Abstract
Objective
Scleroderma is a complex connective tissue disorder characterized by hardening and thickening of skin. One hallmark of scleroderma is excessive accumulation of collagen accompanied by increased levels of pyridinoline collagen cross-links derived from hydroxylysine residues in the collagen telopeptide domains. Lysyl hydroxylase 2 (LH2), an important alternately-spliced enzyme in collagen biosynthesis, acts as a collagen telopeptide hydroxylase. Changes in the pattern of LH2 alternative splicing, favoring increased inclusion of the alternatively-spliced LH2 exon 13A thereby increasing levels of the long transcript of LH2 [LH2(long)], are linked to scleroderma pathology. In this study we have examined the role played by RNA binding protein Fox-2 in regulating exon 13A inclusion that leads to the generation of scleroderma-associated LH2(long) mRNA.
Methods and Results
We report that over-expression of Fox-2 enhances inclusion of exon 13A and increases the generation of LH2(long) mRNA, whereas knockdown of Fox-2 decreases the LH2(long) transcripts. Mutational analysis of an LH2 minigene demonstrated that two of the four Fox binding motifs flanking LH2 exon 13A are required for its inclusion. In early passage fibroblasts derived from patients with systemic scleroderma, the knockdown of Fox-2 protein significantly decreased the endogenous levels of LH2(long) mRNA.
Conclusions
Fox-2 appears to play an integral role in the regulation of LH2 splicing. Knockdown of Fox-2 and other methods to decrease the levels of fibrosis-associated LH2(long) mRNA in primary scleroderma cells may suggest a novel approach to strategies directed against scleroderma.
The biosynthesis and assembly of the triple helical collagen fibrils is a multi-step process that begins inside a cell but is completed in the extracellular matrix (ECM) (1). One of the important groups of enzymes that play a key role in the collagen biosynthesis pathway is the lysyl hydroxylase (LH) family that includes LH1, LH2 and LH3 (also described as PLOD1, 2 and 3; procollagen lysine oxoglutarate dehydrogenases). These enzymes catalyze the conversion of specific lysine residues (Lys) on the nascent collagen chains to hydroxylysines (Hyl). The target sequences of LH are located in both the helical and non-helical telopeptide collagen domains (Gly-X-Lys and Ala/Ser-X-Lys, respectively) (2). The telopeptide hydroxylysines in turn act as precursors that are responsible for the formation of inter-molecular tri-functional pyridinoline cross-links (Pyr). These Pyr cross-links are critical for the mechanical stability and tensile strength of collagen fibrils. Although each member of the LH family hydroxylates collagen lysine residues, only LH2 has been reported to act specifically on the telopeptide lysines of collagen molecules (3). LH2 is also the only member of the LH family that is known to be alternately spliced (4). Its alternative splicing leads to the generation of two mRNA transcripts, LH2(long) and LH2(short), that either include or exclude the 63bp exon 13A (4, 5).
The identification of LH2(long) in dermal fibroblasts and its differential expression in various tissues was first reported by our laboratory (4, 6). Subsequent studies have reported that LH2 (long) mRNA is significantly increased in scleroderma and other fibrotic conditions (7, 8). LH2(long) over-expression is accompanied by an associated increase in the Pyr cross-links present in the accumulated collagen in the scleroderma patients (9–13). Therefore factors affecting the changes in LH2 alternative splicing pattern appear to play an important role in scleroderma. Our current study has established that one of these regulatory factors, Fox-2, promotes the generation of LH2(long) mRNA by enhancing the inclusion of the LH2 alternative exon 13A. We have utilized a new LH2 minigene that recapitulates the endogenous LH2 splicing pattern to identify the relative contributions of the four Fox-binding motifs that are present in the introns flanking the alternately-spliced exon 13A.
Using site-directed mutagenesis and Fox-2 over-expression experiments we have demonstrated that the two Fox motifs located in the upstream intron are required for the efficient inclusion of exon 13A. Targeting the expression of Fox-2 protein in primary cultured cells derived from scleroderma biopsy samples using Fox-2 specific siRNAs significantly reduced the endogenous levels of LH2(long) mRNA that are elevated in scleroderma. Our results not only shed new light on the pathobiology of scleroderma, but may indicate a future approach towards development of new strategies that decrease the levels of fibrosis-associated LH2(long) in scleroderma.
MATERIALS AND METHODS
Phylogenetic comparison
Phylogenetic comparisons were performed using the Clustal W alignment function of the MacVector program (14). The parameters used for sequence analysis are described elsewhere (15).
Cell culture
293 TRex, A431, A375, Huh-7, HeLa cells and the mouse embryonic fibroblasts (MEFs) were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Hyclone). Early passage cells (<5) cultured from biopsies of clinically affected and non-affected skin from SSc patients and healthy controls were cultured under similar conditions. All research was carried out in compliance with the Helsinki Declaration and approved by the Institutional Review Board of the Duke University Health System.
Tetracycline Induction
The 293 TRex cells that had either the Fox-2 cDNA or the EGFP cDNA stably integrated in a single copy at the FRT site were a gift from Prof. Mariano A.Garcia-Blanco, Duke University Medical Center. The tetracycline induction experiments were carried out as described previously (16). After 36 hours, the cells were harvested from three wells for RNA isolation using Trizol reagent (Invitrogen) following manufacturer’s instructions. Total cell lysate was prepared from the fourth well as described later. Cells from the fifth well were used for FACS analysis at the Duke FACS facility.
LH2 Minigene design
An LH2 genomic fragment containing 1163 nucleotides of intron 13, 63 nucleotides of exon 13A and 951 nucleotides of intron 13A was amplified from human genomic DNA using LH2 specific primers. The forward and the reverse primers had XbaI and XhoI overhangs, respectively and their sequences are described in Supplementary data Table S1.
The amplified genomic LH2 fragment was digested with XbaI and XhoI and ligated to pI-11 vector with compatible ends. All sequences were verified by DNA sequencing.
Fox-2 over-expression
Mouse Fox-2 over-expression vector, Fox-2 deletion mutant vector and the over-expression protocol have been described elsewhere (16). RNA was extracted from three of the four wells (Trizol) and total cell lysate was prepared from the fourth well as described below.
Preparation of total cell lysate and western blot
Media was removed from the cells and they were washed twice with PBS (pH 7.4). The cells were then scraped from the wells and re-suspended in PBS followed by centrifugation at 2500 RPM for 5 minutes. The supernatant was discarded and the cells resuspended in cold lysis buffer containing 1% NP-40, incubated for 30 minutes on ice and centrifuged at 13,000 RPM for 2 minutes. The supernatant was then collected.
The total protein concentration of cell lysates was measured by a standard Bradford assay using Bradford Reagent (Bio-Rad) following manufacturer’s instructions. 25ug of protein was loaded in each well of a 7.5% SDS-PAGE gel and, post-electrophoresis, the gel was transferred to a PVDF membrane (Millipore). The following dilutions of primary antibodies were used: αV5 (1: 5000) (Invitrogen), αFox-2 (1:1000) (Abcam), αCA150 (1:2000). The following dilutions of the secondary antibodies conjugated to HRP were used: αMouse IgG-HRP (1:5000) (GE Healthcare), αRabbit IgG-HRP (1:5000) (GE Healthcare). The products of the western blot were detected using a chemiluminiscence detection kit (Denville Scientific) following manufacturer’s instructions.
Site-directed mutagenesis
Mutations were introduced in the LH2 minigene using chimaeric PCRs. All primers used in this study were obtained from Integrated DNA Technologies. Their sequences are included in the Supplementary data, Table S1.
Transfections
The protocols for the generation of stable cell lines and transient transfections have been described elsewhere (17). In brief, one day prior to transfections the cells were plated in antibiotic-free DMEM in a 6-well tissue culture dish and these were transfected the next day with the DNA using lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. For selection of stables, at 48 hours post-transfection, the cells were transferred to a T-25 flask containing DMEM supplemented with 10% FBS and the selection marker Zeocin (200 μg/ml). For transient transfections, the cells were harvested 48 hours post-transfection for the extraction of either RNA or protein. The stably-transfected cells were harvested usually 2–3 weeks after selection. All RNA extractions in this study were performed using Trizol (Invitrogen) following manufacturer’s instructions. Each condition was performed in triplicate and each experiment was repeated twice.
RT-PCR
Reverse Transcription (RT) was performed using 1μg of RNA from each sample, unless mentioned otherwise. The RT was performed using MMLV-RT (Invitrogen) as previously described. Minigene specific PCRs were carried out with T7 and SP6 primers and endogenous LH2 PCRs were carried out using LH2 specific primers and using 32P dCTP as described elsewhere.
Polyacrylamide Gel Electrophoresis and Quantification of RT-PCR
The PCR samples labeled with 32P dCTP were separated on a 5% polyacrylamide gel. The gel was quantified using a phosphorimager (Storm 820) and ImageQuant software as described (18).
Fox-2 siRNA knockdown
All of the knockdowns were performed using the 2-hit siRNA knockdown protocol as described (19). Four wells of a 24-well tissue culture plate were plated for each condition. Cells were transfected twice (on days 1 and 3) with 3μl of 20 nM siRNA using the oligofectamine reagent (Invitrogen) following manufacturer’s instructions. On day 5, the cells were harvested and RNA was extracted from three wells (for each condition) using Trizol and total cell lysate was prepared from the fourth well as described earlier. Sequences of the Fox-2 specific siRNAs (Dharmacon) used were as follows:
Fox-2 siRNA #1: 5’ CCUGGCUAUUGCAAUAUUU dTdT 3’ (20)
Fox-2 siRNA #2: 5’ GUUAUAUGCAGCAUCCAGC dTdT 3’
RESULTS
Phylogenetic sequence analysis of the introns flanking exon 13A reveals high sequence conservation of the upstream Fox binding motifs
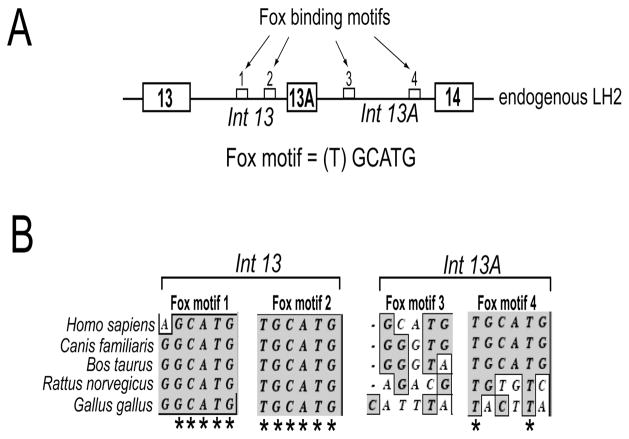
The sequence analysis of introns that flank the alternately spliced human LH2 exon 13A revealed the presence of four RNA binding motifs (U)GCAUG (21). This motif has been shown to regulate the alternate splicing via Fox proteins Fox-1 and Fox-2 in different genes (16, 22–24). As seen in Figure 1, the two Fox binding motifs (1 and 2) present in the upstream intron 13 were found to be highly conserved across the five different species that were selected for our analysis. The other two Fox binding motifs (3 and 4) present in the downstream intron 13A, were not as highly conserved. This suggested that the upstream Fox motifs may play a more important role in regulating LH2 alternative splicing.
Figure 1. Clustal W sequence analysis of the flanking introns of exon 13A reveals that the Fox-2 binding motifs located in the upstream intron are highly conserved.
(A) Schematic (not drawn to scale) showing the genomic arrangement of the alternately spliced exon 13A and the constitutively spliced exons 13 and 14. The introns that flank exon 13A are also shown (Int 13 and Int 13A). The small white boxes represent the four Fox-2 binding motifs, shown by arrows, identified in the introns that flank exon 13A. The DNA sequence of the Fox binding motif is shown. The brackets around (T) indicate that it is the most variable base of the sequence. (B) Phylogenetic comparison of the sequence of the two Fox binding motifs present in intron 13 and two motifs present in intron 13A from five different species (humans, dog, cow, rat and chicken). Only the sequences of the two Fox binding motifs present in intron 13 and two motifs present in intron 13A are shown. The asterisks (*) denote complete sequence conservation across the five species.
Induction of Fox-2 protein increases the inclusion of exon 13A and the generation of endogenous LH2(long) mRNA
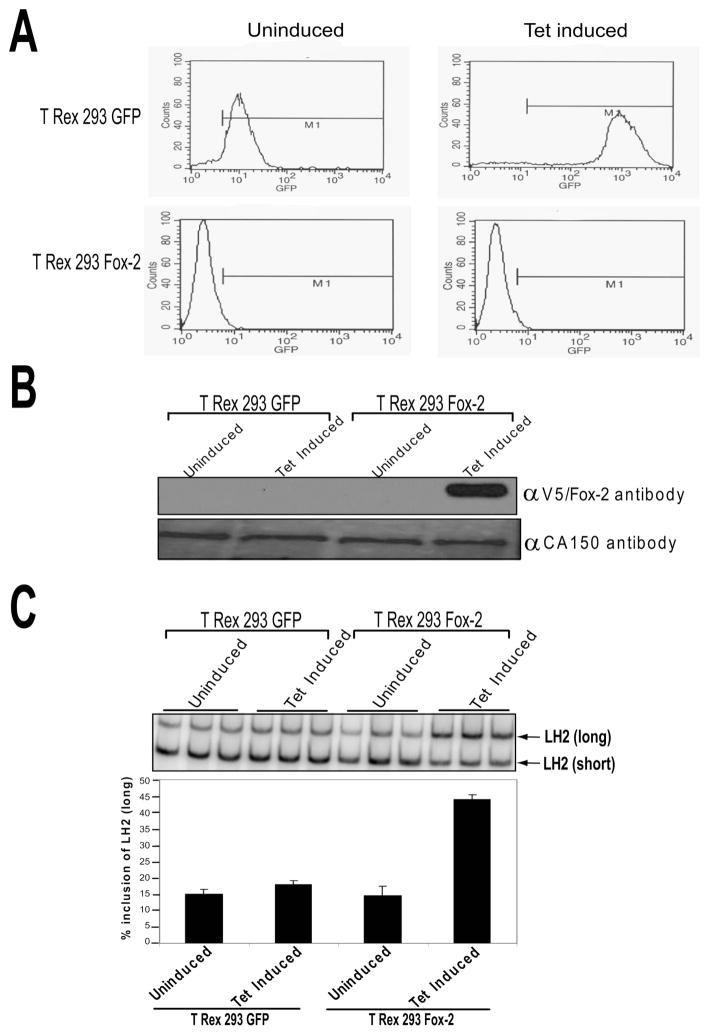
In order to establish the role that Fox-2 protein plays in regulating the inclusion of the LH2 alternately spliced exon 13A, we initially examined the effect of increasing levels of Fox-2 protein on the pattern of LH2 splicing. To accomplish this, we used a tetracycline (tet) inducible system comprised of Fox-2 cDNA sub-cloned under a tet inducible promoter, stably integrated in the FRT site of the 293 T Rex cells. A separate 293 T Rex cell line in which the cDNA of the Enhanced Green Fluorescent Protein (EGFP) had been sub-cloned under the same tet inducible promoter was used as a control for this experiment. This system has been used earlier to test the role played by the Fox-2 protein in regulating the alternative splicing pattern of Fibroblast Growth Factor Receptor 2 (FGFR2) (16). As seen in Figure 2A, FACS analysis was used to show that, whereas GFP was induced by tet in 293 T Rex GFP cells, the GFP levels were unchanged in the 293 T Rex Fox-2 cells. The robust induction of Fox-2 expression in 293 T Rex Fox-2 cells observed upon the addition of tet to the media (Figure 2B), led to a significant increase in the levels of endogenous LH2(long) mRNA (Figure 2C). This striking effect was specific and was not observed in the case of tet induced 293 T Rex GFP cells (Figure 2C).
Figure 2. Induction of Fox-2 protein enhances the inclusion of LH2 exon 13A leading to LH2(long) mRNA.
Analysis of the uninduced and tetracycline-induced 293 TRex GFP control cells and the 293 TRex Fox-2 cells by (A) FACS analysis, (B) Western blot using the V5 antibody to detect the induced Fox-2. CA150 was used as a loading control, and (C) radiolabeled RT-PCR analysis on a 5% SDS-PAGE gel (top) and the quantification of the gel depicting LH2(long) mRNA as a percentage of the total, LH2(long) + LH2(short), as described in Materials and Methods, is shown at the bottom. The graph is the result of independent triplicates with ± Standard Deviation (SD) shown in the error bars.
Validation of a new LH2 minigene and mutagenesis of Fox binding motifs to establish their relative importance
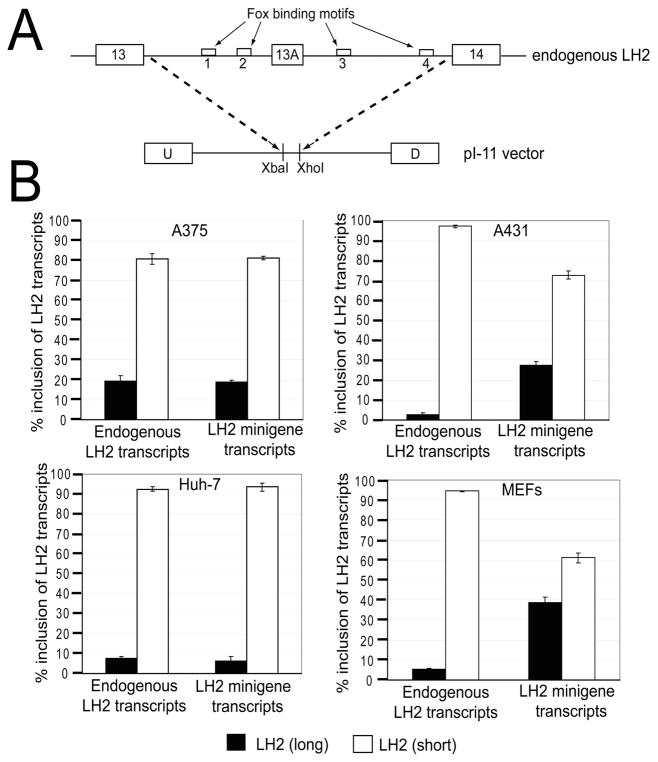
As the previous experiment had shown that increased levels of Fox-2 significantly increased the generation of LH2(long) mRNA, we subsequently identified which of the four (U)GCAUG motifs were important for Fox-2 binding. To examine the relative contribution of each of the four Fox binding motifs to the pattern of LH2 splicing, we mutated each of the four Fox motifs in our newly designed LH2 minigene. This minigene included an LH2 genomic fragment containing 1163 nucleotides of intron 13, 63 nucleotides of the alternately spliced exon 13A, and 951 nucleotides of intron 13A cloned under the CMV promoter of the pI-11 vector (Figure 3A). As shown in Figure 3A, this minigene contains two Fox binding motifs that are located upstream and downstream of exon 13A, respectively. The pI-11 vector has been described in detail elsewhere (25); in brief, it includes two strong exons, the upstream (U) and the downstream (D) adenoviral exons separated by a constitutively-spliced intron containing a multiple cloning site which was used to sub-clone the amplified genomic LH2 fragment. The design of this new LH2 minigene mirrored the endogenous genomic arrangement of LH2 exon 13A in which a “weak” alternately-spliced exon 13A with sub-optimal splice sites is surrounded by “strong” constitutively-spliced exons with optimal splice sites. We then tested and validated this new LH2 minigene in multiple cell lines to test whether the ratio of LH2(long) to LH2(short) mRNA followed a similar trend in transcripts originating either from the endogenous LH2 gene or the transfected LH2 minigene. In Figure 3B, four examples from the tested cell lines (two human skin epithelial cell lines A375 cells and A431, human hepatocellular carcinoma Huh-7 cells and MEFs) are shown. We selected skin cells for the validation experiment as we chose to perform our Fox-2 over-expression experiments in A431 cells due to their high transfection efficiency.
Figure 3. Schematic representation and validation of the human LH2 minigene.
(A) Schematic representation showing the endogenous LH2 at the top, and below, LH2 sequence inserted in the p1–11 vector. The empty white boxes, indicated by arrows, show the four Fox binding motifs that flank the LH2 alternative exon 13A. The constitutive exons 13 and 14 are also shown. The broken lines depict the endogenous LH2 genomic region that was sub-cloned in the XbaI and XhoI sites of the pI-11 vector, between its constitutively expressed adenoviral derived upstream (U) and downstream (D) exons. (B) Validation of the LH2 minigene following transfection in four representative cell lines, skin melanoma epithelial cells (A375) (stables), human hepatocelluler carcinoma cells (Huh-7) (stables), skin epithelial carcinoma cells (A431) (transients) and mouse embryonic fibroblasts (MEFs) (transients) are shown. The ratios of the transcripts that originate from the endogenous LH2 pre-mRNA and the minigene LH2 pre-mRNA, respectively, are shown. The quantification of each LH2 mRNA isoform (long and short) is expressed as a percentage of the total, [LH2(long) + LH2(short)], as described in Materials and Methods. The graph is the result of three independent wells with ± Standard Deviation (SD) shown in the error bars.
We also validated our minigene in Huh-7 liver cells (Figure 3B) and lung cells (data not shown) since these organs also frequently undergo fibrosis. As shown in Figure 3B, in each case the alternately-spliced LH2 minigene transcripts reflected the same trend as the spliced endogenous LH2 transcripts, in which LH2(long) was the minor RNA species and LH2(short) was the major RNA species.
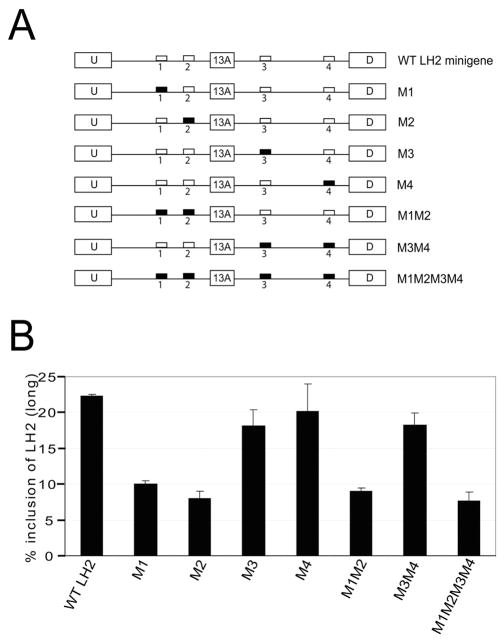
The validated LH2 minigene was then used in mutagenesis experiments in which we mutated all of the four Fox binding motifs either individually or in different combinations (Figure 4A). The wild-type LH2 minigene and its different mutated versions were then stably transfected in the MEFs. We chose to use MEFs for this experiment because the inclusion of exon 13A and hence LH2(long) levels are higher in these cells compared to the other cell lines that we had tested. Since the results from our tet induction experiments with 293 TRex cells stably transfected with the Fox-2 cDNA suggested that Fox-2 protein enhances the inclusion of exon 13A (Figure 2), thereby increasing the levels of LH2(long) mRNA, we predicted that the mutations of Fox binding motifs would lead to decreased inclusion of exon 13A. We therefore performed stable transfections of MEFs with the mutated LH2 minigenes, so that any decrease in LH2(long) caused by mutations in the Fox-2 motifs would be more easily detectable. The individually mutated LH2 minigenes (M1–M4) together with the combination of mutations (M1M2, M3M4, M1M2M3M4) are shown in Figure 4A.
Figure 4. Mutation of the Fox binding motifs located upstream of the exon 13A in the LH2 minigene leads to a significant reduction in the levels of LH2(long).
(A) Schematic representation of the different mutations of the Fox binding motifs that were used for the mutagenesis experiment shown below in 4B. The white boxes represent wild-type Fox binding motifs and the black boxes represent the mutated Fox motifs. The name of each construct is shown on the right hand side.
(B) The bar graph shows the quantification of a stable transfection experiment in MEFs with LH2(long) mRNA shown as a percentage of the total, [LH2(long) + LH2(short)], as described in Materials and Methods. The graph depicts the result of three independent wells with ± Standard Deviation (SD) shown in the error bars. Each experiment was performed twice in triplicate.
We observed a significant decrease in LH2(long) mRNA level when the Fox binding motifs located in the upstream intron 13 were mutated either individually (M1 and M2) or in combination with other Fox motifs (Figure 4B). In contrast, no significant decrease was observed when either of the two Fox motifs located in intron 13A, downstream of exon 13A, were mutated either individually (M3 and M4) or in combination (M3M4).
Our data therefore suggests that both Fox motifs located in intron 13 upstream of LH2 exon 13A play an important role in the efficient inclusion of this exon and hence in determining the level of the LH2(long) mRNA. In contrast, the downstream intronic Fox motifs do not appear to be essential for exon 13A inclusion.
Fox-2 over-expression leads to increased exon 13A inclusion in the LH2 minigene transcripts; both intron 13 Fox-binding motifs are required for LH2(long) expression
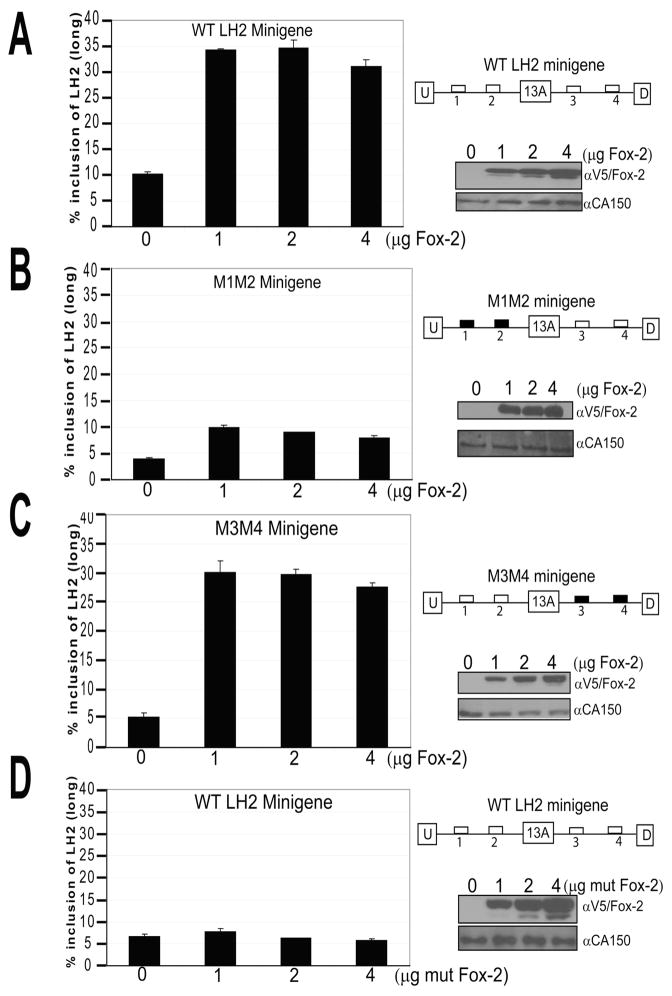
In order to confirm the requirement for Fox-2 protein in the generation of LH2(long) mRNA and the importance of both upstream Fox binding motifs, we performed Fox-2 over-expression experiments using the previously-described LH2 minigene. To accomplish this, A431 cells (human epithelial skin carcinoma) were transiently transfected with increasing amounts of a Fox-2 over-expression vector together with 100ng of the wild-type LH2 minigene. In parallel, A431 cells were transfected with increasing amounts of Fox-2 over-expression vector together with 100ng of the M1M2 minigene in which both Fox motifs (#1 and #2) located in the upstream intron 13 had been mutated (Figure 4A). As shown in Figure 5A and 5B respectively, increased levels of LH2(long) were observed only when increasing amounts of Fox-2 expression vector were co-transfected with the wild-type LH2 minigene but not with its mutated M1M2 version. As shown in Figure 5C, the lack of any effect of downstream Fox binding motifs #3 and #4 was confirmed in a similar Fox-2 over-expression experiment in which the mutated M3M4 minigene (in which both downstream Fox binding motifs had been mutated) still led to a robust increase in the levels of LH2(long) mRNA, similar to that observed in the case of the wild-type LH2 minigene. Comparable results were obtained in HeLa cells (data not shown). We also examined the effect of co-transfection of increasing amounts of a different Fox-2 expression vector that expressed a mutated form of Fox (in which the last 82 amino acids that harbor the RNA Recognition Motif were deleted) together with the wild-type LH2 minigene (16). As shown in Figure 5D, no increase in LH2(long) expression was observed even when the mutated Fox-2 protein was expressed at high levels.
Figure 5. Over-expression of Fox-2 results in significantly increased LH2(long) levels when the upstream Fox-2 binding motifs are intact, irrespective of the status of the downstream motifs.
Bar graphs representing the results of transient transfection experiments in A431 cells in which 100ng of (A) WT LH2 minigene, (B) M1M2 LH2 minigene in which upstream Fox binding motifs were mutated and, (C) M3M4 LH2 minigene in which downstream Fox binding motifs were mutated, were co-transfected together with increasing amounts of WT Fox-2 cDNA, respectively. In (D), 100ng of WT LH2 minigene was co-transfected together with increasing amounts of a mutated Fox-2 cDNA. The quantification of LH2(long) mRNA has been described in Figure 2 and in Materials and Methods. The schematic representation of each minigene used in each experiment is shown on the right. The western blots for each individual experiment are shown below the minigene schematic. V5 antibody was used to detect the induced Fox-2. CA150 was used as a loading control. All the graphs are the result from three independent wells with ± Standard Deviation (SD) shown in the error bars. Each experiment was performed twice in triplicate.
Overall, this data shows that (a) Fox-2 protein plays an integral role in the inclusion of exon 13A, (b) levels of LH2(long) mRNA increased by Fox-2 over-expression requires two intact Fox binding motifs located upstream of the LH2 alternative exon 13A, and (c) the wild-type Fox-2 protein must interact physically with the LH2 pre-mRNA to be able to exert its effect.
Knockdown of Fox-2 protein decreases the inclusion of LH2 exon 13A
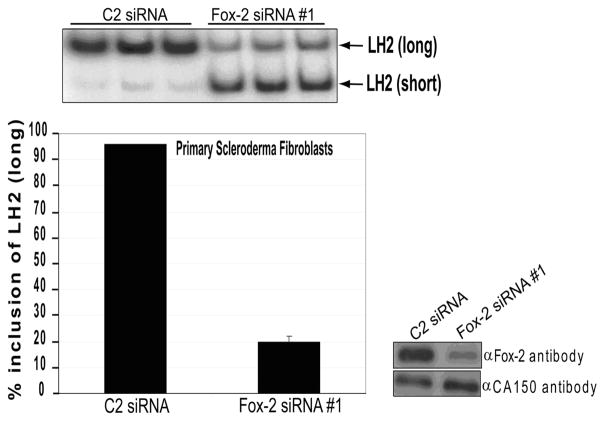
In light of previous studies showing that LH2(long) mRNA is significantly over-expressed in scleroderma (7) and our preliminary results suggesting that this could be regulated by increased levels of Fox-2 (data not shown), we tested the effect of a knockdown of Fox-2 protein on LH2(long) mRNA levels in fibroblasts from scleroderma patients. Initially, we performed this experiment in MEFs and demonstrated that LH2(long) expression was reduced by two-fold upon Fox-2 knockdown (data not shown). Subsequently, we carried out the Fox-2 knockdown in fibroblasts cultured from the biopsies of affected skin from patients with systemic sclerosis (clinically described in Supplementary data Table S2). As shown in cultured fibroblasts from a representative patient in Figure 6, a robust knockdown of Fox-2 protein using a specific siRNA resulted in a significant reduction in the levels of endogenous LH2(long) mRNA. A similar decrease was observed when a comparable experiment was performed in fibroblasts that had been cultured from a biopsy taken from unaffected skin of a scleroderma patient (data not shown). This decrease in LH2(long) levels observed in Figure 6 was even more striking in view of the fact that that these scleroderma primary cells expressed about four-fold higher levels of endogenous LH2(long) mRNA compared to their normal counterparts, as reported earlier by our laboratory (8).
Figure 6. A knock-down of Fox-2 in primary cells derived from a SSc patient leads to a robust decrease in the levels of endogenous LH2(long) mRNA.
The RT-PCR gels of the knockdown experiments are shown above the bar graph. C2 and Fox-2 siRNA are the non-specific control siRNA and Fox-2 specific siRNAs, respectively. The LH2(long) and LH2(short) bands are shown by arrows. The bar graph represents the quantification of the RT-PCR products. The LH2(long) quantification has been described in previous figure legends. The Western blots for these experiments are shown at the right. Fox-2 specific antibody was used to detect the endogenous levels of Fox-2. All the graphs are the result of three independent wells with ± Standard Deviation (SD) shown in the error bars. Each experiment was performed twice in triplicate.
DISCUSSION
Scleroderma is a chronic, degenerative and heterogeneous disorder that has significant autoimmune, vascular and connective tissue components (26–29). In the current study on the regulation of LH2 that modulates the over-accumulation of collagen cross-links associated with scleroderma, we have demonstrated that the regulatory RNA binding protein Fox-2 is required for the generation of the long transcript of LH2 that is significantly increased in scleroderma. This finding is important in view of the evidence that LH2(long) acts as the collagen telopeptide lysyl hydroxylase that hydroxylates the telopeptide lysines prior to the formation of Pyr cross-links (3, 11). As these cross-links are increased in the excessive production of collagen that is a characteristic of scleroderma, both the current and future studies on LH2(long) regulation should provide a novel approach to an understanding of the biology of this complex disease.
While a number of earlier studies have traditionally investigated this complex disease either from an autoimmune or a vascular perspective, the interest in this disease from the RNA biology viewpoint is fairly recent (30–34). Although there have been several reports of increased expression of the LH2(long) transcript in cells from patients with specific fibrotic conditions such as hypertrophic scars, keloids and Dupuytren’s syndrome (9, 11), our study is one of the first to examine LH2 in scleroderma from an alternative splicing perspective. Analysis of factors regulating the alternative splicing pattern of LH2 should broaden our understanding of this relatively understudied but important aspect of scleroderma and aid in defining the molecular mechanisms that are responsible for the increased LH2(long) transcript in scleroderma.
In our previous work we provided evidence for the involvement of the RNA binding protein TIA (T-cell Intracellular Antigen) family members in the regulation of the alternative splicing of LH2 (8). Our current study advances this work in a detailed examination of the effect of the Fox RNA binding proteins, a group of proteins known to regulate alternate splicing, on the splicing of the LH2(long) transcript. The Fox proteins were initially identified as homologs of Caenorhabditis elegans RNA binding protein Feminization on X (35, 36), but subsequently they have been found to play an important role in the alternative splicing of a number of genes including non-muscle myosin chain-B, C-src, fibronectin and FGFR2 (16, 22–24). The Fox-2 protein (or RNA Binding-Motif Protein 9) has different isoforms and is encoded by multiple transcripts (16, 37). It is also regulated by androgens and it is known to regulate its own alternative splicing (16, 38). The importance of the Fox binding motif (U)GCAUG, was initially recognized in the brain but since then this motif has been reported to play a significant role in other tissues (22, 39, 40). A recent genome-wide analysis of the targets of the Fox family proteins predicted thousands of probable targets for Fox-2 and its paralog Fox-1 proteins (41). Although this study did correctly predict and partially validate LH2 as one of the targets of Fox-2 protein, in the current manuscript we have provided an in-depth experimental analysis of the effect of increased Fox-2 expression on both endogenous LH2(long) transcripts and on LH2(long) transcripts generated from a new LH2 minigene.
We have demonstrated the relative importance of the four Fox binding motifs, on the regulation of LH2 splicing. Since phylogenetic analysis suggested that the two highly conserved upstream Fox motifs may play a more influential role in the regulation of LH2 alternate splicing, we tested this in a novel minigene by mutational analysis of the four Fox motifs either singly or in combination. Our mutagenesis experiments confirmed our predictions from the phylogenetic examination and suggested that Fox proteins acts via the two binding motifs in intron 13 upstream of the alternative exon 13A. This finding is in contrast with earlier genome-wide computational studies, demonstrating an over-representation of Fox binding motifs near tissue-specific alternate exons, that suggest that the Fox motifs located in the downstream introns played a more important role in alternative splicing (42, 43). Although the high phylogenetic conservation of the upstream Fox motifs does suggest the importance of these motifs, it is also possible that in the case of LH2, the sequences surrounding the Fox motifs make significant contributions to the relative effect of these Fox motifs on promoting exon inclusion (Seth and Yeowell, unpublished data).
In order to definitively establish the role of Fox-2 in the regulation of alternative splicing of LH2, we performed Fox-2 over-expression experiments in multiple established cell lines. The use of these established cell lines was necessitated because of the very low transfection efficiency of the fibroblasts. To overcome this problem, we not only performed these experiments in multiple cell lines but we also examined the effect of Fox-2 over-expression on both the endogenous LH2 transcripts (in 293 TRex cells) and on the LH2 minigene transcripts (in A431 and HeLa cells). The consistent results from these experiments indicate that the role played by Fox-2 in regulating LH2 alternative splicing is a general phenomenon. As shown in Figure 4, the result of the mutagenesis experiments performed in the murine embryonic fibroblasts suggest that the effect of Fox-2 is also species independent.
In primary fibroblasts cultured from two scleroderma patients, in which LH2 (long) is over-expressed, we successfully decreased the levels of LH2(long) mRNA by knocking down the levels of Fox-2 protein. Although cultured fibroblasts have been reported to have a different expression profile compared to the scleroderma tissue, we did not observe any significant difference in the levels of Fox-2 mRNA between the scleroderma biopsy samples and the fibroblasts cultured from these biopsies (data not shown). Hence, we predict that significantly decreasing Fox-2 in scleroderma fibroblasts would produce the same effect on LH2(long) in scleroderma skin. Consequently, targeting of Fox-2 protein to reduce the level of fibrosis-associated LH2(long) mRNA could provide an effective strategy against decreasing the collagen cross-links that are a characteristic of scleroderma. Moreover, the design of other strategies to reduce LH2(long) may open up exciting new avenues for future research on fibrosis.
Supplementary Material
Acknowledgments
The authors would like to gratefully acknowledge Dr. Mariano A. Garcia-Blanco, MD PhD Center for RNA Biology, Duke University Medical Center, for sharing some of the reagents used in this study. We would also like to thank Linda Walker and Liza McClellan for helpful discussions. We acknowledge the critical reading of this manuscript by Paula Miliani de Marval, PhD and Divya Seth, PhD. This work was supported by NIH Grant RO1 AG10215 from the National Institute on Aging (to HNY) and by the Scleroderma Foundation (to HNY).
Abbreviations
- A2BP1
Ataxin-2 Binding Protein 1
- RBM9
RNA Binding-Motif Protein 9
- Lys
Lysine
- Hyl
Hydroxylysine
- LH
lysyl hydroxylase
- PLOD
procollagen lysine oxoglutarate dehydrogenase
- Pyr
pyridinoline
- ECM
extracellular matrix
- CMV
cytomegalovirus
- tet
tetracycline
- MEFs
murine embryonic fibroblasts
- RRM
RNA recognition motif
- SSc
systemic sclerosis
Footnotes
This data was presented, in part, at the Annual Meeting of the Society for Investigative Dermatology, in Montreal, May 6–9, 2009. J. Invest. Dermatol. 129, S85, 2009
References
- 1.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20(1):33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Kivirikko KI, Myllyla R. Post-translational processing of procollagens. Ann N Y Acad Sci. 1985;460:187–201. doi: 10.1111/j.1749-6632.1985.tb51167.x. [DOI] [PubMed] [Google Scholar]
- 3.Takaluoma K, Lantto J, Myllyharju J. Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix Biol. 2007;26(5):396–403. doi: 10.1016/j.matbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Yeowell HN, Walker LC. Tissue specificity of a new splice form of the human lysyl hydroxylase 2 gene. Matrix Biol. 1999;18(2):179–87. doi: 10.1016/s0945-053x(99)00013-x. [DOI] [PubMed] [Google Scholar]
- 5.Valtavaara M, Papponen H, Pirttila AM, Hiltunen K, Helander H, Myllyla R. Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle. J Biol Chem. 1997;272(11):6831–4. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- 6.Walker LC, Overstreet MA, Yeowell HN. Tissue-specific expression and regulation of the alternatively-spliced forms of lysyl hydroxylase 2 (LH2) in human kidney cells and skin fibroblasts. Matrix Biol. 2005;23(8):515–23. doi: 10.1016/j.matbio.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 7.van der Slot AJ, Zuurmond AM, Bardoel AF, Wijmenga C, Pruijs HE, Sillence DO, et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278(42):40967–72. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 8.Yeowell HN, Walker LC, Mauger DM, Seth P, Garcia-Blanco MA. TIA nuclear proteins regulate the alternate splicing of lysyl hydroxylase 2. J Invest Dermatol. 2009;129(6):1402–11. doi: 10.1038/jid.2008.386. [DOI] [PubMed] [Google Scholar]
- 9.van der Slot AJ, Zuurmond AM, van den Bogaerdt AJ, Ulrich MM, Middelkoop E, Boers W, et al. Increased formation of pyridinoline cross-links due to higher telopeptide lysyl hydroxylase levels is a general fibrotic phenomenon. Matrix Biol. 2004;23(4):251–7. doi: 10.1016/j.matbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 10.van der Slot AJ, van Dura EA, de Wit EC, De Groot J, Huizinga TW, Bank RA, et al. Elevated formation of pyridinoline cross-links by profibrotic cytokines is associated with enhanced lysyl hydroxylase 2b levels. Biochim Biophys Acta. 2005;1741(1–2):95–102. doi: 10.1016/j.bbadis.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Brinckmann J, Notbohm H, Tronnier M, Acil Y, Fietzek PP, Schmeller W, et al. Overhydroxylation of lysyl residues is the initial step for altered collagen cross-links and fibril architecture in fibrotic skin. J Invest Dermatol. 1999;113(4):617–21. doi: 10.1046/j.1523-1747.1999.00735.x. [DOI] [PubMed] [Google Scholar]
- 12.Pornprasertsuk S, Duarte WR, Mochida Y, Yamauchi M. Lysyl hydroxylase-2b directs collagen cross-linking pathways in MC3T3-E1 cells. J Bone Miner Res. 2004;19(8):1349–55. doi: 10.1359/JBMR.040323. [DOI] [PubMed] [Google Scholar]
- 13.Pornprasertsuk S, Duarte WR, Mochida Y, Yamauchi M. Overexpression of lysyl hydroxylase-2b leads to defective collagen fibrillogenesis and matrix mineralization. J Bone Miner Res. 2005;20(1):81–7. doi: 10.1359/JBMR.041026. [DOI] [PubMed] [Google Scholar]
- 14.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 15.Mistry N, Harrington W, Lasda E, Wagner EJ, Garcia-Blanco MA. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. Rna. 2003;9(2):209–17. doi: 10.1261/rna.2470903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26(4):1209–22. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol Cell Biol. 2003;23(24):9327–37. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seth P, Miller HB, Lasda EL, Pearson JL, Garcia-Blanco MA. Identification of an intronic splicing enhancer essential for the inclusion of FGFR2 exon IIIc. J Biol Chem. 2008;283(15):10058–67. doi: 10.1074/jbc.M800087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner EJ, Garcia-Blanco MA. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol Cell. 2002;10(4):943–9. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 20.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25(22):10005–16. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8(13):1561–74. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 22.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271(30):17613–6. [PubMed] [Google Scholar]
- 23.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17(11):6537–45. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18(7):3900–6. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carstens RP, McKeehan WL, Garcia-Blanco MA. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18(4):2205–17. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakkas LI, Platsoucas CD. Is systemic sclerosis an antigen-driven T cell disease? Arthritis Rheum. 2004;50(6):1721–33. doi: 10.1002/art.20315. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T. Scleroderma--pathophysiology. Eur J Dermatol. 2009;19(1):14–24. doi: 10.1684/ejd.2008.0570. [DOI] [PubMed] [Google Scholar]
- 28.Kahaleh MB. Raynaud phenomenon and the vascular disease in scleroderma. Curr Opin Rheumatol. 2004;16(6):718–22. doi: 10.1097/01.bor.0000138677.88694.a4. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T. The bleomycin-induced scleroderma model: what have we learned for scleroderma pathogenesis? Arch Dermatol Res. 2006;297(8):333–44. doi: 10.1007/s00403-005-0635-z. [DOI] [PubMed] [Google Scholar]
- 30.Tan FK, Hildebrand BA, Lester MS, Stivers DN, Pounds S, Zhou X, et al. Classification analysis of the transcriptosome of nonlesional cultured dermal fibroblasts from systemic sclerosis patients with early disease. Arthritis Rheum. 2005;52(3):865–76. doi: 10.1002/art.20871. [DOI] [PubMed] [Google Scholar]
- 31.Whitfield ML, Finlay DR, Murray JI, Troyanskaya OG, Chi JT, Pergamenschikov A, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100(21):12319–24. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pendergrass SA, Whitfield ML, Gardner H. Understanding systemic sclerosis through gene expression profiling. Curr Opin Rheumatol. 2007;19(6):561–7. doi: 10.1097/BOR.0b013e3282f00375. [DOI] [PubMed] [Google Scholar]
- 33.Sargent JL, Milano A, Connolly MK, Whitfield ML. Scleroderma gene expression and pathway signatures. Curr Rheumatol Rep. 2008;10(3):205–11. doi: 10.1007/s11926-008-0034-5. [DOI] [PubMed] [Google Scholar]
- 34.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3(7):e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388(6638):200–4. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- 36.Skipper M, Milne CA, Hodgkin J. Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics. 1999;151(2):617–31. doi: 10.1093/genetics/151.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33(7):2078–89. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman AP, Friedlich DL, Harmison G, Howell BW, Jordan CL, Breedlove SM, et al. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem Biophys Res Commun. 2001;282(2):499–506. doi: 10.1006/bbrc.2001.4617. [DOI] [PubMed] [Google Scholar]
- 39.Deguillien M, Huang SC, Moriniere M, Dreumont N, Benz EJ, Jr, Baklouti F. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood. 2001;98(13):3809–16. doi: 10.1182/blood.v98.13.3809. [DOI] [PubMed] [Google Scholar]
- 40.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. Embo J. 2003;22(4):905–12. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22(18):2550–63. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29(11):2338–48. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33(2):714–24. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.