Abstract
Background
To develop less costly methods to virologically monitor patients receiving antiretroviral therapy, we evaluated methods that use pooled blood samples and quantitative information available from viral load assays to monitor a cohort of patients on first-line antiretroviral therapy for virologic failure.
Methods
We evaluated 150 blood samples collected after 6 months of therapy from participants enrolled in a San Diego primary infection program between January 1998 and January 2007. Samples were screened for virologic failure with individual viral load testing, 10 × 10 matrix pools and minipools of five samples. For the pooled platforms (matrix and minipools), we used a search and retest algorithm based on the quantitative viral load data to resolve samples that remained ambiguous for virologic failure. Viral load thresholds were more than 500 and more than 1500 copies/ml for the matrix and more than 250 and more than 500 copies/ml for the minipool. Efficiency, accuracy and result turnaround times were evaluated.
Results
Twenty-three percent of cohort samples were detectable at more than 50 HIV RNA copies/ml. At an algorithm threshold of more than 500 HIV RNA copies/ml, both minipool and matrix methods used less than half the number of viral load assays to screen the cohort, compared with testing samples individually. Both pooling platforms had negative predictive values of 100% for viral loads of more than 500 HIV RNA copies/ml and at least 94% for viral loads of more than 250 HIV RNA copies/ml.
Conclusion
In this cohort, both pooling methods improved the efficiency of virologic monitoring over individual testing with a minimal decrease in accuracy. These methods may allow for the induction and sustainability of the virologic monitoring of patients receiving antiretroviral therapy in resource-limited settings.
Keywords: antiretroviral therapy, drug resistance, HIV, monitoring, pooling, viral load
Introduction
In settings in which resources are available, diagnosis of the failure of antiretroviral therapy (ART) to suppress HIV replication is made by the regular monitoring of HIV RNA levels (viral loads) in the blood while a patient is being treated [1]. Monitoring for virologic failure during ART is important to limit the development and transmission of HIV drug resistance [2–4], as both can significantly diminish the effectiveness of ART and resistance that develops to one antiretroviral drug in a regimen often confers resistance to other drugs of the same class [5–7]. The current clinical guideline for monitoring viral loads during ART is every 3 months, and this monitoring strategy has proven cost-effective in resource-wealthy settings [8]. Because of capability and cost, the use of viral loads to monitor for virologic failure during ART is not recommended or performed in most resource-limited settings [9–13], and in efforts to save resources, some have proposed detailed historical, hematological, immunological and clinical monitoring instead [9,14–16]. Recent studies, however, have demonstrated that measures that do not monitor for active viral replication are not sufficient to detect virologic failure and foster the spread of drug resistance within a population [11,13,14,17]. Less expensive methods to monitor for virologic failure during ART will thus be required in resource-limited settings to make virologic monitoring feasible.
Pioneering efforts to use HIV RNA detection among people presenting for HIV testing found that a pooling strategy which had previously been used for syphilis testing [18] would identify considerable numbers of individuals who were acutely infected with HIV despite a negative HIV antibody test [19–21]. Because testing for HIV RNA in each blood sample would be expensive, the innovation was to pool blood samples and perform one HIV RNA assay on a pooled sample. If the sample was negative for HIV RNA, then most likely all individuals in the pool were negative for HIV infection [21–23]. This strategy was found to be a highly efficient and affordable means of identifying individuals with acute HIV infection [21]. Currently, HIV RNA and hepatitis C virus RNA nucleic acid testing (NAT) on blood plasma pooled from blood donors is used to screen the United States blood supply for HIV and hepatitis C virus contamination [22]. In a similar fashion, we demonstrate that viral load monitoring in pooled blood samples can be used to monitor for active HIV replication in patients receiving ART, that is, virologic failure [24]. We applied two pooling algorithms to screen for virologic failure after 6 months of first-line ART for a cohort of patients in the United States. One method was based on a 10 × 10 pooling matrix to maximize assay efficiency in terms of number of assays performed per number of individual samples screened. The second strategy used nonoverlapping minipools of five samples per screen to decrease result turnaround time while also improving assay efficiency compared with viral load testing on each individual sample. Because of the potential cost savings of these methods, these data could have substantial consequences for the clinical management of HIV-infected patients receiving ART in resource-limited settings.
Methods
Study population and drug resistance testing
This study was approved by our local ethics committee. We included all participants of the San Diego Acute Infection and Early Disease Research Program (AIEDRP) between January 1998 and January 2007, who had received their first ART regimen, containing at least three agents, for 6 months (±2 weeks). The timing of study participation was determined so as to include 150 eligible samples. Eligibility for the AIEDRP required that all participants have laboratory-documented evidence of HIV infection within 12 months of enrollment, as previously described [25], though timing of ART in relation to duration of infection was not an eligibility requirement for this study. Blood plasma samples collected during the AIEDRP study had been aliquoted and stored at −80°C without previous thaws. As part of their participation in the San Diego site of AIEDRP, most participants received baseline drug resistance testing (Geneseq; Monogram Biosciences, South San Francisco, California, USA).
Nucleic acid testing
NAT was performed using the ultrasensitive Amplicor HIV-1 monitor viral load assay (Roche Molecular Diagnostics, Pleasanton, California, USA), which has a lower level of viral detection of 50 HIV RNA copies/ml. We compared three NAT screening methods to evaluate the identification of virologic failure during ART among our cohort: testing samples individually, 10 × 10 matrix pools and minipools of five samples. Samples were individually tested and placed within the matrix and minipool platforms based on chronological collection in the AIEDRP cohort (1–150). Individual sample testing and reformatting into minipools or matrices were performed by a single technician who was blinded to the clinical information of the individuals. The average time to perform a viral load assay by the technician was measured as well as the technician time required to constitute the minipools and matrix pools. Algorithm thresholds for resolving ambiguities within the two pooling platforms were defined a priori as more than 500 and more than 1500 HIV RNA copies/ml for the matrix and more than 250 and more than 500 HIV RNA copies/ml for the minipool. A search algorithm that combined quantitative viral load information from the pooled samples and viral load information from individual samples was used to resolve ambiguities based on the algorithm thresholds [26]. See Supplementary methods for details.
Matrix
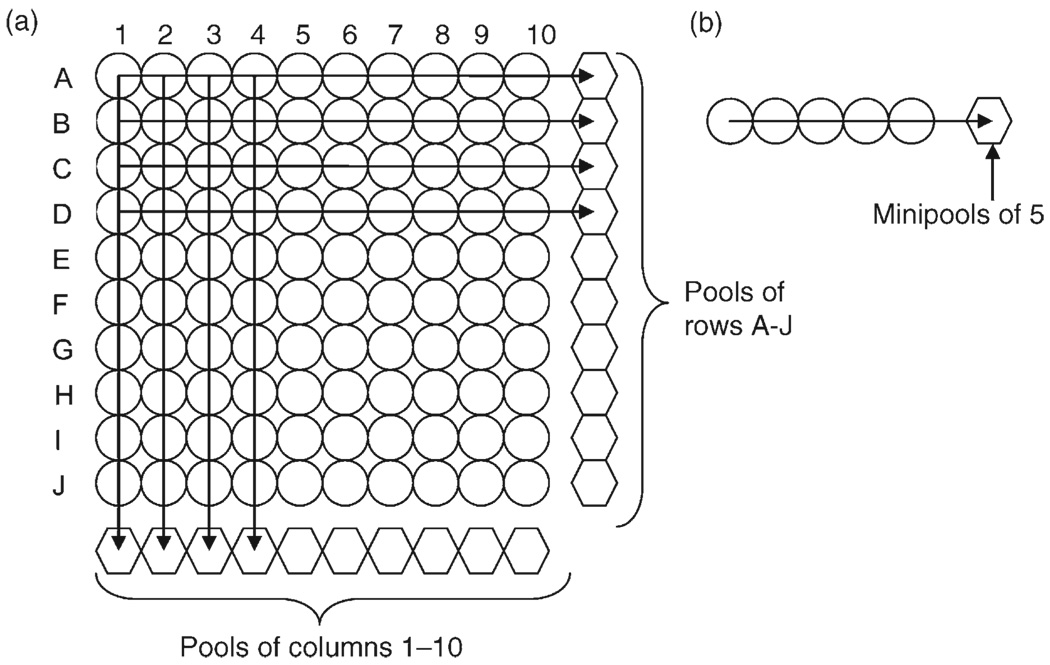
On the basis of previous research [23,24], a 10 × 10 matrix platform was used for these analyses (Fig. 1a). Eligible blood plasma samples were thawed once, and 50µl was pooled from 10 samples for a total of 500µl for each row (letters A–J) and column (numbers 1–10). Matrix pooled blood plasma samples were assayed for viral loads (Amplicor; Roche Molecular Diagnostics). On the basis of this platform, each sample was tested twice, once in a row pool and once in a column pool for each matrix. Using the 150 eligible samples, three matrix pools were tested, such that each sample was included in two different matrices (Matrix 1: samples 1–100, Matrix 2: 51–150, Matrix 3: 1–50, 101–150). We used a search algorithm that combines quantitative viral load information from the pooled samples and viral load information from individual samples to resolve ambiguities, as described in the Supplementary methods [24].
Fig. 1. A 10 × 10 matrix and minipools of five.
Individual samples (circles) were pooled (hexagons) by row (A–J) and by column (1–10) in the 10 × 10 matrix (a) and by five samples in the minipool (b) approach. Viral load tests were then performed on the pooled samples only (hexagons). When ambiguity was generated as to which individual samples in the matrix method (A) were above the threshold of HIV RNA detection and represented virologic failure, then individual samples (circles) that were ambiguous were assayed using the search and retest algorithm as described in the methods section. Similarly, when a minipool (hexagon) was tested and was above the threshold of HIV RNA detection, a simple procedure was used to resolve which individual sample(s) (circles) in the minipool harbored virologic failure.
Minipools of five samples
Similarly, a platform of minipools of five samples was used for these analyses (Fig. 1b). Separate aliquots of blood plasma collected from eligible participants after 6 months of ART used in the 10 × 10 matrix and individual testing experiments above were thawed once and 100µl were pooled from five samples for a total of 500µl for each minipool. Pooled blood plasma samples were assayed for HIV RNA levels, as described above. Using the 150 eligible samples, 30 minipools were tested. Samples were placed within minipools based on chronological collection in the AIEDRP cohort (1–150) and assayed for HIV RNA levels. Similar to the matrix analyses, we used a search algorithm to resolve samples with ambiguous viral loads, as described in the Supplementary methods [24].
Results
Participants
Between January 1998 and January 2007, 168 study participants started ART during their participation in the cohort and had blood plasma samples available for analysis after 6 months of therapy. Eighteen of these participants were not eligible for the study because they had discontinued their first ART regimen before completing 6 months of ART. Twelve of these had changed or stopped their first-line regimen secondary to medication intolerance. The remaining six individuals changed or stopped their ART regimen secondary to virologic failure after virologic suppression, which was identified between 2 and 5 months after the start of their initial regimen. The average viral load at time of virologic failure for these six individuals was 3.2 log10HIV RNA copies/ml (range 64–6780 HIV RNA copies/ml). The majority of the remaining 150 participants were White men who reported sex with men as their HIV risk factor (91%). At the time of starting ART, the average estimated duration of infection was 266 days (range 8–1918 days), the median CD4 cell count was 466 cells/µl and mean viral load was 3.9 log10HIV RNA copies/ml (Table 1).
Table 1.
Comparisons of virologic outcomes of study cohort.
| Demographic | Total (n = 150) | Virologic success (n = 116) | Virologic failure (n = 34) |
|---|---|---|---|
| Male sex; n (%) | 143 (95) | 110 (95) | 33 (97) |
| Age; mean years (range) | 42 (23–67) | 42 (23–67) | 40 (27–58) |
| Ethnicity; n (%) | 149 (100) | ||
| White | 106 (71) | 85 (74) | 21 (62) |
| African–American | 8 (5) | 5 (4) | 3 (9) |
| Latino/a | 24 (16) | 16 (14) | 8 (24) |
| Other | 11 (7) | 9 (8) | 2 (6) |
| HIV risk factor; n (%) | |||
| MSM | 136 (91) | 104 (90) | 32 (94) |
| MSM + IDU | 3 (2) | 3 (2) | 0 (0) |
| Heterosexual | 11 (7) | 9 (8) | 2 (6) |
| Viral load at start of ART; mean log10HIV RNA copies/ml (range) |
3.9 (1.1–6.4) N = 147 | 3.8 (1.1–6.4) N = 114 | 4.1 (2.6–5.8) N = 33 |
| CD4 cell count at start of ART; median cells/µl (range) |
466 (85–1332) N = 149 | 466 (85–1332) N = 115 | 470 (153–920) N = 34 |
| CD4 cell count after 6 months of ART; median cells/µl (range) |
583 (203–1301) | 610 (203–1301) | 511 (254–827) |
| Nonclade B; n (%) | 3 (2) | 3 (3) | 0 (0) |
| Baseline drug resistancea | |||
| Total missing; n (%) | 17 (11) | 11 (9) | 6 (18) |
| Any drug; n (%) | 17 (13) | 13 (12) | 4 (14) |
| NRTI; n (%) | 6 (5) | 5 (5) | 1 (4) |
| NNRTI; n (%) | 12 (9) | 9 (9) | 3 (11) |
| PI; n (%) | 5 (4) | 3 (3) | 2 (7) |
| ART regimen | |||
| PI based; n (%) | 71 (47) | 52 (45) | 19 (56) |
| NNRTI based; n (%) | 57 (38) | 48 (41) | 9 (26) |
| PI and NNRTI; n (%) | 11 (7) | 7 (6) | 2 (6) |
| NRTI only; n (%) | 11 (7) | 9 (8) | 2 (6) |
Virologic success defined as viral load less than 50 HIV RNA copies/ml after 6 months of ART. ART, antiretroviral treatment; IDU, injection drug users; MSM, men who have sex with men; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Rates of HIV-1 clade and drug resistance were calculated on available genotypic data.
Viral load testing of individual eligible samples collected after 6 months of ART revealed that 23% of samples were detectable (>50 HIV RNA copies/ml), and the mean viral load of these samples was 2.3 log10HIV RNA copies/ml (range: 51–5180 HIV RNA copies/ml). Although participants receiving protease inhibitor-based ART regimens had a nonsignificantly higher percentage of virologic failure than those receiving nonnucleoside reverse transcriptase inhibitor-based ART regimens (27 versus 16%; P > 0.1, Exact test). There was also no difference between those with virologic failure and those without when comparing demographics (age and sex) and other baseline characteristics including reported HIV risk factor, baseline plasma viral load, CD4 cell counts and drug resistance (P > 0.2, Exact and Wilcoxon tests) (Table 1).
Matrix and minipool testing characteristics for the study cohort
One hundred and fifty samples were used to evaluate each method for relative efficiency comparing the number of viral load assays performed with the number of individual samples screened and ability to identify individual samples harboring virologic failure during ART at various levels of viral loads. For algorithm thresholds of 500 and 1500 HIV RNA copies/ml, the average relative efficiency of 10 × 10 matrix pools was 0.56 and 0.70, respectively. This means that with a relative efficiency of 0.70, the matrix approach uses 70% fewer assays than individual testing (Table 2). At the 500 HIV RNA copies/ml threshold, the average negative predictive values for individual samples with viral loads more than 500, more than 250, more than 100 and more than 50 copies HIV RNA copies/ml was 100, 97, 90 and 86% for the matrix approach, respectively. At the 1500 HIV RNA copies/ml threshold, the average negative predictive values for individual samples with viral loads more than 500, more than 250, more than 100 and more than 50 copies HIV RNA copies/ml were 100, 95, 89 and 83%for the matrix approach, respectively (Table 2). For the minipool method, the average relative efficiency was 0.41 for the 250 HIV RNA copies/ml threshold and 0.56 for the 500 HIV RNA copies/ml threshold (Table 2). At the 250 HIV RNA copies/ml threshold, the average negative predictive values for individual samples with viral loads more than 500, more than 250, more than 100 and more than 50 copies HIV RNA copies/ml were 100, 98, 93 and 88% for the minipool approach, respectively. At the 500 HIV RNA copies/ml threshold, the average negative predictive values for individual samples with viral loads more than 500, more than 250, more than 100 and more than 50 copies HIV RNA copies/ml were 100, 94, 88 and 82% for the minipool approach, respectively (Table 2). In summary, the negative predictive value was always 100% for either the minipool or matrix platform at the thresholds evaluated when an individual sample had a viral load more than 500 HIV RNA copies/ml, that is, in this study population, the pooling platforms correctly identified all samples without virologic failure (< 50 HIV RNA copies/ml) as confirmed by viral load testing of the individual samples.
Table 2.
Assays performed, costs and result turnaround time.
| Algorithm thresholds (HIV RNA copies/ml) |
Assays for 100 samples (average) |
Negative predictive value at HIV RNA copies/ml |
Cost per samplea (average in $) |
Turnaround time for all results (average days) |
||||
|---|---|---|---|---|---|---|---|---|
| >500 (%) | >250 (%) | >100 (%) | >50 (%) | |||||
| Individual testing | NA | 100 | 100 | 100 | 100 | 100 | 75 | 5 |
| Minipool | 250 | 59 | 100 | 98 | 93 | 88 | 45 | 7 |
| 500 | 44 | 100 | 94 | 88 | 82 | 33 | 5 | |
| Matrix | 500 | 44 | 100 | 97 | 90 | 86 | 33 | 17b |
| 1500 | 30 | 100 | 95 | 89 | 83 | 23 | 9b | |
Costs are in US dollars. NA, not applicable.
Only accounting for cost of assay performed at commercial laboratory, not additional costs to constitute the matrix or minipool.
Although the matrix pooling platform required an average of 17 and 9 days to resolve all samples below the threshold, ≥66% of all individual samples with viral loads more than 50 HIV RNA copies/ml were resolved in the first day.
The calculated time for one technician to perform 100 viral load assays on 100 individual samples was 5 days. As we used a search and retest method to identify samples within the matrix or minipools that required individual viral load testing and the results of those viral loads determined the next sample to be tested, there was a turnaround time for all ambiguous samples to be resolved for both pooling methods. The time to screen and resolve 100 individual samples in our cohort for virologic failure using the 10 × 10 matrix and the minipool method with the same threshold of 500 HIV RNA copies/ml was an average of 17 days and 5 days, respectively (Table 2). Although the matrix platform required an average of 17 days to resolve all samples above the 500 HIV RNA copies/ml threshold, 66% of all individual samples with viral loads more than 50 HIV RNA copies/ml were resolved in the first day.
Discussion
In one of the largest and most expensive public health endeavors ever, over 3 million people worldwide are receiving potent ART [10,27–29]. The continued success of this intervention is limited when ART fails to suppress HIV replication. HIV drug resistance can then develop and be transmitted [4,11,15,30,31]. To improve clinical outcomes and limit the development of drug resistance, clinical programs in resource-wealthy settings regularly monitor patients receiving ART with viral load assays. Many resource-limited settings, however, have minimized laboratory monitoring in exchange for wider availability of HIV treatment [11,13,16,32,33], and virologic monitoring is not recommended or performed in most resource-limited settings [10,11,27,34]. Some have proposed historical, hematological, immunological and clinical monitoring instead [15], but these have proven suboptimal [13,14,35,36]. Although expensive, virologic monitoring will provide optimal clinical outcomes for individual patients and for populations; less expensive methods to monitor for viral replication during ART will be needed to make such monitoring feasible in resource-limited settings [10,13,17,27]. To this end, we demonstrate here using existing clinical data and banked samples how NAT applied to pooled blood samples can maintain accuracy while being used to reduce the number of viral load tests needed to screen a population of patients receiving ART.
Building on methods used to screen individuals for acute HIV infection [21], we demonstrate that pooling strategies can also be used to virologically monitor patients receiving ART with excellent accuracy and greatly improved efficiency, as compared with performing viral loads on individual samples. The use of NAT on blood samples pooled from patients receiving ART to identify instances when ART is failing, however, is more challenging than using NAT on pooled blood samples to identify instances of acute HIV infection. First, during acute HIV infection, the level of HIV RNA is much higher than when ART fails to fully suppress HIV replication. Because the threshold level of HIV RNA detection is much higher for NAT when blood samples are pooled, the number of samples that can be pooled and still detect a clinically meaningful level of viral replication during ART is much smaller than what is often needed for the detection of acute HIV infection. Second, among people being tested for HIV or donating blood, the prevalence of acute HIV infection is rare, while for patients receiving ART the prevalence of virologic failure would be much greater and therefore require more resolution testing of pooled samples than would be expected for acute HIV infection screening. To overcome these obstacles, we implemented NAT pooling strategies combined with a search and retest algorithm using the quantitative information available from the viral load assay.
Overall, these methods demonstrated excellent accuracy compared with performing viral loads on individual samples. Theoretically, the minimum detectable viral load on a pool of 10 samples (matrix method) is 500 HIV RNA copies/ml if all samples without virologic failure are assumed to have a viral load of zero HIV RNA copies/ml; however, the presented matrix method could detect individual samples of more than 250 and more than 100 HIV RNA copies/ml with 95 and 89% negative predictive values, respectively. Similarly, the minipool method demonstrated excellent accuracy across all viral load thresholds tested. These methods could detect virologic failure at levels less than the ‘theoretical’ lower level of detection of the assay in the pool because the search and retest algorithm used to resolve ambiguous samples involves testing of individual samples in the pool. Almost all of the levels of detection evaluated were well below the required viral load that is necessary to perform most commercial drug resistance assays, generally more than 1000 HIV RNA copies/ml [37].
In addition to maintaining accuracy to detect virologic failure at the lower viral load levels, the matrix pooling and minipool methods demonstrated a considerable reduction in the number of viral load assays required to screen individuals receiving ART. Using the lowest level viral load thresholds, the matrix pooling method used an average of 44 assays and the minipool method used an average of 59 assays to screen 100 study participants for virologic failure while receiving ART (Table 2). The cost of a single ultrasensitive viral load at UCSD Medical Center is $75 (USD); therefore, these methods reduce the cost to $23 (USD) per assay, that is, 70% savings (Table 2). Although labor costs associated with constituting the matrix could offset any savings, the technician time required to perform the additional viral load assays in our experiments was greater than that required to constitute the matrices. Of course, all costs will vary by clinical setting and therefore must be evaluated locally.
Although we used a particular commercially available viral load platform (Ultrasensitive Amplicor; Roche), the methods presented here are not platform specific and most likely could be translated to other viral load platforms. In resource-limited settings with diverse HIV-1 subtypes, adaptation of assays in current clinical use would allow the efficiency gains described here without sacrificing sensitivity to locally relevant subtypes. Their use in conjunction with lower cost technologies being developed for the measurement of HIV RNA could reduce the cost even further than is demonstrated here. In fact, these methods could potentially be used for a wide variety of clinical diagnostics in which measurement data are continuous. Common examples of these types of laboratory measurements include viral load measurements of hepatitis B and C viruses and cancer diagnostics such as prostate-specific antigen for prostate cancer, alpha-feta protein for hepatocellular carcinoma or carcinoembryonic antigen for colon cancer. These methods would need to be validated in clinical populations.
Although less expensive methods are needed to bring virologic monitoring to resource-limited settings, whether or not these proposed methods will be sufficient needs to be evaluated in local clinical and laboratory settings. The investigations presented here were performed in a well equipped laboratory by well trained technicians, which may not always be available in resource-limited settings. The proposed strategies may be too cumbersome or require unacceptably long turnaround times to be useful in some resource-limited clinical settings, especially if the most efficient matrix algorithms are used. To assist in the implementation of the proposed methods at remote sites, we developed a freely available web-based application, ‘Measurement Enhanced Pooling Assay Calculator’ (http://mepac.ucsd.edu) (see Supplementary methods). The application tracks and supports the implementation of the search and retest algorithm based on the viral load values of the pooled and individual samples, such that it determines which sample to test next in the algorithm and provides the relative efficiency of each experiment. Previously published simulations also provide data on how various pool sizes and platforms can be tested based on the theoretical prevalence of virologic failure in the population [24]. Additionally, other methods used in conjunction with this proposed strategy may prove even more useful, such as self-reported adherence to ART or risk scores to identify a subset of patients with a high likelihood of harboring virologic failure that could be tested separately [38,39]. Similar to the pooled NAT strategy overall, the use of adherence measures and the Measurement Enhanced Pooling Assay Calculator (MEPAC) application will need to be evaluated under local conditions, especially in resource-limited settings, to determine feasibility and cost-effectiveness.
Several factors, in addition to cost, efficiency and accuracy, must be considered before instituting a method to screen for virologic failure during ART. Turnaround time has a significant impact on the clinical management of patients. The turnaround time for viral load results was faster for the minipool than for individual testing; however, the more efficient matrix platform had the longest turnaround time. Nevertheless, on average, 66% of the samples were resolved in the first day using the matrix platform. The time required to obtain enough samples for each pooling method will impact their turnaround time. The size of the clinical population receiving ART and the frequency at which screening occurs will determine the rate of specimens available for screening [24]. For these studies, we screened our population retrospectively; so all samples were immediately available. We also chose the time point of 6 months after the start of each patient’s initial ART regimen for screening, which would most likely limit the frequency of virologic failure in this study population followed regularly with viral load monitoring. The prevalence of virologic failure in our setting was 23%, and with a higher or lower prevalence, both matrix and minipool methods can be expected to be less or more efficient, respectively [26]. The prevalence of virologic failure is likely to be high and the range of viral loads wide in resource-limited populations among patients that have not previously received virological monitoring during ART [13,14]. With regular virologic monitoring during ART, however, as with our proposed NAT methods, virologic failure can be identified and therapy changed earlier, likely reducing the prevalence of virologic failure and the progressive accumulation of antiviral drug resistance. Examples of other factors that should be considered locally when choosing a particular pooling method to screen for virologic failure during ART are presented in Table 3.
Table 3.
Considerations for choosing viral load strategy to detect virologic failure.
| Factors | Costs | Clinical considerations |
|---|---|---|
| Assay type and level of detection | Cost per assay | Accuracy |
| Inherent error of assay type | Cost per assay | Accuracy |
| Laboratory space to avoid contamination during processing | Space cost | Quality assurance |
| Personnel availability | Personnel cost | Turnaround time |
| Personnel training | Personnel cost | Quality assurance |
| Clinic population size | Cost per pool | Turnaround time |
| Rate of virologic failure | Cost per pool | Accuracy |
| Rate of screening | Cost per pool | Accuracy and turnaround time |
In summary, both of the pooling methods, matrix and minipool, demonstrated improved efficiency over individual testing to retrospectively screen a study cohort receiving ART for virologic failure, while maintaining excellent accuracy. Ultimately, these methods may prove to be a cost-effective way to monitor patients receiving ART in resource-limited settings; thereby, limiting the development and transmission of HIV drug resistance and preserving ART options for patients and communities.
Acknowledgements
This work was supported by National Institutes of Health grants MH083552, AI077304, AI69432, MH62512, AI27670, AI38858, AI43638, AI43752, AI047745, NS51132, UCSD Centers for AIDS Research Viral Pathogenesis Core (AI36214), AI29164, AI47745, AI064086, AI57167 and the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Healthcare System (10-92-035).
This study was approved by our local ethics committee. Written informed consent was obtained from all patients, and the human experimentation guidelines of the US Department of Health and Human Services and the individual institutions were followed in conducting this research.
We would like to thank Dr Robert Schooley and Dr Anthony Gamst for insightful comments and Demetrius dela Cruz for his administrative assistance.
Footnotes
Authors contributions and potential conflicts of interest: All authors have read and approved the paper, have met the criteria for authorship as established by the International Committee of Medical Journal Editors, believe that the article represents honest work and are able to verify the validity of the results reported. All authors have seen and approved the final version of the manuscript.
Author contributions: D.M.S. conceived study design, assisted in the collection, analysis and interpretation of data and in the writing of the report. S.M. assisted in study design, assisted in the analysis and interpretation of data and in the writing of the report. J.P.-S. assisted in the analysis and interpretation of data and in the writing of the report. M.C.S. assisted in study design, assisted in the analysis and interpretation of data and in the writing of the report. C.C.I. assisted in the collection, analysis and interpretation of data and in the writing of the report. R.H.H. assisted in study design, assisted in the interpretation of data and in the writing of the report. C.A.B. assisted in study design, assisted in the interpretation of data and in the writing of the report. D.D.R. assisted in study design, assisted in the collection and interpretation of data and in the writing of the report. S.J.L. assisted in study design, assisted in the collection and interpretation of data and in the writing of the report.
Possible author conflicts of interest (including financial and other relationships) for each author include the following: D.M.S. has served as a consultant and board member for Symmunity LLC. S.M. has no reported conflicts. J.P.-S. no reported conflicts. M.C.S. has served as an employee and board member of Symmunity LLC. C.C.I. has no reported conflicts. R.H.H. received speaking honoraria or consultant fees from Abbott, Ardea, Boehringer Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Merck, Pfizer, Schering, Roche, Tibotec and has received research support from Abbott, Glaxo-SmithKline, Pfizer and Tibotec. C.A.B. has consulted for GlaxoSmithKline, Merck and Pfizer, has participated in the Data Safety and Monitoring Board for Achillion and Johnson & Johnson, Ltd – Australia and has received research support from Gilead. Her spouse has consulted for GlaxoSmithKline, Vertex, Monogram Biosciences, Achillion, Gilead, Pfizer, Taimed, Inhibitex, Myriad and Tobira and has received research support from Gilead. D.D.R. has consulted for Chimerix, Geneprobe, Pfizer, Merck, Bristol Myers Squibb, Gilead, Idenix, Roche, Monogram Biosciences. S.J.L. has no reported conflicts.
References
- 1.Bagnarelli P, Menzo S, Valenza A, Paolucci S, Petroni S, Scalise G, et al. Quantitative molecular monitoring of human immunodeficiency virus type 1 activity during therapy with specific antiretroviral compounds. J Clin Microbiol. 1995;33:16–23. doi: 10.1128/jcm.33.1.16-23.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katzenstein DA, Holodniy M. HIV viral load quantification, HIV resistance, and antiretroviral therapy. AIDS Clin Rev. 1995–1996:277–303. [PubMed] [Google Scholar]
- 3.Cook J, Dasbach E, Coplan P, Markson L, Yin D, Meibohm A, et al. Modeling the long-term outcomes and costs of HIV antiretroviral therapy using HIV RNA levels: application to a clinical trial. AIDS Res Hum Retroviruses. 1999;15:499–508. doi: 10.1089/088922299311024. [DOI] [PubMed] [Google Scholar]
- 4.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 5.Richman DD. HIV chemotherapy. Nature. 2001;410:995–1001. doi: 10.1038/35073673. [DOI] [PubMed] [Google Scholar]
- 6.Deeks SG, Wrin T, Liegler T, Hoh R, Hayden M, Barbour JD, et al. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N Engl J Med. 2001;344:472–480. doi: 10.1056/NEJM200102153440702. [DOI] [PubMed] [Google Scholar]
- 7.Zolopa AR, Shafer RW, Warford A, Montoya JG, Hsu P, Katzenstein D, et al. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendavid E, Young SD, Katzenstein DA, Bayoumi AM, Sanders GD, Owens DK. Cost-effectiveness of HIV monitoring strategies in resource-limited settings: a southern African analysis. Arch Intern Med. 2008;168:1910–1918. doi: 10.1001/archinternmed.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips AN, Pillay D, Miners AH, Bennett DE, Gilks CF, Lundgren JD. Outcomes from monitoring of patients on antiretroviral therapy in resource-limited settings with viral load, CD4 cell count, or clinical observation alone: a computer simulation model. Lancet. 2008;371:1443–1451. doi: 10.1016/S0140-6736(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 10.Schooley RT. Viral load testing in resource-limited settings. Clin Infect Dis. 2007;44:139–140. doi: 10.1086/510090. [DOI] [PubMed] [Google Scholar]
- 11.Calmy A, Ford N, Hirschel B, Reynolds SJ, Lynen L, Goemaere E, et al. HIV viral load monitoring in resource-limited regions: optional or necessary? Clin Infect Dis. 2007;44:128–134. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- 12.Gilks CF, Crowley S, Ekpini R, Gove S, Perriens J, Souteyrand Y, et al. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006;368:505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 13.Mee P, Fielding KL, Charalambous S, Churchyard GJ, Grant AD. Evaluation of the WHO criteria for antiretroviral treatment failure among adults in South Africa. AIDS. 2008;22:1971–1977. doi: 10.1097/QAD.0b013e32830e4cd8. [DOI] [PubMed] [Google Scholar]
- 14.Moore DM, Awor A, Downing R, Kaplan J, Montaner JS, Hancock J, et al. CD4R T-cell count monitoring does not accurately identify HIV-infected adults with virologic failure receiving antiretroviral therapy. J Acquir Immune Defic Syndr. 2008;49:477–484. doi: 10.1097/QAI.0b013e318186eb18. [DOI] [PubMed] [Google Scholar]
- 15.Colebunders R, Moses KR, Laurence J, Shihab HM, Semitala F, Lutwama F, et al. A new model to monitor the virological efficacy of antiretroviral treatment in resource-poor countries. Lancet Infect Dis. 2006;6:53–59. doi: 10.1016/S1473-3099(05)70327-3. [DOI] [PubMed] [Google Scholar]
- 16.Petti CA, Polage CR, Quinn TC, Ronald AR, Sande MA. Laboratory medicine in Africa: a barrier to effective healthcare. Clin Infect Dis. 2006;42:377–382. doi: 10.1086/499363. [DOI] [PubMed] [Google Scholar]
- 17.Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Piyavong B, Chumpathat N, Chantratita W. Options for a second-line antiretroviral regimen for HIV type 1-infected patients whose initial regimen of a fixed-dose combination of stavudine, lamivudine, and nevirapine fails. Clin Infect Dis. 2007;44:447–452. doi: 10.1086/510745. [DOI] [PubMed] [Google Scholar]
- 18.Dorfman R. The detection of defective members of large populations. Ann Math Stat. 1943;14:436–440. [Google Scholar]
- 19.Klausner JD, Grant RM, Kent CK. Detection of acute HIV infections [with reply from Pilcher CD and Leone PA] N Engl J Med. 2005;353:631–633. doi: 10.1056/NEJM200508113530620. [DOI] [PubMed] [Google Scholar]
- 20.Pilcher CD, Eron JJ, Jr, Galvin S, Gay C, Cohen MS. Acute HIV revisited: new opportunities for treatment and prevention. J Clin Invest. 2004;113:937–945. doi: 10.1172/JCI21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilcher CD, Fiscus SA, Nguyen TQ, Foust E, Wolf L, Williams D, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352:1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
- 22.Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, Wright DJ, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 23.Westreich DJ, Hudgens MG, Fiscus SA, Pilcher CD. Optimizing screening for acute HIV infection with pooled nucleic acid amplification tests. J Clin Microbiol. 2008;46:1785–1792. doi: 10.1128/JCM.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May S, Gamst A, Haubrich R, Benson C, Smith D. Pooled nucleic acid testing to identify antiretroviral treatment failure during HIV infection. J Acquir Immune Defic Syndr Hum Retrovirol. 2009 doi: 10.1097/QAI.0b013e3181ba37a7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hecht FM, Wang L, Collier A, Little S, Markowitz M, Margolick J, et al. A multicenter observational study of the potential benefits of initiating combination antiretroviral therapy during acute HIV infection. J Infect Dis. 2006;194:725–733. doi: 10.1086/506616. [DOI] [PubMed] [Google Scholar]
- 26.May S, Gamst A, Haubrich R, Benson C, Smith D. UW Biostatistics Working Paper Series. Working Paper 343. The Berkeley Electronic Press; 2009. Pooled nucleic acid testing to identify antiretroviral treatment failure during HIV infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DM, Schooley RT. Running with scissors: using antiretroviral therapy without monitoring viral load. Clin Infect Dis. 2008;46:1598–1600. doi: 10.1086/587110. [DOI] [PubMed] [Google Scholar]
- 28.Landman R, Schiemann R, Thiam S, Vray M, Canestri A, Mboup S, et al. Once-a-day highly active antiretroviral therapy in treatment-naive HIV-1-infected adults in Senegal. AIDS. 2003;17:1017–1022. doi: 10.1097/00002030-200305020-00010. [DOI] [PubMed] [Google Scholar]
- 29.Coetzee D, Hildebrand K, Boulle A, Maartens G, Louis F, Labatala V, et al. Outcomes after two years of providing antiretroviral treatment in Khayelitsha, South Africa. AIDS. 2004;18:887–895. doi: 10.1097/00002030-200404090-00006. [DOI] [PubMed] [Google Scholar]
- 30.Smith DM, Wong JK, Shao H, Hightower GK, Mai SH, Moreno JM, et al. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J Infect Dis. 2007;196:356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 31.Kamya MR, Mayanja-Kizza H, Kambugu A, Bakeera-Kitaka S, Semitala F, Mwebaze-Songa P, et al. Predictors of long-term viral failure among Ugandan children and adults treated with antiretroviral therapy. J Acquir Immune Defic Syndr. 2007;46:187–193. doi: 10.1097/QAI.0b013e31814278c0. [DOI] [PubMed] [Google Scholar]
- 32.Ashton M, Sopwith W, Clark P, McKelvey D, Lighton L, Mandal D. An outbreak no longer: factors contributing to the return of syphilis in Greater Manchester. Sex Transm Infect. 2003;79:291–293. doi: 10.1136/sti.79.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koenig SP, Kuritzkes DR, Hirsch MS, Leandre F, Mukherjee JS, Farmer PE, del Rio C. Monitoring HIV treatment in developing countries. BMJ. 2006;332:602–604. doi: 10.1136/bmj.332.7541.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Cock KM. Prevention of mother-to-child HIV transmission in resource-poor countries: translating research into policy and practice. JAMA. 2000;283:1175–1182. doi: 10.1001/jama.283.9.1175. [DOI] [PubMed] [Google Scholar]
- 35.Moore DM, Mermin J, Awor A, Yip B, Hogg RS, Montaner JS. Performance of immunologic responses in predicting viral load suppression: implications for monitoring patients in resource-limited settings. J Acquir Immune Defic Syndr. 2006;43:436–439. doi: 10.1097/01.qai.0000243105.80393.42. [DOI] [PubMed] [Google Scholar]
- 36.Bagchi S, Kempf MC, Westfall AO, Maherya A, Willig J, Saag MS. Can routine clinical markers be used longitudinally to monitor antiretroviral therapy success in resource-limited settings? Clin Infect Dis. 2007;44:135–138. doi: 10.1086/510072. [DOI] [PubMed] [Google Scholar]
- 37.Dunne AL, Mitchell FM, Coberly SK, Hellmann NS, Hoy J, Mijch A, et al. Comparison of genotyping and phenotyping methods for determining susceptibility of HIV-1 to antiretroviral drugs 11. AIDS. 2001;15:1471–1475. doi: 10.1097/00002030-200108170-00003. [DOI] [PubMed] [Google Scholar]
- 38.Powers KA, Miller WC, Pilcher CD, Mapanje C, Martinson FE, Fiscus SA, et al. Improved detection of acute HIV-1 infection in sub-Saharan Africa: development of a risk score algorithm. AIDS. 2007;21:2237–2242. doi: 10.1097/QAD.0b013e3282f08b4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bangsberg DR. Preventing HIV antiretroviral resistance through better monitoring of treatment adherence. J Infect Dis. 2008;197 Suppl 3:S272–S278. doi: 10.1086/533415. [DOI] [PubMed] [Google Scholar]