Abstract
A common pattern observed in molecular evolution is that reproductive genes tend to evolve rapidly. However, most previous studies documenting this rapid evolution are based on genes expressed in just a few male reproductive organs. In mammals, sperm become motile and capable of fertilization only after leaving the testis, during their transit through the epididymis. Thus, genes expressed in the epididymis are expected to play important roles in male fertility. Here, we performed evolutionary genetic analyses on the epididymal transcriptome of mice. Overall, epididymis-expressed genes showed evidence of strong evolutionary constraint, a finding that contrasts with most previous analyses of genes expressed in other male reproductive organs. However, a subset of epididymis-specialized, secreted genes showed several signatures of adaptive evolution, including an increased rate of nonsynonymous evolution. Furthermore, this subset of genes was overrepresented on the X chromosome. Immunity and protein modification functions were significantly overrepresented among epididymis-specialized, secreted genes. These analyses identified a group of genes likely to be important in male reproductive success.
Keywords: reproduction, epididymis, evolution, selection
Introduction
Traits involved in reproduction are directly tied to organismal fitness. Genes that underlie reproductive traits often evolve rapidly, a pattern that is commonly interpreted as evidence for continual functional turnover in response to natural and/or sexual selection. Rapid evolution of reproductive genes has been observed in animals as diverse as Drosophila (Coulthart and Singh 1988a, 1988b; Aguadé 1999; Begun et al. 2000; Wagstaff and Begun 2005a, 2005b), crickets (Andres et al. 2006), abalone (Lee et al. 1995; Swanson et al. 2001; Swanson and Vacquier 2002), sea urchins (Metz and Palumbi 1996), and mammals (Wyckoff et al. 2000; Torgerson et al. 2002; Waterston et al. 2002; Swanson et al. 2003; Castillo-Davis et al. 2004; Dorus et al. 2004; Gibbs et al. 2004; Clark and Swanson 2005; Nielsen et al. 2005).
Although the pattern of rapid evolution among reproductive genes appears general, much of our thinking has been shaped by studies on a subset of reproductive tissues. In Drosophila, for example, many studies have focused on genes expressed in the male accessory glands (Tsaur et al. 1998; e.g., Aguadé 1999; Begun et al. 2000; Wagstaff and Begun 2005a). Accessory gland proteins are present in an ejaculate and have been associated with many reproductive phenotypes that are likely involved in coevolutionary interactions (Wolfner 1997; Chapman et al. 2001; Heifetz et al. 2001; Wigby and Chapman 2005). In contrast to accessory gland proteins, proteins that are in or on mature Drosophila sperm show evidence of strong evolutionary constraint, suggesting that different compartments of the male reproductive tract experience different evolutionary dynamics (Dorus et al. 2006). In mammals, most evolutionary genetic studies have focused on genes expressed in the testis, finding rapid protein evolution associated with biological function (Swanson et al. 2003; Nielsen et al. 2005), knockout phenotypes (Torgerson et al. 2005), and developmental timing of gene expression (Good and Nachman 2005).
Another potential signature of adaptive evolution in reproductive genes is their preferential location on the X chromosome (Vicoso and Charlesworth 2006). Genes with male-specific benefits are expected to accumulate on the X chromosome, especially if those same genes confer a cost in females (Rice 1984). In the mammalian testis, this prediction is complicated by X inactivation when genes are silenced about midway through spermatogenesis (Khil et al. 2004). An excess of X linkage has been shown for prostate-specific genes (Lercher et al. 2003), but the generality across mammalian male reproductive tissues remains unknown.
Although the testis is the site of spermatogenesis, other male reproductive organs play central roles in male fertility, such as the epididymis and several accessory glands including the seminal vesicles, prostate, coagulating glands, and bulbourethral glands. A recent study of ejaculated proteins found evidence of extensive positive selection in primates (Clark and Swanson 2005), but in many cases, the tissue of origin of these proteins was unknown. Therefore, it remains unclear how frequent adaptive evolution is across mammalian male reproductive tissues.
There are 2 general functions of the epididymis that may cause genes to be subject to intense positive selection. First, many epididymal proteins interact directly with maturing sperm and are necessary for male fertility (reviewed by Yanagimachi 1994; Jones 1998). Sperm membrane proteins can be added, removed, or modified during a sperm's 9-day transit through the epididymis (Kohane, Gonzalez Echeverria, et al. 1980; Olson and Orgebin-Crist 1982; Jones et al. 1983; Eddy et al. 1985; Cooper 1986; Bedford and Hoskins 1990; Tulsiani et al. 1998; Dacheux et al. 2003). Such modifications may proceed in a kind of assembly-line process, as several features vary across the 10 morphologically distinct epididymal segments in mice (fig. 1), including patterns of gene expression (Johnston et al. 2005; Zhang et al. 2006; Jelinsky et al. 2007), protein content (Kohane, Cameo, et al. 1980; Dacheux et al. 2003, 2005), enzymatic activity (Tulsiani et al. 1993), and lumen morphology (Maneely 1959). Second, the epididymis plays a central role in immune defense (Yenugu et al. 2004). Multiple pathogen defense genes that are expressed in the epididymis have experienced positive natural selection (Maxwell et al. 2003), possibly driven by interactions with pathogenic bacteria. Additionally, epididymal immunity genes may alter the immune response of the female reproductive tract, either by promoting immune response to potential incoming infections (Mueller et al. 2007) or by protecting sperm from the female immune response (Robertson 2007).
Fig. 1.
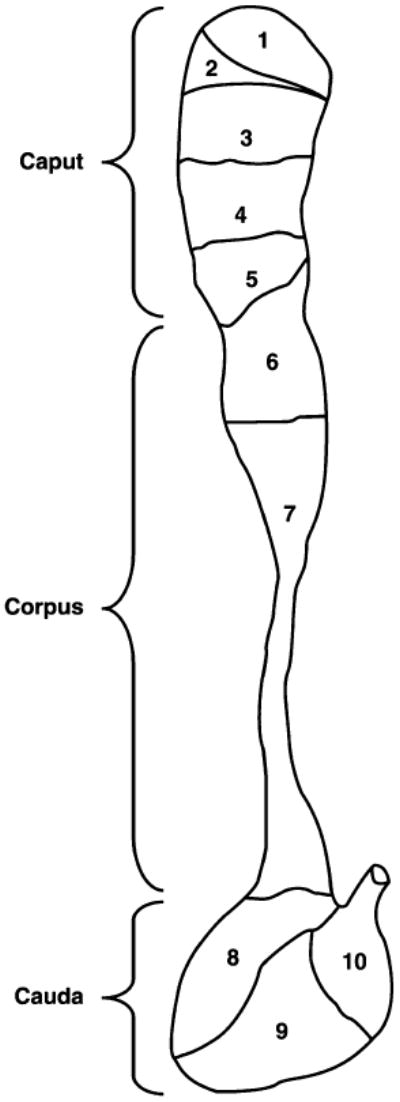
An illustration of the epididymis (reprinted with permission from Biology of Reproduction) showing the 10 morphologically defined segments that were interrogated for patterns of gene expression.
Although epididymal function is relatively well characterized, we know virtually nothing about the evolutionary forces acting on genes expressed in this specialized male reproductive organ. Here, we reanalyzed mouse epididymal transcriptome data (Johnston et al. 2005) in an evolutionary genomics context. We report 4 main findings: 1) genes expressed in the epididymis show unusually strong evolutionary constraint and exhibited less nonsynonymous evolution compared to other genes in the genome, 2) a subset of epididymis-specialized and secreted genes showed signatures of adaptive evolution, 3) epididymis-specialized and secreted genes were significantly overrepresented on the X chromosome as predicted by some theory, and 4) these genes were enriched for immunity and protein modification functions.
Materials and Methods
Expression Data
Johnston et al. (2005) interrogated gene expression patterns from 23 tissues taken from adult C57BL/6 mice with Affymetrix microarrays. They isolated RNA from the whole epididymis, from the 10 morphologically distinct segments within the epididymis (fig. 1), and from 22 non-epididymal tissues. Modifying their definitions slightly, we defined 4 categories: epididymis-expressed genes were detected in the epididymis at a minimum threshold of 100 units, epididymis-selective genes were mostly expressed in the epididymis (expression in the epididymis at least 3-fold greater than all 22 nonepididymal tissues assayed), epididymis-exclusive genes were only expressed in the epididymis (no expression greater than 50 signal units in any of 22 nonepididymal tissues), and segmentally regulated genes were differentially expressed across the epididymis (significant variation in gene expression among segments, with expression in 1 segment at least 4-fold greater than at least one other segment). These 4 groups of genes were defined solely from the Johnston et al. (2005) data. We use the term “epididymis-specialized” to refer to epididymis-selective, epididymis-exclusive, and segmentally regulated genes as a group. These terms summarize patterns of expression and do not necessarily reflect knowledge of biological function. A total of 16,312 genes were included.
Due to proprietary restrictions, detailed expression data for nonepididymal tissues were not available from the Johnston et al. (2005) study. To identify a control set of nonepididymal tissue–selective genes, we analyzed the data of Su et al. (2002), who assayed >36K transcripts from 61 tissues in the mouse, including testis. We filtered the Su et al. (2002) data to mirror the 22 tissues assayed by Johnston et al. (2005); we assumed that the term “retina” was equivalent to “eye,” “7.5-day embryo” was equivalent to “embryo,” and “large intestine” was equivalent to “colon.” Furthermore, we averaged expression data across 9 brain tissues (Su et al. 2002) to approximate the term “brain” (Johnston et al. 2005). The other 18 tissue types were named identically across the 2 data sets.
Gene Annotation and Features
Expression data were linked to gene annotations using the BioMart tool (Ensembl version 39, Mouse Genome Build 36, www.ensembl.org). Any Affymetrix probe sets that hit more than one gene, or hit a pseudogene, were discarded as the expression patterns may be spurious. Probes hitting more than one transcript from the same gene were retained.
What Proportion of the Epididymal Transcriptome Codes for Secreted Proteins?
The presence of a secretory signal was of interest because such proteins may enter the lumen of the epididymis and interact directly with maturing sperm. Presence of a secretory signal was determined using SIGNALP version 3.0 (Nielsen et al. 1997; Bendtsen et al. 2004) and TARGETP version 1.1 (Emanuelsson et al. 2000). A gene was considered secreted if at least one of its transcripts contained a secretory signal.
Which Sperm Membrane Protein Genes Were Expressed in the Epididymis?
Proteins on the membranes of mature sperm may be associated with gamete recognition and fertility success. Stein et al. (2006) identified 114 unique proteins from purified membranes and acrosome vesicles isolated from mature sperm in the caudal end of the epididymis. We were able to associate 98 of these proteins with Ensembl gene annotation and the epididymal transcriptome data.
Molecular Evolution of the Epididymal Transcriptome
Estimating Rates of Nonsynonymous Change (dN/dS)
To estimate rates of evolution, we calculated dN/dS (the number of nonsynonymous substitutions per nonsynonymous site normalized by the number of synonymous substitutions per synonymous site, Goldman and Yang 1994) for the 12,203 mouse genes that had a one-to-one ortholog in rat based on the Ensembl annotation. Protein sequences were aligned using ClustalW version 1.83 (Thompson et al. 1994) and then associated with their coding DNA sequences using REVTRANS version 1.5 (Wernersson and Pedersen 2003). We estimated dN/dS using the CODEML package in PAML version 3.15 (Yang 1997). For genes with multiple transcripts, we estimated dN/dS for all possible pairwise comparisons between mouse and rat and then chose the pair with the lowest estimated dS to represent that gene. Under an assumption of selective neutrality of synonymous sites, dS is a rough estimate of alignment quality. We excluded any genes with fewer than 100 codons, an estimated dN > 1, or an estimated dS ≥ 0.398 (twice the median dS value across the 12,203 genes). We constructed 95% confidence intervals (CIs) of the estimated median by sampling 10,000 bootstrap replicates with R (www.r-project.org).
Testing for Recurrent Positive Selection
To test for recurrent positive selection acting on genes, we used a maximum likelihood framework implemented in CODEML (Yang 1997). Using the same pair of sequences chosen in the above mouse–rat comparisons, we retrieved all one-to-one orthologs in human, cow, and dog. A total of 6,110 genes had one-to-one orthologs across these 5 species. For genes with multiple transcripts in any of these latter 3 species, we chose the longest transcript. Alignments were made as described above. Using the unrooted phylogeny ((human, (mouse, rat), cow, dog), we fit the data to 3 alternative models of molecular evolution (the M7, M8a, and M8 models as described by: Yang et al. 2000; Swanson et al. 2003). In essence, M7 and M8a represent different null hypotheses, as neither allows for codons within a sequence to experience recurrent positive selection, whereas model M8 relaxes this constraint.
The 3 CODEML models consider dS to be invariant across codons. However, synonymous substitution rate may vary across a gene, potentially leading to spurious comparisons of dN and dS (Kosakovsky Pond and Muse 2005). We used a 2-rate fixed-effects likelihood (FEL) model developed by Kosakovsky Pond and Frost (2005), as implemented in the program HYPHY (Kosakovsky Pond et al. 2005) version 0.9920070619beta to compare dN and dS in a likelihood framework. This model allows dS to vary among codons.
We took a conservative approach and considered a gene to have experienced recurrent positive selection if all 5 of the following criteria were met: 1) M8 fit the data significantly better than M7 at P < 0.01, using a likelihood ratio test; 2) M8 fit the data significantly better than M8a at P < 0.01; 3) the additional class of dN/dS estimated by M8 was greater than 1.1; 4) at least 1% of the codons belonged to this additional class of dN/dS; and 5) at least one codon showed significant evidence (P < 0.10) of positive selection (dN/dS > 1.1) in an FEL framework. As further quality control, we estimated pairwise dS between mouse and each of the 4 other species using the runmode = −2 option in CODEML. We excluded any genes that had fewer than 100 codons or produced pairwise dS of mouse–rat ≥ 0.384, mouse–human ≥ 1.190, mouse–dog ≥ 1.368, or mouse–cow ≥ 1.442 (each representing greater than twice the median dS estimated from these respective genome pairs).
Because our expression definitions were based on data collected within a single inbred strain of mice, but evolutionary rates were estimated across a diversity of mammalian species, we expected any association between them to be conservative. Nevertheless, we made a more direct link using a free-ratio model implemented in CODEML (Yang et al. 2000). This model estimated a separate dN/dS ratio for each branch in the above phylogeny. We then tested whether evidence of recurrent positive was associated with increased dN/dS along the lineage leading to Mus, excluding estimates of “infinity” which occur when dS = 0.
Functional Analyses
To better understand the biological processes associated with various gene groups, we performed analyses of functional overrepresentation. We downloaded Mouse Genome Informatics (MGI) terms from Ensembl, excluding any transcripts with more than one MGI term, as well as any MGI terms associated with more than one gene. We tested for overrepresentation of Gene Ontology terms (Ashburner et al. 2000) using ONTOLOGIZER version 2.0 (Robinson et al. 2004). We used the “Term-for-Term” calculation method and considered functional terms with Bonferroni-corrected P < 0.05 to be significantly overrepresented in gene groups.
Results
Expression Data
Of 16,312 genes, 6,739 were epididymis expressed, 209 were epididymis selective, 59 were epididymis exclusive, and 1,115 were segmentally regulated (table 1, fig. 2). For the remainder of the manuscript, we use the term “epididymis-specialized” to refer to epididymis-selective, epididymis-exclusive, and segmentally regulated genes as a group (N = 1,137 genes). Statistical statements did not change whether we analyzed epididymis-specialized genes together or separately for the 3 included groups. It should be noted that the epididymis-specialized group consists mostly of segmentally regulated genes (fig. 2).
Table 1. Number of Probesets, Transcripts, and Genes Analyzed.
| Group | Probe Sets | Transcripts | Genes |
|---|---|---|---|
| Whole genome | 26,147 | 21,321 | 16,312 |
| Epididymis expressed | 12,519 | 9,070 | 6,739 |
| Epididymis specialized | 1,967 | 1,480 | 1,137 |
| Epididymis selective | 317 | 262 | 209 |
| Epididymis exclusive | 89 | 74 | 59 |
| Segmentally regulated | 1,934 | 1,451 | 1,115 |
Fig. 2.
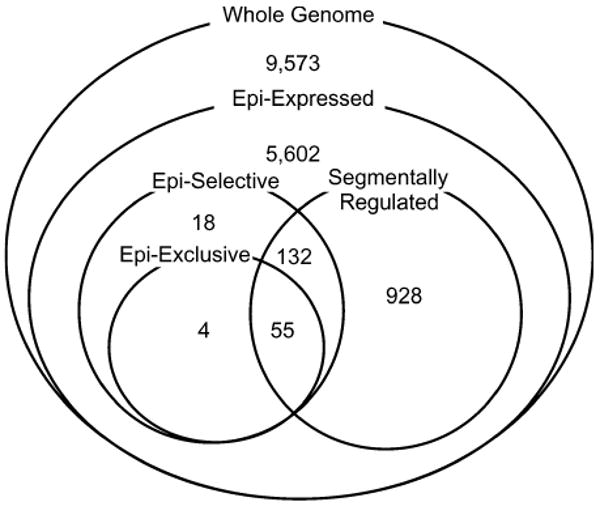
The distribution of the 16,312 genes included in this study with respect to expression definition. Epi-expressed: the number of genes expressed in the epididymis. Epi-selective: the number of genes that were mostly expressed in the epididymis compared with 22 other tissues. Epi-exclusive: the number of genes that were only expressed in the epididymis compared with 22 other tissues. Segmentally regulated: the number of genes that were mostly expressed in one or a few epididymal segments.
What Proportion of the Epididymal Transcriptome Codes for Secreted Proteins?
In general, gene products that are secreted into the lumen of the epididymis might be more likely to interact directly with maturing sperm. We used the entire genome as our null expectation so that different subsets of genes could be compared with the same null distribution. Comparing a subset of genes with the whole genome makes our results conservative. Epididymis-expressed genes showed a general paucity of secreted genes (table 2). Specifically, 1,337 of 6,739 (20%) epididymis-expressed genes were secreted compared with 3,984 of 16,312 (24%) genes from the whole genome (Fisher's exact test [FET], P < 10−13). Thus, most of the epididymal transcriptome probably does not interact directly with maturing sperm.
Table 2. Number of Secreted Genes.
| Group | Secreted | Not Secreted | % Secreted |
|---|---|---|---|
| Whole genome | 3,984 | 12,328 | 24 |
| Epididymis expressed | 1,337 | 5,402 | 20 |
| Epididymis specialized | 396 | 741 | 35 |
| Epididymis selective | 98 | 111 | 47 |
| Epididymis exclusive | 35 | 24 | 59 |
| Segmentally regulated | 390 | 725 | 35 |
In contrast, epididymis-specialized genes were significantly more likely to encode secreted proteins. Of the 1,137 epididymis-specialized genes, 396 (35%) were secreted, significantly more than the whole genome (FET, P < 10−6). For segmentally regulated genes, there was no difference in the proportion of secreted genes among the 10 morphologically distinct epididymal segments (fig. 1).
Which Sperm Membrane Protein Genes Were Expressed in the Epididymis?
Of 98 sperm membrane protein genes (Stein et al. 2006), 56 (57%) were epididymis-expressed genes (fig. 3), including 25 epididymis-specialized, 12 epididymis-selective, 2 epididymis-exclusive, and 22 segmentally regulated genes (these latter 3 categories are not mutually exclusive, fig. 2). To compare these findings to those from testis-expressed genes, we used the data of Su et al. (2002). There were 50 sperm membrane protein genes that were testis-expressed, including 18 testis-selective genes. Of 98 sperm membrane protein genes, 30 (30.6%) showed transcription in the epididymis and not in the testis. Although additional empirical work would be needed to demonstrate the relationship between transcription and protein acquisition, this pattern suggests that a large fraction of sperm membrane proteins derives from the epididymis.
Fig. 3.
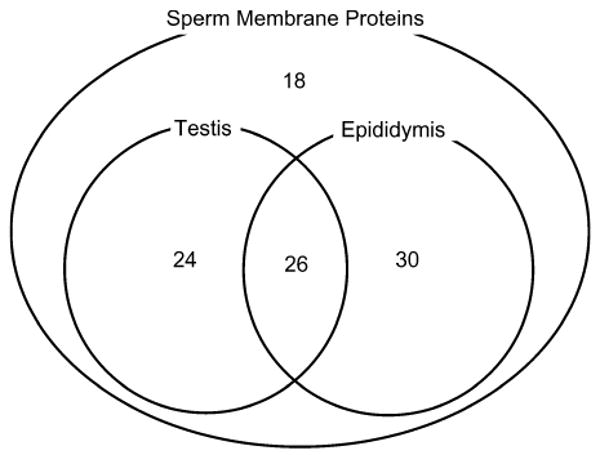
The number of sperm membrane protein genes with transcripts detected in the epididymis and/or testis.
Molecular Evolution of the Epididymal Transcriptome
Strong Evolutionary Constraint Acting on Epididymis-Expressed Genes
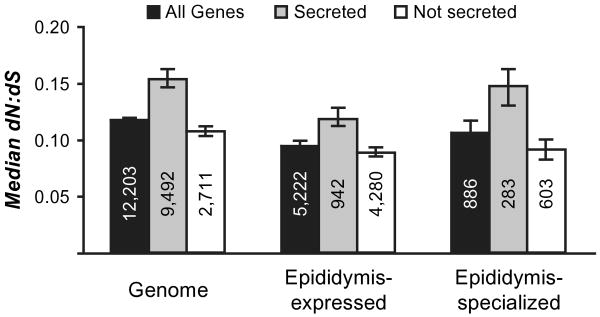
Based on nonoverlapping 95% CIs (fig. 4), epididymis-expressed genes exhibited significantly lower dN/dS (95% CI = 0.090–0.097) compared with the genome (0.114– 0.119). Epididymis-specialized genes showed significantly higher dN/dS (0.097–0.117) than epididymis-expressed genes, but their dN/dS was still lower than the genome. Within epididymis-specialized genes, dN/dS was positively correlated with mean expression in the whole epididymis (P < 10−15, r = 0.27). Secreted genes have undergone significantly more nonsynonymous evolution than non-secreted counterparts in epididymis-specialized genes (secreted: 0.133–0.169 vs. nonsecreted: 0.085–0.101), epididymis-expressed genes (0.114–0.133 vs. 0.085– 0.092), and the whole genome (0.148–0.164 vs. 0.104– 0.110). Secreted genes have been shown previously to experience elevated rates of nonsynonymous evolution (Winter et al. 2004; Julenius and Pedersen 2006).
Fig. 4.
Median pairwise estimates of dN/dS between mouse and rat one-to-one orthologs. Numbers within bars indicate the number of genes. Error bars represent 95% CI around the median, constructed from 10,000 bootstrap replicates.
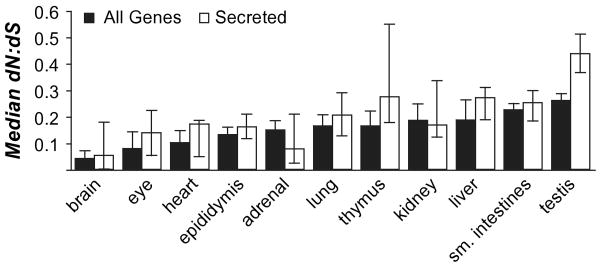
To place estimates of dN/dS in context with other tissues, we identified genes selectively expressed in nonepididymal tissues using the data of Su et al. (2002). Compared with other tissues, epididymis-selective genes showed a relatively high degree of evolutionary constraint, with the third lowest dN/dS among 11 tissues from which at least 20 selectively expressed genes were identified (fig. 5).
Fig. 5.
Rank order of dN/dS among tissue-specialized genes. Tissues were included if at least 20 genes were selectively expressed in them.
For segmentally regulated genes, there was no significant difference in dN/dS among segment of upregulation as their CIs overlapped broadly (N = 204, 43, 52, 33, 53, 74, 105, 75, 100, and 168 genes in the 10 epididymal segments, respectively). This result held even after pooling segmentally regulated genes into the 3 major regions of caput, corpus, and cauda. Among segmentally regulated genes, there was a very small but statistically significant positive correlation (P < 0.05, r = 0.07) between dN/dS and the degree of segmental regulation, defined as the expression in the segment of upregulation divided by the sum of expression across all 10 segments.
Epididymis-Specialized, Secreted Genes Have Undergone Recurrent Positive Selection
Inferring patterns of selection based on the above pair-wise estimates is difficult because high dN/dS may result from relaxed evolutionary constraint or from increased frequency of positive selection. To distinguish between these alternatives, we performed codon-based maximum likelihood estimates among several mammalian species. In the whole genome, 205 of 6,110 genes (3.4%) showed statistically significant evidence of recurrent positive selection. We would expect approximately 75 false positives in our data set assuming a binomial distribution (described in Castillo-Davis et al. 2004). Thus, the majority of these 205 genes are unlikely to be spurious, and our main conclusions should be robust.
Two interesting patterns emerge from these tests. First, a smaller proportion of epididymis-expressed genes experienced recurrent positive selection compared with the whole genome (table 3). Specifically, 77 of 2,786 (2.8%) epididymis-expressed genes showed statistical evidence of positive selection compared with 205 of the 6,110 (3.8%) genes from the whole genome. This difference was not statistically significant (FET, P = 0.15). Second, epididymis-specialized, secreted genes showed a higher incidence of positive selection than the whole genome (table 3). Specifically, 13 of 164 (7.9%) epididymis-specialized, secreted genes showed evidence of positive selection compared with the 205 of 6,110 positively selected in the whole genome (FET, P < 0.01) (table 3). All 13 of these positively selected genes were classified as segmentally regulated, and all occurred on autosomes. One was also classified as epididymis-selective.
Table 3. Proportion of Genes Subject to Recurrent Positive Selection.
| Group | All | Secreted | Not Secreted |
|---|---|---|---|
| Whole genome | 205/6110 (3.4%)a | 73/1446 (5.0%) | 132/4664 (2.8%) |
| Epididymis expressed | 77/2786 (2.8%) | 23/567 (4.1%) | 54/2219 (2.4%) |
| Epididymis specialized | 22/461 (4.8%) | 13/164 (7.9%) | 9/297 (3.0%) |
| Epididymis selective | 4/77 (5.2%) | 1/35 (2.9%) | 3/42 (7.1%) |
| Epididymis exclusive | 1/21 (4.8%) | 0/12 (0.0%) | 1/9 (11.1%) |
| Segmentally regulated | 22/451 (4.9%) | 13/161 (8.1%) | 9/290 (3.1%) |
This ratio was taken as the null expectation in all statistical comparisons, so that different gene groups could be compared with the same null.
We might expect higher rates of evolution among this class of genes simply because secreted proteins and genes with tissue-selective patterns of expression evolve rapidly (Winter et al. 2004; Julenius and Pedersen 2006). However, further investigation showed that molecular evolution among epididymis-specialized, secreted genes is higher than expected based on these features. Of 437 genes that were selectively expressed in a nonepididymal tissue, 20 (4.6%) showed evidence of recurrent positive selection. From 137 genes that were both selectively expressed in a nonepididymal tissue and possessed a secretory signal, 7 (5.1%) showed significant evidence of positive selection. Although not statistically significant, the frequency of positive selection in epididymis-specialized, secreted genes is higher than expected.
Given that we defined epididymis-specialized, secreted genes with mouse data, the link to positive selection, which was inferred across a diverse mammalian phylogeny, should be taken as highly conservative. Nevertheless, a free-ratio model showed that along the lineage leading to Mus, epididymis-specialized, secreted genes have significantly elevated dN/dS (median = 0.13, 95% CI = 0.10–0.16) compared to all genes in the genome (median = 0.09, 0.087–0.093).
Epididymis-Specialized Genes Were Overrepresented on the X Chromosome
X linkage may reflect adaptive evolution because selection can operate more efficiently in the hemizygous sex if new mutations are on average recessive (Rice 1984; Charlesworth et al. 1987). In addition, genes that are favored in one sex but disfavored in the other (i.e., sexually antagonistic) are expected to accumulate on the X chromosome under a much broader array of conditions than on the autosomes (Rice 1984). Of 6,739 epididymis-expressed genes, 237 (3.5%) were X linked, a nonsignificant difference from the genome, where 555 of 16,312 (3.4%) were X linked (FET, P = 0.51). In contrast, epididymis-specialized genes were significantly overrepresented on the X chromosome. Of 1,137 epididymis-specialized genes, 53 (4.7%) were X linked (FET, P = 0.03). Nine of these 53 genes were also secreted.
Functional Analyses
Immunity and Protein Modification Functions Were Overrepresented among Epididymis-Specialized, Secreted Genes
With their increased frequency of positive selection, we were most interested in the functions represented in epididymis-specialized, secreted genes. Within this group, immune response and various modification functions, including transferase and metabolic activities, were significantly overrepresented (table 4; for complete hierarchical relationships among overrepresented terms, see supplementary fig. 1, Supplementary Material online).
Table 4. Overrepresented Functions among Epididymis-Specialized, Secreted Genes.
| Function (GO term) | Number of Genes Observeda | Number of Genes Expectedb | Pc |
|---|---|---|---|
| Biological process | |||
| Defense response (0006952) | 26 | 10.3 | <0.03 |
| Response to bacterium (0009617)d | 10 | 1.4 | <0.01 |
| Lipid metabolism (0006629) | 36 | 14.5 | <10−3 |
| Cell adhesion (0007155) | 38 | 13.7 | <10−4 |
| Signaling (0007167) | 22 | 6.7 | <0.01 |
| Molecular function | |||
| Peptidase activity (0008233) | 33 | 15.1 | <0.05 |
| Transferase activity (0016757)d | 23 | 5.4 | <10−5 |
| Structural (0005201) | 10 | 1.9 | <0.04 |
| Phospholipid binding (0005543)d | 20 | 5.5 | <0.01 |
| Enzyme inhibition (0004857)d | 17 | 5.0 | <0.03 |
The number of epididymis-specialized, secreted genes that are associated with a function.
The number of genes expected to have that function, based on the genome.
The Bonferroni-corrected probability of observing this deviation by chance alone.
This term has additional daughter terms which are significantly overrepresented (supplementary fig. 1, Supplementary Material online).
Discussion
Many reproductive genes show signatures of recurrent positive selection, suggesting that continual functional turnover is favored due to sperm competition among males or to conflicting reproductive interests between males and females (e.g., sexual antagonism). However, our understanding of the evolutionary dynamics of reproductive genes draws mostly from studies of Drosophila (reviewed by Clark et al. 2006). In mammals, evolutionary genetic analyses of reproductive genes come mostly from the testis (Torgerson et al. 2002; Torgerson and Singh 2003, 2006) or seminal fluid proteins (Clark and Swanson 2005). Whether natural selection acts differently on genes expressed in other reproductive organs remains an open question. Here, we investigated the evolutionary dynamics of the epididymal transcriptome.
Strong Evolutionary Constraint Acting on Epididymis-Expressed Genes
Previous studies of genes expressed in male reproductive organs commonly revealed recurrent positive selection in terms of increased dN/dS, increased frequency of positive selection, and increased birth/death of genes (reviewed by Clark et al. 2006). In contrast to previous studies of male reproductive tissues, epididymis-expressed genes exhibited lower dN/dS and reduced frequency of recurrent positive selection compared with the genome. Both patterns indicate strong evolutionary constraints suppressing the fixation of nonsynonymous mutations (fig. 4). However, genes that show expression specialization within the epididymis and are secreted (an indication their proteins may interact directly with maturing sperm) may experience more frequent functional turnover.
Epididymis-Specialized, Secreted Genes Have Undergone Recurrent Positive Selection
In contrast to epididymis-expressed genes, epididymis-specialized, secreted genes have been subject to recurrent positive selection, as evidenced by high pairwise estimates of dN/dS, high frequency of recurrent positive selection, and elevated rates of dN/dS along the phylogenetic lineage leading to Mus. The high rates of molecular evolution (fig. 4 and table 3) were not due to the overrepresented class of immunity genes. Although immunity genes are thought to participate in coevolutionary interactions, none had a one-to-one ortholog in rat based on the Ensembl annotation and therefore were not included in analyses of molecular evolution. Furthermore, a greater proportion of epididymis-specialized, secreted genes showed evidence of recurrent positive selection compared to selective and/or secreted genes of other tissues. Recurrent positive selection was not concentrated among particular epididymal segments; rather, targets of selection were distributed across different developmental stages of sperm maturation.
Epididymis-Specialized, Secreted Genes Were Overrepresented on the X Chromosome
In addition to high rates of nonsynonymous evolution, epididymis-specialized, secreted genes showed a subtle signature of adaptive evolution in their increased frequency on the X chromosome. In spite of the theory predicting that male-specific genes will accumulate on the X (Rice 1984; Charlesworth et al. 1987), this pattern has not been widely observed. Male-specific genes are virtually absent from the X chromosome in Drosophila (Reinke et al. 2000) and Caenorhabditis (Reinke et al. 2000). Several hypotheses have been proposed to explain this paucity (Wu and Xu 2003; Oliver and Parisi 2004). One hypothesis states that selection disfavors X linkage of male-biased genes because genes on the X become inactivated during spermatogenesis (Hense et al. 2007). Consistent with this hypothesis, only genes expressed prior to X inactivation in testis germ cells are overrepresented on the X chromosome in mammals (Wang et al. 2001; Khil et al. 2004).
If X inactivation explains the dearth of testis-expressed genes on the X, then we might expect an excess of X linkage in somatic male-specific tissues. This prediction is supported in mammals, where prostate-specific genes (Lercher et al. 2003) and genes specific to the somatic cells of the testis (Khil et al. 2004) are overrepresented on the X. However, Drosophila show a very different pattern: Genes expressed in somatic accessory glands (Mueller et al. 2005) as well as Drosophila sperm proteome genes (Dorus et al. 2006) were virtually absent from the X chromosome. The present study supports an emerging generality that genes expressed in male-specific somatic tissue accumulate on the mammalian X chromosome, in contrast to Drosophila. Interestingly, we found only a single sperm membrane protein gene (out of 98) on the X chromosome, and its transcript was detected in both testis and epididymis. Of the 53 epididymis-specialized, secreted genes that were X linked, none overlapped with the 13 that showed signs of recurrent positive selection in the maximum likelihood analyses.
Immunity and Protein Modification Functions Were Overrepresented among Epididymis-Specialized, Secreted Genes
Surprisingly, reproduction or gamete development functions were not overrepresented among epididymis-specialized, secreted genes. This conclusion should be taken cautiously as functional characterization of most genes is probably incomplete. No biological functions were significantly overrepresented among the 13 epididymis-specialized, secreted genes that showed evidence of recurrent positive selection (supplementary table 1, Supplementary Material online).
Immune response and various protein modification functions were significantly overrepresented among epididymis-specialized, secreted genes (table 4, supplementary fig. 1, Supplementary Material online). How do these functions fit in the context of epididymal biology? Several bacteria, including gonorrhea and chlamydia, can cause epididymitis and lead to male infertility in humans (Schoysman 1981). Innate immunity proteins identified in the lumen of the epididymis bind to sperm and may protect them during the maturation process (Dacheux et al. 2003; Yenugu et al. 2003, 2004; Zanich et al. 2003; Shayu et al. 2006). Immunity proteins may also influence the female immune system, perhaps allowing sperm to escape detection as a foreign body (Robertson 2007). To our knowledge, no studies have characterized epididymal infections among natural populations of house mice, but some bacterial infections have been identified from male accessory glands in laboratory strains (reviewed in Casey and Irving 1982). Given that multiple mating is common in house mice (Dean et al. 2006), immunity genes may protect against sexually transmitted diseases, as suggested for primates (Nunn et al. 2000; Anderson et al. 2004). Future studies of naturally occurring epididymal infections are needed.
There are several protein modification functions that were overrepresented among this group of genes and these may be related to remodeling of sperm during the maturation process (reviewed by Yanagimachi 1994). Transferase activity is one such modification function that is an important process in sperm maturation (Brown et al. 1983; Jones 1989; Tulsiani et al. 1998). One example is the sperm membrane protein β-1,4-galactosyltransferase, which must be properly glycosylated to bind to the ZP3 glycoprotein found in egg zona pellucida and undergo the acrosome reaction (Macek and Shur 1988; Miller et al. 1992; Nixon et al. 2001).
Our understanding of the function of genes will inevitably benefit from evolutionary genetic studies such as those presented here. We have shown that epididymis-specialized, secreted genes experience recurrent evolutionary turnover. Such turnover may be indicative of the actions of natural and/or sexual selection and suggest that these genes play important roles in male-male as well as male-female interactions.
Supplementary Material
Acknowledgments
We are grateful to Daniel S. Johnston and Scott A. Jelinsky of Wyeth Research as well as Terry T. Turner from the University of Virginia Health Science System for making their epididymal transcriptome data available and for many helpful discussions. M. Worobey provided computing resources. Members of the Nachman laboratory, E. Kelleher, E. Holmes, and 2 anonymous reviewers provided constructive comments on the manuscript. This research was supported by National Science Foundation and National Institutes of Health (NIH) grants to M.W.N. and NIH postdoctoral fellowship F32GM070246-02 to M.D.D.
Footnotes
Supplementary Material: Supplementary figure 1 and table 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Literature Cited
- Aguadé M. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics. 1999;152:543–551. doi: 10.1093/genetics/152.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Hessel JK, Dixson AF. Primate mating systems and the evolution of immune response. J Reprod Immunol. 2004;61:31–38. doi: 10.1016/j.jri.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Andres JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. Molecular evolution of seminal proteins in field crickets. Mol Biol Evol. 2006;23:1574–1584. doi: 10.1093/molbev/msl020. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford JM, Hoskins DD. The mammalian spermatozoon: morphology, biochemistry, and physiology. In: Lamming GE, editor. Marshall's physiology of reproduction. London: Churchill Livingstone; 1990. pp. 379–568. [Google Scholar]
- Begun DJ, Whitley P, Todd BL, Waldrip-Dail HM, Clark AG. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brown CR, von Glos KI, Jones R. Changes in plasma membrane glycoproteins of rat spermatozoa during maturation in the epididymis. J Cell Biol. 1983;96:256–264. doi: 10.1083/jcb.96.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey HW, Irving GW., 3rd . Bacterial, mycoplasmal, mycotic, and immune-mediated diseases of the urogenital system. In: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. New York: Academic Press; 1982. pp. 43–53. [Google Scholar]
- Castillo-Davis CI, Kondrashov FA, Hartl DL, Kulathinal RJ. The functional genomic distribution of protein divergence in two animal phyla: coevolution, genomic conflict, and constraint. Genome Res. 2004;14:802–811. doi: 10.1101/gr.2195604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Herndon LA, Heifetz Y, Partridge L, Wolfner MF. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proc R Soc Lond B Biol Sci. 2001;268:1647–1654. doi: 10.1098/rspb.2001.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. The epididymis, sperm maturation, and fertilization. Berlin (Germany): Springer-Verlag; 1986. [Google Scholar]
- Coulthart MB, Singh RS. Differing amounts of genetic polymorphism in testes and male accessory glands of Drosophila melanogaster and Drosophila simulans. Biochem Genet. 1988a;26:153–164. doi: 10.1007/BF00555496. [DOI] [PubMed] [Google Scholar]
- Coulthart MB, Singh RS. High level of divergence of male-reproductive-tract proteins, between Drosophila melanogaster and its sibling species, D. simulans. Mol Biol Evol. 1988b;5:182–191. doi: 10.1093/oxfordjournals.molbev.a040484. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Castella S, Gatti JL, Dacheux F. Epididymal cell secretory activities and the role of proteins in boar sperm maturation. Theriogenology. 2005;63:319–341. doi: 10.1016/j.theriogenology.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Gatti JL, Dacheux F. Contribution of epididymal secretory proteins for spermatozoa maturation. Microsc Res Tech. 2003;61:7–17. doi: 10.1002/jemt.10312. [DOI] [PubMed] [Google Scholar]
- Dean MD, Ardlie KG, Nachman MW. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus) Mol Ecol. 2006;15:4141–4151. doi: 10.1111/j.1365-294X.2006.03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–1445. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36:1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- Eddy EM, Vernon RB, Muller CH, Hahnel AC, Fenderson BA. Immunodissection of sperm surface modifications during epididymal maturation. Am J Anat. 1985;174:225–237. doi: 10.1002/aja.1001740305. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences: a maximum likelihood approach. J Mol Evol. 1994;40:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Heifetz Y, Tram U, Wolfner MF. Male contributions to egg production: the role of accessory gland products and sperm in Drosophila melanogaster. Proc R Soc Lond B Biol Sci. 2001;268:175–180. doi: 10.1098/rspb.2000.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. X Chromosome Inactivation during Drosophila Spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, Dicandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005;73:404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Jones R. Membrane remodelling during sperm maturation in the epididymis. Oxf Rev Reprod Biol. 1989;11:285–337. [PubMed] [Google Scholar]
- Jones R. Plasma membrane structure and remodelling during sperm maturation in the epididymis. J Reprod Fertil. 1998;53(Suppl):73–84. [PubMed] [Google Scholar]
- Jones R, von Glos KI, Brown CR. Changes in the protein composition of rat spermatozoa during maturation in the epididymis. J Reprod Fertil. 1983;67:299–306. doi: 10.1530/jrf.0.0670299. [DOI] [PubMed] [Google Scholar]
- Julenius K, Pedersen AG. Protein evolution is faster outside the cell. Mol Biol Evol. 2006;23:2039–2048. doi: 10.1093/molbev/msl081. [DOI] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Kohane AC, Cameo MS, Piñeiro L, Garberi JC, Blaquier JA. Distribution and site of production of specific proteins in the rat epididymis. Biol Reprod. 1980;23:181–187. doi: 10.1095/biolreprod23.1.181. [DOI] [PubMed] [Google Scholar]
- Kohane AC, Gonzalez Echeverria FM, Piñeiro L, Blaquier JA. Interaction of proteins of epididymal origin with spermatozoa. Biol Reprod. 1980;23:737–742. doi: 10.1095/biolreprod23.4.737. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- Lee YH, Ota T, Vacquier VD. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol Biol Evol. 2003;20:1113–1116. doi: 10.1093/molbev/msg131. [DOI] [PubMed] [Google Scholar]
- Macek MB, Shur BD. Protein-carbohydrate complementarity in mammalian gamete recognition. Gamete Res. 1988;20:93–109. doi: 10.1002/mrd.1120200109. [DOI] [PubMed] [Google Scholar]
- Maneely RB. Epididymal. structure and function: a historical and critical review. Acta Zool. 1959;40:1–20. [Google Scholar]
- Maxwell AI, Morrison GM, Dorin JR. Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol Immunol. 2003;40:413–421. doi: 10.1016/s0161-5890(03)00160-3. [DOI] [PubMed] [Google Scholar]
- Metz EC, Palumbi SR. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Macek MB, Shur BD. Complementarity between sperm surface beta-1,4-galactosyltransferase and egg-coat ZP3 mediates sperm-egg binding. Nature. 1992;357:589–593. doi: 10.1038/357589a0. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Page JL, Wolfner MF. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics. 2007;175:777–783. doi: 10.1534/genetics.106.065318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Ravi Ram K, McGraw LA, Bloch Qazi MC, Siggia ED, Clark AG, Aquadro CF, Wolfner MF. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics. 2005;171:131–143. doi: 10.1534/genetics.105.043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nixon B, Lu Q, Wassler MJ, Foote CI, Ensslin MA, Shur BD. Galactosyltransferase function during mammalian fertilization. Cells Tissues Organs. 2001;168:46–57. doi: 10.1159/000016805. [DOI] [PubMed] [Google Scholar]
- Nunn CL, Gittleman JL, Antonovics J. Promiscuity and the primate immune system. Science. 2000;290:1168–1170. doi: 10.1126/science.290.5494.1168. [DOI] [PubMed] [Google Scholar]
- Oliver B, Parisi M. Battle of the Xs. Bioessays. 2004;26:543–548. doi: 10.1002/bies.20034. [DOI] [PubMed] [Google Scholar]
- Olson GE, Orgebin-Crist MC. Sperm surface changes during epididymal maturation. Ann N Y Acad Sci. 1982;383:372–392. doi: 10.1111/j.1749-6632.1982.tb23179.x. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Robertson SA. Seminal fluid signaling in the female reproductive tract: lessons from rodents and pigs. J Anim Sci. 2007;85:E36–44. doi: 10.2527/jas.2006-578. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Wollstein A, Bohme U, Beattie B. Ontologizing gene-expression microarray data: characterizing clusters with Gene Ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- Schoysman R. Epididymal causes of male infertility: pathogenesis and management. In: Bollack C, Clavert A, editors. Progress in reproductive biology, pathology and pathophysiology of the epididymis. Basel (Switzerland): Karger; 1981. pp. 102–109. [Google Scholar]
- Shayu D, Chennakesava CS, Rao AJ. Differential expression and antibacterial activity of WFDC10A in the monkey epididymis. Mol Cell Endocrinol. 2006;259:50–56. doi: 10.1016/j.mce.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Stein KK, Go JC, Lane WS, Primakoff P, Myles DG. Proteomic analysis of sperm regions that mediate sperm-egg interactions. Proteomics. 2006;6:3533–3543. doi: 10.1002/pmic.200500845. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Aquadro CF, Vacquier VD. Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol Biol Evol. 2001;18:376–383. doi: 10.1093/oxfordjournals.molbev.a003813. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Kulathinal RJ, Singh RS. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol Biol Evol. 2002;19:1973–1980. doi: 10.1093/oxfordjournals.molbev.a004021. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol Biol Evol. 2003;20:1705–1709. doi: 10.1093/molbev/msg193. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity. 2006;96:39–44. doi: 10.1038/sj.hdy.6800749. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Whitty BR, Singh RS. Sex-specific functional specialization and the evolutionary rates of essential fertility genes. J Mol Evol. 2005;61:650–658. doi: 10.1007/s00239-005-0007-5. [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Ting CT, Wu CI. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: divergence versus polymorphism. Mol Biol Evol. 1998;15:1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- Tulsiani DR, Orgebin-Crist MC, Skudlarek MD. Role of luminal fluid glycosyltransferases and glycosidases in the modification of rat sperm plasma membrane glycoproteins during epididymal maturation. J Reprod Fertil. 1998;53(Suppl):85–97. [PubMed] [Google Scholar]
- Tulsiani DR, Skudlarek MD, Holland MK, Orgebin-Crist MC. Glycosylation of rat sperm plasma membrane during epididymal maturation. Biol Reprod. 1993;48:417–428. doi: 10.1095/biolreprod48.2.417. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Wagstaff BJ, Begun DJ. Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol Biol Evol. 2005a;22:818–832. doi: 10.1093/molbev/msi067. [DOI] [PubMed] [Google Scholar]
- Wagstaff BJ, Begun DJ. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics. 2005b;171:1083–1101. doi: 10.1534/genetics.105.043372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S, Chapman T. Sex peptide causes mating costs in female Drosophila melanogaster. Curr Biol. 2005;15:316–321. doi: 10.1016/j.cub.2005.01.051. [DOI] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner MF. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- Wu CI, Xu EY. Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R. Mammalian fertilization. In: Knobil E, Neill JD, editors. The physiology of reproduction. New York: Raven Press; 1994. pp. 189–317. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenugu S, Hamil KG, Birse CE, Ruben SM, French FS, Hall SH. Antibacterial properties of the sperm-binding proteins and peptides of human epididymis 2 (HE2) family; salt sensitivity, structural dependence and their interaction with outer and cytoplasmic membranes of Escherichia coli. Biochem J. 2003;372:473–483. doi: 10.1042/BJ20030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenugu S, Hamil KG, French FS, Hall SH. Antimicrobial actions of the human epididymis 2 (HE2) protein isoforms, HE2alpha, HE2beta1 and HE2beta2. Reprod Biol Endocrinol. 2004;2:61. doi: 10.1186/1477-7827-2-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanich A, Pascall JC, Jones R. Secreted epididymal glycoprotein 2D6 that binds to the sperm's plasma membrane is a member of the beta-defensin superfamily of pore-forming glycopeptides. Biol Reprod. 2003;69:1831–1842. doi: 10.1095/biolreprod.103.018606. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Liu Q, Li YM, Hall SH, French FS, Zhang YL. Genome-wide profiling of segmental-regulated transcriptomes in human epididymis using oligo microarray. Mol Cell Endocrinol. 2006;250:169–177. doi: 10.1016/j.mce.2005.12.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.