Bottini N Italian Collaborative, G. Hum. Mol. Genet. 2009;18:569. doi: 10.1093/hmg/ddn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaskovich MA. Curr. Med. Chem. 2009;16:2095. doi: 10.2174/092986709788612693. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZY. Curr. Opin. Chem. Biol. 2001;5:416. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZY. Annu. Rev. Pharmacol. Toxicol. 2002;42:209. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Sun JP, He YT, Guo XL, Liu SJ, Zhou B, Hudmon A, Zhang ZY. Proc. Natl. Acad. Sci. USA. 2007;104:19767. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karver MR, Krishnamurthy D, Kulkarni RA, Bottini N, Barrios AM. J. Med. Chem. 2009;52:6912. doi: 10.1021/jm901220m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu SD, Bottini M, Rickert RC, Mustelin T, Tautz L. ChemMedChem. 2009;4:440. doi: 10.1002/cmdc.200800375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy D, Karver MR, Fiorillo E, Orru V, Stanford SM, Bottini N, Barrios AM. J. Med. Chem. 2008;51:4790. doi: 10.1021/jm800101w. [DOI] [PubMed] [Google Scholar]
- 23.Xie YL, Liu YD, Gong GL, Rinderspacher A, Deng SX, Smith DH, Toebben U, Tzilianos E, Branden L, Vidovic D, Chung C, Schurer S, Tautz L, Landry DW. Bioorg. Med. Chem. Lett. 2008;18:2840. doi: 10.1016/j.bmcl.2008.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman HE, Blair ER, Johndrow JE, Bishop AC. J. Am. Chem. Soc. 2005;127:2824. doi: 10.1021/ja043378w. [DOI] [PubMed] [Google Scholar]
- 25.Bishop AC, Zhang XY, Lone AM. Methods. 2007;42:278. doi: 10.1016/j.ymeth.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XY, Bishop AC. J. Am. Chem. Soc. 2007;129:3812. doi: 10.1021/ja069098t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang XY, Chen VL, Rosen MS, Blair ER, Lone AM, Bishop AC. Bioorg. Med. Chem. 2008;16:8090. doi: 10.1016/j.bmc.2008.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X-Y, Bishop AC. Biochemistry. 2008;47:4491. doi: 10.1021/bi800014c. [DOI] [PubMed] [Google Scholar]
- 29.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 30.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J. Am. Chem. Soc. 2002;124:6063. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 31.Iversen LF, Moller KB, Pedersen AK, Peters GH, Petersen AS, Andersen HS, Branner S, Mortensen SB, Moller NP. J. Biol. Chem. 2002;277:19982. doi: 10.1074/jbc.M200567200. [DOI] [PubMed] [Google Scholar]
- 32.Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, Hansen SK. Nat. Struct. Mol. Biol. 2004;11:730. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 33.Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Biochemistry. 2005;44:7704. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- 34.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Mol. Cell. Biol. 2001;21:7117. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barr AJ, Ugochukwu E, Lee WH, King ONF, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Cell. 2009;136:352. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. Nature. 2000;407:395. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 37.Griffin BA, Adams SR, Jones J, Tsien RY. Meth. Enzymol. 2000;327:565. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- 38.Liu SJ, Zhou B, Yang HY, He YT, Jiang ZX, Kumar S, Wu L, Zhang ZY. J. Am. Chem. Soc. 2008;130:8251. doi: 10.1021/ja711125p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang ZY, Maclean D, Thieme-Sefler AM, Roeske RW, Dixon JE. Anal. Biochem. 1993;211:7. doi: 10.1006/abio.1993.1224. [DOI] [PubMed] [Google Scholar]
- 40.Krishnan B, Gierasch LM. Chem. Biol. 2008;15:1104. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat. Chem. Biol. 2007;3:779. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop AC, Buzko O, Shokat KM. Trends Cell Biol. 2001;11:167. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 43.Madani F, Lind J, Damberg P, Adams SR, Tsien RY, Graslund AO. J. Am. Chem. Soc. 2009;131:4613. doi: 10.1021/ja809315x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montalibet J, Skorey KI, Kennedy BP. Methods. 2005;35:2. doi: 10.1016/j.ymeth.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Wang T, Yan P, Squier TC, Mayer MU. ChemBioChem. 2007;8:1937. doi: 10.1002/cbic.200700209. [DOI] [PubMed] [Google Scholar]