Abstract
Background
Pooling strategies have been used to reduce the costs of polymerase chain reaction-based screening for acute HIV infection in populations in which the prevalence of acute infection is low (less than 1%). Only limited research has been done for conditions in which the prevalence of screening positivity is higher (greater than 1%).
Methods and Results
We present data on a variety of pooling strategies that incorporate the use of polymerase chain reaction-based quantitative measures to monitor for virologic failure among HIV-infected patients receiving antiretroviral therapy. For a prevalence of virologic failure between 1% and 25%, we demonstrate relative efficiency and accuracy of various strategies. These results could be used to choose the best strategy based on the requirements of individual laboratory and clinical settings such as required turnaround time of results and availability of resources.
Conclusions
Virologic monitoring during antiretroviral therapy is not currently being performed in many resource-constrained settings largely because of costs. The presented pooling strategies may be used to significantly reduce the cost compared with individual testing, make such monitoring feasible, and limit the development and transmission of HIV drug resistance in resource-constrained settings. They may also be used to design efficient pooling strategies for other settings with quantitative screening measures.
Keywords: AIDS, efficiency, matrix
INTRODUCTION
In high-resource settings, the failure of antiretroviral therapy (ART) to suppress HIV replication (ie, virologic failure) while a patient is receiving ART is detected by the regular monitoring of HIV RNA levels in the blood (viral loads).1–3 Viral loads are not preformed regularly in resource-constrained settings, where changes in CD4 cell count are used as a surrogate, although CD4-based criteria have been shown to be insensitive in detecting virologic failure during ART.4–8 It is, therefore, imperative to develop and implement less expensive methods to monitor for viral replication during ART in resource-constrained settings to limit the development and transmission of drug-resistant HIV.9–13
Various efforts demonstrated that screening for HIV RNA among people presenting for HIV testing or blood donation can be used efficiently to identify individuals who were acutely infected with HIV despite a negative HIV antibody test, because individuals testing during the window period between acute HIV infection and seroconversion will escape routine detection.14–17 Because testing for HIV RNA in each blood sample would be expensive, a commonly used strategy is to pool blood samples from a group of individuals and perform one HIV RNA assay on a pooled sample.14–17 If the pool tests positive, individual samples or pools of smaller size might be tested again to identify affected individuals. Performance and advantages of different pooling strategies for screening purposes have been extensively investigated. In a landmark paper, Dorfman18 calculated the optimal pool size for a prevalence ranging from 1% to 30% for a single-stage pooling algorithm. Hammick and Gastwirth19 suggest obtaining two samples per individual and form two independent sets of groupings to estimate the prevalence of HIV while preserving confidentiality. An advantage of this procedure is that it can be efficient for a prevalence of up to approximately 30%. Brookmeyer20 developed an estimator of disease prevalence and its variance for multistage pooling studies. Pooling approaches for HIV-infected patients have been investigated by others as well.21–26 Westreich et al27 investigated the performance of three different pooling strategies (two hierarchical and one matrix-based approach) to identify acute HIV infection. Kim et al28 derive and compare operating characteristics of hierarchical as well as square array-based testing algorithms in the presence of testing error.19,20,27,28 Nevertheless, all of the existing work is based on settings where the outcome is binary (disease versus no disease). Also, the testing for HIV RNA in blood samples pooled from patients receiving ART to identify instances when ART is failing to suppress viral replication is considerably different from nucleic acid testing on pooled blood samples to identify instances of acute HIV infection. Specifically, more efficient algorithms are needed because of the higher prevalence of instances of virologic failure, the inherent variability of the viral load assay, and the need to identify lower viral load levels. In this report, we demonstrate how screening for HIV RNA among pooled blood samples can be used to monitor for active HIV replication during ART to identify individuals who are experiencing virologic failure. Because of the potential cost savings of these pooling methods, these results could have a profound impact on the ability to virologically monitor HIV-infected patients receiving ART in resource-constrained settings and ultimately limit the development and transmission of drug-resistant HIV strains.
METHODS
Simulations were used to generate sample values of viral loads that represent a population of patients on ART. These simulations allow the evaluation of the different approaches for a variety of prevalences. The distribution of viral load values of the simulated population was based on a natural history cohort of HIV-infected individuals. Briefly, for viral load values below 500 copies/mL, 85%, 5%, and 10% of values were generated to be uniformly distributed below 50 copies/mL, between 50 and 100 copies/mL, and between 100 and 500 copies/mL, respectively. For values above 500 copies/mL, simulated viral load values were based on two gamma distributions with modes at 1000 and approximately 6000 copies/mL (on log10 scale, the gamma location parameter was 2.7, scale parameter was 0.5, and shape parameter was 1.6 for the first and 3.18 for the second gamma distribution resulting in a mode, mean, and standard deviation of 3.0, 3.5, and 0.63 for the first and 3.8, 4.3, and 0.89 for the second gamma distribution, respectively). The proportion of values generated for the first and second gamma distribution were 93% and 7%, respectively. Both individual and pooled values were generated to include measurement error from viral load assay variability based on normally distributed measurement error with mean zero. Assay standard deviation was assumed to be zero, 0.12, and 0.20 on the log10 scale. The value of zero represented a best case scenario, the value of 0.12 was based on published data,29 and 0.20 represented a more conservative alternative.30 Virologic failure during ART was defined using cut points of 500, 1000, or 1500 copies/mL. These were chosen based on clinically meaningful thresholds at which drug resistance testing could be performed and decisions to amend therapy might be expected.31–33
Three algorithms were evaluated (Figs. 1 and 2). The first was based on Dorfman’s18 two-stage “minipool” approach in which a fixed number of samples would be combined into one pool. If the pooled samples yielded an assay value above the lower limit of detection for the pool (eg, 50 copies/mL or greater), then all individual samples would be tested in the second stage. Samples of pools with assay values below the lower limit of detection would be considered virologically suppressed and no further tests would be performed. For the second algorithm, minipools would be formed in the same manner as for Dorfman’s approach; however, if the pooled samples yielded a viral load value above the lower limit of detection, then individual samples would be tested consecutively (in random order) and subtracted from the pooled sample estimate until the lower limit of detection for the pool was achieved. We call this approach “minipool + algorithm.” The third approach applied a similar algorithm to a k by k matrix. For this approach, samples would be organized in a matrix structure and pools formed over k rows and k columns. Samples of pools would be considered virologically suppressed if either the row or column pools yielded values below the cut point defined as virologic failure. After obtaining (k + k) values for pools across rows and columns, individual samples would be consecutively tested based on the order of highest column and row value, that is, the individual sample that resides in the matrix at the intersection of the row and column pools with the highest viral load values would be tested individually. Viral load values from the individual test would then be subtracted from the appropriate column and row pool totals, and individual testing would proceed one sample at a time until no further row and column pools had values above the lower level of interest. If a single row (or column) showed a value above the lower limit of interest, but none of the columns (or rows) did, all samples from such a row (or column) would be considered virologically suppressed.
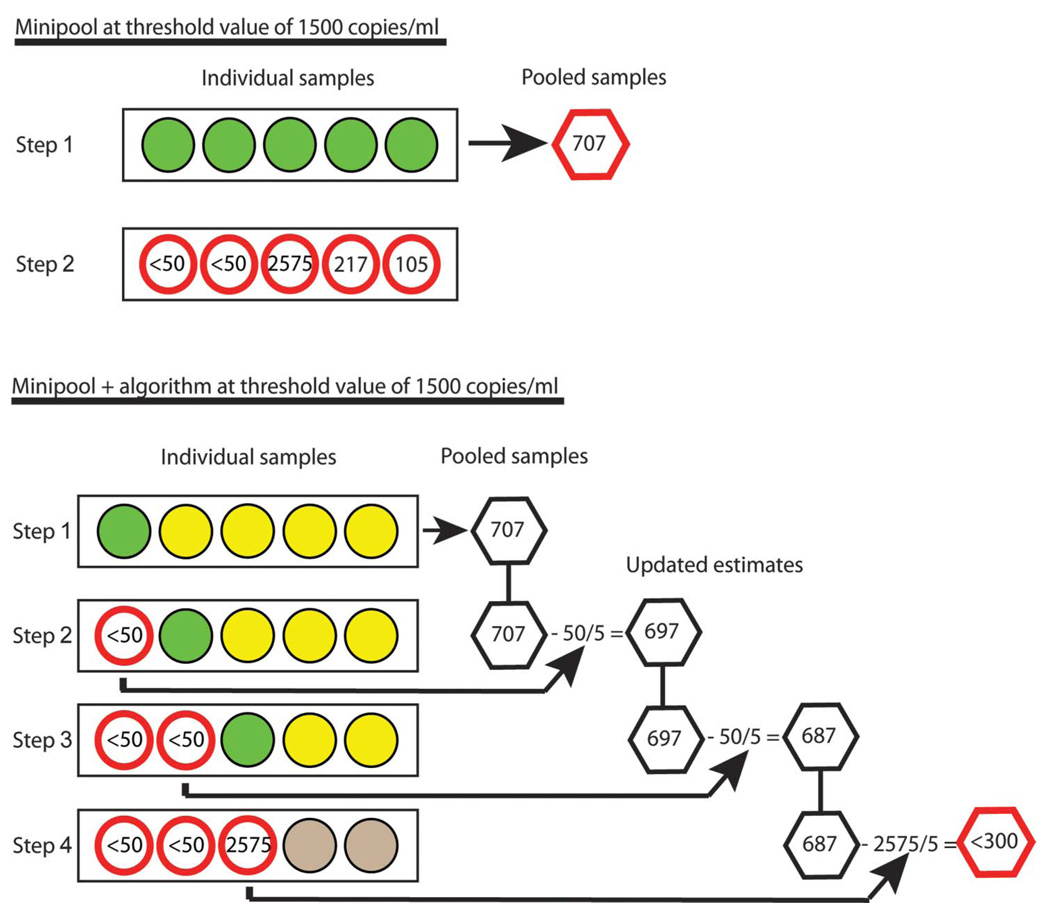
FIGURE 1.
Schematic overview of minipool and minipool + algorithm approaches. Each circle represents an individual sample; each hexagon represents a pooled sample. The numbers within the nonshaded circles are viral load values measured in the individual samples. Green circles indicate individual samples that will have viral loads measured in the next step. Yellow circles indicate samples that might have viral loads measured depending on the outcome of the measures for the green circles. In step 1 for the minipool approach (A), individual samples (green circles) are pooled (hexagon) and a viral load measured in the pooled sample. In this example, because the pooled sample is greater than the threshold of interest (here 300 copies/mL or less because there are five individual samples in a pool), then all individual samples are tested individually in step 2. In step 1 for the minipool + algorithm approach (B), individual samples (green and yellow circles) are pooled (hexagon) and a viral load measured in the pooled sample. In steps 2 through 4, because the viral load of the pooled sample is greater than the threshold of interest, individual samples of pools with values above the threshold are consecutively tested and individual estimates (divided by pool size) are subtracted from pool total (hexagon) until the threshold value is reached (red hexagon). Remaining samples (gray circles) that have not been tested individually are assumed to be below the cut point defining virologic failure (here 1500 copies/mL or less). Note, estimated values for individual samples will typically not add up to estimated values for pooled samples because of assay variability.
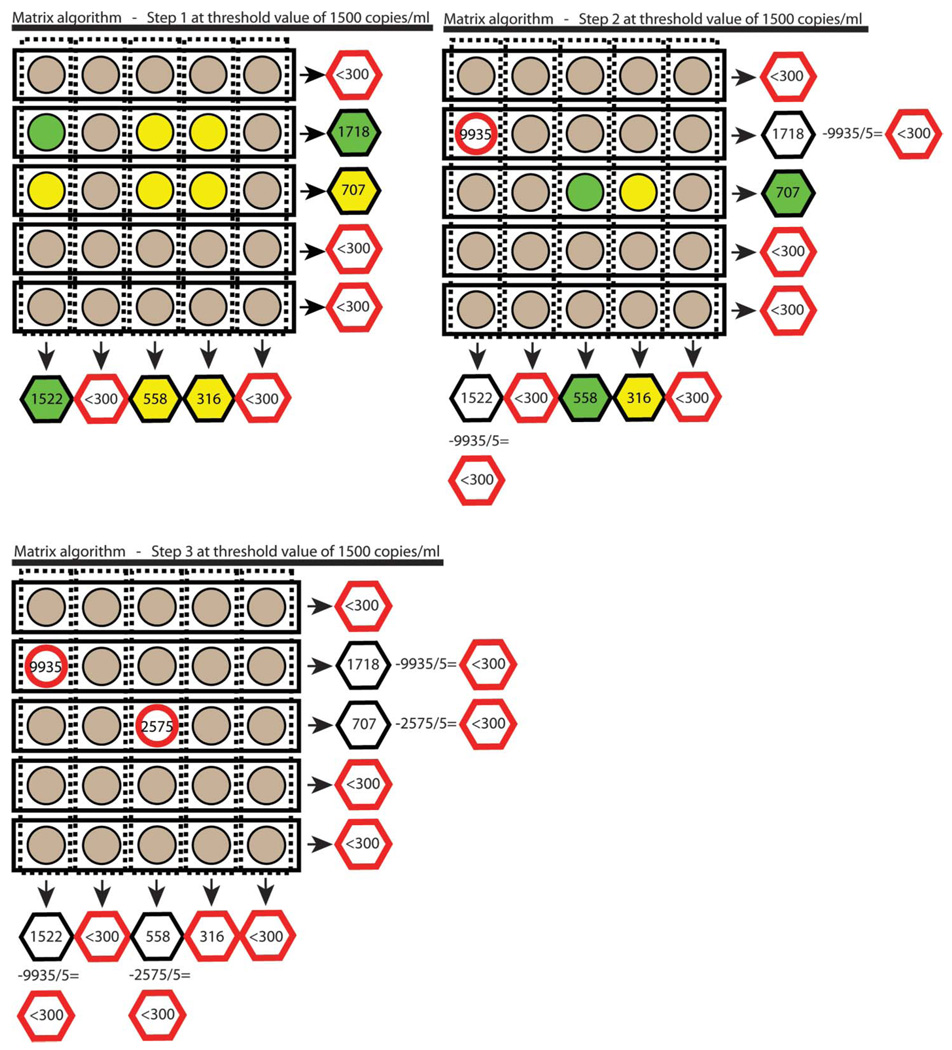
FIGURE 2.
Schematic overview of matrix approach. Each circle represents an individual sample and each hexagon a pooled sample. The numbers within the nonshaded circles are viral load values measured on individual samples. Viral load values for gray circles are assumed to be below the cut point defining virologic failure (here 1500 copies/mL or less). Green circles indicate samples that will be measured in the next step. Yellow circles indicate samples that might be measured depending on the outcome of the measures for the individual samples marked by the green circles. Red circles and hexagons indicate that no further testing/estimation will be performed for the sample or pool. Note, estimated values for individual samples will typically not add up to estimated values for pooled samples because of assay variability. In step 1 of the matrix approach, individual samples (circles) are pooled across rows and columns (hexagons), and viral loads are then measured in each pool. All individual samples are considered to be below the cut point defining virologic failure (gray circle) if their respective row or column pool has a viral load below the corresponding threshold level of interest. Individual samples that belong to a row and a column pool that have viral loads above the threshold remain ambiguous as to whether they contain viral loads above the cut point defining virologic failure and might be tested individually (green and yellow circles). In step 2, the individual sample that belongs to the intersection of the row and column pools with the highest viral load values (green circle) has its viral load measured. The viral load of this individual sample is divided by the size of the pools (here five), and this value is subtracted from the respective row and column pool totals (hexagons), and this subtraction can resolve previously ambiguous samples (yellow circles) to be considered below the viral load level of interest (gray circles). In step 3, this continues until all individual samples belong to a row or column pool that has reached the threshold level. In this example, the first individual sample that is tested has a viral load of 9935. This value divided by the pool size (9935/5 = 1987) is subtracted from column (1522 – 1987 < 0) and row estimate (1718 – 1987 < 0). Samples or rows and columns with pool estimates below the threshold (here 300 or less) are not tested individually and are assumed to be below the cut point defining virologic failure (here 1500 or less).
Based on various prevalences of and cut points for defining virologic failure, we compared relative efficiency and negative predictive value for four approaches (individual samples, minipool, minipool + algorithm, and matrix approaches) and various pool sizes within each of the pooling approaches (minipool, minipool + algorithm, and matrix approaches). Relative efficiency was defined as one minus the average number of assays performed divided by the number of samples. We use a different definition of efficiency than others27 because of the natural interpretation that the best algorithm will have the highest values of efficiency relative to individual testing. With this definition, an algorithm with efficiency zero has no advantage over individual testing and an algorithm with efficiency of 0.25 uses 25% fewer samples than would be used with individual testing.
Negative predictive values were compared for the various approaches to identify individual samples that had viral load values below the same range of cut points (500, 1000, and 1500 HIV RNA copies/mL) similar to the comparisons of relative efficiency. For our purposes, the negative predictive value was the proportion of individuals who were virologically suppressed below the cut point defining virologic failure (true- and test-negative) among those who were considered suppressed by each proposed method (test-negative). We focus here on achieving high negative predictive values, because of the clinical importance of missing virologic failure among patients receiving ART by the testing approaches.
Additionally, we evaluated the various approaches in the context of the following clinically relevant factors: 1) level of viral load for defining virologic failure; 2) standard deviation of the viral load assay; and 3) prevalence of virologic failure in the sampled population. Screening approaches for higher prevalence of virologic failure and assays with larger standard deviation were expected to be less efficient than for lower prevalence of virologic failure or assays with smaller standard deviation.
Pool sizes of three, four, five, six, seven, eight, 10, and 20 were considered. Because the lowest possible level of viral load detection depends on the dilution factor of the pool, the level of detection was considered in relation to the size of the pool. For example, with a lower detection limit for a viral load assay of 50 copies/mL and a pool size of 10, the on average lowest level of virologic failure that we can expect to detect for any individual sample based on the pooled analysis was 500 copies/mL. When pools of 20 individual samples were considered, the lowest detectable level of virologic failure was 1000 copies/mL. All simulations were run using the statistical software Stata34 and were repeated 1000 times for all conditions.
RESULTS
Relative Efficiency
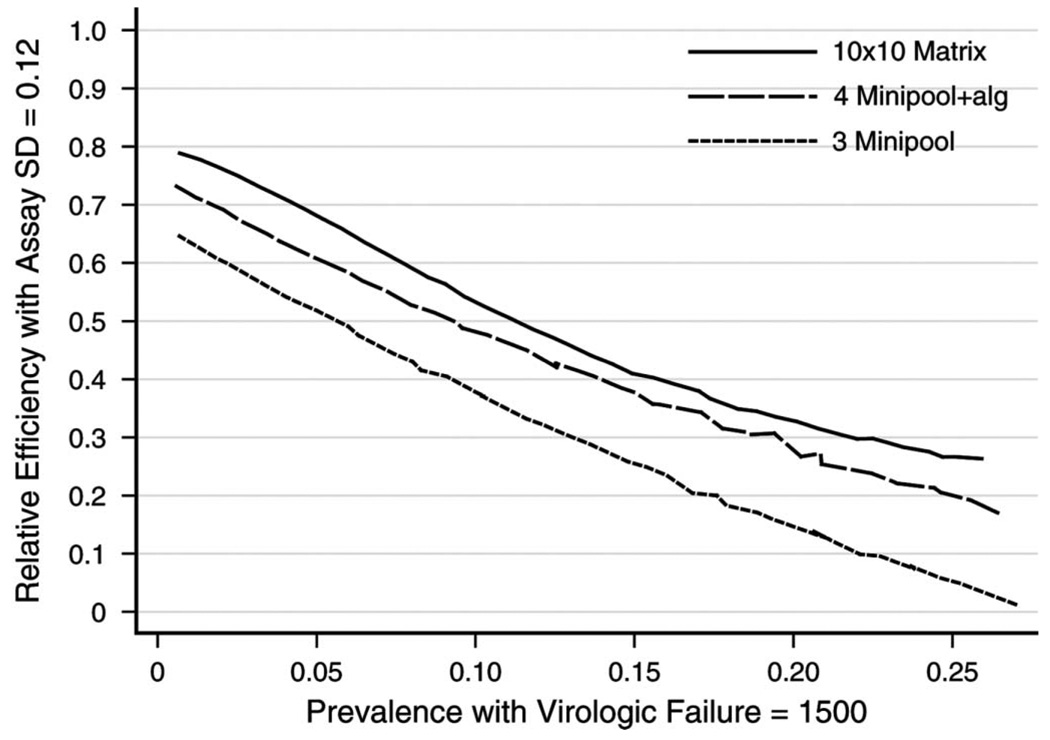
Of the three pooling approaches, most demonstrated relatively high efficiency (greater than 0.70) when the prevalence of virologic failure was low (less than 3%) (Fig. 3), that is, less than 30% of viral load assays would be used in a pooling approach relative to performing viral loads for each individual sample. When the prevalence of virologic failure was higher (greater than 3%), relative efficiency varied markedly; Dorfman’s18 minipools showed lower relative efficiency than algorithms that used the quantitative information available. A matrix approach with intermediate pool size (eight or 10) appeared to be the most efficient approach when: 1) virologic failure was defined as 1500 copies/mL or greater; 2) the standard deviation of the assay was 0.12; and 3) the prevalence of virologic failure of the sampled population was between 5% and 20%. Specifically, the relative efficiency for the matrix approach with a pool size of 10 ranged from 0.68 to 0.33 and for a matrix approach with a pool size of eight, it ranged from 0.65 to 0.32 when the prevalence of virologic failure was 5% and 20%, respectively. Among the approaches that use the minipools + algorithm, the one with five samples per pool appeared most efficient under similar conditions with relative efficiency from 0.63 to 0.26 for a prevalence of virologic failure of 5% and 20%, respectively. Thus, the relative efficiency of the five minipool + algorithm was close (no more than 0.07 difference) to the most efficient matrix approaches. For minipool and minipool + algorithm approaches of the same size, there was an advantage for using the quantitative information when the prevalence of virologic failure was higher than 5%. According to Dorfman’s studies,18 the optimal pool size in the minipool approach is five for a prevalence of 7%, which can achieve a 50% reduction in the number of samples being tested. For our setting, the minipool of size five achieved a 50% reduction in samples being tested for a prevalence of 6%. When quantitative information was used (minipool + algorithm), the same reduction could be achieved for a prevalence of 9% and with the matrix approach of size 10, the same reduction could be achieved for a prevalence of 11%. Similarly, an efficiency of 30% can be achieved for a prevalence of 11%, 18%, and 21% for the minipool, minipool + algorithm, and matrix approaches, respectively. As a result, the same efficiency can be achieved at approximately double the prevalence when using quantitative information and the matrix approach compared with when quantitative information is ignored like in the minipool approach. The relative efficiency over the various approaches changed only slightly when the standard deviation of the assay was 0.20. For example, when the prevalence of virologic failure was 5% in the sampled population, the relative efficiency for the matrix approach with a pool size of 10 was: 1) 0.66 when the standard deviation of the viral load assay was 0.20; 2) 0.68 when the standard deviation was 0.12; and 3) 0.73 when the standard deviation was zero. For Dorfman’s minipool approach, the most efficient pool size was three (compare Table 1 of Dorfman18) with relative efficiency ranging from 0.52 to 0.13 when the prevalence of virologic failure was 5% and 20%, respectively.
FIGURE 3.
Relative efficiency of minipool, minipool + algorithm, and matrix approaches for various pool sizes using a definition of virologic failure of HIV RNA 1500 copies/mL or greater and assay standard deviation (SD) 0.12 copies/mL (on log10 scale).
Overall, pool sizes of three and four had the highest relative efficiency among the minipool and minipool + algorithm approaches and a pool size of 10 had the highest relative efficiency among the matrix approaches when the prevalence of virologic failure was between 5% and 20% (Fig. 4). Results were not substantially different when virologic failure was defined as 500 or greater, 1000 copies/mL or greater, or 1500 copies/mL or greater. For example, when using the matrix approach with a pool size of 10, assay standard deviation of 0.12, and a prevalence of virologic failure of 20%, the relative efficiency was 0.68 when virologic failure was defined at 1500 copies/mL or greater, 0.66 at 1000 copies/mL or greater, and 0.65 at 500 copies/mL or greater.
FIGURE 4.
Relative efficiency for minipool, minipool + algorithm, and matrix approaches with highest (among different pool sizes) relative efficiency compared with individual testing using a definition of virologic failure of viral load 1500 copies/mL or greater and assay standard deviation (SD) 0.12 copies/mL (on log10 scale).
Negative Predictive Values
The results for negative predictive values were uniformly good. For a prevalence of virologic failure up to 10%, negative predictive values were at least 99% and 97% for the matrix approach with assay standard deviations of 0.12 and 0.20 copies/mL, respectively. Similarly, the corresponding negative predictive values were 99% and 98% for all minipool + algorithm approaches. For a prevalence of virologic failure up to 20%, negative predictive values were always at least 92% for all matrix approaches and always above 96% for the minipool + algorithm approach when the standard deviation of the assay was 0.12. These results changed only minimally for an assay standard deviation of 0.20. Negative predictive values were always higher than 99% for Dorfman’s minipool approach (without algorithm). These results remained virtually unchanged for different definitions of virologic failure (500, 1000, and 1500 HIV RNA copies/mL).
Other Factors
In addition to relative efficiency and negative predictive value, there are other factors that can influence the clinical usefulness of these approaches. One is the local need of turnaround time of the results, which will depend on availability of the assays, personnel performing the assays, and size of the clinical population receiving ART. The proposed pooling strategies will vary in the time from sample collection to the time individual viral load results are available to the clinician for management of their patients. For example, if a laboratory has the capability of performing 20 viral loads a day, then 20 individuals could be screened daily using individual viral load testing and it would take 5 days to screen 100 individuals. If minipools of size five are used, then 100 individuals could be screened in the first day, but some pools would require additional testing and, therefore, additional days would be required before results are available. The number of additional days would depend on the prevalence of virologic failure in the sampled population and the specific pooling approach. When 100 patients are screened using minipools + algorithm of size five, virologic failure is defined as 1500 HIV RNA copies/mL or greater and the prevalence of virologic failure is 10%, there would be an average of 2 days until the viral loads of all samples have been resolved. Similarly, the initial screening of the same 100 patients using the matrix approach with a pool size of 10 would require 1 day, but the complete resolution of all samples would require on average 28 days (1 day for testing all row and column pools and an average of approximately 27 days for 27 individual assays that must be tested consecutively); however, on average, 65% of the individual samples would have been resolved on the first day. From a financial perspective, when 100 patients are screened, virologic failure is defined as 1500 HIV RNA copies/mL or greater, assay standard deviation is 0.12, the prevalence of virologic failure is 10%, and one assay costs $75 (as an example), then the average cost per patient tested would be $75 for individual testing and approximately $49, $38, and $35 for using the minipool of size five, minipool + algorithm of size five, and 10 by 10 matrix approaches, respectively, to screen 100 patients.
DISCUSSION
Because commercial viral load assays cost between U.S. $50 and $150 per test (diagnostics pricing; Clinton Foundation; available at: http://www.clintonfoundation.org/cf-pgm-hs-ai-work3.htm; accessed May 23, 2008),8 the costs of virologic monitoring during ART may exceed those of ART itself, but the development and transmission of drug-resistant HIV may ultimately compromise the effectiveness of ART in many populations.9,31,32,35,36 In resource-constrained settings, less expensive methods to monitor viral replication during ART are necessary to make such monitoring feasible and ART sustainable. In this report, we demonstrate how nucleic acid testing on pooled blood samples can be used to reduce the overall number of viral load tests needed to screen patients receiving ART compared with individual testing. By taking advantage of the quantitative information available, efficiency can be achieved for approximately double the prevalence than for methods that use qualitative information only (eg, disease/no disease). To our knowledge, none of the previous work on pooling methods has exploited quantitative information.
Because the prevalence of virologic failure in the sampled population will impact the relative efficiency and accuracy of the presented pooling and testing methods, other strategies that can identify individuals with virologic failure before nucleic acid testing could greatly increase the usefulness of the proposed methods. Such strategies include the measurement of adherence to ART4,37–39 or longitudinal trajectory of CD4 counts4; however, these methods are by no means perfect predictors of virologic failure4–8 and will need to be evaluated in individual clinical settings. Additionally, the definition of virologic failure is an obvious factor in determining the prevalence of virologic failure in a population. Although some research considers the consequences of different definitions of virologic failure,40–42 it remains unclear which level of HIV RNA viral load constitutes the most important clinical cut point to define virologic failure. Differences regarding negative predictive values among different definitions of virologic failure (500, 1000, and 1500 HIV RNA copies/mL) appear to be very small (2% or less) and reasonably small (7% or less) for prevalence of virologic failure up to 10% and 20%, respectively. Differences in relative efficiency for different definitions of virologic failure (500, 1000, and 1500 HIV RNA copies/mL) were also small. In essence, results regarding relative efficiency and negative predictive value did not depend on the definition of virologic failure. It might be possible to further improve the efficiency of the matrix approach. The current approach for using the quantitative information is simple and straightforward, but more sophisticated methods, for example, for choosing the order in which individual samples are tested, might further improve efficiency.
Because the goal of virologic monitoring during ART is to achieve and maintain a low prevalence of virologic failure in the population of patients receiving ART, another option to achieve low prevalence would be to monitor the population for virologic failure at shorter intervals. Recent HIV treatment guidelines recommend monitoring viral loads every 3 to 4 months1; however, these recommendations are based solely on experts’ interpretation of the published literature,1 and more frequent monitoring has been proposed based on prospective clinical trial data.2 More likely, the frequency of virologic monitoring needed to optimize clinical outcomes will vary by clinical setting, which would include factors such as the potency, tolerability, and durability of the available ART regimens; support available for patient adherence; and prevalence of transmitted drug resistance. Taken together, the methods used to monitor for virologic failure during ART will need to be evaluated based on the needs of each clinical setting. We have focused here on relative efficiency and negative predictive value, but depending on the setting, the number of false-positive results might be considered as well. Associated costs could offset some or all of any efficiency achieved through the pooling, like unnecessary resistance testing performed or patients being unnecessarily switched to a new regimen.
Similarly, each clinical program will most likely have unique requirements for assay characteristics (overall costs, turnaround time of results, and monitoring accuracy). For example, in some areas, the overall cost of performing a viral load may be the greatest limiting factor for monitoring for virologic failure; therefore, a clinical program in this setting might best be served by a method that has the highest relative efficiency for the assays independent of the turnaround time of individual results or maximal accuracy. Alternatively, other programs might require more rapid turnaround times for obtaining viral load results such as programs with smaller patient populations receiving ART. These settings would require more time to obtain enough samples to constitute a pool and more frequent testing or smaller pool sizes or even individual viral load testing might be the most cost-effective. Another factor that must be considered for each clinical setting is quality assurance, which will mostly depend on the expertise of the personnel performing the assays (sample handling, processing, and technical consistency with the viral load assay), which could lead to errors in pooling, resolution testing, and inconsistency in calculating viral load results. To further address quality assurance, retesting of samples that are assumed or estimated to be virologically suppressed can be performed using pooling techniques in an efficient manner.43 Similar to choosing a method to screen for virologic failure, the extent and nature of technical training required to perform various pooling approaches and maximize quality assurance will need to be evaluated in each local laboratory and clinical setting.
The proposed approaches are not limited to HIV research but could be used in other settings where quantitative measures are available for screening. In settings where confidentiality is essential, the proposed methods could be applied in two different ways. First, the methods could be used to estimate prevalence.19 Second, if individual identification of samples is important, the estimate of prevalence could be used to guide the choice of pooling method.
We present characteristics for a variety of algorithms and pool sizes that can incorporate available quantitative viral load data and data on how pooling methods can be used efficiently and accurately to monitor for virologic failure in patients receiving ART. These data could be used by individual laboratory and clinical settings to make choices about optimal local virologic monitoring strategies. They may also be used to design efficient pooling strategies for other settings where screening involves quantitative measures. Although promising, further investigation in resource-constrained settings is required to determine if these methods are feasible and cost-effective with respect to the factors that could not be included in the simulations like turnaround time of results, additional personnel costs in local settings, and individual and public health costs of a patient population with virologic failure during ART.
ACKNOWLEDGMENTS
We thank two reviewers and Drs. Susan Little, Robert Schooley, and Matthew C. Strain for insightful comments.
This work was supported by grants AI080353, AI27670, AI43638, the UCSD Center for AIDS Research AI 36214, AI29164, AI47745, AI57167, AI55276, K24-AI064086, (CHRP) CH05-SD-607-005, MH62512 from the National Institutes of Health, and the San Diego Veterans Affairs Healthcare System.
REFERENCES
- 1.Hammer SM, Saag MS, Schechter M, et al. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society–USA panel. JAMA. 2006;296:827–843. doi: 10.1001/jama.296.7.827. [DOI] [PubMed] [Google Scholar]
- 2.Haubrich RH, Currier JS, Forthal DN, et al. A randomized study of the utility of human immunodeficiency virus RNA measurement for the management of antiretroviral therapy. Clin Infect Dis. 2001;33:1060–1068. doi: 10.1086/322636. [DOI] [PubMed] [Google Scholar]
- 3.Hughes MD, Johnson VA, Hirsch MS, et al. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. ACTG 241 Protocol Virology Substudy Team. Ann Intern Med. 1997;126:929–938. doi: 10.7326/0003-4819-126-12-199706150-00001. [DOI] [PubMed] [Google Scholar]
- 4.Bisson GP, Gross R, Bellamy S, et al. Pharmacy refill adherence compared with CD4 count changes for monitoring HIV-infected adults on antiretroviral therapy. PLoS Med. 2008;5:e109. doi: 10.1371/journal.pmed.0050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petti CA, Polage CR, Quinn TC, et al. Laboratory medicine in Africa: a barrier to effective health care. Clin Infect Dis. 2006;42:377–382. doi: 10.1086/499363. [DOI] [PubMed] [Google Scholar]
- 6.Bisson GP, Gross R, Strom JB, et al. Diagnostic accuracy of CD4 cell count increase for virologic response after initiating highly active antiretroviral therapy. AIDS. 2006;20:1613–1619. doi: 10.1097/01.aids.0000238407.00874.dc. [DOI] [PubMed] [Google Scholar]
- 7.Moore DM, Mermin J, Awor A, et al. Performance of immunologic responses in predicting viral load suppression: implications for monitoring patients in resource-limited settings. J Acquir Immune Defic Syndr. 2006;43:436–439. doi: 10.1097/01.qai.0000243105.80393.42. [DOI] [PubMed] [Google Scholar]
- 8.Fiscus SA, Cheng B, Crowe SM, et al. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller V, Larder BA. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir Ther. 2001;6 Suppl 3:25–44. [PubMed] [Google Scholar]
- 10.Phillips AN, Pillay D, Miners AH, et al. Outcomes from monitoring of patients on antiretroviral therapy in resource-limited settings with viral load, CD4 cell count, or clinical observation alone: a computer simulation model. Lancet. 2008;371:1443–1451. doi: 10.1016/S0140-6736(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 11.Calmy A, Ford N, Hirschel B, et al. HIV viral load monitoring in resource-limited regions: optional or necessary? Clin Infect Dis. 2007;44:128–134. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- 12.Smith DM, Schooley RT. Running with scissors: using antiretroviral therapy without monitoring viral load. Clin Infect Dis. 2008;46:1598–1600. doi: 10.1086/587110. [DOI] [PubMed] [Google Scholar]
- 13.Durant J, Clevenbergh P, Halfon P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 14.Pilcher CD, McPherson JT, Leone PA, et al. Real-time, universal screening for acute HIV infection in a routine HIV counseling and testing population. JAMA. 2002;288:216–221. doi: 10.1001/jama.288.2.216. [DOI] [PubMed] [Google Scholar]
- 15.Pilcher CD, Fiscus SA, Nguyen TQ, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352:1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
- 16.Busch MP, Glynn SA, Stramer SL, et al. A new strategy for estimating risks of transfusion-transmitted viral infections based on rates of detection of recently infected donors. Transfusion. 2005;45:254–264. doi: 10.1111/j.1537-2995.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 17.Patterson KB, Leone PA, Fiscus SA, et al. Frequent detection of acute HIV infection in pregnant women. AIDS. 2007;21:2303–2308. doi: 10.1097/QAD.0b013e3282f155da. [DOI] [PubMed] [Google Scholar]
- 18.Dorfman R. The detection of defective members of large populations. Annals of Mathematical Statistics. 1943;14:436–440. [Google Scholar]
- 19.Hammick PA, Gastwirth JL. Group-testing for sensitive characteristics: extension to higher prevalence levels. International Statistical Review. 1994;62:319–331. [Google Scholar]
- 20.Brookmeyer R. Analysis of multistage pooling studies of biological specimens for estimating disease incidence and prevalence. Biometrics. 1999;55:608–612. doi: 10.1111/j.0006-341x.1999.00608.x. [DOI] [PubMed] [Google Scholar]
- 21.Behets F, Bertozzi S, Kasali M, et al. Successful use of pooled sera to determine HIV-1 seroprevalence in Zaire with development of cost-efficiency models. AIDS. 1990;4:737–741. doi: 10.1097/00002030-199008000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Cahoon-Young B, Chandler A, Livermore T, et al. Sensitivity and specificity of pooled versus individual sera in a human immunodeficiency virus antibody prevalence study. J Clin Microbiol. 1989;27:1893–1895. doi: 10.1128/jcm.27.8.1893-1895.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gastwirth JL, Hammick PA. Estimation of the prevalence of a rare disease, preserving the anonymity of the subjects by group-testing—application to Estimating the prevalence of AIDS antibodies in blood-donors. Journal of Statistical Planning and Inference. 1989;22:15–27. [Google Scholar]
- 24.Tu XM, Litvak E, Pagano M. On the Informativeness and accuracy of pooled testing in estimating prevalence of a rare disease—application to HIV screening. Biometrika. 1995;82:287–297. [Google Scholar]
- 25.Quinn TC, Brookmeyer R, Kline R, et al. Feasibility of pooling sera for HIV-1 viral RNA to diagnose acute primary HIV-1 infection and estimate HIV incidence. AIDS. 2000;14:2751–2757. doi: 10.1097/00002030-200012010-00015. [DOI] [PubMed] [Google Scholar]
- 26.Kline RL, Brothers TA, Brookmeyer R, et al. Evaluation of human immunodeficiency virus seroprevalence in population surveys using pooled sera. J Clin Microbiol. 1989;27:1449–1452. doi: 10.1128/jcm.27.7.1449-1452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westreich DJ, Hudgens MG, Fiscus SA, et al. Optimizing screening for acute HIV infection with pooled nucleic acid amplification tests. J Clin Microbiol. 2008;46:1785–1792. doi: 10.1128/JCM.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HY, Hudgens MG, Dreyfuss JM, et al. Comparison of group testing algorithms for case identification in the presence of test error. Biometrics. 2007;63:1152–1163. doi: 10.1111/j.1541-0420.2007.00817.x. [DOI] [PubMed] [Google Scholar]
- 29.Brambilla D, Reichelderfer PS, Bremer JW, et al. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. The Women Infant Transmission Study Clinics. Virology Quality Assurance Program. AIDS. 1999;13:2269–2279. doi: 10.1097/00002030-199911120-00009. [DOI] [PubMed] [Google Scholar]
- 30.Jagodzinski LL, Wiggins DL, McManis JL, et al. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J Clin Microbiol. 2000;38:1247–1249. doi: 10.1128/jcm.38.3.1247-1249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrigan PR, Hogg RS, Dong WW, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. J Infect Dis. 2005;191:339–347. doi: 10.1086/427192. [DOI] [PubMed] [Google Scholar]
- 32.Haupts S, Ledergerber B, Boni J, et al. Impact of genotypic resistance testing on selection of salvage regimen in clinical practice. Antivir Ther. 2003;8:443–454. [PubMed] [Google Scholar]
- 33.Hirsch MS, Brun-Vezinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society–USA Panel. Clin Infect Dis. 2003;37:113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 34.StataCorp. College Station, TX: StataCorp; Stata Statistical Software: Release 10. 2007
- 35.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 36.Vijayaraghavan A, Efrusy MB, Mazonson PD, et al. Cost-effectiveness of alternative strategies for initiating and monitoring highly active antiretroviral therapy in the developing world. J Acquir Immune Defic Syndr. 2007;46:91–100. doi: 10.1097/QAI.0b013e3181342564. [DOI] [PubMed] [Google Scholar]
- 37.Haubrich RH, Little SJ, Currier JS, et al. California Collaborative Treatment Group. The value of patient-reported adherence to antiretroviral therapy in predicting virologic and immunologic response. AIDS. 1999;13:1099–1107. doi: 10.1097/00002030-199906180-00014. [DOI] [PubMed] [Google Scholar]
- 38.Gifford AL, Bormann JE, Shively MJ, et al. Predictors of self-reported adherence and plasma HIV concentrations in patients on multidrug antiretroviral regimens. J Acquir Immune Defic Syndr. 2000;23:386–395. doi: 10.1097/00126334-200004150-00005. [DOI] [PubMed] [Google Scholar]
- 39.Paterson DL, Swindells S, Mohr J, et al. Adherence to protease inhibitor therapy and outcomes in patients with HIV infection. Ann Intern Med. 2000;133:21–30. doi: 10.7326/0003-4819-133-1-200007040-00004. [DOI] [PubMed] [Google Scholar]
- 40.Raboud JM, Seminari E, Rae SL, et al. Comparison of costs of strategies for measuring levels of human immunodeficiency virus type 1 RNA in plasma by using Amplicor and Ultra Direct assays. J Clin Microbiol. 1998;36:3369–3371. doi: 10.1128/jcm.36.11.3369-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raboud JM, Rae S, Hogg RS, et al. Suppression of plasma virus load below the detection limit of a human immunodeficiency virus kit is associated with longer virologic response than suppression below the limit of quantitation. J Infect Dis. 1999;180:1347–1350. doi: 10.1086/314998. [DOI] [PubMed] [Google Scholar]
- 42.Macias J, Palomares JC, Mira JA, et al. Transient rebounds of HIV plasma viremia are associated with the emergence of drug resistance mutations in patients on highly active antiretroviral therapy. J Infect. 2005;51:195–200. doi: 10.1016/j.jinf.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Johnson WO, Gastwirth JL. Dual group screening. Journal of Statistical Planning and Inference. 2000;83:449–473. [Google Scholar]