Abstract
T-box (Tbx)3, a known transcriptional repressor, is a member of a family of transcription factors, which contain a highly homologous DNA binding domain known as the Tbx domain. Based on the knowledge that mutation of the Tbx3 gene results in limb malformation, Tbx3 regulates osteoblast proliferation and its expression increases during osteoblast differentiation, we predicted that Tbx3 is an important regulator of osteoblast cell functions. In this study, we evaluated the consequence of transgenic overexpression of Tbx3 on osteoblast differentiation. Retroviral overexpression increased Tbx3 expression > 100-fold at the mRNA and protein level. Overexpression of Tbx3 blocked mineralized nodule formation (28 ± 8 vs. 7 ± 1%) in MC3T3-E1 cells. In support of these data, alkaline phosphatase (ALP) activity was reduced 33–70% (P < 0.05) in both MC3T3-E1 cells and primary calvaria osteoblasts overexpressing Tbx3. In contrast, Tbx3 overexpression did not alter ALP activity in bone marrow stromal cells. Tbx3 overexpression blocked the increase in expression of key osteoblast marker genes, ALP, bone sialoprotein, and osteocalcin that occurs during normal osteoblast differentiation, but had little or no effect on expression of proliferation genes p53 and Myc. In addition, Tbx3 overexpression abolished increased osterix and runx2 expression observed during normal osteoblast differentiation, but the change in Msx1 and Msx2 expression over time was similar between control and Tbx3 overexpressing cells. Interestingly, osterix and runx2, but not Msx1 and Msx2, contain Tbx binding site in the regulatory region. Based on these data and our previous findings, we conclude that Tbx3 promotes proliferation and suppresses differentiation of osteoblasts and may be involved in regulating expression of key transcription factors involved in osteoblast differentiation.
Keywords: BONE, DIFFERENTIATION, OSTEOBLAST, TRANSCRIPTION FACTOR, T-BOX3
The process of bone formation is dependent on the proliferation and subsequent differentiation of mesenchymal stem cells into osteoblast cells and osteoblast function. These processes are tightly regulated by several known growth factors and transcription factors. Although several transcription factors have been identified as key regulators of osteoblasts and bone formation in the past decade, several new transcription factors are being identified that are involved in bone formation [Karsenty, 2001; Lian and Stein, 2003; Stains and Civitelli, 2003], as well as several other developmental processes. Identification of novel transcription factors and their mechanisms of action in bone will enable us to fully understand the bone formation process.
The T-box (Tbx) proteins are a family of transcription factors, which contain a highly homologous DNA binding domain (the T-box). In general, the Tbx genes are essential for early development and function as activators and/or repressors of transcription [Smith, 1999]. These proteins have been identified in several key developmental processes, including limb and digit formation and identity, heart development, cancer cell cycle and growth, mammary gland development, skeletal development, and osteogenesis [Papaioannou and Silver, 1998; Carlson et al., 2001; Moorman et al., 2004]. Of the family of at least 18 genes, Tbx3 has been shown to be critical for embryonic development in mice and humans as demonstrated by embryonic lethality when a homozygous mutation of Tbx3 is present [Davenport et al., 2003]. Mutation of Tbx3 is also associated with human ulnar mammary syndrome (UMS), which is characterized by posterior forelimb deficiency or duplication, apocrine/mammary gland hypoplasia or dysfunction, and hair, genital and dental defects [Bamshad et al., 1995; Bamshad et al., 1997].
Although it has been well established that Tbx3 is critical for embryonic development, little is known about the role of this gene in postnatal development. Recent findings demonstrate that Tbx3 is important in regulating cell cycle as demonstrated by increased cell growth and inhibition of senescence with overexpression of Tbx3 in MEF cells [Fan et al., 2004]. In support of these data, inhibition of Tbx3 in bladder epithelial cells increases apoptosis and senescence [Ito et al., 2005]. In terms of a role for Tbx3 in bone development, we recently demonstrated that Tbx3 is expressed in mouse bone, Tbx3 expression is regulated by GH, a known mediator of growth and bone development, and Tbx3 is required for osteoblast proliferation [Govoni et al., 2006b]. Lee et al. [2007] also demonstrated that blocking Tbx3 expression inhibits the differentiation of stromal cells into osteoblasts. In addition, other Tbx genes are also known to be involved in regulating bone development, thus suggesting that these genes which are critical for embryonic development, may also play an important role in postnatal bone development. Specifically, Tbx2 overexpression in NIH3T3 cells increases expression of osteoblast and chondroblast lineage genes [Chen et al., 2001] and Tbx15 null mice display reduced bone size and length, as well as, delayed endochondral bone development [Singh et al., 2005].
Based on the important role of other Tbx proteins in bone development, that Tbx3 is required for embryonic limb development, and that Tbx3 regulates osteoblast proliferation, we hypothesized that Tbx3 must be an important transcriptional regulator of osteoblast function and thus critical for optimal bone development and maintenance. Based on our previous findings that Tbx3 expression increases during osteoblast differentiation [Govoni et al., 2006b], we sought to determine the role of Tbx3 in regulating osteoblast differentiation. To this end, we evaluated the consequence of transgenic overexpression of Tbx3 in osteoblast cells and demonstrated a critical role for Tbx3 in inhibiting osteoblast differentiation.
MATERIALS AND METHODS
TISSUE CULTURE REAGENTS
α-Minimal essential medium (α-MEM), penicillin–streptomycin suspension, and calf serum were purchased from Gibco BRL (Gaithersburg, MD). Normocin was purchased from Invivogen (San Diego, CA). Bovine serum albumin (BSA) was purchased from Fluka (Buchs, Switzerland). Recombinant human bone morphogenic protein (BMP)-2 was obtained from Genetics Institute (Cambridge Park, MA). All other chemicals were purchased from Fisher Scientific (Tustin, CA) or Sigma (St. Louis, MO).
ISOLATION OF PRIMARY CALVARIA OSTEOBLAST CELLS
Primary mouse calvaria osteoblasts were cultured as previously described [Govoni et al., 2007]. Briefly, calvaria were removed from 4- to 8-day-old pups and digested with collagenase A (2 mg/ml; Sigma) and hyaluronidase (1 mg/ml; Sigma) for 20 min. The first digestion was discarded and the second and third digestions (30 min each) were harvested and plated in α-MEM + 10% FBS + 100 U/ml penicillin, and 100 µg/ml streptomycin + 1% fungizone or α-MEM + 10% FBS + 1% normocin at a density of 50,000 cells/well in 6-well plates for transduction.
ISOLATION OF MOUSE BONE MARROW STROMAL CELLS (BMSC)
Mouse BMSC were isolated as previously described [Govoni et al., 2006b]. Brifely cells were isolated from 4 to 6 weeks old mice and plated in α-MEM + 10% FBS + 100 U/ml penicillin, and 100 µg/ml streptomycin + 1% fungizone at a density of 4 million cells/well in 6-well plates. Once cells reached approximately 50% confluency cells were transduced as described below.
CLONING OF TBX3 INTO MLV VECTOR
Tbx3 was amplified by PCR using Mus musculus bone cDNA (50 ng) as a template. Cloning was accomplished using PCR selection kit-high fidelity (Invitrogen, Carlsbad, CA). The reaction mixture consisted of 5 µl of 10× Pfx50 PCR mix, 1 µl each of forward and reverse primers, 2.5 µl of 10 mM dNTPs, 32.5 µl of DNA water, 2.5 µl DMSO, and 0.5 µl Pfx50 DNA polymerase. The 5′ end of the forward primer contains a Kozak consensus sequence that initiates protein translation. The primer sequences are (5′ to 3′): Tbx3 forward: 5′-GGG TCG ACG CCA CCA TGA GCC TCT CCA TGA GAG ATC CGG T-3′ and Tbx3 reverse: 5′-CGC GGA TCC TTA AGG GGA CCC GCT GCA AGA CCT-3′. Amplification was carried out at 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 1 min, and 72°C for 3 min using Pfx50 DNA polymerase (Invitrogen) for PCR cloning due to its high specificity and high fidelity. The PCR product was purified with GeneClean spin kit (Qbiogene, Carlsbad, CA) according to the manufacturer’s instructions and then digested overnight at 37°C with BamHI and SalI. The digested product was run on a gel and purified again using GeneClean spin kit. Finally the PCR product was subcloned into the corresponding restriction sites of the MLV expression vector [Peng et al., 2001]. All plasmids were transformed into E. coli XL1 BLUE cells and the plasmids were isolated using EndoFree® Plasmid Maxi Kit (Qiagen, Inc., Valencia, CA). The Tbx3 sequence was verified by DNA sequencing using a 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
MLV-BASED VECTOR PRODUCTION AND MLV TRANSDUCTION
We used the VSV-G protein as the envelope for our MLV-based vector and the vectors were generated by transient transfection in 293T cells as described previously [Peng et al., 2001]. Briefly, a 10-cm plate of 293T cells was transfected with a mixture of 20 µg of retroviral expression vector (TBX3 or the control vectors), 10 µg of MLV-GP expression vector, and 1 µg of VSV-G expression vector by CaPO4 precipitation. The conditioned medium containing the viral vectors was collected 48 h after the transfection. The viral titer was determined by the end-point dilution assay for the marker gene (β-gal or eGFP) expression in HT1080 cells.
MC3T3-E1 (mouse pre-osteoblast cell line), mouse primary calvaria osteoblast cells or BMSC were seeded at a density of 100,000, 50,000, or 4,000,000 cells/well, respectively in 6-well plates. When MC3T3-E1 cells were about 70% confluent (approximately 1–2 days after plating), calvaria osteoblasts were 40–50% confluent (approximately 4–5 days after plating), and BMSC were 50% confluent, the cells were transduced as previously described [Peng et al., 2001] with Tbx3, β-galactosidase (β-gal) or green-fluorescent protein (GFP) constructs. The titer of β-gal cells was 1.7 × 108 tfu/ml. A volume of 200, 100, and 60 µl was added to MC3T3-E1, primary osteoblast cells and BMSC, respectively. For our dose–response experiment, 0, 2, 6.3, 20, and 63 µl of Tbx3 and β-gal virus was used to transduce MC3T3-E1 cells. Twenty-four hours after transduction, cells were trypsinized and plated in α-MEM + 10% FBS + 1% normocin. Cells for Tbx3 mRNA expression were plated at 100,000 cells/well in a 6-well dish. MC3T3-E1 cells for Tbx3 protein quantification were plated at 300,000 cells/dish in 10 cm plates. Cells for all other experiments were plated as described below. To determine the efficiency of transduction, fluorescence in GFP transduced cells was visualized and quantified using a FACSCalibur System (BD Biosciences, San Jose, CA).
WESTERN BLOT ANALYSIS
Equal amounts of protein (20 µg) were resolved on a 10% SDS–polyacrylamide gel. Proteins were transferred to a 0.2 µM nitrocellulose membrane at 300 mA for 40 min at 4°C (BioRad, Hercules, CA). The membrane was blocked in 5% milk in TBST overnight with rotation at 4°C. The following day, the membrane was probed with specific antibody to anti-Tbx3 (1:1,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1 h. After washing, the membrane was incubated with horseradish peroxidase conjugated anti-goat antibody (1:5,000; Sigma–Aldrich, St. Louis, MO) for 1 h and then washed. Detection of Tbx3 was visualized using Super Signal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
RNA EXTRACTION
RNA was extracted from the cells using Trizol (Invitrogen) and an RNeasy Mini Kit (Qiagen, Inc.) according to the manufacturer’s instructions. Following extraction, up to 10 µg of RNA was DNase treated with a DNA-free kit (Ambion, Austin, TX) to remove any residual DNA. RNA quality was determined using a 2100 Bioanalyzer (Agilent, Palo Alto, CA) and RNA was quantified using a NanoDrop Spectrophotometer (Wilmington, DE).
GENE EXPRESSION ANALYSIS
Quantitative real-time RT-PCR analysis was used to determine the expression levels of Tbx3, alkaline phosphatase (ALP), bone sialoprotein (BSP), osteocalcin (Oc), Osterix, Runx2, Msx1, Msx2, p53, Myc, and PPIA (peptidylprolyl isomerase A; endogenous control) as previously described [Govoni et al., 2006a]. Primers used were validated as previously described [Govoni et al., 2006a]. Delta CT values were determined (CT value for gene of interest minus CT value for control gene) and comparisons of the CT values were used for relative quantification of gene expression [Dorak, 2006].
OSTEOBLAST DIFFERENTIATION
MC3T3-E1 cells were plated at 100,000 cells/well in 6-well plates and cultured with α-MEM containing 10% calf serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. After 3 days, media was changed to a differentiation media containing 300 µg/ml ascorbic acid 2 phosphate and 10 mM β-glycerophosphate. This was considered day 0. The culture media was changed every 3 days for 28 days. At day 28 cells were stained with Alizarin Red to identify mineralized nodules. These experiments were repeated two times. Mineralization was quantified using Image Pro Plus 4.5 software (Media Cybernetics, Inc., Bethesda, MD) and expressed as % of area. For gene expression analysis, RNA was extracted as described above from cells at day 0, 3, and 6 after addition of differentiation media.
ALP ACTIVITY ASSAY
After transduction, cells (MC3T3-E1 or calvaria osteoblasts) were trypsinized and plated at a density of 5,000 cells/well in a 96-well dish in α-MEM + 10% FBS + 1% normocin + 50 µg/ml ascorbic acid 2 phosphate and 10 mM β-glycerophosphate. Six hours after plating, media was changed to serum free (α-MEM + 0.1% BSA + 1% normocin + 50 µg/ml ascorbic acid 2 phosphate and 10 mM β-glycerophosphate). Eighteen hours later cells were treated with BMP-2 (30 ng/ml) or 0.1% BSA for 3 days. BMSC were left on plates and serum depleted for 18 h prior to treatment. ALP activity was measured as previously described [Farley et al., 1994]. Protein concentration was measured by the Bradford method. ALP activity was standardized based on cellular protein content and expressed as milliunits per milligram of protein (mU/mg protein).
STATISTICAL ANALYSIS
Data were analyzed by ANOVA and or Student’s t-test where appropriate. Posthoc analysis was performed using Newman–Keuls analysis. Data were analyzed using Statistica 6 software (StatSoft, Tulsa, OK). Data are presented as mean ± SE and a significant difference was determined at P ≤ 0.05.
RESULTS
EFFICIENCY OF TBX3 TRANSDUCTION IN OSTEOBLASTS
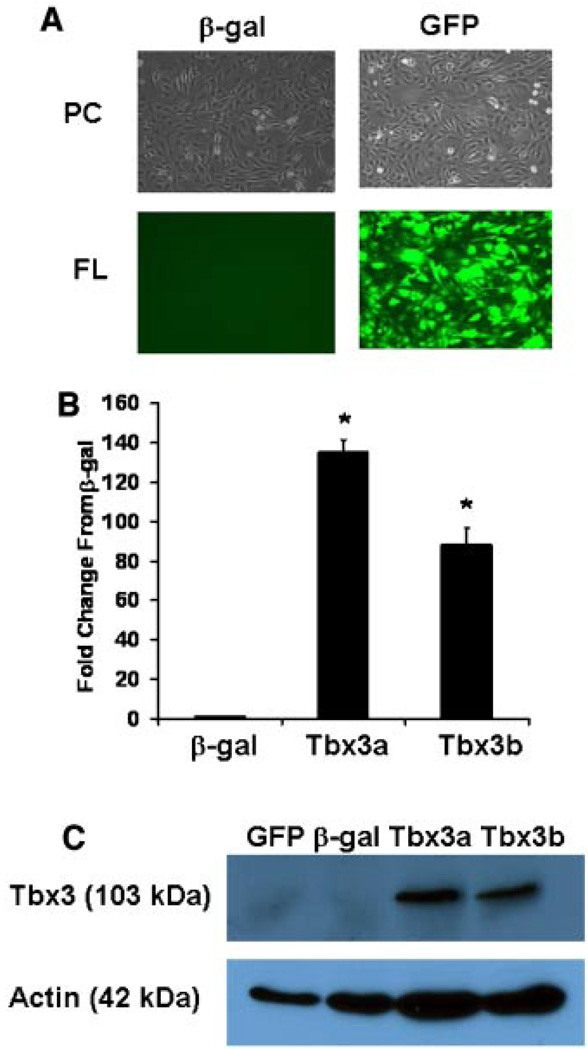
To determine the efficiency of transduction, MC3T3-E1 and calvaria osteoblast cells were transduced with GFP and evaluated by examining fluorescence (Fig. 1A). We observed >90% transduction efficiency for both cell types by fluorescence (Fig. 1A representative of MC3T3-E1 cells), and confirmed by FACS analysis in which >90% of the cells were positive for GFP in both cell types (data not shown).
Fig. 1.
Increased Tbx3 expression and protein levels in osteoblasts transduced with MLV-based vector. A: Image of β-gal and GFP transduced MC3T3-E1 cells under phase contrast (PC) and fluorescent (FL) lighting. B: mRNA expression of Tbx3 in β-gal and Tbx3 transduced MC3T3-E1 cells (n = 3/treatment). *indicates a significant difference from control (β-gal) cells. C: Protein expression of Tbx3 in transduced MC3T3-E1 cells as determined by western immunoblot. Actin served as a control for protein concentration. Tbx3a and Tbx3b are two different transductions, which yielded similar overexpression of Tbx3. Similar findings for transduction efficiency (fluorescence and mRNA expression) were observed for primary calvaria osteoblasts. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To determine the efficiency of Tbx3 overexpression, we evaluated Tbx3 mRNA by real-time RT-PCR and determined a > 100-fold increase in Tbx3 compared with β-gal cells in multiple transductions in both cell types. Representatives of two of our transductions in MC3T3-E1 cells (Tbx3a and Tbx3b) are presented in Figure 1B. To determine if Tbx3 protein was also increased we evaluated Tbx3 protein in control and Tbx3 overexpressing MC3T3-E1 cells. Although endogenous levels of Tbx3 were not detectable in control cells, a significant amount of protein of anticipated size (103 kDa) was present in Tbx3 overexpressing cells (Fig. 1C). These findings demonstrate that our model effectively overexpresses the Tbx3 gene and induces increased Tbx3 protein in osteoblasts.
TBX3 INHIBITS OSTEOBLAST DIFFERENTIATION
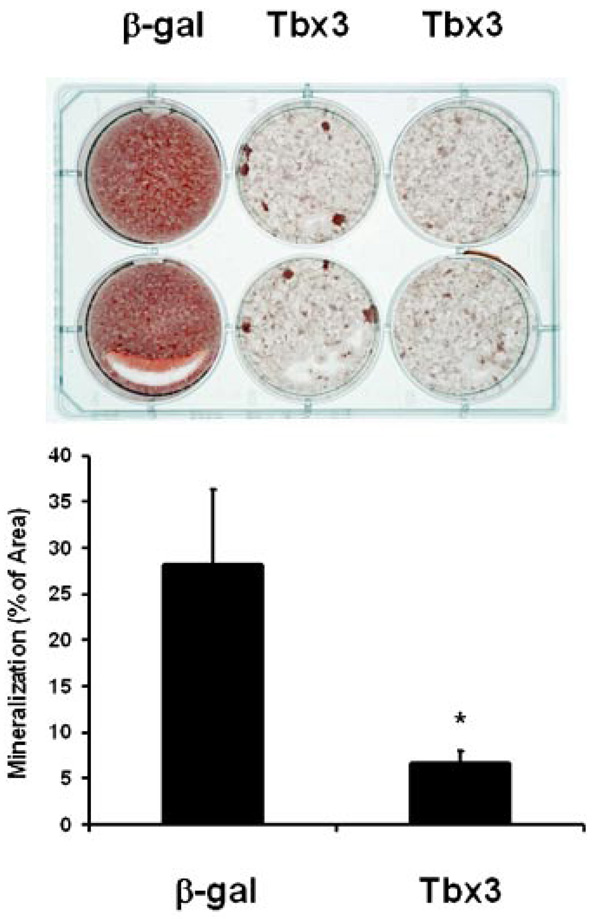
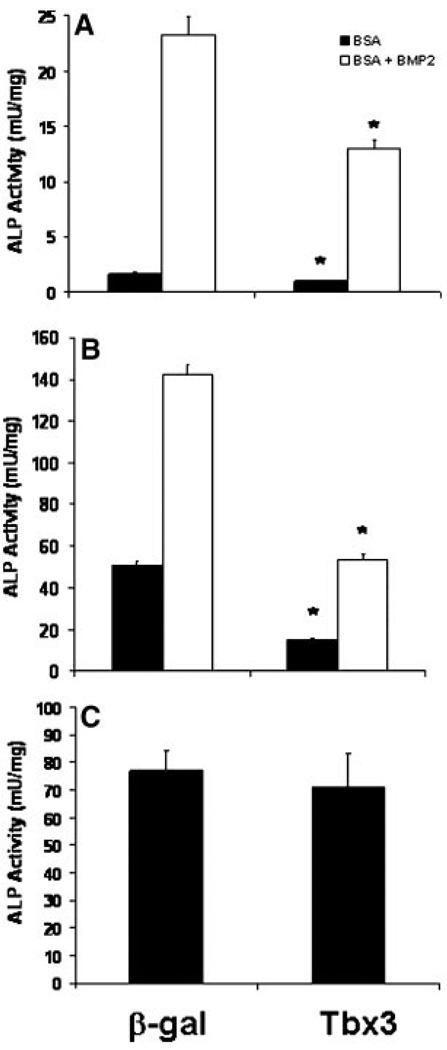
We previously determined that Tbx3 expression increases during differentiation of mouse bone marrow stromal cells into osteoblasts [Govoni et al., 2006b]. To determine if Tbx3 plays a role in regulating osteoblast differentiation, we evaluated the consequence of overexpressing Tbx3 on the differentiation of MC3T3-E1 cells. Tbx3 overexpression in MC3T3-E1 cells caused a 76% reduction in the area of Alizarin Red stained mineralized nodules compared to β-gal overexpressing control cultures (Fig. 2). Consistent with these findings, Tbx3 overexpression reduced ALP activity 40% and 35%, respectively in cells treated without or with BMP-2 (Fig. 3A). To confirm that our findings were not specific to the cell line we used, we investigated the consequence of Tbx3 overexpression on ALP activity in primary osteoblasts. Accordingly, Tbx3 overexpression reduced ALP activity 70% and 63%, respectively in primary osteoblast cells treated without or with BMP-2 (Fig. 3B). Interestingly, overexpression of Tbx3 in undifferentiated BMSC did not reduce ALP activity compared with β-gal control cells (Fig. 3C).
Fig. 2.
Tbx3 inhibits mineralization of osteoblast cells. A: Image is a representation of two experiments in which cells were grown in differentiation media (α-MEM + 10% calf serum + 100 U/ml penicillin + 100 µg/ml streptomycin + 300 µg/ml ascorbic acid 2 phosphate + 10 mM β-glycerophosphate) for 28 days. Mineralization was determined by Alizarin Red staining and quantified with Image Pro Plus 4.5 software. B: Data are presented as mean ± SE and expressed as % of area. β-gal (n = 7). Tbx3 (n = 14). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Fig. 3.
Tbx3 inhibits ALP activity in osteoblasts. ALP activity was determined in MC3T3-E1 (A), primary calvaria osteoblasts (B) and bone marrow stromal cells (C) transduced with Tbx-3 and treated with or without BMP-2. n = 6 to 16/treatment. Data are presented as mean ± SE. *indicates a significant difference from β-gal transduced cells at P < 0.01. BMP-2 increase in ALP activity was significant (P > 0.05) in MC3T3-E1 and primary osteoblast cells.
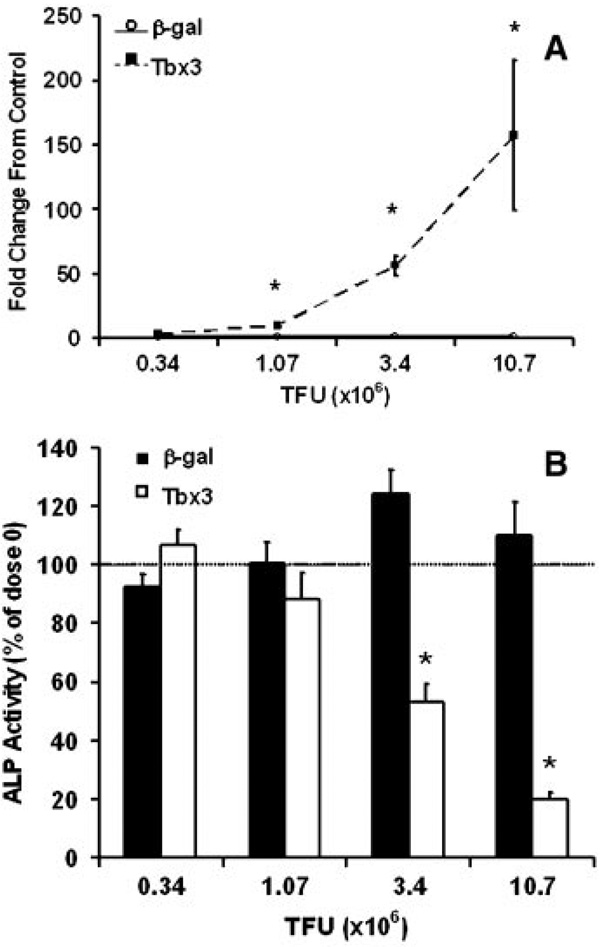
To determine if the reduced ALP activity and mineralization was due to the high expression of Tbx3 (> 100-fold) in our model we performed a dose–response with different levels of β-gal and Tbx3 virus added to MC3T3-E1 cells. Figure 4A shows that as expected Tbx3 expression increased with increasing amount of MLV-Tbx3 used for transfection in Tbx3 overexpressing cells. In contrast, MLV-β-gal infection did not cause a change in Tbx3 expression at the same doses (Fig. 4A). More importantly, we observed a dose-dependent reduction in ALP activity in Tbx3 overexpressing cells, but no difference with different doses of β-gal virus (Fig. 4B).
Fig. 4.
Dose-dependent decrease in ALP activity in response to Tbx3 overexpression. MC3T3-E1 cells were transduced with four doses of β-gal and Tbx3 virus. A: mRNA expression of Tbx3. *indicates a significant difference between β-gal and Tbx3 cells at P < 0.01. No change in Tbx3 expression was observed in β-gal control cells with increased dose (P ≥ 0.40), however a dose dependent increase in Tbx3 was observed in Tbx3 cells (P < 0.01). B: ALP activity. *indicates a significant difference from dose 0 for respective treatment groups at P < 0.01. n = 6 to 8/treatment. TFU = transforming units.
TBX3 REGULATES EXPRESSION OF KEY OSTEOBLASTS MARKERS DURING OSTEOBLAST DIFFERENTIATION
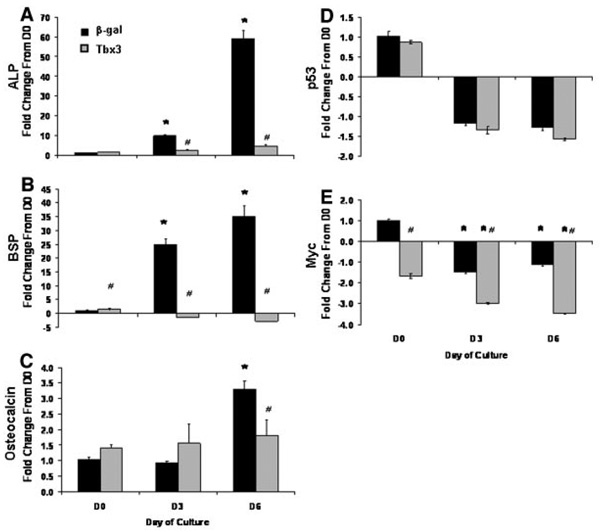
Consistent with reduced ALP activity and mineralization in Tbx3 overexpressing cells, expression of markers of osteoblast differentiation were reduced (Fig. 5A–C). Specifically, expression of ALP, BSP, and Oc increased 59-, 35-, and 3-fold, respectively during differentiation in β-gal control cells; however overexpression of Tbx3 blocked the increase in ALP, BSP, and Oc expression between day 0 and day 6.
Fig. 5.
Tbx3 inhibits expression of osteoblast marker genes during osteoblast differentiation. day 0 represents the day differentiation media was added. mRNA expression of alkaline phosphatase (ALP; A), bone sialoprotein (BSP; B), osteocalcin (C), p53 (D), and Myc (E) were determined by real-time RT-PCR. n = 5 to 6/treatment. Data are presented as mean ± SE and expressed as fold change from control (β-gal control cells) at day 0. *indicates a significant difference from Day 0 for respective treatment groups at P < 0.01. #indicates a significant difference from β-gal control cells at P < 0.05.
Tbx3 is a known regulator of cell cycle. We, therefore, evaluated the expression of key cell cycle genes (p53 and Myc) in cultures overexpressing Tbx3 or β-gal. We found that the expression of p53 was not different between Tbx3 overexpressing and β-gal control cells at all three time points (P ≥ 0.30; Fig. 5D). Although expression of Myc was lower in Tbx3 overexpressing cells at all three time points (P ≤ 0.01; Fig. 5E), β-gal control cells and Tbx3 overexpressing cells follow a similar pattern of reduced Myc expression over time. Consistent with these data, we found that Tbx3 overexpression did not influence cell proliferation as evaluated by cell number in this model under culture conditions used in this study (data not shown).
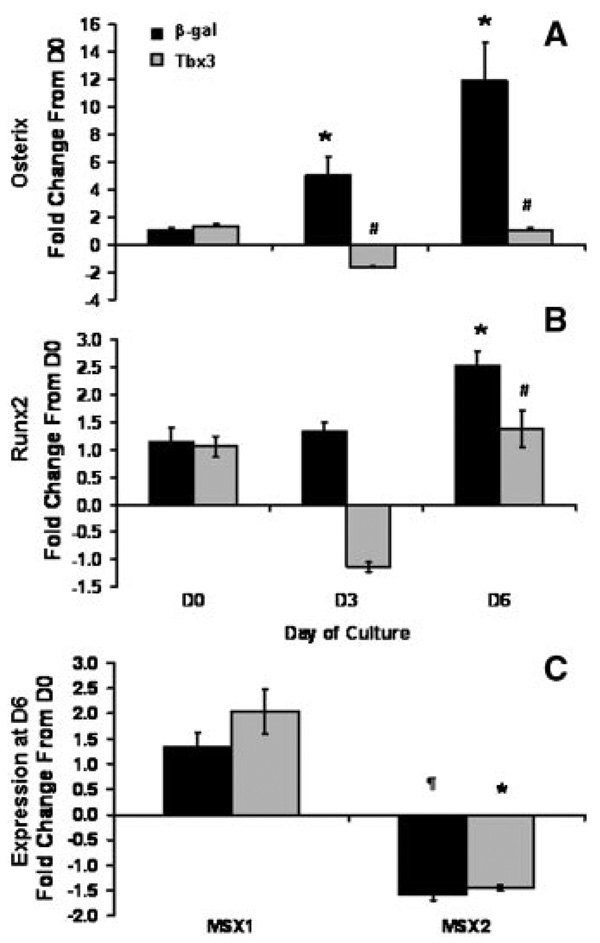
Based on our findings that Tbx3 inhibits osteoblast differentiation and the knowledge that Tbx3 is a known transcriptional repressor, we questioned whether Tbx3 blocks the expression of transcriptions factors that are key regulators of osteoblast differentiation (osterix and runx2/cbfa1). As expected, expression of osterix and runx2 increased 12- and 2.5-fold, respectively in β-gal control cells (P < 0.01; Fig. 6A,B). However in Tbx3 overexpressing cells expression of osterix and Runx2 did not change significantly during differentiation (Fig. 6A,B).
Fig. 6.
Tbx3 inhibits expression of osterix and Runx2, but does not alter Msx1 and 2 expression during osteoblast differentiation. day 0 represents the day differentiation media was added. mRNA expression of Osterix (A), Runx2 (B), Msx1 and 2 (C) were determined by real-time RT-PCR. Msx1 and 2 data are expression at day 6 of culture, presented as mean ± SE and expressed as fold change from control (β-gal control cells) at day 0. n = 5 to 6/treatment. Data are presented as mean ± SE and expressed as fold change from control (β-gal control cells) at day 0. *indicates a significant difference from Day 0 for respective treatment groups at P < 0.01. #indicates a significant difference in mRNA from β-gal control cells at P < 0.05. ¶indicates a tendency (P ≥ 0.06) for a difference from day 0.
Recently, it was demonstrated that Tbx3 binds to transcription factors Msx1 and Msx2, known regulators of osteoblast differentiation, in cardiac cells [Boogerd et al., 2008]. To determine if Tbx3 regulates the expression of these genes in osteoblasts, we looked at their expression in our model. Expression of Msx1 and Msx2 were not different between Tbx3 overexpressing and β-gal control cells (Fig. 6C).
DISCUSSION
Our findings in MC3T3-E1 and primary osteoblast cells overexpressing Tbx3 demonstrate that Tbx3 is a negative regulator of osteoblast differentiation. These findings were in contrast to the study by Lee et al. [2007] in which blocking Tbx3 expression with siRNA inhibited the differentiation of human adipose tissue stromal cells into osteoblasts and reduced expression of Runx2 and osteopontin. One potential explanation for our findings could be the dose of Tbx3 in our experiments. However, our dose–response data in which low doses of Tbx3 were able to reduce ALP activity in MC3T3-E1 osteoblasts suggest that the negative effects of Tbx3 cannot be explained on the basis of the Tbx3 dose. A more likely explanation for our unexpected findings could be that Tbx3 acts in a stage-dependent manner to regulate osteoblast differentiation [Aubin, 1998], as demonstrated by the lack of effect of Tbx3 overexpression on ALP activity in the BMSC. In support of this concept, Lee et al. utilized mesenchmyl stem cells, which are not already commited to the osteoblast lineage. Therefore, Tbx3 may have a different role in regulating differentiation depending on the stage of the cell in the osteoblast lineage. Consistent with these findings, a previous report in C2C12 myoblasts demonstrated that overexpression of Tbx3 blocked myoblast differentiation [Carlson et al., 2002]. Again, these cells were committed to a specific cell lineage, suggesting that during the later stages of differentiation, Tbx3 may have an inhibitory role. In support of an osteoblast stage-dependent role of Tbx3, it was previously shown that FGF2 and noggin act at different stages of osteoblast differentiation to regulate osteoblastogenesis [Kalajzic et al., 2003] and that stage-specific expression of Dlx5 regulates osteoblast differentiation [Ryoo et al., 1997]. Further studies are needed to determine if the effects of Tbx3 are indeed dependent on the stage of differentiation.
In vivo data for the role of Tbx3 are limited to phenotype data from humans with UMS and a mutant mouse model during embryonic development. Both models demonstrate limb malformation, in particular aplasia of the ulna and deficiencies or duplication of the fourth and fifth digit [Davenport et al., 2003; Klopocki et al., 2006]. Although the mechanism of Tbx3 action during critical stages of embryogenesis are not fully known, it has been suggested that lack of Tbx3 interferes with apoptosis or differentiation [Bamshad et al., 1997; Carlson et al., 2002; Davenport et al., 2003]. In addition, expression patterns in the mouse mutant model suggest an interaction with Shh signaling [Davenport et al., 2003]. However the mechanism of Tbx3 action during postnatal skeletal development is still not known. In support of a role of for Tbx genes in regulating postnatal bone development, a recent study using a Tbx15 mutant mouse demonstrated that Tbx15 is required for proliferation of mesenchymal precursor cells, but did not affect prehypertrophic chondrocytes in postnatal bone development [Singh et al., 2005]. Our previous findings that Tbx3 regulates osteoblast proliferation and is responsive to GH administration in mice along with our current findings suggest that Tbx3 may regulate bone formation by stimulating osteoblast proliferation and blocking the differentiation of cells to mature osteoblasts. Consistent with this model is the role of PTHrP, a gene known to regulate chondrocytes during endochondral bone formation, in which a mutation of the PTHrP gene causes cells to undergo premature entry into differentiation and ultimately affect bone growth by decreasing the number of proliferating cells at the proliferating zone [Kronenberg, 2006]. Similarly, Tbx3 may regulate bone formation by stimulating the proliferation of cells through the osteoblast lineage and blocking the differentiation of these cells into mature osteoblasts. Further studies in vitro during the various stages of differentiation and using transgenic mouse models are needed to determine the specific role of Tbx3 in regulating postnatal bone formation.
In terms of the potential mechanisms of Tbx3 action, previous findings have demonstrated that Tbx genes can form homodimers as well as bind to other transcription factors to regulate gene expression [Hiroi et al., 2001; Fan et al., 2003; Garg et al., 2003; Krause et al., 2004; Plageman and Yutzey, 2004, 2005]. Specifically, Tbx genes bind a well-conserved DNA binding domain, which contains a partially palindromic sequence (TCACxCCx) [Kispert and Herrmann, 1993; Smith, 1999; Carlson et al., 2001]. Since Tbx3 has been shown to bind this sequence and is known to bind and regulate promoter regions of Nppa, Cx40, and p19ARF in other tissues [Brummelkamp et al., 2002; Lingbeek et al., 2002], we hypothesized that Tbx3 may regulate osteoblast differentiation by binding to key transcription factors. Accordingly, we determined that the expression of osterix and runx2, two transcription factors that are essential for osteoblast differentiation [Karsenty, 2001; Lian and Stein, 2003; Shui et al., 2003; Stains and Civitelli, 2003; Stein et al., 2004; Xing et al., 2007], were suppressed in Tbx3 overexpressing cells. Whereas expression of Msx1 and 2, which play a role in osteoblast function, were similar between control and overexpressing cells. Consistent with our hypothesis that Tbx3 binds other transcription factors to regulate their expression, we identified the partially palindromic DNA binding site in osterix and runx2 regulatory regions, but not in Msx1 and Msx2. However, a recent report demonstrated that Tbx3 binds to Msx2 to regulate Connexin43 promoter activity in cardiac cells [Boogerd et al., 2008]. These findings demonstrate that in addition to regulating expression of key transcription factors, Tbx3 may also function as a regulator of osteoblast function by forming complexes with other transcription factors and regulating their activity. Future studies will identify the molecular mechanism through which Tbx3 regulates osterix and runx2 expression.
Since we and others have established that Tbx3 is required for cell proliferation in numerous cell types [Carlson et al., 2001; Brummelkamp et al., 2002; Fan et al., 2004; Suzuki et al., 2004; Ito et al., 2005; Lee et al., 2007; Platonova et al., 2007], our study focused on the role of Tbx3 during osteoblast differentiation. To demonstrate the specificity of our model we evaluated the role of Tbx3 overexpression on the expression of cell cycle genes (p53 and Myc) in our differentiating cells. As expected, there was little to no change in the expression of p53 between Tbx3 and control cells. Surprisingly, we observed a slight reduction in Myc expression during the differentiation as well as slightly lower expression in Tbx3 overexpressing cells. These findings suggest that Tbx3 did partially regulate cell cycle in our model. However, based on the abundant reduction in ALP activity when adjusted for protein, we are confident that Tbx3 is also an important regulator of differentiation in osteoblasts.
In conclusion, using a well established mouse osteoblast cell line, as well as primary osteoblasts, we demonstrated that Tbx3 is an inhibitor of osteoblast differentiation and may function by binding to key osteoblast transcription factors and altering their function and/or expression during key stages in the differentiation process. These findings, combined with our previous data demonstrate that Tbx3 is indeed necessary for optimal osteoblast function and therefore an important gene in the bone formation process. Further studies to identify key binding partners for Tbx3 in osteoblasts and the role of Tbx3 in vivo during postnatal bone development will advance our understanding of the bone formation process.
ACKNOWLEDGMENTS
All work was performed in facilities provided by the Department of Veterans Affairs. The authors thank Joe Rung-Aroon and Catrina Alarcon for technical assistance. This study was supported by National Institute of Health (AR048139 to S.M.).
Grant sponsor: National Institute of Health; Grant number: AR048139.
REFERENCES
- Aubin JE. Advances in the osteoblast lineage. Biochem Cell Biol. 1998;76:899–910. [PubMed] [Google Scholar]
- Bamshad M, Krakowiak PA, Watkins WS, Root S, Carey JC, Jorde LB. A gene for ulnar-mammary syndrome maps to 12q23-q24.1. Hum Mol Genet. 1995;4:1973–1977. doi: 10.1093/hmg/4.10.1973. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Boogerd KJ, Wong LY, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AF, Barnett P. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc Res. 2008;78:486–493. doi: 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem. 2002;277:6567–6572. doi: 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: Relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ. Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation. Oncogene. 2002;21:3827–3835. doi: 10.1038/sj.onc.1205476. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhong Q, Wang J, Cameron RS, Borke JL, Isales CM, Bollag RJ. Microarray analysis of Tbx2-directed gene expression: A possible role in osteogenesis. Mol Cell Endocrinol. 2001;177:43–54. doi: 10.1016/s0303-7207(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Dorak MT. REAL-TIME PCR. 2006 http://dorakmt.tripod.com/genetics/realtime.html.
- Fan C, Liu M, Wang Q. Functional analysis of TBX5 missense mutations associated with Holt-Oram syndrome. J Biol Chem. 2003;278:8780–8785. doi: 10.1074/jbc.M208120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3 + 2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- Farley JR, Hall SL, Tanner MA, Wergedal JE. Specific activity of skeletal alkaline phosphatase in human osteoblast-line cells regulated by phosphate, phosphate esters, and phosphate analogs and release of alkaline phosphatase activity inversely regulated by calcium. J Bone Miner Res. 1994;9:497–508. doi: 10.1002/jbmr.5650090409. [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Govoni KE, Amaar YG, Kramer A, Winter E, Baylink DJ, Mohan S. Regulation of insulin-like growth factor binding protein-5, four and a half lim-2, and a disintegrin and metalloprotease-9 expression in osteoblasts. Growth Horm IGF Res. 2006a;16:49–56. doi: 10.1016/j.ghir.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, Mohan S. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab. 2006b;291:E128–E136. doi: 10.1152/ajpendo.00592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govoni KE, Lee SK, Chung YS, Behringer RR, Wergedal JE, Baylink DJ, Mohan S. Disruption of insulin-like growth factor-I expression in type IIalphaI collagen-expressing cells reduces bone length and width in mice. Physiol Genomics. 2007;30:354–362. doi: 10.1152/physiolgenomics.00022.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219:105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Kalajzic Z, Hurley MM, Lichtler AC, Rowe DW. Stage specific inhibition of osteoblast lineage differentiation by FGF2 and noggin. J Cell Biochem. 2003;88:1168–1176. doi: 10.1002/jcb.10459. [DOI] [PubMed] [Google Scholar]
- Karsenty G. Minireview: Transcriptional control of osteoblast differentiation. Endocrinology. 2001;142:2731–2733. doi: 10.1210/endo.142.7.8306. [DOI] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993;12:3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Neumann LM, Tonnies H, Ropers HH, Mundlos S, Ullmann R. Ulnar-mammary syndrome with dysmorphic facies and mental retardation caused by a novel 1.28 Mb deletion encompassing the TBX3 gene. Eur J Hum Genet. 2006;14:1274–1279. doi: 10.1038/sj.ejhg.5201696. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. PTHrP and skeletal development. Ann NY Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- Lee HS, Cho HH, Kim HK, Bae YC, Baik HS, Jung JS. Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells. Mol Cell Biochem. 2007;296:129–136. doi: 10.1007/s11010-006-9306-4. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS. Runx2/Cbfa1: A multifunctional regulator of bone formation. Curr Pharm Des. 2003;9:2677–2685. doi: 10.2174/1381612033453659. [DOI] [PubMed] [Google Scholar]
- Lingbeek ME, Jacobs JJ, van Lohuizen M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J Biol Chem. 2002;277:26120–26127. doi: 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Soufan AT, Hagoort J, de Boer PA, Christoffels VM. Development of the building plan of the heart. Ann NY Acad Sci. 2004;1015:171–181. doi: 10.1196/annals.1302.014. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, Silver LM. The T-box gene family. Bioessays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Peng H, Chen ST, Wergedal JE, Polo JM, Yee JK, Lau KH, Baylink DJ. Development of an MFG-based retroviral vector system for secretion of high levels of functionally active human BMP4. Mol Ther. 2001;4:95–104. doi: 10.1006/mthe.2001.0423. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279:19026–19034. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. T-box genes and heart development: Putting the “T” in heart. Dev Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- Platonova N, Scotti M, Babich P, Bertoli G, Mento E, Meneghini V, Egeo A, Zucchi I, Merlo GR. TBX3, the gene mutated in ulnar-mammary syndrome, promotes growth of mammary epithelial cells via repression of p19ARF, independently of p53. Cell Tissue Res. 2007;328:301–316. doi: 10.1007/s00441-006-0364-4. [DOI] [PubMed] [Google Scholar]
- Ryoo HM, Hoffmann HM, Beumer T, Frenkel B, Towler DA, Stein GS, Stein JL, van Wijnen AJ, Lian JB. Stage-specific expression of Dlx-5 during osteoblast differentiation: Involvement in regulation of osteocalcin gene expression. Mol Endocrinol. 1997;11:1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- Shui C, Spelsberg TC, Riggs BL, Khosla S. Changes in Runx2/Cbfa1 expression and activity during osteoblastic differentiation of human bone marrow stromal cells. J Bone Miner Res. 2003;18:213–221. doi: 10.1359/jbmr.2003.18.2.213. [DOI] [PubMed] [Google Scholar]
- Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Smith J. T-box genes: What they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- Stains JP, Civitelli R. Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biol. 2003;4:222. doi: 10.1186/gb-2003-4-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T. Tbx genes specify posterior digit identity through Shh and BMP signaling. Dev Cell. 2004;6:43–53. doi: 10.1016/s1534-5807(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J Biol Chem. 2007;282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]