Abstract
Neuronal nicotinic acetylcholine receptors are activated by both endogenous acetylcholine and exogenous nicotine, making sequence variations in these receptors likely candidates for association with tobacco phenotypes. Previous studies have found evidence for significant association between SNPs in the genomic region containing the CHRNA6 and CHRNB3 genes and tobacco behaviors (Bierut et al, 2007; Greenbaum et al, 2006; Saccone et al, 2007; Zeiger et al, 2008). In this study, we provide support for an association between these genes and tobacco dependence in the National Youth Survey Family Study wave 10, a nationally representative sample of households. Eight single nucleotide polymorphisms (SNPs) in the CHRNA6 and CHRNB3 genomic region were genotyped in 1051 subjects, approximately half of whom are members of sibling pairs. Genetic association with DSM-IV dependence was assessed using a family-based approach as implemented in the statistical package PBAT. Individual SNPs were tested for association with quit attempts and overall dependence. Variation in CHRNA6 was found to be associated with tobacco dependence (p=0.007 in Caucasians). SNPs in CHRNB3 were found to be associated with the number of quit attempts (p=0.0024). Together these results further implicate the region downstream of CHRNA6 and the region upstream of CHRNB3 in risk of nicotine dependence.
Keywords: Nicotinic receptors, SNP, Genetic association, Tobacco use, Nicotine Dependence
Introduction
Tobacco use is a major health problem in the United States and worldwide. Despite the increasing awareness of health related consequences, about 25% of American adults continue to smoke (Center for Disease Control 2000). Some individuals continue despite knowledge of the risks, and others do not understand the risks, but the majority of individuals continue smoking because they are nicotine dependent (Hymowitz et al, 1997; Jarvis, 2004). That is, despite having the desire to quit, a large percentage of smokers are unsuccessful in stopping. Studies have shown that genes influence level of nicotine dependence with heritability estimates indicating genetic factors account for between 40–70% of the phenotypic variance (Li et al, 2003; Maes et al, 2004; Rhee et al, 2003; Swan et al, 1990). Determining which genes are contributing to this heritability is an area of active research.
Neuronal nicotinic acetylcholine receptors (nAChRs) are likely candidate genes to be involved in mediating nicotine dependence. These pentameric ion channels are present in the peripheral and central nervous systems, including dopaminergic neurons, and are activated by endogenous acetylcholine and exogenous nicotine (Mineur and Picciotto, 2008). The CHRNA6 and CHRNB3 genes coding for the α6 and β3 receptor subunits are located contiguously in a tail to tail configuration on chromosome 8. Their protein products are co-localized in nicotinic receptors in the substantia nigra, ventral tegmental area, striatum, and locus coeruleus (Gotti et al, 2006). Located on chromosome 8 at position 8p11.21, CHRNA6 spans 16.01 kb (NCBI Build 35) and has at least three alternatively spliced transcripts. CHRNB3 is located on chromosome 8 at position 8p11.2, and spans 39.99 kb. Six exons can be alternatively spliced into at least three different transcripts (GeneCards: http://www.genecards.org (Rebhan et al, 1998)).
Both α6- and β3- containing receptors have been found to have a significant role in dopaminergic neurotransmission, thus contributing to reward and reinforcement of behavior (Cui et al, 2003). Receptors containing α6β2β3 as well as α6α4β2β3 containing receptors in the striatum mediate αConotoxin MII-sensitive dopamine release. In contrast, α6β2 containing receptors in the superior colliculus seem to be involved in GABA release (Champtiaux et al, 2003; Gotti et al, 2006; Salminen et al, 2004). The β3 subunit is believed to be important for assembly and stability of α6 containing nAChRs (Cui et al, 2003).
Recent genetic association studies have provided evidence that polymorphisms (SNPs) in CHRNA6 and CHRNB3 are associated with nicotine dependence and subjective response to nicotine. Bierut et al (2007) and Saccone et al (2007) found SNPs upstream of CHRNB3 as well as in exon 6 (3’ UTR) of CHRNA6 to be associated with nicotine dependence in a case-control sample (Bierut et al, 2007; Saccone et al, 2007). Greenbaum et al (2006) also found association of a SNP in α6 (intron 2) with nicotine dependence in a sample of female Israeli students (Greenbaum et al, 2006). Additionally, Zeiger et al have found significant associations between SNPs located in the upstream region of CHRNB3 and in exon 6 of the CHRNA6 gene and subjective responses to nicotine, in separate samples of selected and community young adults (Zeiger et al, 2008). To the best of our knowledge, association of CHRNA6 and CHRNB3 with dependence measured by DSM-IV has not been reported, nor has quit attempts been examined. In an effort to strengthen these recent findings as relevant to the general U.S. population, we present our results from testing a subset of SNPs in these genes in a population-representative sample using the DSM-IV definition of dependence.
Materials and methods
Subjects
NYS- FS Study Participants and Phenotypic Assessments
Polymorphisms in the CHRNA6 and CHRNB3 subunit genes were examined in a genetic supplemental sample participating in the National Youth Survey Family Study (NYSFS; Institute for Behavioral Science, and Institute for Behavioral Genetics, University of Colorado) wave 10. The National Youth Survey is a nationally representative probability sample of adolescents aged 11–17 in 1976 and living in the United States in 1977. A follow-up interview was conducted in 2002 (age range 35–44 years) where behavioral data and buccal cell swab samples were obtained on a strictly voluntary basis (Elliott et al, 1989). Of the 1051 individuals who agreed to follow-up interviews and provided buccal swab DNA samples, 856 of them had tried cigarettes. Although there has been some selective/disproportionate loss in some waves by sex, or age, or race/ethnicity, or social class across the first 10 waves of the NYS, these differences are quite small and often do not significantly affect the original distribution of these variables as established at the first wave of the NYS, which has been shown to be representative of individuals born in the continental U.S. in 1959–1965. One of the few significant and the largest of these is a change from 53.2% to 49.2% male (46.8% to 50.8% female) from the 1st to 10th survey. There has been no significant disproportionate loss over the years by initial alcohol use, marijuana use, or other illicit drug use or by general delinquency or more serious Uniform Crime Rates (UCR) offending. The distributions of these variables are not significantly different between respondents who completed interviews in a given year and those who did not. In addition, the distributions of sex, age, race/ethnicity, and social class of waves 7–10 are usually not significantly different from those observed in Wave 1 (1976) or Wave 6 (1983). Thus, overall, the NYS sample appears to remain reasonably representative of the U.S. population born in 1959–1965. The sample consisted of 227 families with sibships ranging from 2 to 5 offspring per family (592 individuals) and 479 individuals without siblings in the sample. 48.1% of subjects were male and 51.9% female, frequencies of minorities were reflective of the US population in 1976 with 80.3% White, 12.3% African American, and 5.4% of Hispanic or other ancestry. Additional summary statistics are given in Table 1.
Table 1.
Sample characteristics for the NYS-FS sample.
| Sample | Male (%) | Female (%) | Ever Tobacco (%) |
Ever Alcohol (%) |
|---|---|---|---|---|
| Pooled (1051) | 506 (49) | 545 (51) | 856 (81) | 990 (94) |
| Caucasian (860) | 410 (48) | 450 (52) | 727 (85) | 824 (96) |
| Hispanic (28) | 11 (39) | 17 (61) | 23 (82) | 27 (96) |
| African-American (132) | 67 (51) | 65 (49) | 82 (62) | 109 (83) |
Alcohol and tobacco use behaviors were assessed during a face-to-face structured interview including an adaptation of the Composite International Diagnostic Interview – Substance Abuse Module (CIDI-SAM) (Cottler and Keating, 1990). Nicotine dependence was assessed using DSM-IV criteria. The seven symptoms with their respective endorsement rates are given in Table 2. An individual was considered dependent if they endorsed three or more symptoms. Two phenotypes were analyzed: nicotine dependence as defined by three or more DSM-IV dependence symptoms, and reported number of unsuccessful quit attempts. The number of unsuccessful quit attempts was used because it is a quantitative measure and was hypothesized to be a closer biological measure of addiction (than overall dependence). Phenotypes were examined for ethnic and sex differences using ANOVA in SPSS.
Table 2.
Phenotype descriptions.
| DSM-IV dependence symptoms | Endorsement rate % (n) |
|
|---|---|---|
| Tolerance: | Increased amounts needed for same effect, or decreased effect. |
39.8% (426) |
| Withdrawal | Use to avoid symptoms, or symptoms when discontinue use | 25.2% (270) |
| Increased use | Increasing either quantity or duration | 26.4% (283) |
| Quit failure | Loss of control inability to quit or cut down despite desire to. | 18.3% (196) |
| Important activities |
Activities given up due to substance use | 1.9% (20) |
| Time | A great deal of time is spent obtaining, using, or recovering from effects of the substance. |
8.7% (94) |
| Continued use | Continued use despite substance causing or exacerbating physical or psychological problems. |
18.8% (201) |
Three subsets of the sample were used, described in Table 3. The initial analysis was done only with “Current frequent smokers” which included individuals reporting having used tobacco at least 180 days (almost every day) in the six months prior to the interview. This is more stringent (restricting the sample to current smokers) but similar to the inclusion criteria used by Bierut et al of having smoked at least 100 cigarettes (Bierut et al, 2007). Secondary analyses were done with 1) sets of “smokers” who were individuals who reported having used tobacco almost every day for at least a month at some point in their life, and 2) the entire sample of 1051 individuals.
Table 3.
Inclusion criteria.
| Subsample | Description |
|---|---|
| All | All subjects interviewed and genotyped. |
| Smokers | Individuals reporting having ever used tobacco almost everyday for at least a month. |
| Current frequent smokers | Individuals who reported using tobacco 180 days in the six months prior to interview (almost every day). |
Selection of SNPs and genotyping
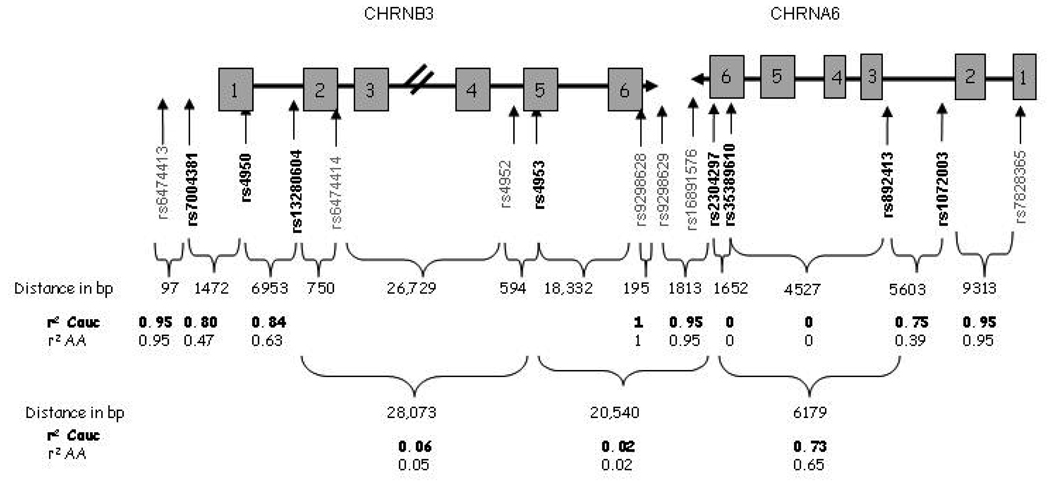
Candidate polymorphisms for the CHRNA6 and CHRNB3 genes were identified using the SNPbrowser Software version 3.5 from Applied Biosystems (http://www.appliedbiosystems.com), the public database, dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and other published research. SNPs were chosen to span the two genes, have moderate minor allele frequencies, and assay reliably. The majority of SNPs chosen were identical to those used in Zeiger et al except for a few additional SNPs added to examine regions where association had recently been reported. The structures of the CHRNA6 and CHRNB3 genes, and the SNPs selected, are shown in Figure 1, with SNPs examined in this study shown in bold. Other recently published SNPs are also included in grey to show proximity and spacing.
Figure 1.
Cartoon representation of the CHRNA6-CHRNB3 locus. Boxes represent exons separated by intronic regions (not drawn to scale). Eight SNPs were genotyped shown in bold, with their reference sequence numbers and gene locations indicated. The number of nucleotide base pairs (bp) between each SNP, and the linkage disequilibrium measured by r2, is also indicated.
Genomic DNA was isolated from buccal cell swabs and preamplified using the method of Zhang et al (Zhang et al, 1992). Data obtained using this DNA are high-quality; these methods have been shown to be reliable for genotyping (Anchordoquy et al, 2003; Haberstick and Smolen, 2004). A Biomek® 3000 Laboratory Automation Workstation (Beckman Coulter) was used to automate DNA genotyping assay preparation in a 384 well plate format. TaqMan® assays for allelic discrimination (Applied Biosystems) were used to determine SNP genotypes, per instructions of the manufacturer under standard conditions using an ABI PRISM® 7900 instrument. In total, eight SNPs located in the CHRNA6 – CHRNB3 region of chromosome 8 were genotyped in 1051 subjects.
Analytic Methods
Linkage disequilibrium (LD) and genotype quality were analyzed using Haploview (Barrett et al, 2005). This freely available software package allows for estimation of several LD statistics, including pairwise LD correlations, or r2. All SNPs were checked for Hardy Weinberg Equilibrium within ethnic groups using Haploview and further verified in SPSS version 15.0.
Analyses were conducted separately on the subset of genetically related individuals (227 families totaling 529 individuals). Single marker analyses were performed using PBAT (Lange et al, 2004). This statistical genetics program takes full advantage of the family structure, making use of both the between and within-family information (Fulker et al, 1999). All analyses were first conducted on the subset of “current frequent smokers” using the ethnic group information as covariates, and then conducted separately for the Caucasians. Three pairs of Monozygotic twins were removed and 10 suspected half siblings were removed from the data set prior to genetic analysis. Secondarily, because subject inclusion and recruitment criteria vary widely among genetic association studies, following the initial analysis, post hoc exploratory analyses were done to examine the effects of the inclusion criteria on the significance of the observed genetic associations. The inclusion criteria are described in Table 3.
Following the analysis of the siblings, subsequent analyses were conducted using the singleton individuals (i.e. those with no genetic data from other family members). The analysis of the singletons (which could not be included in the PBAT analysis due to lack of family genotypic information) was done in SPSS version 15.0 and SAS version 9.1 using Chi Square tests for binary traits and ANOVA/regression models for continuous traits. In SPSS and SAS ethnicity was included as a covariate when possible, and also the subset of Caucasians was examined separately. Thus two sub-samples were generated from the genetic sample: the group of individuals with sibships who were analyzed with PBAT, and the singletons group, analyzed using SAS and SPSS. Significance levels from the two samples were combined using Fisher’s Method where appropriate.
Results
Phenotype Analysis in the NYS-FS
The mean number of DSM-IV symptoms for each represented ethnic group, as well as each phenotypic subgroup is given in Table 4, along with the significance of group differences tested by ANOVA. Overall responses to the DSM-IV items were available for 1051 individuals with a mean symptom count of 1.4 ± 1.8. Within “current frequent smokers” the mean symptom count was 3.3 ± 1.6. No significant differences in DSM-IV symptoms were seen between the sexes, nor between ethnicities. Hispanic individuals tended to endorse fewer symptoms than Caucasians and African Americans, but this was not significant. As expected, number of quit attempts and nicotine dependence were correlated with an r2 = 0.35 (p < 0.001) in the whole sample and r2 = 0.30 (p < 0.001) within current frequent smokers.
Table 4.
Phenotype summary by ethnicity.
| Mean DSM-IV dependence symptom counts (n)* | |||||
|---|---|---|---|---|---|
| Sample | all | CEU | AA | Hispanic | American Indian |
| All | 1.4±1.8 (1051) |
1.5±1.9 (860) |
1.2±1.7 (132) |
0.9±1.5 (28) |
1.3±1.7 (12) |
| Smokers | 2.9±1.7 (488) |
3.0±1.7 (412) |
2.7±1.7 (56) |
2.1±1.8 (12) |
2.3±1.7 (7) |
|
Current frequent smokers |
3.3±1.6 (295) |
3.4±1.6 (245) |
2.9±1.7 (40) |
1.8±0.8 (5) |
2.8±1.8 (5) |
CEU = Caucasian, AA = African American
Individual SNP frequencies in different ethnic groups
The allele frequencies in the NYS were similar to those reported in the literature (Table 5). There was evidence for differences in SNP minor allele frequencies among ethnic groups, to the extent that the minor allele was different in Caucasians and African Americans. This dramatic difference in allele frequencies was seen in rs7004381, rs4950, rs13280604, rs35389610, rs892413, and rs1072003 (Chi-Square, all p<0.01). Only rs4953 showed no evidence of differences between allele frequencies in African Americans versus Caucasians (p = 0.30). The representation of other ethnic groups, such as Hispanics, is too small in this sample to obtain accurate allele frequencies and test for frequency differences. One SNP, rs35389610 was too rare to be informative in a sample of this size (MAF = 0.006) and was excluded from all genetic association analyses.
Table 5.
Allele frequencies and ethnic differences in the NYS-FS
| Reference sequence (rs) |
Location | Gene | Position Chr. 8 |
Relative position |
MAF sample |
MAF CEU* |
MAF AA* |
Minor Allele CEU* |
Minor Allele AA* |
P-value Eth. diff |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7004381 | Upstream | CHRNB3 | 42670318 | 0 | 0.285 | 0.227 | 0.399 | A | G | 0.0084 |
| rs4950 | 3’ UTR | CHRNB3 | 42671790 | 1472 | 0.291 | 0.215 | 0.258 | G | A | <0.0001 |
| rs13280604 | Intron 1 | CHRNB3 | 42678743 | 8425 | 0.298 | 0.218 | 0.299 | G | A | <0.0001 |
| rs4953 | Exon 5 | CHRNB3 | 42706816 | 36498 | 0.041 | 0.03 | 0.06 | C | C | 0.3 |
| rs2304297 | Exon 6 | CHRNA6 | 42725148 | 57038 | 0.295 | 0.237 | 0.271 | C | G | <0.0001 |
| rs35389610 | Intron 5 | CHRNA6 | 42726954 | 58690 | 0.006 | 0.007 | 0 | T | C | - |
| rs892413 | Intron 2 | CHRNA6 | 42727356 | 63217 | 0.267 | 0.212 | 0.308 | A | C | <0.0001 |
| rs1072003 | Intron 2 | CHRNA6 | 42729008 | 68840 | 0.236 | 0.186 | 0.345 | G | C | <0.0001 |
CEU = Caucasian, AA = African American
Block structure and interSNP correlations in the NYS were similar to those available from HapMap. Due to the large differences in allele frequencies, LD between SNPs was examined separately in African Americans and Caucasians. The patterns of LD and correlations are shown in Figure 1. In CHRNA6, rs892413 and rs1072003 are in high LD with an r2 of 0.75 in Caucasians and r2 of 0.39 in African Americans. These SNPs also form a weak block with rs2304297. Although rs35389610 is located between rs2304297 and rs89214, it is only polymorphic in Caucasians and so rare that the estimated r2 between it and any other SNP is effectively 0. SNPs rs7004381 and rs4950 upstream of CHRNB3 also showed high LD with r2 of 0.80 in Caucasians and 0.47 in African Americans. All SNPs were in Hardy-Weinberg Equilibrium (HWE) within self-reported ethnic groups.
Genetic Association
Primary Analysis
Association tests focusing on overall nicotine dependence and number of quit attempts were done using PBAT on the group of “current frequent smokers”. Significant associations in the NYS-FS are shown in Table 6. In cases where the p-value within Caucasians is different than across ethnic groups it is given. In examining overall nicotine dependence, rs2304297 showed association in Caucasians (p=0.0066). In the case of number of quit attempts, three SNPs at the upstream end of CHRNB3 showed significant association under a recessive model (rs7004381, p=0.0024; rs4950, p=0.021; rs13280604, p=0.024). Marker rs2304297 in the 3’ region of CHRNA6 was associated also with number of quit attempts in Caucasians (p=0.0044). Using a Bonferroni correction for multiple testing (8 SNPs and 2 phenotypes = 16 tests) the cutoff for a 5% significance level would be a p-value < 0.003. Only the association of rs7004381 with quit attempts is significant at this level. However, it is worth noting that this cutoff is overly conservative due to both correlations between the SNPs (LD) and between the two phenotypes.
Table 6.
Association results for significant SNP-phenotype associations “current frequent smokers”*.
| Gene | SNP | 3 or more dependence symptoms† |
quit attempts p-value† | ||||
|---|---|---|---|---|---|---|---|
| # inform Families |
risk allele |
additive | recessive | additive | recessive | ||
| CHRNB3 | rs7004381 | 58 | G | 0.60 | 0.39 | 0.011 | 0.0024 |
| CHRNB3 | rs4950 | 47 | A | 0.85 | 0.98 | 0.030 | 0.021 |
| CHRNB3 | rs13280604 | 49 | A | 0.65 (0.037) | 0.72 (0.09) | 0.074 (0.036) | 0.024 |
| CHRNB3 | rs4953 | 13 | G | 0.24 | 0.25 | 0.030 | 0.031 |
| CHRNA6 | rs2304297 | 43 | G | 0.91 (0.0066) | 0.56 (0.018) | 0.55 (0.0044) | 0.43 (0.013) |
| CHRNA6 | rs892413 | 46 | - | 0.72 | 0.68 | 0.96 | 0.64 |
p-values for association within current frequent smokers reporting at time of interview at least 180 days of smoking in past six months.
p-value within Caucasians given in parenthesis.
Examination of inclusion criteria
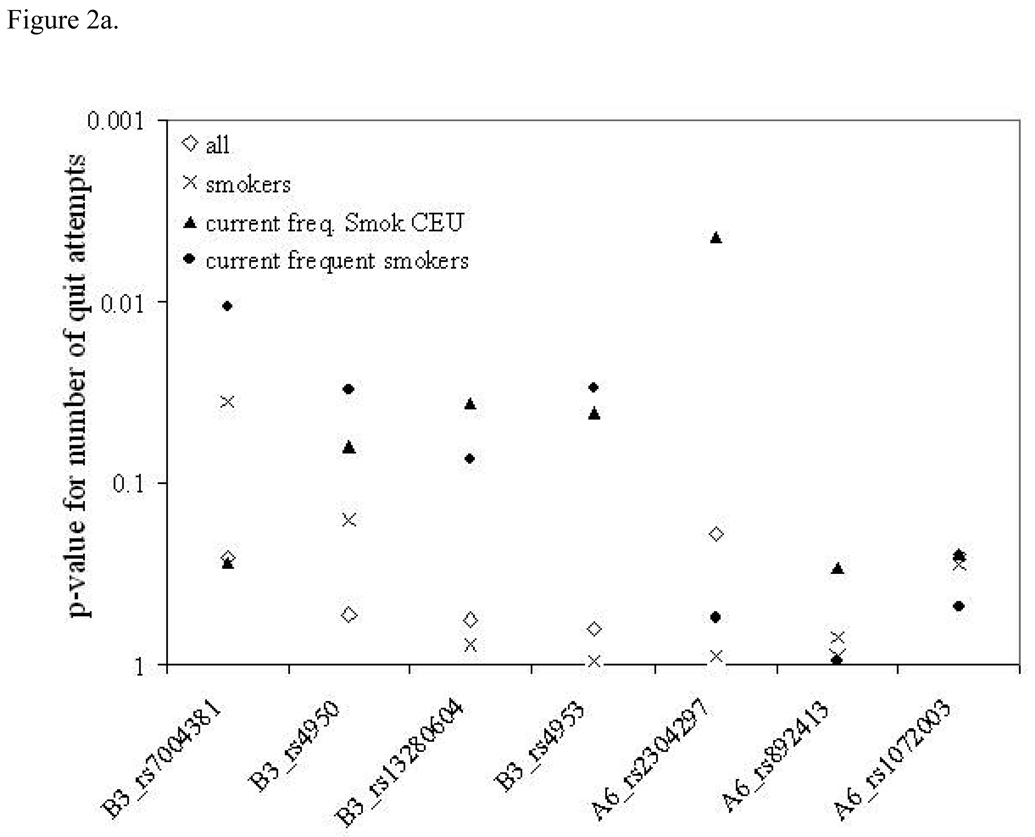
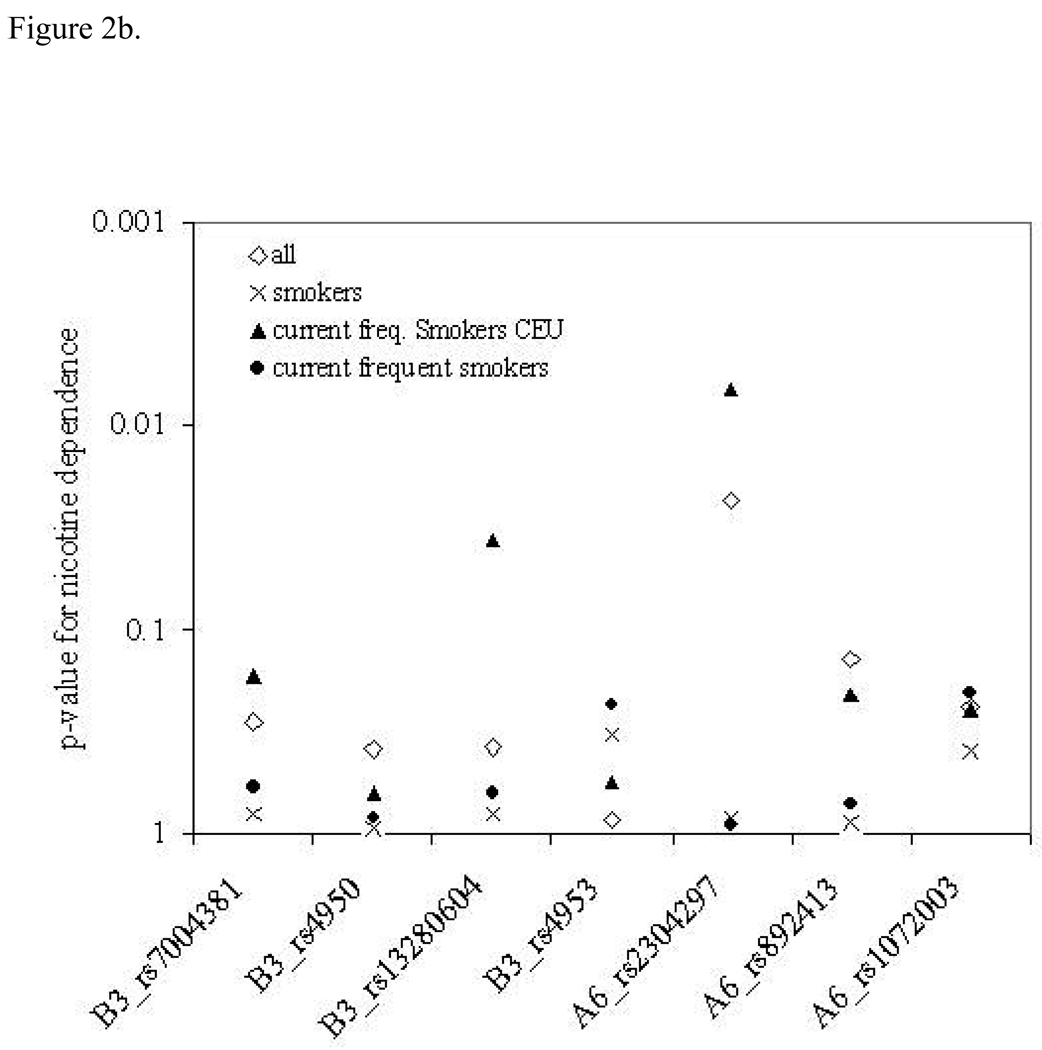
Since subject inclusion and recruitment criteria vary widely among genetic association studies we carried out further analyses to examine the effects of the inclusion criteria on the significance of the observed genetic associations. Figure 2 clearly shows that p-values are different within different subsets of the sample. In some cases, the more stringent inclusion criteria yielded stronger genetic associations, whereas in other cases association was most significant in the whole sample. This set of analyses was done last and conducted separately to examine the patterns of p-values across inclusion criteria. Individual p-values from these plots are not adjusted for multiple testing, because the goal was to examine the pattern, not to extract genetic significance of a single SNP.
Figure 2.
Association results for number of quit attempts (a) and nicotine dependence (b) and across the CHRNA6 CHRNB3 region. The y-axis shows p-values for each SNP across the x-axis for each of the three different sample subsets.
Singletons
Unrelated individuals who could not be included in the PBAT analyses were used to confirm the results. These singletons were examined for genetic association of SNPs with quit attempts and with overall dependence, using simple Chi Square and ANOVA analysis in SPSS. The left -most two columns of Table 7 show the results of these analyses in the current frequent smokers. rs7004381, rs4950, and rs13280604 in CHRNB3 were found to be associated with overall nicotine dependence using a recessive model (p=0.039, p=0.007, p=0.006), with corresponding odds ratios of: 3.36 (1.01 –11.11) for rs7004381, 4.62 (1.44 – 14.84) for rs4950, and 4.67 (1.46 – 14.95) for rs13280604. Fisher’s method was used to combine the p-values from the above-described PBAT sibpairs analysis with the results from the singletons, shown in the second set of columns in Table 7 (Combined). In addition to boosting the significance of the CHRNB3 SNP rs4950 to p=0.022, rs2304297 in CHRNA6 was also nominally associated with tobacco dependence (p=0.023). Results of analysis of all subjects are shown in the right half of Table 7. In the singletons, there was modest evidence for rs2304297 using an additive model (p=0.028), and modest evidence for rs13280604 and rs892413 with a recessive model (p=0.044 and p=0.024). After combining p-values with the PBAT sibpair results, there is some evidence for association of rs2304297 in CHRNA6 (p=0.011).
Table 7.
Association results for tobacco dependence within singletons and combined.
| Gene | SNP | Current frequent smokers | all | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Singletons | Combined | Singletons | Combined | ||||||||
| Add. | Rec. | N | Add. | Rec. | Add. | Rec. | N | Add. | Rec. | ||
| B3 | rs7004381 | 0.059 | 0.039 | 109 | 0.055 | 0.060 | 0.30 | 0.056 | 410 | 0.299 | 0.118 |
| B3 | rs4950 | 0.071 | 0.007 | 101 | 0.185 | 0.022 | 0.34 | 0.45 | 375 | 0.393 | 0.307 |
| B3 | rs13280604 | 0.661 | 0.006 | 105 | 0.114 | 0.004 | 0.50 | 0.044 | 388 | 0.500 | 0.044 |
| B3 | rs4953 | ns | ns | ns | ns | ns | ns | ns | ns | ||
| A6 | rs2304297 | 0.232 | 0.188 | 107 | 0.011 | 0.023 | 0.028 | 0.89 | 392 | 0.005 | 0.107 |
| A6 | rs892413 | 0.095 | 0.091 | 110 | 0.097 | 0.099 | 0.051 | 0.024 | 398 | 0.042 | 0.015 |
ns – not significant
Number of quit attempts was not normally distributed so the data were log transformed in an attempt to achieve the normality and satisfy homoscedasticity assumptions necessary for ANOVA, which were then sufficiently, but not perfectly, satisfied. rs7004381, rs4950, and rs13280604 showed a trend toward association with number of quit attempts in the singletons. For example, for rs7004381 individuals with AA genotype averaged 1.38 ±1.16 quit attempts versus 5.77 ± 2.22 in GG individuals; similarly at rs4950 individuals with GG genotype averaged 0.47 ± 0.27 quit attempts versus 4.38 ± 1.74 in AA individuals, and at rs13280604 individuals with CC genotype averaged 1.2 ± 0.82 quit attempts versus 5.81 ± 2.81 in AA genotype individuals. However, these failed to be significant due to a low response rate for this question which greatly reduced power (fewer than 100 individuals within current frequent smokers).
Discussion
Here we provide additional support for a role of the CHRNA6 and CHRNB3 genes in mediating nicotine dependence. This study has used a population-representative sample to show that three SNPs, rs7004381, rs4950, and rs2304297 are associated with tobacco dependence. To our knowledge, this is the first study to show evidence for association of variation in CHRNA6 and CHRNB3 with dependence measured by DSM-IV symptom endorsement. Therefore, results presented here suggest that association of tobacco dependence with these genes is present using different diagnostic criteria across samples, thus strengthening the evidence that they are likely to contribute to nicotine related behaviors.
In the context of this study, it is useful to compare results for specific SNPs, phenotypes, and samples to those that have been previously reported. To facilitate this process, Supplementary Table 1 summarizes results from the four other publications to compare with new data presented here. The fourth column gives the allele of each SNP present in the most common haplotype using data from the caucasian HapMap sample. It is important to recognize that some of the studies included subjects from other ethnic backgrounds, including hispanics in the CADD sample and African Americans in the Add Health sample, where allele frequencies were found to differ. Therefore, although the risk alleles for individual SNPs across studies are in agreement for much of this region, there are some instances where they differ. It is possible that certain alleles of specific SNPs exert different effects among different ethnic groups, consistent with the fact that in animal models, the effect of a gene deletion may differ depending on genetic background (Crabbe et al, 2006). In this NYS sample of adults, results from the DSM-IV analyses in the sibpairs sample suggest that SNP rs2304297 in CHRNA6 is associated with nicotine dependence (in Caucasians) and number of unsuccessful quit attempts. This SNP has been examined for its involvement in nicotine dependence in all of the other studies. Greenbaum et al (2006) did not find any evidence for association, as measured by FTND. However, results from a genome-wide association (Beirut et al, 2007), as well as a candidate gene study (Saccone et al, 2007) in the same sample, revealed a highly significant association between FTND-defined nicotine dependence and the same risk allele associated in this paper (G). Although Zeiger et al (2008) did not find evidence for association with nicotine dependence, there was suggestive evidence for association between rs2304297 and early subjective response to tobacco in both samples studied. Each of these samples consisted of adolescents/young adults (mean ages of 18.21 ± 1.5 and 22.4 ± 1.7), as was the relatively small sample in the Greenbaum et al (2006) paper (mean age 23.3 ± 2.3), which may include individuals who are still in an “experimental” phase of smoking and haven’t yet tried to quit. The NYS sample studied here, as well as the NICSNP sample used in Saccone et al (2007) and Bierut et al (2007), include older adults (mean ages 39 ± 3 and 45 ± 8, approximated from table 6 p45 in the Saccone et al (2007) paper). Therefore, of the five samples where rs2304927 was investigated, two have found evidence for early subjective response to tobacco (Zeiger et al, 2008), two have found evidence for nicotine dependence (Saccone et al, 2007; current study), and one did not find any evidence of association (Greenbaum et al, 2006).
In the CHRNB3 gene, SNP rs7004381 was found to be associated with number of unsuccessful quit attempts, which is correlated with, and a component of, nicotine dependence measured by DSM-IV. rs7004381 is in close proximity to rs6474413 and rs6474412 (97 bp and 663bp downstream respectively), rs6474413 was one of the top associations in the Saccone et al (2007) candidate genes study and rs6474412 was within the top 40 associations out of 2,400,000 markers in the Bierut et al (2007) study. This SNP of CHRNB3 is further upstream of the gene than any of those included the Greenbaum et al (2006) or the Zeiger et al (2008) studies. SNPs rs4950 and rs13280604, which are moderately associated with number of unsuccessful quit attempts in this NYS sample, have only been examined in the current study and the Zeiger et al (2008) study. Although that study did not find evidence for association with dependence, these two SNPs were significantly associated with early subjective response to tobacco. It is possible that the lack of association with nicotine dependence may be related to the fact that both samples were composed of adolescents and young adults. Number of unsuccessful quit attempts was also modestly associated with rs4953. While rare, this SNP was also found to be moderately associated with dependence in the Saccone et al (2007) study and the Zeiger et al (2008) study.
Although CHRNA6 and CHRNB3 are adjacent to each other it is biologically plausible that the α6 and β3 subunits may affect slightly different components of nicotine addiction. The β3 subunit is found in receptors lacking α6, and has no acetylcholine binding site. nAChR receptors with β3 localize with α3 and β4 in the interpeduncular nucleus and medial habenula, whereas receptors with β3, α6 and β2 are found in the substantia negra, VTA, striatum, and locus coeruleus (Gotti et al, 2006). Likewise, α6β2 containing receptors (lacking β3) are found in the superior colliculus and have been shown to be involved in GABA release (Gotti et al, 2006). Analysis of quit attempts suggests that this measure may have a strong underlying genetic component that can be explained in part by the CHRNA6 and CHRNB3 genes (Table 6).
In the singletons sample, the most significant associations with dependence were found for three SNPs in the CHRNB3 gene (rs7004381, rs4950, and rs13280604). Evidence for an association with the CHRNA6 gene was less consistent in the singletons (Table 7). However, although the sample size for the quit attempt variable in singletons was quite small, directional trends emerged that were consistent with findings in the families. The consistency of the results across this region further supports the findings in the larger set of families.
Our initial analysis focused on the subset of “current frequent smokers” which included individuals who reported using tobacco every day in the six months prior to interview. Our hypothesis for selection criteria seems to be supported because the strongest genetic association was detected in the comparison of dependence symptoms among daily smokers. Specifically, the mean number of symptoms in current frequent smokers with the genotype AA at rs7004381 is 2.2 ± 0.37 versus 3.4 ± 0.17 symptoms in GG individuals.
Reports in the literature support this idea that comparing exposed, regular users may be one way to tease out genes which contribute to dependence. For example, in the recent studies by Bierut et al (2007) and Saccone et al (2007) who used a minimum of smoking 100 or more times, they found highly significant associations with CHRNB3 SNPs and nicotine dependence (Bierut et al, 2007; Saccone et al, 2007). However, other studies that used more inclusive criteria did not find results that were as significant (Greenbaum et al, 2006). Replication of associations between specific SNPs and complex traits has been a challenge for researchers investigating and interpreting these studies. While lack of replication may be due to the initial finding being a false positive, there are many other sources of inconsistency between findings, including the definition of the phenotype as well as inclusion criteria of the sample. However, it is important to keep in mind that using highly stringent inclusion criteria can greatly reduce power due to reduced sample size and phenotypic heterogeneity. For this reason, we conducted a secondary analysis of our data whereby different subject inclusion criteria were evaluated to determine the effect of sample selection on the ability to detect association.
It is important to comment on the differences in allele frequencies that are present in Caucasians and African Americans, because ethnicity may be an important factor in the biological relevance of these SNPs. Even though all ethnic groups could be analyzed together when using the family-based PBAT analysis which controls for stratification, the association of rs2304297 became more significant when Caucasians were analyzed separately. This may be due to differences in the size of the LD blocks in Caucasians versus African Americans. There is only a small probability that rs2304297 is “the functional” allele, so the strength of association will depend on the level of LD between rs2304297 and the actual disease allele. Based on data from the HapMap project NCBI Build 35 (2003) as well as the results of genotyping presented here, the LD block which contains rs2304297 extends beyond exon 4 of CHRNA6 in Caucasians but is weaker in African Americans. In addition, as mentioned above, the differences between the results within ethnicities may be affected by genetic background, a well documented factor in genetically engineered mouse studies (Crabbe et al, 2006), but relatively unexamined in humans.
Based on the local sequence around these SNPs, there are several putative functional elements that may be affected by different alleles. The 5’ UTR of CHRNB3 shows evidence for a variety of putative transcription factor binding sites (as predicted by Transcriptional Element Search System: http://www.cbi.upenn.edu/tess). In particular rs4950 appears to be in close proximity to a binding site of the ubiquitous trans-activator AP1 (ID:T00029). In the case of CHRNA6 there are numerous predicted transcription factor binding sites, particularly near the SNPs in intron 2, but none of note near rs2304297. Polymorphisms such as rs2304297 would be more likely to effect expression through trafficking of mRNA, miRNA binding, and/or rate of degradation since it is the 3’ UTR (Brant-Zawadzki et al, 2007; Maziere and Enright, 2007).
We have provided evidence for the involvement of α6 and β3 in nicotine dependence measured by DSM-IV in a population-representative sample, further supporting the results in the literature for association of dependence defined by the Fagerstrom Test for Nicotine Dependence (FTND) with these genes in selected samples (Bierut et al, 2007; Greenbaum et al, 2006; Saccone et al, 2007). Furthermore, in our secondary analysis using different subsets of the sample based on phenotype definition, we also showed that sample selection can greatly affect the ability to detect association. Given the collection of studies that have now provided evidence for an association between these genes and nicotine behaviors, it will be important for future studies to focus on understanding the molecular functional consequences of different SNPs. This should include work aimed at identifying all of the variation in the region, followed by in vitro and in vivo approaches to study the effects of these variants. Ultimately, it will take a careful integration of results from human genetic studies with laboratory based methods in order to tease apart the complex underlying genetic mechanisms which contribute to risk for nicotine dependence.
Supplementary Material
Acknowledgements
This work was supported by NIH grants AA015336 (MAE), AA007464 (NRH), DA017637 (IS, NRH), and AA11949-03 (NYS-FS).
Footnotes
Disclosures and Potential Conflicts of Interest
Dr. Nicole R. Hoft, Isabel R. Schlaepfer, Dr. Robin P. Corley, Dr. Matthew B. McQueen, Dr. David Huizinga, and Dr. Marissa A. Ehringer do not have any potential conflicts of interest, financial or otherwise, relevant to the subject matter of this work.
References
- The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of three candidate genes after whole-genome preamplification of DNA collected from buccal cells. Behav Genet. 2003;33:73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant-Zawadzki PB, Schmid DI, Jiang H, Weyrich AS, Zimmerman GA, Kraiss LW. Translational control in endothelial cells. J Vasc Surg. 2007;45 Suppl A:A8–A14. doi: 10.1016/j.jvs.2007.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champtiaux N, Gotti C, Cordero-Erausquin M, David DJ, Przybylski C, Lena C, et al. Subunit composition of functional nicotinic receptors in dopaminergic neurons investigated with knock-out mice. J Neurosci. 2003;23:7820–7829. doi: 10.1523/JNEUROSCI.23-21-07820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottler LB, Keating SK. Operationalization of alcohol and drug dependence criteria by means of a structured interview. Recent Dev Alcohol. 1990;8:69–83. [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, et al. The beta3 nicotinic receptor subunit: a component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23:11045–11053. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DS, Huizinga D, Menard SW. Multiple problem youth : delinquency, substance use, and mental health problems. New York: Springer-Verlag; 1989. [Google Scholar]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Riganti L, Vailati S, Clementi F. Brain neuronal nicotinic receptors as new targets for drug discovery. Curr Pharm Des. 2006;12:407–428. doi: 10.2174/138161206775474486. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, et al. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11:312–322. doi: 10.1038/sj.mp.4001774. 223. [DOI] [PubMed] [Google Scholar]
- Haberstick BC, Smolen A. Genotyping of three single nucleotide polymorphisms following whole genome preamplification of DNA collected from buccal cells. Behav Genet. 2004;34:541–547. doi: 10.1023/B:BEGE.0000038492.50446.25. [DOI] [PubMed] [Google Scholar]
- Hymowitz N, Cummings KM, Hyland A, Lynn WR, Pechacek TF, Hartwell TD. Predictors of smoking cessation in a cohort of adult smokers followed for five years. Tob Control. 1997;6 Suppl. 2:S57–S62. doi: 10.1136/tc.6.suppl_2.s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MJ. Why people smoke. Bmj. 2004;328:277–279. doi: 10.1136/bmj.328.7434.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Maziere P, Enright AJ. Prediction of microRNA targets. Drug Discov Today. 2007;12:452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Mineur YS, Picciotto MR. Genetics of nicotinic acetylcholine receptors: Relevance to nicotine addiction. Biochem Pharmacol. 2008;75:323–333. doi: 10.1016/j.bcp.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, et al. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65:1526–1535. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Rosenman RH, Fabsitz RR, Christian JC. Smoking and alcohol consumption in adult male twins: genetic heritability and shared environmental influences. J Subst Abuse. 1990;2:39–50. doi: 10.1016/s0899-3289(05)80044-6. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17:724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.