Abstract
The synthesis of oligomers containing two or three acridine units linked through 2-aminoethylglycine using solid-phase methodology is described. Subsequent studies on cell viability showed that these compounds are not cytotoxic. Binding to several DNA structures was studied by competitive dialysis, which showed a clear affinity for DNA sequences that form G-quadruplexes and parallel triplexes. The fluorescence spectra of acridine oligomers were affected strongly upon binding to DNA. These spectral changes were used to calculate the binding constants (K). Log K were found to be in the order of 4–6.
1. Introduction
Small organic molecules with specific interactions with DNA have become antitumor, antiviral, and antibiotic drugs [1, 2]. Duplex DNA-binding drugs interact in two main ways, through groove binding and through intercalation. Medicinal chemistry has made a considerable effort in searching for and testing of a large number of drugs with increased selectivity to a range of DNA sequences or structures. More recently, some of this interest has moved to the search of new ligands for G-quadruplexes [3]. This structure motif is formed by the planar association of four guanines in a cyclic Hoogsteen hydrogen bonding tetrad.
Guanine-rich sequences form G-quadruplex structures and have been found in telomeres [4] and in transcriptional regulatory regions of critical oncogenes such as c-myc and c-kit [5, 6]. Ligands that selectively bind and stabilize these structures have become anticancer drugs of interest [7].
The G-quadruplex stabilization occurs in most cases by π-π stacking and electrostatic interaction. G-quadruplex ligands are normally planar aromatic molecules that are prone to stacking with G-tetrads. Some of them are also positively charged or have hydrophilic groups to favor electrostatic interaction [8].
Although there is a long way to go in the development of potent drugs that target G-quadruplexes, some promising lead compounds have been achieved [9]. Several ligand structures have been studied, such as anthraquinones, cationic porphyrins, perylene derivatives, and a large number of compounds [9]. Among the acridine compounds, 3,6,9-trisubstituted acridines have inhibitory activity in the nanomolar range and they have entered preclinical studies [8, 10, 11].
In previous studies we described the preparation of sequence specific oligomers of DNA-intercalating drugs using protocols based on solid-phase synthesis in an attempt to facilitate the preparation of compounds with improved DNA-binding selectivity [12, 13]. It has been proposed that bis- and tris-intercalating drugs show promising activity and selectivity [14, 15]. Here we described solid-phase synthesis protocols for the preparation of several acridine oligomers linked through 2-aminoethylglycine units as well as their DNA-binding properties. Although the acridine derivatives described in this study are not cytotoxic, they show a clear affinity for several DNA G-quadruplex structures, especially those sequences found in the promoter regions of c-myc [16] and bcl-2 [17, 18] oncogenes.
2. Materials and Methods
2.1. Chemicals
The phosphoramidites and ancillary reagents used during oligonucleotide synthesis were obtained from Applied Biosystems (USA) and Link Technologies Ltd. (Scotland). The rest of the chemicals were purchased from commercial sources. The Slide-A-Lyzer Mini Dialysis Units 3500 MWCO were purchased from Pierce. Acridine-9-carboxylic acid was obtained from Aldrich. 2-(Acridine-9-carboxamide)acetic acid was prepared by reaction of acridine-9-carboxylic acid with glycine methyl ester and subsequent saponification of the methyl ester as described [10]. Boc-(2-aminoethyl)glycine(Fmoc) (Boc-Aeg(Fmoc)-OH) was obtained from Iris Biotech and Fmoc-glycine (Fmoc-Gly-OH) was obtained from Bachem. Boc-6-aminohexyl hemisuccinate was prepared by reaction of Boc-6-aminohexanol with succinic anhydride.
2.2. Oligonucleotide Synthesis
Oligonucleotide sequences (Table 1) were prepared on an automatic Applied Biosystems 3400 DNA synthesizer on 1 μmoL (CPG resin) scale using commercially available 2-cyanoethyl phosphoromidites. After the assembly of the sequences, oligonucleotide-supports were deprotected using 32% aqueous ammonia at 55°C for 16 h. Ammonia solutions were concentrated to dryness and the residue was desalted by a NAP-10 (Sephadex G-25) column.
Table 1.
Sequences of oligonucleotides.
| No. | Name | Sequence (5′–3′) |
|---|---|---|
| 1 | T20 | TTT TTT TTT TTT TTT TTT TT |
| 2 | 24bclc | CCC GCC CCC TTC CTC CCG CGC CCG |
| 3 | Dickerson | CGC GAA TTC GCG |
| 4 | Ds26 | CAA TCG GAT CGA ATT CGA TCC GAT TG |
| 5 | GA triplex | GAA AGA GAG GAG GCC TTT TTG GAG GAG AGA AAG + CCT CCT CTC TTT C |
| 6 | TC triplex | CCT CCT CTC TTT CCC TTT TTC TTT CTC TCC TCC + GAA AGA GAG GAG G |
| 7 | TG4T | TGG GGT |
| 8 | TBA | GGT TGG TGT GGT TGG |
| 9 | HT24 | TAG GGT TAG GGT TAG GGT TAG GGT |
| 10 | 24bcl | CGG GCG CGG GAG GAA GGG GGC GGG |
| 11 | cmyc | GGG GAG GGT GGG GAG GGT GGG GAA GGT GGG G |
2.3. Solid-Phase Synthesis of Acridine Oligomers
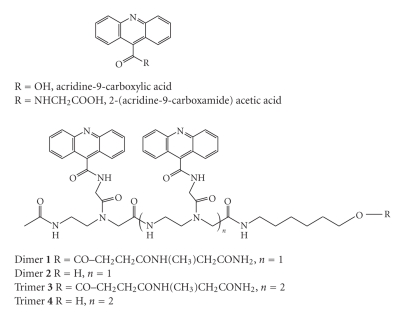
Acridine dimers and trimers (1–4, Figure 1) were prepared with the 2-aminoethylglycine scaffold, which allows the growth of a polyamide skeleton on solid-phase and the following incorporation of acridine unit.
Figure 1.
Structure of the acridine derivatives prepared.
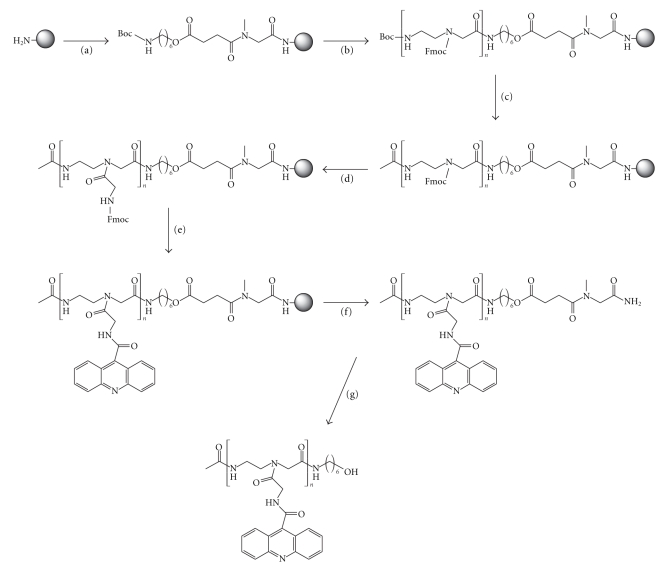
The assembly of 2-aminoethylglycine derivatives was carried out on methylbenzhydrylamine (MBHA) polystyrene-1%-divinylbenzene solid support applying an Fmoc/Boc hybrid strategy using Boc-Aeg(Fmoc)-OH, Fmoc-Gly-OH, and acridine-9-carboxylic acid as building blocks (Figure 2).
Figure 2.
Solid-phase synthesis of dimer and trimer acridine derivatives. (a) (i) Fmoc-Sar-OH, PyBOP, DIEA; (ii) 20% piperidine, DMF; (iii) Boc–NH–(CH2)6–OCH2CH2COOH, PyBOP, DIEA; (b) (i) 40% TFA, DCM; (ii) Fmoc-Aeg(Boc)-OH, PyBOP, DIEA; (iii) Repeat steps (i) and (ii) n times; (c) (i) 40% TFA, DCM; (ii) Ac2O, DIEA, DMF; (d) (i) 20% piperidine, DMF; (ii) Fmoc-Gly-OH, PyBOP, DIEA; (e) (i) 20% piperidine, DMF; (ii) acridine-9-carboxylic acid, PyBOP, DIEA; (f) anhydrous HF (0°); (g) 32% aqueous NH3.
Fmoc-Sarcosine-OH (5 eq) was coupled to the resin using standard coupling conditions (5 eq. PyBOP and 10 eq. DIEA, 1 h), then the Fmoc group was removed (20% of piperidine in DMF, 30 min), and Boc-6-aminohexyl hemisuccinate (Boc–NH–(CH2)6–OCOCH2CH2COOH) was coupled (5 eq R-COOH, 5 eq. PyBOP and 10 eq. DIEA, 1 h) to the support. The residual unreacted amino groups were acetylated with 5 eq. of acetic anhydride and 5 eq. of DIEA.
Next, the Boc group was removed (40% trifluoroacetic acid in dichloromethane) and the 2-aminoethylglycine skeleton was synthesized by repetitive couplings of Boc-Aeg(Fmoc)-OH until reaching the desired dimer or trimer compound. The last Boc group of the sequence was removed and the resulting amino group was acetylated (5 eq. acetic anhydride and 5 eq. DIEA).
Once the aminoethylglycine backbone was built, the Fmoc-protecting groups of the side chains were removed (20% of piperidine in DMF, 30 min), and Fmoc-Gly-OH followed by 9-acridine carboxyl acid was coupled to the support. The progress of the coupling reactions was followed by ninhydrine test and by UV monitoring of the 9-methylene-9H-fluorene released during deprotection, which allowed optimization of the coupling conditions.
The acridine dimer 1 and trimer 3 were obtained by treatment of the appropriate solid supports with HF anhydrous at 0°C. Finally the acridine dimer 2 and trimer 4 were obtained by treatment of dimer 1 and trimer 3 respectively with 32% aqueous ammonia (1 h, 55°C).
Good yields and purities were obtained for the products (around 85% for 1 and 2, 75% for 3 and 4). HPLC and MALDI-TOF spectra are shown in Supplementary Material available onlie at doi: 10.4061/2010/489060.
The compounds were analyzed by MALDI-TOF, 1 [M+Na+] = 1080.3 (expected 1054.2), 2 [M] = 885.2 (expected 884.0), 3 [M] = 1418.9 (expected 1416.5), and 4 [M] = 1247.4 (expected 1246.4). MALDI-TOF spectra were obtained using a Perseptive Voyager DETMRP mass spectrometer, equipped with nitrogen laser at 337 nm using a 3 ns pulse. The matrix used contained 2,5-dihydroxybenzoic acid (DHB, 10 mg/mL in water).
Analytical HPLC was performed using XBridge OST C18 (Waters), 2.5 μm, 4.6 × 50 mm column using a 10-minute linear gradient from 9% to 45% B, flow rate 1 mL/min; solution A was 5% ACN in 0,1 M aqueous TEAA, and B 70% ACN in 0.1M aqueous TEAA. HPLC chromatograms and MS spectra can be found in Supplementary Material.
Fluorescence spectra were recorded using a Jasco FP-6200 spectrofluorometer equipped with a Peltier temperature controller, λ em = 435 nm (λ exc = 252 nm).
UV spectra were recorded using a Jasco spectrophotometer V-650, λ max = 252 nm, 360 nm.
2.4. Cell Viability Assays
The in vitro cytotoxicity of the compounds (Figure 1) was evaluated by colorimetric assays with tetrazole salts (MTT) on Jurkat clone E6-1 (human leukemia), GLC-4 clone (human lung carcinoma) cell lines, and one mouse fibroblast cell line (NIHT-3T3).
GLC4 and Jurkat cell lines were cultured in RPMI and NIH3T3 in DMEM and supplemented with 10% fetal calf serum, 10000 u/mL penicillin, 10 μg/mL streptomycin and 200 mM L-glutamine. Cells were grown in a humidified atmosphere of air containing 5% CO2 at 37°. Cells were plated in triplicate wells (1.5·104 cells well) in 100 μL of growth medium in 96-well plates and proliferate for 24 h and then treated with increasing concentrations of acridine oligomers. After 72 h of incubation, 10 μM of MTT (5 mg/mL in Phosphate buffer saline 10%) was added for an additional 4 h. The absorbance at 570 nm was measured on a multiwell plate reader after addition of 100 μl of isopropanol : 1N HCl (24 : 1). Cell viability was expressed as a percentage of control and IC50 was determined as the concentration of drug that produced a 50% reduction of absorbance at 570 nm.
2.5. Competitive Dialysis Assays
100 μL of a 50 μM oligonucleotide (Table 1) in potassium phosphate buffer (185 mM NaCl, 185 mM KCl, 2 mM NaH2PO4, 1 mM Na2EDTA, 6 mM Na2HPO4 at pH 7) was introduced into a separated dialysis unit. A blank sample containing only buffer without oligonucleotide was also prepared. All 12 dialysis units were then placed in the beaker containing the 1 μM solution of the appropriate acridine derivative. The samples were allowed to equilibrate with continuous stirring at room temperature overnight. After the equilibration period, DNA samples were removed to an Eppendorf tube. SDS is usually added to denature the DNA sample and release the acridine oligomer, but in our case the presence of K+ ions induced the formation of a white precipitate, which interfered with the measurement of the fluorescence spectra of the samples.
In order to measure the compound retained in the dialysis unit, samples were treated with snake-venom phosphodiesterase to degrade the DNA and release the acridine oligomer. 350 μL of potassium phosphate buffer (without EDTA), buffer at pH 8.5, 50 μL of 100 mM MgCl2, and 1 μm of snake venom phosphodiesterase solution were added for an additional overnight incubation at 37°C. Finally, the fluorescence of each sample was measured (λ ex and λ em were set to 252 and 435 nm, resp.).
2.6. Fluorescence Assays
The study of the interaction equilibrium of acridine derivatives and oligonucleotides consists of recording the fluorescence spectra of a 0.2 μM solution of the acridine derivative after the addition of increasing amounts of oligonucleotide (from 0 to 10 μM) in potassium phosphate buffer (185 mM NaCl, 185 mM KCl, sodium phosphate, 1 mM EDTA, pH 7). These experiments were carried out by adding small volumes of an oligonucleotide stock solution to the 0.2 μM solution of the acridine derivative. After 24 h the emission spectra of the resulting solutions were recorded from 300 to 500 nm at 252 nm excitation wavelength at 25°C.
The macroscopic binding constant (K) corresponding to the reaction
| (1) |
was calculated from the multivariate analysis of fluorescence data recorded in the range 300–390 nm using the hard-modeling program Equispec [19]. This program performs a nonlinear least squares optimization of K and of the pure fluorescence spectra corresponding to each of the species considered (DNA, ligand, and interaction complex). A 1 : 1 stoichiometry DNA : ligand for the interaction complex was assumed. The logarithms of the binding constants calculated are given as their weighted means with twice their standard errors (units of the least significant digit). Results are shown in Table 2.
Table 2.
Logarithm of the binding constants (log K) calculated from data recorded throughout fluorescence titrations using Equispec program assuming a 1 : 1 stoichiometry DNA : ligand for the interaction complex (details in materials and methods). Compounds 1–4 correspond to the acridine dimers and trimers prepared in this study. HT24, 24bcl, cmyc, and Dickerson correspond to oligonucleotide sequences shown in Table 1.
| 1 | 2 | 3 | 4 | |
|---|---|---|---|---|
| HT24 | 4.9 ± 0.4 | 4.6 ± 0.5 | 5.3 ± 0.3 | n.d. |
| 24bcl | 6.8 ± 0.5 | 5.1 ± 0.1 | 5.3 ± 0.2 | n.d. |
| cmyc | 5.5 ± 0.3 | 4.9 ± 0.2 | 4.8 ± 0.5 | 5.3 ± 0.2 |
| Dickerson | n.d. | n.d. | n.d. | n.d. |
n.d. not determined.
2.7. Circular Dichroism
An increasing amount of 1 (from 0 to 8 μM) in potassium phosphate buffer (185 mM NaCl, 185 mM KCl, sodium phosphate, 1 mM EDTA, pH 7) was added to a 1 μM solution of the oligonucleotide. The CD spectra were recorded after 24 h on a Jasco J-810 spectropolarimeter attached to a Julabo F/25HD circulating water bath in 1 cm path-length quartz cylindrical cells, using a 50 nm/min scan rate, a spectral band width of 1 nm, and a time constant of 4 s. All the spectra were corrected with the buffer blank, normalized to facilitate comparisons and noise-reduced using Matlab software. CD spectra are shown as supplementary data.
3. Results and Discussion
3.1. Synthesis of Acridine Oligomers
The synthesis of acridine dimer and trimers has been described previously [13] using a Boc-(2-aminoethyl)glycine derivative carrying the 2-(acridine-9-carboxamide)acetyl residue. The synthesis of this monomer was long and yields were low.
An alternative method developed is to first assemble the Boc-(2-aminoethyl)glycine backbone on solid-phase, and then the intercalating agent is added. This strategy is more convenient for rapid synthesis, as it is unnecessary to construct each intercalating monomer.
Thus, acridine oligomers were assembled using the methylbenzhydrylamine (MBHA) resin by applying an Fmoc/Boc hybrid strategy and using Boc-(2-aminoethyl)-(Fmoc)glycine [Boc-Aeg(Fmoc)-OH] as building block (Figure 2). The Boc group was used to protect the aminoethyl group of each unit, which thus facilitated elongation of the backbone. Fmoc was the semipermanent protecting group for the amino group of glycine through which the intercalating compound was introduced. The succinyl linker was selected to connect the solid support and the oligomer. This linker is used in oligonucleotide synthesis and it is labile to ammonia. Unfortunately, the linker is not compatible with Fmoc chemistry. It has been described that an intramolecular side reaction can lead to premature loss of the oligomers during the base treatment used to remove the Fmoc group [20]. Thus, N-methylglycine (sarcosine) was incorporated between the amino-support and the succinyl linker. The presence of the N-methyl group prevents the potential side reaction [20]. 6-Aminohexanol was used to connect the succinyl linker and the oligomer backbone, as described for the synthesis of peptide nucleic acid (PNA) oligomers [21].
The (2-aminoethyl)glycine backbone was assembled by consecutive additions of Boc-Aeg(Fmoc)-OH to obtain the dimer and trimer sequence. Acetylation of the N-terminal position was carried out using acetic anhydride and a base.
Next, the removal of the Fmoc group allowed the addition of a Fmoc-glycine unit as a spacer, which was followed by the addition of acridine-9-carboxylic acid. The acridine dimer and trimer were synthesized in this way (Figure 2).
After assembly of the oligomers, the resulting supports were treated with ammonia. The desired oligomers were not released from the support even after prolonged time and high temperatures. We therefore treated the supports with anhydrous HF to yield the acridine oligomers 1 and 3, which contain the sarcosyl succinyl linker. At this point HPLC spectra showed a major peak that had the expected molecular weight for the oligomers 1 and 3 carrying the sarcosyl succinyl linker (see supplementary data). This observation indicates that the simultaneous incorporation of all acridines proceeded with excellent yields. Ammonia treatment of acridines 1 and 3 in solution now yielded the desired acridine dimer 2 and trimer 4 in excellent yields and purity (see supplementary data). Compounds 1–4 were fully characterized and their properties were analyzed.
The pKa of the acridine ring of acridine-9-carboxylic acid and 2-(acridine-9-carboxamide)acetic acid was measured by UV titration. pKa of acridine-9-carboxylic acid was 5.5 ± 0.1 and 2-(acridine-9-carboxamide)acetic acid 4.1 ± 0.2. We therefore estimated that the acridine rings of compounds 1–4 are mainly unprotonated at pH 7.0.
3.2. Cell Viability Assay
The in vitro cytotoxicity of the compounds was evaluated by colorimetric assays with tetrazole salts (MTT). This assay is based on the capacity of living cells to incorporate and reduce MTT. This reaction can be followed by the change of absorbance of the reduced and oxidized forms. This reaction is done by the action of the mitochondrial enzyme succinatehydrogenase, which is active only in living cells. The intensity of color is directly correlated with the number of living cells in the sample. No cytotoxicity activity was observed in compounds 1–4 at concentrations up to 50 μM.
3.3. Competitive Dialysis Studies
In order to evaluate the selectivity of the compounds for DNA structures, a competitive dialysis experiment was performed using 11 oligonucleotides (Table 1) representing several nucleic acid structures [22]. The more acridine accumulated in the dialysis unit indicates a higher binding affinity to the oligonucleotide present in the dialysis unit. As model for single stranded structures we used T20 and the C-rich complementary strand of bcl-2 (24bclc). This last oligonucleotide folds in an i-form quadruplex structure at acidic pH but has no structure in the conditions used in the dialysis (pH 7) [18]. As duplexes we used the self-complementary sequences Dickerson-Drew dodecamer (Dickerson) and a 26 mer (ds26). A parallel triplex (TC triplex) and an antiparallel triplex (GA triplex) were also prepared by mixing a hairpin Watson-Crick sequence and the corresponding triplex-forming sequence. Finally, the following G-quadruplex sequences were prepared: the tetramolecular parallel G-quadruplex TG4T [23], the antiparallel thrombin-binding aptamer (TBA) [24], the human telomere sequence (HT24) [25], and the promoter sequences of c-myc (cmyc) [16] and bcl-2 (24bcl) [17, 18].
We used the protocol described by Ren and Chaires [26] with a few modifications. The buffer solution was similar to that described in [26] but K+ was included (185 mM) to ensure the formation of the most stable G-quadruplex structures. At the end of the dialysis experiment, the amount of acridine derivative bound to the DNA was analyzed by fluorescence measurement of the acridine compound. After dialysis, we observed that the spectra of the acridine derivatives of some samples (especially those from G-quadruplexes) differed greatly from the fluorescence spectra of the initial compounds. This difference is attributed to the interaction of the acridine derivatives with the G-quadruplex. In order to release the acridine oligomer, the addition of SDS is recommended [26, 27]. Using SDS, the presence of K+ ions resulted in the formation of a white precipitate with SDS which did not allow the measurements of the fluorescence spectra [27]. In order to solve this problem, the oligonucleotide was digested with snake venom phosphodiesterase at the end of the dialysis experiment. Thus, the fluorescence spectra of the acridine derivatives were recorded with high accuracy without the interference of DNA and without the use of SDS.
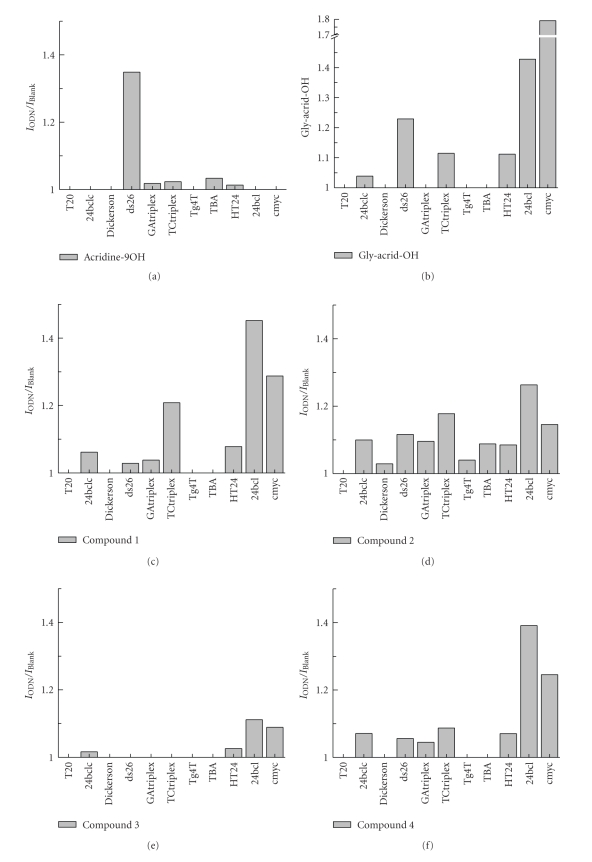
We measured the binding preferences of compounds 1–4 and the acridine monomers acridine-9-carboxylic acid and 2-(acridine-9-carboxamide)acetic acid (Figure 1). Acridine-9-carboxylic acid showed affinity only for duplex ds26 (Figure 3). Unexpectedly, the addition of the glycine residue, used as spacer, induced a change in the affinity. 2-(Acridine-9-carboxamide)acetic acid showed the highest affinity for the G-quadruplex sequences cmyc and 24bcl and less affinity for duplex ds26.
Figure 3.
Results obtained by the competitive dialysis assay. The amount of ligand bound to each DNA structure is shown as a bar graph. The fluorescence of each sample was measured using an excitation wavelength of 252 nm and an emission wavelength of 435 nm, respectively.
Dimer 1 (dimer with the sarcosylsuccinyl linker) has a similar profile as 2-(acridine-9-carboxamide)acetic acid. In this case, the G-quadruplex 24bcl is preferred to the cmyc sequence. Some affinity for the TC triplex is observed but no affinity for duplex ds26. Surprisingly, dimer 2 (without the sarcosylsuccinyl linker) lost most of the selectivity although some residual higher affinity for G-quadruplex 24bcl was observed.
In contrast, trimer 3 (trimer with the sarcosylsuccinyl linker) presented lower binding affinity than the other compounds. However, trimer 4 (without the sarcosylsuccinyl linker) recovered most of the affinity for 24bcl and cmyc showing a similar profile to that observed for dimer 1.
3.4. Measurement of G-Quadruplex-Affinity Constants by Fluorescence Spectroscopy
Dialysis experiments suggest that some of the acridine derivatives prepared have special affinity for 24bcl and cmyc G-quadruplexes. In order to confirm this observation, binding constants were estimated using mole-ratio experiments monitored with fluorescence spectroscopy. Hence, increasing amounts of oligonucleotides 24bcl, HT24, cmyc, and Dickerson were added to a solution with a fixed concentration of the acridine derivatives, and the fluorescence spectra were recorded at excitation wavelengths 252 and 360 nm.
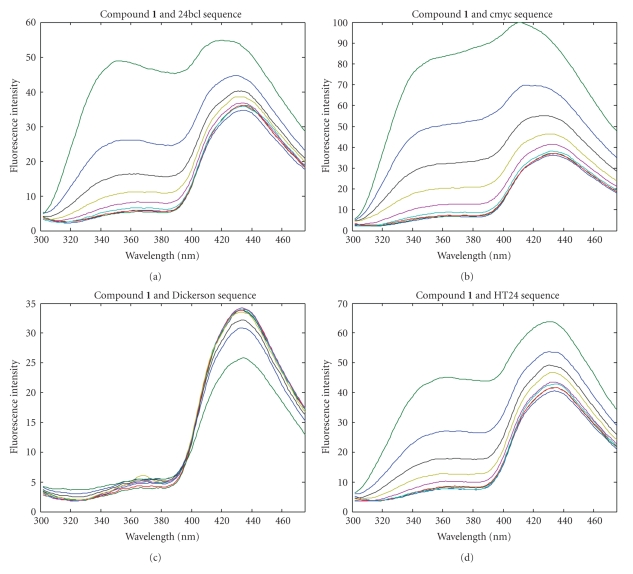
At both wavelengths, changes in the fluorescence spectra upon the addition of oligonucleotides were observed. Figure 4 shows the changes in the fluorescence spectra of acridine dimer 1 at excitation wavelengths 252 nm when oligonucleotides 24bcl, HT24, cmyc, and Dickerson were added. A dramatic increase in fluorescence intensity was observed around 360 nm upon addition of G-quadruplex DNA sequences (24bcl, HT24, and cmyc). The greatest changes were seen with 24bcl and cmyc. Similar results were found with compounds 2, 3, and 4 as well as 2-(acridine-9-carboxamide)acetic acid (see supplementary data). Interestingly, when the Dickerson dodecamer was added, no changes in the fluorescence spectra were detected. The progressive modification of the fluorescence spectrum of these compounds reflects their interaction with the G-quadruplex.
Figure 4.
Fluorescence titration spectra. Fluorescence spectra of a 0.2 μM solution of the acridine derivative after the addition of increasing amounts of oligonucleotide (from 0 to 10 μM) in potassium phosphate buffer. Excitation wavelength is 252 nm.
Fluorescence data obtained at an excitation wavelength of 252 nm were analyzed with the hard-modelling EQUISPEC program in order to calculate the corresponding binding constants (Table 2). The values of the logarithm of the binding constant (log K) obtained lie in the range 4–6, suggesting a weak interaction with DNA. Of all the compounds, dimer 1 showed the highest binding constants, thereby suggesting a stronger interaction with DNA than the other compounds studied. However these values were slightly lower than the binding constants calculated for other similar ligands, such as the acridine monomers BRACO-19 (log K = 7.4) and BSU6048 (log K = 6.5) [28] or a hemicyanine-peptide ligand (log K = 7.1 [29]), when interacting with human telomere quadruplex.
Changes in the fluorescence spectra at an excitation wavelength of 360 nm were also recorded. Fluorescent emission at this wavelength 360 nm was much lower than that recorded at 252 nm; so the fluorescent signal was low. Upon addition of the oligonucleotide to a solution of compounds 1–4, the formation of a new maximum at 442 nm was observed (see supplementary data). Although the fluorescent intensity was low, we could estimate the binding constant of the stronger interactions of compound 1 with 24bcl (7.2 ± 0.4) and cmyc (5.5 ± 0.2) sequences (see supplementary data). These values are in agreement with those recorded at an excitation wavelength of 252 nm (Table 1).
Finally, CD spectra of the DNA : ligand mixtures showed no significant differences in relation to those of DNA (see supplementary data). This observation suggests that the DNA G-quadruplex structure is not altered significantly upon binding of the acridine derivatives.
4. Conclusions
In summary, here we have described a new optimized protocol for the synthesis of acridine oligomers with a (2-aminoethyl)glycine backbone. In this method, the Boc-(2-aminoethyl)glycine backbone is first assembled on solid-phase, and then the intercalating agent is assembled on the backbone. This strategy is faster and more efficient than the one described previously [13] and yields the desired oligomers with good yields. A succinyl linker was used to connect the oligomers to the solid support. The succinyl linker attached to sarcosine was unexpectedly too stable and oligomers could not be directly released from the support by a single ammonia treatment. Instead a two-step protocol was used obtaining the desired compounds and an intermediate oligomer carrying a long succinyl sarcosine chain at the C-terminal position.
Competitive dialysis experiments have shown differences on the affinity of acridine oligomers to G-quadruplexes. Higher affinities are found in G-quadruplex sequences present on the promoter regions of c-myc and bcl-2 oncogenes. This affinity is modulated by the number of acridines and the presence of the succinyl sarcosine chain at the C-terminal position, dimer 1 and trimer 4 being the more relevant compounds for G-quadruplex binding. The monomer 2-(acridine-9-carboxamide)acetic acid also shows binding properties of interest and it is the simplest compound to be prepared. Unfortunately, the compounds synthesized in this study did not have antiproliferative activity in spite of their affinity to quadruplex. This observation contrasts with other reported quadruplex-binding acridine derivatives, such as BRACO-19 [11, 30] which shows anticancer activity. The lower affinity to telomere G-quadruplex sequence and the larger size of the acridine derivatives described in the present study may hinder cellular uptake and may explain the absence of antiproliferative activity.
The acridine nucleus is described to interact to G-quadruplex. Depending on the substituents, the acridine nitrogen can be charged when bound to DNA, and with the ring stacked on a G-tetrad, the charge will occupy a position similar to that of the potassium cation that stabilizes the G-quadruplex [28]. The introduction of protonable side chains on the acridine ligand enhances binding by electrostatic interactions [28]. In our case the acridines had no protonable substituents and the acridine nitrogen was not charged when bound to DNA. For this reason, the affinity of oligomeric acridines to G-quadruplexes is due to the multimeric nature of the compounds as well as the addition of a glycine to acridine-9-carboxylic acid. An interesting possibility for future development is the introduction of protonable sites at the oligomeric acridines, which may increase solubility in water, affinity to target, and cellular uptake. The method described here will contribute to accelerating the preparation of potential active oligomeric compounds.
Supplementary Material
Supplementary data contains HPLC profiles and MALDI-TOF spectra of compounds 1- 4; fluorescent titrations spectra of compounds 1-4 with oligonucleotides; acid-base titration of acridine derivatives and circular dichroism of cmyc sequence with compound 1.
Acknowledgments
This study was supported by the Dirección General de Investigación Científica y Técnica (Grant BFU2007-63287) and the Generalitat de Catalunya (2009/SGR/208). R. Ferreira acknowledges a predoctoral fellowship from the Spanish Ministry of Science.
Abbreviations
- ACN:
Acetonitrile
- Ac2O:
acetic anhydride
- Aeg:
(2-aminoethyl)glycine
- Boc:
t-butoxycarbonyl
- CPG:
controlled pore glass
- DIEA:
diisopropylethylamine
- DCM:
dichloromethane
- DMEM:
Dulbecco's Modified Eagle's Medium
- DMF:
N,N-dimethylformamide
- EDTA:
Ethylenediaminetetraacetic acid
- Fmoc:
(9-fluorenyl)methoxycarbonyl
- MALDI-TOF:
Matrix-assisted laser desorption ionization time-of-flight
- MBHA:
methylbenzhydrylamine
- MTT:
Thiazolyl blue tetrazolium bromide
- PNA:
peptide nucleic acid
- PyBOP:
(benzotriazol-1-yloxy) trispyrrolidino-phosphonium hexafluorophosphate
- Sar:
sarcosine
- SDS:
sodium dodecasulphate
- TEAA:
Triethylammonium acetate
- TFA:
trifluoroacetic acid
- UV:
Ultraviolet.
References
- 1.Palchaudhuri R, Hergenrother PJ. DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action. Current Opinion in Biotechnology. 2007;18(6):497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Neto BAD, Lapis AAM. Recent developments in the chemistry of deoxyribonucleic acid (DNA) intercalators: principles, design, synthesis, applications and trends. Molecules. 2009;14(5):1725–1746. doi: 10.3390/molecules14051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Research. 2007;35(22):7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 5.Huppert JL. Hunting G-quadruplexes. Biochimie. 2008;90(8):1140–1148. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochemical Society Transactions. 2001;29(6):692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 7.Han H, Hurley LH. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends in Pharmacological Sciences. 2000;21(4):136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 8.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. Journal of the American Chemical Society. 2008;130(21):6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 9.Monchaud D, Teulade-Fichou M-P. A hitchhiker’s guide to G-quadruplex ligands. Organic and Biomolecular Chemistry. 2008;6(4):627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 10.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. Journal of Molecular Biology. 2003;326(1):117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 11.Moore MJB, Schultes CM, Cuesta J, et al. Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3,6- and 9-side chains on quadruplex binding, telomerase activity, and cell proliferation. Journal of Medicinal Chemistry. 2006;49(2):582–599. doi: 10.1021/jm050555a. [DOI] [PubMed] [Google Scholar]
- 12.Aviñó A, Navarro I, Farrera-Sinfreu J, et al. Solid-phase synthesis of oligomers carrying several chromophore units linked by phosphodiester backbones. Bioorganic and Medicinal Chemistry Letters. 2008;18(7):2306–2310. doi: 10.1016/j.bmcl.2008.02.074. [DOI] [PubMed] [Google Scholar]
- 13.Farrera-Sinfreu J, Aviñó A, Navarro I, et al. Design, synthesis and antiproliferative properties of oligomers with chromophore units linked by amide backbones. Bioorganic and Medicinal Chemistry Letters. 2008;18(7):2440–2444. doi: 10.1016/j.bmcl.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 14.Arya DP, Willis B. Reaching into the major groove of B-DNA: synthesis and nucleic acid binding of a neomycin-Hoechst 33258 conjugate. Journal of the American Chemical Society. 2003;125(41):12398–12399. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 15.Fechter EJ, Olenyuk B, Dervan PB. Design of a sequence-specific DNA bisintercalator. Angewandte Chemie—International Edition. 2004;43(27):3591–3594. doi: 10.1002/anie.200454231. [DOI] [PubMed] [Google Scholar]
- 16.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai J, Dexheimer TS, Chen D, et al. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. Journal of the American Chemical Society. 2006;128(4):1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.del Toro M, Bucek P, Aviñó A, et al. Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie. 2009;91(7):894–902. doi: 10.1016/j.biochi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Dyson RM, Kaderli S, Lawrance GA, Maeder M. Second order global analysis: the evaluation of series of spectrophotometric titrations for improved determination of equilibrium constants. Analytica Chimica Acta. 1997;353(2-3):381–393. [Google Scholar]
- 20.Stengele K-P, Pfleiderer W. Improved synthesis of oligodeoxyribonucleotides. Tetrahedron Letters. 1990;31(18):2549–2552. [Google Scholar]
- 21.Will DW, Breipohl G, Langner D, Knolle J, Uhlmann E. The synthesis of polyamide nucleic acids using a novel monomethoxytrityl protecting-group strategy. Tetrahedron. 1995;51(44):12069–12082. [Google Scholar]
- 22.Granotier C, Pennarun G, Riou L, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Research. 2005;33(13):4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gros J, Rosu F, Amrane S, et al. Guanines are a quartet’s best friend: iImpact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Research. 2007;35(9):3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38(49):16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 27.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43(4):313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White EW, Tanious F, Ismail MA, et al. Structure-specific recognition of quadruplex DNA by organic cations: influence of shape, substituents and charge. Biophysical Chemistry. 2007;126(1–3):140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Green JJ, Ladame S, Ying L, Klenerman D, Balasubramanian S. Investigating a quadruplex—ligand interaction by unfolding kinetics. Journal of the American Chemical Society. 2006;128(30):9809–9812. doi: 10.1021/ja0615425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burger AM, Dai F, Schultes CM, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Research. 2005;65(4):1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data contains HPLC profiles and MALDI-TOF spectra of compounds 1- 4; fluorescent titrations spectra of compounds 1-4 with oligonucleotides; acid-base titration of acridine derivatives and circular dichroism of cmyc sequence with compound 1.