Abstract
Macrocyclic scaffolds are particularly attractive for designing selective G-quadruplex ligands essentially because, on one hand, they show a poor affinity for the “standard” B-DNA conformation and, on the other hand, they fit nicely with the external G-quartets of quadruplexes. Stimulated by the pioneering studies on the cationic porphyrin TMPyP4 and the natural product telomestatin, follow-up studies have developed, rapidly leading to a large diversity of macrocyclic structures with remarkable-quadruplex binding properties and biological activities. In this review we summarize the current state of the art in detailing the three main categories of quadruplex-binding macrocycles described so far (telomestatin-like polyheteroarenes, porphyrins and derivatives, polyammonium cyclophanes), and in addressing both synthetic issues and biological aspects.
1. Introduction
G-rich DNA strands are naturally involved in duplex-DNA architecture through association with their complementary C-rich DNA strands by the canonical Watson-Crick pairing [1–3]. However, a growing body of evidence currently testifies that this canonical association is not the unique mode of stabilization of G-rich DNA in cells. Indeed, given that four guanine residues can self-associate in a planar arrangement through a Hoogsteen-type hydrogen-bonding network [4] to form a structure called G-quartet, G-rich strands can adopt a peculiar three-dimensional arrangement called G-quadruplex DNA [5–9] resulting from the stacking of several contiguous G-quartets (Figure 1).
Figure 1.
Schematic representation of the folding of an oligonucleotide that mimics the human telomeric sequence (d[AG3(T2AG3)3]). The polymorphism of the quadruplex is represented through the various possible structures, namely, the hybrid (left), antiparallel (centre), and parallel (right) forms; these structures differ by strand orientation (grey dashed arrows) and loop arrangement (represented in orange, yellow, and red).
The formation of G-quadruplex DNA is easily conceivable in DNA sequences that are present as single strands in cells, such as the telomeric overhang. The structural and functional integrity of this overhang is based on its association with a constellation of specific proteins, some of them belonging to the shelterin complex [10]. Altogether, this nucleoproteic assembly caps the chromosomes, protects their integrity, and is also deeply involved in the telomeric replication process [11–13]. Numerous studies currently suggest that quadruplex formation in this overhang alters the structure and function of telomeres, inducing a damage response and rapid apoptosis in particular in cancer cells [14–24]. Thus, over the past decade telomeric G-quadruplex DNA has been thoroughly studied with an initial focus on the possible interferences with telomerase activity [25].
The existence of G-quadruplex DNA is also heavily implied in the promoter region of genes and oncogenes, and is thus assumed to play an important regulatory role in their transcription [26]. It has been indeed demonstrated that the involvement of G-rich sequences in duplex architecture is compatible with their folding into quadruplex structures, thanks to the breathing of duplex DNA [27]. Nevertheless, we have to keep in mind that these G-quadruplex-forming sequences although transiently single-stranded are a priori not as easily accessible as the telomeric sequences, due to the presence of the transcription machinery [28].
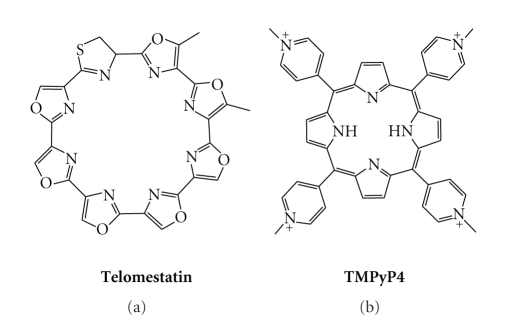
Stabilization of quadruplex architecture by small molecules is thus emerging as a potential anticancer approach since it is thought to interfere with oncogenic expression and telomeric maintenance in cancer cells [11–13]. Interestingly, several classes of small molecules have been developed that efficiently target G-quadruplex DNA [29–32]. Among them, macrocycles rapidly became popular for recognition of quadruplexes motivated by the fact that telomestatin is one of the most active known G-quadruplex ligands [33]. This natural 8-ring 24-membered macroheterocycle (Figure 2) displays high affinity for quadruplex and most importantly has no affinity for duplex DNA (vide infra). Nevertheless, apart from several reports on telomestatin-like molecules, true polyheteroaryl analogues are still scarce, essentially because of the difficulty to synthesise such molecules. Another well-known G-quadruplex ligand—and arguably one of the most studied—is TMPyP4 [34, 35], a tetramethylpyridinium porphyrin (Figure 2). This molecule has been widely used, essentially due to its great affinity for several quadruplex targets, as well as its commercial availability. However, the interest in employing TMPyP4 is somewhat counterbalanced by its lack of quadruplex selectivity (vide infra).
Figure 2.
Chemical structures of telomestatin and TMPyP4 (with p-CH3(C6H4)SO3 − as counterions).
These two examples illustrate the main reasons that made the macrocyclic scaffold particularly interesting for targeting G-quadruplex DNA: (i) a broad aromatic surface that favours the stacking interactions with the external G-quartets of the quadruplex, (ii) a rigid structure that maximizes the quartet overlap and, combined with their large size, impedes intercalation into duplex DNA, and (iii) in the case of TMPyP4, a cationic charge that promotes the electrostatic interactions with the negatively charged biopolymer. Unfortunately, the high cationic charge of TMPyP4 represents both an advantage and a drawback due to the nonspecificity of electrostatic interactions which promote association with any form of DNA, but in particular duplex DNA, thereby decreasing the binding selectivity (vide infra).
A great deal of research effort around the macrocyclic scaffold was thus motivated by these two examples. The macrocyclic systems described up to now as G-quadruplex ligands can be divided in three different categories: (i) telomestatin like (i.e., neutral and rigid macrocycles), (ii) porphyrin (TMPyP4) like (i.e., cationic and rigid macrocycles), and (iii) the less studied family of flexible polyammonium macrocycles (i.e., cationic and non planar), which will be described herein also in the companion paper, “One Ring to Bind Them All”—Part II, by A. Granzhan et al.) in the present issue.
2. Telomestatin-Like Macrocycles
Telomestatin is the benchmark compound in terms of G-quadruplex recognition (Figure 2). This natural compound was isolated from Streptomyces anulatus in 2001 by Shin-ya's group [33], and has been extensively studied due to its outstanding selectivity for G-quadruplex and highly promising biological properties.
One general method to quantify the DNA affinity of a given molecule is to perform thermal denaturation experiments, either in a UV-monitored melting assay [36] or in the so-called “FRET-melting” assay. Here the thermal unfolding of the quadruplex-forming oligonucleotide F21T (FAM-G3[T2AG3]3-Tamra), doubly labelled with a complementary pair of FRET donor and acceptor (FAM: 6-carboxyfluorescein and Tamra: 6-carboxytetramethylrho-damine), is monitored via FRET (fluorescence resonance energy transfer) [37]. Semiquantitative evaluation of ligand-binding affinity is obtained by measuring the increase in melting temperature induced by the ligand (ΔT 1/2). The quadruplex-over-duplex-DNA selectivity may then be established via competitive FRET melting which is carried out in presence of competitive duplex DNA (ds26). In the presence of telomestatin, the increase in melting temperature is very large (ΔT 1/2 = 24°C), and since this value is unaffected by the presence of up to 50 equiv. of competitive duplex DNA (herein ds26, the self-complementary sequence 5′-CA2TCG2ATCGA2T2CGATC2GAT2G-3′), telomestatin stands among the most selective of G-quadruplex ligands. This property has been subsequently confirmed by independent studies using various evaluation techniques.
The enthusiasm for telomestatin was also justified by its ability to inhibit telomerase [38], with the reported IC50 value lying, impressively, in the nanomolar range (IC50-TRAP = 5 nM) [33]. However, the relationship between TRAP assay and telomerase inhibition was recently challenged on the grounds that this assay does not actually reflect the influence of the ligand on the activity of the enzyme but rather the ability of the ligand to inhibit the PCR amplification step [25]. The direct assay, a more constraining method based on the telomerase elongation of a telomeric primer with incorporation of [α-32P]dGTP, provides a more reliable estimation of the telomerase inhibition ability [25]. Interestingly, telomestatin is still highly active, staying among the best reported inhibitors, with an IC50 value of 58 nM.
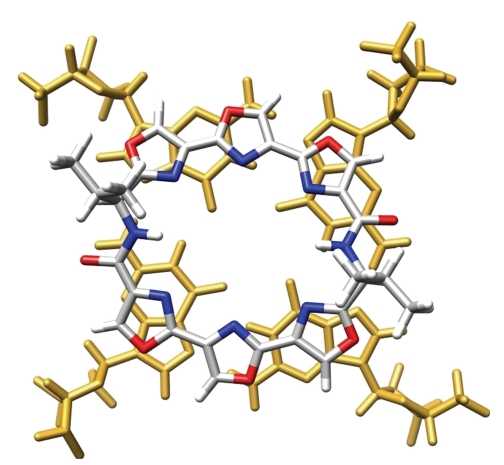
The structure of telomestatin is quite unusual since it is a neutral polycyclic compound, composed of five oxazole, two methyloxazole, and one thiazoline rings, whose overall flatness is impeded by the presence of an s p 3 carbon within the thiazoline ring (Figure 2). Whether this molecule is aggregated in water is yet not known; however, it is likely that the strong hydrophobic character of the molecule should reinforce the hydrophobic forces that contribute to stacking interactions, and so explain its exceptional effectiveness in binding G-quadruplex. This efficiency is also assumed to rely on a perfect shape adaptation between the macrocycle and a G-quartet (Figure 3) [39].
Figure 3.
Qualitative in silico superposition of telomestatin and a G-tetrad (extracted from PDB entry: 2A5R); guanine residues appear in gold; the carbon, nitrogen, oxygen, sulphur, and hydrogen atoms of telomestatin appear in grey, blue, red, yellow, and white, respectively.
Abundant biophysical and biological investigations have been performed with telomestatin. For example, it exhibits an antiproliferative activity against a wide variety of cancer cell lines with IC50 values between 0.1 and 5 μM, including human pancreatic carcinoma [40], myeloid leukaemia [41, 42], breast cancer [43], cervical [43], tumoral HT1080 [44], telomerase-transformed SW39 [45], ALT-transformed SW26 [45], immortalized EcR293 [46], and a panel of myeloma [47] and neuroblastoma lines [48]. Remarkably, telomestatin does not affect normal cell lines, with no effect observed at 5 μM on fibroblast MRC-5 cells [43]. This observation has recently been substantiated by the finding that telomestatin does not interfere with telomere replication in normal cells [49]. Telomestatin was also a tool of choice to investigate the biological role of G-quadruplex ligands in cells by suggesting that effects of ligands might originate essentially from the disruption of the telomeric structure with subsequent displacement of protective proteins (shelterin complex) rather than in telomerase inhibition [44, 46]. Telomestatin was used to investigate the role of putative quadruplex formation within the promoter region of genes like VEGF [50] and hTERT [51] or oncogenes like c-Myc [52], Bcl-2 [53], and RET [54], whose corresponding proteins are overexpressed in some cancers. Recently, telomestatin was shown to interfere with the ability of helicases to unwind G-quadruplex structures [55], and to affect the growth of telomerase-negative ALT (Alternative Lengthening of Telomeres) cell lines via an indirect interaction with a proteinic complex comprised of the shelterin component TRF2, the helicase BLM, and the enzyme Topoisomerase IIIα [56]. The reason why telomestatin—or more generally G-quadruplex ligands—selectively affects tumour cell lines is yet not fully understood. However, differences in plasmic membrane permeability between normal and cancer cells or in the accessibility of the telomere and in particular variations in the composition of the shelterin complex have been proposed; even if the accessibility is identical, one can imagine that the cellular responses are different.
Unexpectedly, only two studies have addressed in detail the actual binding mode of telomestatin to G-quadruplex using in silico investigations [57, 58]. This is particularly surprising, given that there are no structural characterizations of this interaction (NMR, X-ray crystallography) available to date. These studies support concomitant double endstacking on quartets, with a preference for the parallel conformation of the human telomeric quadruplex. This conclusion is somewhat in contradiction with those of other studies based on CD spectroscopy [59] or 125I-radioprobing [60], underlining that further efforts are required to clarify the interaction mode of telomestatin with quadruplexes.
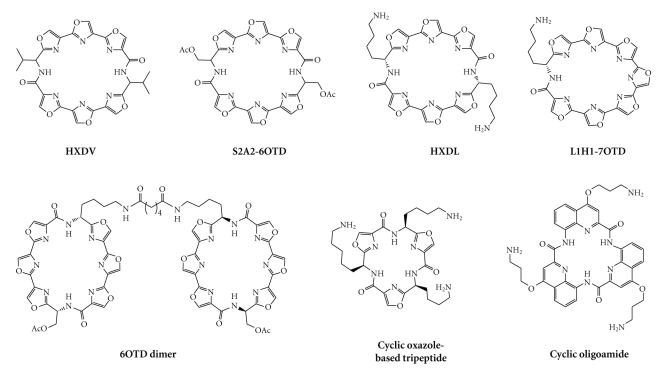
Nevertheless, one major concern is that telomestatin is difficult to synthesise. Despite considerable synthetic efforts to obtain either fragments of telomestatin or telomestatin-like arrays of polyoxazoles [61–65], its total synthesis has been reported only recently [66], and the complexity of the proposed pathway is incompatible with large-scale preparation. This can explain why, whilst the excellence of telomestatin towards quadruplex-recognition can hardly be exaggerated, relatively few reports on telomestatin-like compounds have appeared in the literature. Among them, two hexaoxazole macrocyclic ligands have been independently reported by Rice et al. [67, 68] and by Shin-ya, Nagasawa et al. [69]. The structure of these macrocycles was based on the dimeric association of two symmetrical trioxazole moieties through an amino-acid linker, either a valine (for HXDV, Figure 4) or a protected serine (for S2A2-6OTD, Figure 4), respectively.
Figure 4.
Chemical formulae of telomestatin-related macrocycles.
HXDV has been widely investigated and found to profoundly stabilize the telomeric G-quadruplex structure by UV-melting assay (ΔT m = 24°C), without any significant binding to duplex DNA [67, 68]. Its association with a quadruplex-forming sequence mimicking the human telomeric sequence has recently been confirmed to be based on external stacking, with occupancy of both external G-quartets. Interestingly, the high quadruplex selectivity of HXDV is assumed to originate from its particular telomestatin-like concave shape since the two planes defined by the trioxazole moieties present an angle of ~30°, along with the disposition of the two amino-acid residues on the same face of the cycle, thereby optimizing stacking interaction with quartets (Figure 5) [39]. The biological effects of HXDV have been investigated on human lymphoblastoma (RPM1) and murine leukaemia (P388) cell lines, with IC50 values for inhibition of the population doubling in the low micromolar range (0.4 and 0.5 μM, resp.) [67, 68]. Very interestingly, it has also been shown recently that HXDV elicits antiproliferative effect on both telomerase-positive (HeLa, A875, PC3-1 with IC50 between 0.2 and 0.6 μM) and telomerase-negative cell lines (like SAOS2 or GM847 with IC50 between 0.4 and 0.5 μM) mainly through an M-phase inhibitory effect, thereby contributing to the elucidation of the actual biological effects of G-quadruplex ligands [70].
Figure 5.
Qualitative in silico superposition of HXDV and a G-tetrad (extracted from PDB entry: 2A5R); guanine residues appear in gold, the carbon, nitrogen, oxygen, and hydrogen atoms of HXDV in grey, blue, red and white, respectively.
Interestingly, to circumvent the solubility problems, various HXDV derivatives have been prepared based on the modification of the nature of the side chain(s), from two valine residues (for HXDV), to only one valine [71], one valine and one protected lysine (HXLV-AC) [72], and two lysine residues (HXDL, Figure 4) [73]. Of all these derivatives, HXDL appeared as the most promising, whilst it is not neutral at physiological pH (due to protonation of both terminal amine groups, whose pK a values are estimated as 9.9 and 10.5) [74] and the characteristics of its association with quadruplex are impressive in terms of both affinity (ΔT m = 49°C) and selectivity, since only residual stabilization of salmon testes duplex-DNA is observed. This gain in efficiency is assumed to originate in a higher solubility and in enhanced interactions with quadruplex-DNA, mixing both tetrad-stacking and electrostatic interactions.
The success obtained with HXDL was concomitantly—and independently—confirmed by Nagasawa et al., who developed a series of G-quadruplex binding macrocyclic hexaoxazoles (6OTD for 6-Oxazole Telomestatin Derivatives). The very first example of this series, S2A2-6OTD (Figure 4), [69] was rapidly surpassed by more active compounds, in which the serine residues of the side chain were replaced by lysine (L2H2-6OTD, also called HXDL by Rice et al.) or arginine residues (L2G2-6OTD) [75]. Both compounds were found to be active in vitro in a range similar to that of telomestatin, notably via PCR stop assay (IC50 down to 0.64 μM), TRAP assay (IC50 down to 20 nM), and inhibition of the growth of HeLa cells (IC50 down to 0.5 μM). Current strategies for improvement include the dimerisation of the 6OTD scaffold (6OTD dimer, Figure 4) [76], using a bisamide linker between the two 6OTD moieties, which is long enough to span the thickness of the quadruplex architecture (~15 Å). Unexpectedly the affinity of the dimer so obtained is not significantly increased as compared with the corresponding monomer, as judged by FRET-melting data (ΔT m = 25.1 versus 25.0°C, resp.) and PCR stop assays (IC50 = 3.0 versus 2.9 μM, resp.). On the other hand, the dimer is more selective than the monomer since a ~10-fold improvement is obtained when comparing the inhibitory IC50 values measured by PCR stop assay allowing the authors to claim a 800-fold selectivity. The fact that the dimer is able to form a unique 1 : 1 complex with G-quadruplex, as shown by ESI-MS analysis, added to a proven inability to interact with duplex-DNA, implies that this dimeric scaffold is optimized for quadruplex recognition. Further improvement was found by replacing the hexaoxazole scaffold by the more planar heptaoxazole analogue (L1H1-7OTD, Figure 4) [77]. This compound was shown to interact strongly with quadruplex DNA (as judged by PCR stop assay, IC50 = 0.67 μM), with a fair quadruplex-over-duplex DNA selectivity (~8-fold). Furthermore, it displays a selective cytotoxicity toward telomerase-positive cells (HeLa, IC50 = 2.2 μM) as compared to telomerase-negative cells (Saos-2, IC50 > 30 μM). It is thus highly probable that a dimeric version of the 7OTD scaffold will appear in a near future and that it will help this scaffold to improve its quadruplex-over-duplex DNA selectivity.
Other series of oxazole-based macrocycles have been investigated as G-quadruplex binders and notably macrocycles whose structure is based on a trisoxazole [78], trisfuran, or tetrafuran scaffolds [79] (Figure 4). Amino-terminated chains have been introduced to improve water solubility and favour electrostatic interactions with DNA (the pK a values of the amino side chains depicted in Figure 4 standing in the 8.8–10.7 range) [74], with the chain length finely tuned to reach the quadruplex grooves. The intrinsic qualities of these compounds have been evaluated through FRET-melting assay, and whilst modest results were obtained with the oxazole-based tripeptide (ΔT 1/2 = 6.4°C), moderate (ΔT 1/2 = 10°C) to good (ΔT 1/2 = 17°C) results were obtained with furan-based tri- and tetrapeptides, respectively. Interestingly, when performed with duplex DNA, no stabilization occurred in any case, thus inferring a high quadruplex-selectivity of this series of ligands. But a real breakthrough was achieved in increasing the aromatic surface of the ligands, since stabilizations higher than 30°C were obtained with the aminoquinoline-based cyclic oligoamide depicted in Figure 4 [80, 81]. Once more, this compound retains a high selectivity for quadruplex DNA, since no stabilization was detected when a FRET-melting assay was performed with duplex hairpin-DNA. It is worth noting that this compound is also able to discriminate various types of quadruplex DNA (human telomeric versus oncogenic c-kit), thus opening perspectives for discovery of a novel generation of selective and specific G-quadruplex ligands. Additionally, the very attractive in vitro biophysical results were also complemented by the low general cytotoxicity of the cyclic oligopeptides.
Despite the real viability of the neutral macrocyclic scaffold for G-quadruplex recognition, the equilibrium between hydrosolubility and activity is difficult to achieve. One improvement has been the introduction of amino side chains that increase water-solubility and electrostatic attraction with the DNA target without altering the duplex-versus-quadruplex selectivity of the ligand, which relies on the shape and rigidity of the polyhetroaryl cyclic scaffold itself. Such molecular designs have thus to be further investigated, in order to gain more insights in terms of bioavailability and selective cytotoxicity.
3. Porphyrin- (TMPyP4-) Like Macrocycles
Along with neutral and cationic cyclic polyheteroarenes, cationic porphyrins are probably the most widely used macrocyclic G-quadruplex ligands. TMPyP4 (for 5,10,15, 20-tetra(N-methyl-4-pyridyl)porphyrin) [34, 35] is a representative example of this family of ligands (Figures 2 and 6) [39]. The ability of TMPyP4 to interact with DNA was first mentioned 30 years ago [82–84] whilst its use as a G-quadruplex ligand was reported for the first time twenty years later [34, 35, 85]. Since these seminal reports, this tetracationic porphyrin has been extensively studied: it has demonstrated a high affinity for G-quadruplex DNA (as monitored by FRET-melting assay, ΔT 1/2 = 17°C) [86], but with low selectivity [86–91]. TMPyP4 has been particularly used as a tool to investigate the possibility of downregulating the expression of genes due to quadruplex formation or induction (like c-myc [92–100], RET [54], HIF-1α [101], VEGF [50, 102, 103], Bcl-2 [53, 104, 105], KRAS [106], PDGF-A [107], c-kit [108], and even hTERT genes [51]). The biological behaviour of TMPyP4 has also been investigated in vitro against a broad variety of tumor cell lines (with IC50 between 23 and 310 μM), including human pancreatic [40], breast [85], and prostate carcinomas [85], lymphoma [85], retinoblastoma [109], leukaemia [110] and human gastrointestinal stromal tumor [108] cell lines, as well as telomerase-transformed SW39 [45], ALT-transformed SW26 [45], transformed human breast [85] and HeLa cell lines [111]. TMPyP4 has also been tested on normal cell lines (human fibroblast and breast cell lines) [85], in which it demonstrated moderate to acute cytotoxicity (IC50 down to 13 μM). It has also been used as an in vitro tool for more prospective and fundamental studies [112–115], sometimes exhibiting some unexpected behaviour, including quadruplex-unfolding activity on both quadruplex-forming DNA (d(CGG) repeats [116], or antithrombin aptamer) [117] and RNA sequences (r(CGG) repeats) [118].
Figure 6.
Interaction of TMPyP4 and a G-tetrad (adapted from PDB entry: 2A5R); guanine residues appear in gold; the carbon, nitrogen, oxygen, and hydrogen atoms of TMPyP4 appear in grey, blue, red, and white, respectively.
Interestingly, despite the fact that it was quickly acknowledged that TMPyP4 has poor selectivity for quadruplex structures [86–91], this ligand is still the focus of an intense research interest. Notably, a lot of structural studies are currently undertaken to investigate its binding mode with quadruplex DNA; interestingly, TMPyP4 displays various quadruplex-interactions, ranging from intercalation between adjacent G-quartets (as suggested via Raman spectroscopy studies) [119] to the more expected stacking on the external G-quartet (demonstrated by NMR studies) [120], passing by a mode totally devoid of direct contacts with G-quartets (shown by X-ray analysis of obtained crystal structures) [121], without excluding the possibility of combining several binding modes during a single recognition process (vide infra). These efforts are also currently completed by investigations dedicated to the understanding of the nature of the interaction between TMPyP4 and G-quadruplex, either in silico [100, 122, 123] or via established biophysical methods, UV-Vis [105], steady-state or time-resolved fluorescence [124–127], CD [128–130] or Raman spectroscopies [119], as well as ITC [99, 131–133], ESI-MS [134], DSC [135], SPR [90], or even HPLC methods [136], on a broad variety of quadruplex architectures, for example, telomeric [90, 119, 125, 127, 128, 130–133, 135], c-myc [99], c-kit [108], Bcl-2 [105, 134], TBA [136], the bimolecular (G4T4G4)2 [124], or the tetramolecular (TG4T)4 quadruplexes [126], and even G-wires [129]. Nevertheless, it is worth pointing out that a vast majority of these studies have been carried out in in vitro conditions that have to be considered as “dilute” as compared to the crowded environment within a living cell (due to the presence of naturally occurring proteins, nucleic acids, sugars, etc.). Interestingly, when the conditions are artificially crowded (using molecular crowding agents such as ethylene glycol [130] or poly(ethylene glycol) [125, 132], TMPyP4 displays a higher affinity (for quadruplex-DNA) [125, 130] and selectivity (with regard to duplex DNA) [132] than in dilute conditions, on the basis of a different recognition process (the crowded conditions influencing both the quadruplex DNA structures and the TMPyP4 stacking interactions, becoming multiple but stepwise [125, 132]). Given that crowded conditions are more biologically relevant, these results indicate that TMPyP4 may be more selective than anticipated in cells, and these observations will certainly revitalize TMPyP4-related researches.
Thus, the current status of TMPyP4 is the opposite of telomestatin; it is both commercially and synthetically accessible and its interaction with quadruplexes, although multiple, is fully characterized. Finally, the simple geometric comparison allowed by the overlaps shown in Figures 3 and 6 strongly suggests that the tetrapyrrolic macrocycle is much less suitable than the octacyclic ring for optimizing aromatic-aromatic interactions with a G-quartet. In conclusion, although TMPyP4 remains an interesting tool that has contributed to the understanding of the ligand–quadruplex interactions and to their complexity, its biological use for probing the involvement of quadruplex in biological assays is questionable.
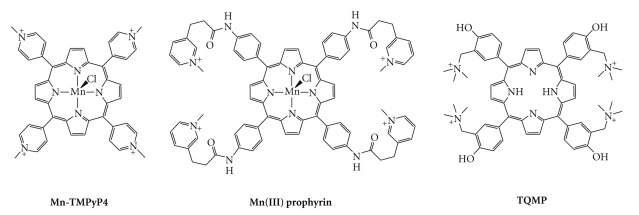
With an inner cavity particularly suited to coordination of a metal, TMPyP4 (and other porphyrins, vide infra) has been also widely exploited to form metal complexes. Given the well-known modulation of DNA association by metallation, this modification was thus performed in order to improve the DNA recognition [82–84]. TMPyP4 has now been used to prepare Pt(II), Cu(II), In(III), Zn(II), Co(II), Fe(III), Ni(II), Mn(III), Mg(II), and Pd(II) complexes [34, 137–142]. Among them, Mn-TMPyP4 (Figure 7) deserves particular attention since it showed a strong improvement in terms of quadruplex-versus-duplex selectivity.
Figure 7.
Chemical formulae of porphyrin-based G-quadruplex ligands.
The porphyrin core, as clearly demonstrated with TMPyP4, has been extensively explored as a macrocyclic scaffold for the design of G-quadruplex ligands. Among the numerous examples that have been reported, either as metal free or in a complexed state, the most interesting ligands are presented in Figure 7. Even if the methods for analyzing G-quadruplex affinity and selectivity are not yet standardised, the nature of the side arms that surround the porphyrin core is unmistakably crucial. Indeed, while the above mentioned Mn-TMPyP4 was able to interact with G-quadruplex 10 times faster than with duplex DNA, a 10,000-fold difference was obtained by moving the N-methylpyridinium moieties from the close vicinity of the porphyrin core to the termini of flexible arms [143]. This modification is assumed to greatly improve the electrostatic interaction with quadruplex grooves, while impeding intercalation into duplex DNA. This kinetic difference was evaluated through biosensor-surface plasmon resonance (SPR) measurements. The SPR technique, based on the use of surface-immobilized oligonucleotides, enables a quantitative analysis of the binding parameters, that is, thermodynamic (equilibrium constant, Gibbs energy of binding, and stoichiometry) and kinetic (k on/k off) parameters of the interaction of small molecules with the target DNA [90, 144]. In the present case, the affinity constant determined with duplex DNA lies in the 104 M−1 range, reaching up to 108 M−1 with quadruplex-DNA. Even if more modest, similar observations were made with TQMP, a porphyrin in which N-methylpyridinium substituents were replaced by (trimethylammonium)methyl-phenol arms (Figure 7) [91]. In this case, an ~100-fold difference in the kinetics of the association with duplex (104 M−1 · s−1 range) and quadruplex DNA (106 M−1 · s−1 range) was obtained. Finally, knowing that the nature and number of the surrounding side arms can be finely tuned in the case of TMPyP4 [145, 146], interesting perspectives exist to modify porphyrin-based G-quadruplex ligands in this way. It is, however, worth noting that the equilibrium between affinity and selectivity is difficult to access with the TMPyP4 scaffold, as demonstrated by a recent study in which a TMPyP4-related compound was included in a high-order supramolecular architecture (ruthenium coordination cubes) [147], on the basis of the elegant idea to use the metal itself to build the macrocyclic scaffold [148]. These cubic complexes were indeed shown to strongly bind not only quadruplex-DNA but also duplex-DNA, despite their peculiar molecular volume. This lack of selectivity may originate in their highly cationic nature (eight positive charges).
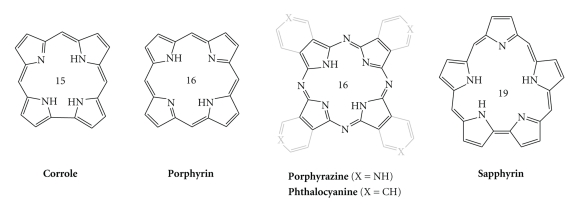
A large number of structural analogues of TMPyP4 have been recently described in the literature, differing notably by the nature of the side-arms (N-methylpyridinium groups have, for example, been replaced by N-propylpyridinium [126], N-methylquinolinium [149], and phenyl groups with short [150] or long aminoalkyl arms [151], and even endowed with fluorescent reporters) [152]. The extensive family of ring-size analogues of the 16-membered porphyrins are exemplified in Figure 8, including the 15-membered corrole, 16-membered phthalocyanine or porphyrazine, and the 19-membered sapphyrin, which have all been employed for the construction of G-quadruplex ligands.
Figure 8.
Polyheterocyclic macrocyclic cores used in the structures of G-quadruplex ligands. The number in the center corresponds to the number of atoms in the central ring.
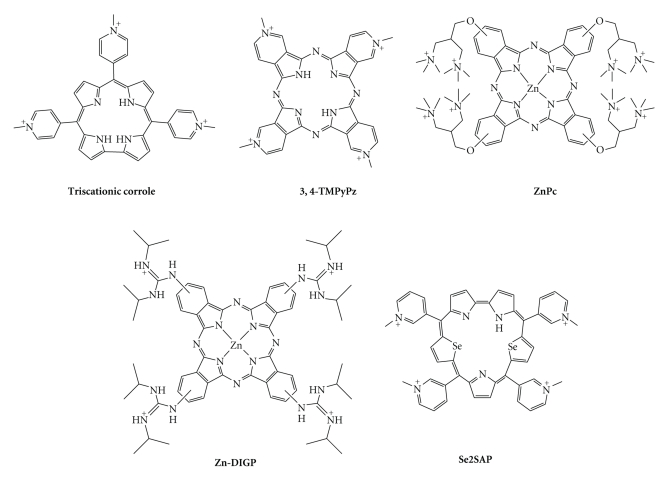
Corroles are tetrapyrrolic macrocyclic structures, lacking one bridging s p 2 carbon with respect to porphyrins. Interestingly, the resulting TMPyP4 analogue, which is substituted with N-methylpyridium groups (Figure 9) [153], is only trisubstituted, with a peculiar spatial disposition of pyridinium moieties relative to each other (Figure 10) [39]. This corrole efficiently stabilizes quadruplex DNA, as judged from CD-melting experiments (T m increase of 36°C), with a fair selectivity, judged by SPR measurements (affinity constants in the range of 105 and 106 M−1 for duplex- and quadruplex-DNA, resp.). Copper corroles have also been studied and proved to be good stabilizing agents for quadruplexes formed from sequences of both the human telomere and the c-myc promoter [154]. Porphyrazine, the polyaza tetra(pyrrolopyridine) analogue of porphyrin (Figures 8 and 9), has also been widely used in the design of G-quadruplex ligands, because of its steric properties (close to those of the porphyrin), but also because its synthesis opens novel perspectives for structural diversity. It enables, for example, to fuse the pyridinium moieties with the cyclic scaffold, thereby leading to a flat, electron-deficient and structurally frozen macrocycle, with an excellent overlap of a G-tetrad (Figure 10) [39]. 3,4-TMPyPz, used either as metal-free or as Zn complex, shows a high affinity and selectivity for quadruplex DNA, as demonstrated by UV-Vis and SPR experiments (affinity constants in the range of 106 M−1 for quadruplex DNA and <105 for duplex-DNA), with a refined binding mode, since a unique binding site was determined using both of these techniques [155]. It is worth noting that 3,4-TMPyPz has also been used as a tool to investigate the role of G-quadruplex ligands in a totally telomere-independent mechanism, that is, an innovative gene-silencing strategy based on the control by a G-quadruplex ligand of the dicing of quadruplex-forming short hairpin RNAs that actively participate to the RNA interference (RNAi) pathway [156].
Figure 9.
Chemical structures of corrole-, porphyrazine-, phthalocyanine-, and sapphyrin-based G-quadruplex ligands.
Figure 10.
Qualitative in silico superposition of 3,4-TMPyPz (a) and corrole (b) and a G-tetrad (extracted from PDB entry: 2A5R); guanine residues appear in gold; the carbon, nitrogen, oxygen, and hydrogen atoms of the ligands in grey, blue, red, and white, respectively.
Phthalocyanines are structurally close to porphyrazines, with the pyridine rings that compose the porphyrazine scaffold replaced by benzene rings. The zinc phthalocyanine complex ZnPc (Figure 9) [157, 158] carries the classical amino side chains in the close vicinity of the macrocyclic scaffold. The results are interesting, since they demonstrate that a highly charged compound (up to 8 positive charges), whilst presenting an expected high degree of quadruplex affinity, can concomitantly retain a satisfactory level of selectivity (on the basis of SPR and CD-melting data), the difference in kinetic parameters with duplex-versus-quadruplex DNA being in the range of one order of magnitude. When the aminoalkyl side arms of ZnPc are replaced by guanidinium moieties, the corresponding Zn-DIGP (for tetrakis-(diisopropylguanidinio) zinc phthalocyanine, Figure 9) elicits quite exceptional properties, both in terms of quadruplex affinity (K d < 2 nM for c-myc quadruplex) and quadruplex-versus-duplex DNA selectivity (Zn-DIGP displays a 5000-fold higher affinity for c-myc quadruplex than for calf thymus DNA). In addition, Zn-DIGP exhibits “turn-on” fluorescent properties, making it a particularly interesting quadruplex-selective dye [159]. Very recent studies have substantiated the interest in this series of molecules, confirming their good cellular uptake and inhibition of promoter-quadruplex-mediated genes (KRAS) [160].
The expanded diselenosapphyrin Se2SAP (Figure 9) represents a novel class of chemically challenging compounds but with a very interesting ability to interact only with its preferential target, the oncogenic c-myc-derived quadruplex [52, 59]. Indeed, Se2SAP discerns, firstly, among the DNAs of various nature (duplex and quadruplex DNA, with a selectivity factor of ~600 as judged by a competitive Taq polymerase assay), and secondly, among quadruplex DNA of various structures (Se2SAP binds specifically c-myc, as compared to human telomeric quadruplex, but displays also a higher affinity for the quadruplex found in the promoter region of VEGF) [102]. The substitution of nitrogen by selenium atoms in the backbone of the ligand was motivated by the decrease of the known photocytotoxicity of the porphyrins, as confirmed by the low general cytotoxicity of Se2SAP in HeLa cells (no effect at 200 μM dose). One major drawback to the use of Se2SAP in cells stands in its limited bioavailability, which remains to be improved for further biological investigations.
A further noteworthy family of porphyrins is hemin and related compounds. Hemin, also known as Fe(III)-protoporphyrin IX (Figure 11), is the oxidized version of heme (Fe(II)-protoporphyrin IX), widely known as a cofactor of hemoglobin (and myoglobin) that actively participates in the reversible binding of dioxygen. Since the pioneering work of Sen et al. [161–165], hemin is known to bind tightly to quadruplex architecture; however, hemin has not been investigated as a G-quadruplex ligand per se, but has been thoroughly investigated for its DNAzyme activity when bound to quadruplex. Indeed, the hemin/quadruplex complex acquires the ability to catalyze the H2O2-mediated oxidation of precursors such as ABTS (2,2′-azino-bis(3-ethylbenzothiozoline-6-sulfonic acid) [161–165], luminol [166–168], or TMB (3,3′,5,5′-tetramethylbenzidine) [169, 170], therefore causing an easily detectable color change. This particular activity has found numerous applications, including the detection of telomerase activity [167, 168], single-stranded DNA [171], SNP (single-nucleotide polymorphism) [172], DNA [173] and DNA analytes [174], proteins (like nucleolin [175], lysozyme [176], or thrombin [177]), various cations (such as Cu2+ [169], Hg2+ [178, 179], Pb2+ [180], or K+) [181], or enzyme cofactors (like L-histidine) [180], as well as a use for constructing DNA logic gates [182, 183], and it has been also implied in the development of novel in vitro assays for screening G-quadruplex ligands [184, 185].
Figure 11.
Chemical formulae of Hemin (a) and NMM (b).
A hemin-related porphyrin, NMM (or N-methyl mesoporphyrin IX) has also been thoroughly investigated for its ability to interact with G-quadruplex. NMM (Figure 11) is interesting as the unique representative of the negatively charged macrocyclic G-quadruplex ligands (its two carboxylic groups being deprotonated at physiological pH) [74, 186–188]. NMM was initially studied since it behaves as stable transition-state analogue for ferrochelatase enzyme, which catalyzes the insertion of Fe2+ ions into protoporphyrin in the final step of the heme biosynthesis [161–165]. Whilst NMM became rapidly known as highly quadruplex selective [87, 189], it was only sparingly studied due to a moderate affinity for its DNA target, despite an excellent overlap with the G-tetrad (Figure 12). However, in recent studies in yeast, it has shown a very interesting ability to control the regulation of genes in direct connection with the probability of quadruplex formation in their promoters [190, 191]. Additionally, given that the effect of NMM is felt at both telomeres and loci throughout the genome, this study provides novel lines of evidence of the possible in vivo roles of G-quadruplexes. Finally, NMM has also been used as a novel tool for isolating G-quadruplex DNA from a mixture of nucleic acids derived from in vivo sources, via its grafting on a sepharose resin, thus offering an innovative way to determine whether or not the in silico detected putative quadruplex-forming sequences (QFSs) [192–194] exist in the cellular context as quadruplex DNA [195].
Figure 12.
Qualitative in silico superposition of NMM and a G-tetrad (extracted from PDB entry: 2A5R); guanine residues appear in gold; the carbon, nitrogen, oxygen, and hydrogen atoms of NMM in grey, blue, red, and white, respectively.
4. Polyammonium Cyclophane-Type Macrocycles
As compared to the telomestatin, porphyrins, and related compounds, polyammonium cyclophane-type macrocycles have not been extensively studied as G-quadruplex ligands. Mainly two types of cyclophane-like macrocycles have been studied for their interaction with quadruplexes: the cyclobisintercalator family (CBI, Figure 13) and the neomycin-capped aromatic platforms (exemplified by NCQ, Figure 13).
Figure 13.
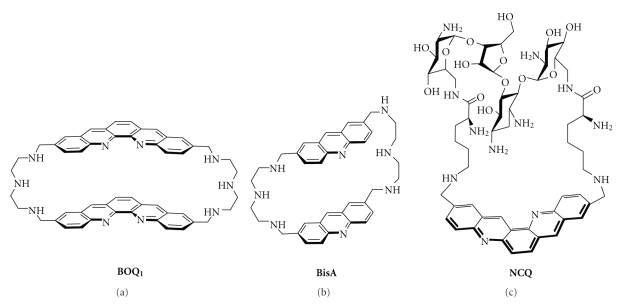
Chemical formulae of nonplanar macrocyclic G-quadruplex ligands: B O Q 1 (a), BisA (b), and the neomycin-capped scaffold NCQ (c).
The CBI macrocycles contain two flat aromatic units, usually derived from simple intercalators linked together by polyamine chains. This particular scaffold is highly soluble at physiological pH thanks to protonation of the four benzylic nitrogen atoms of the linkers [196]. With regard to DNA binding, the CBIs present unique features that are (i) a strong association with DNA bases via aromatic π-π interactions and (ii) a very low affinity for duplex-DNA. This family of compounds has already been studied for its recognition of various unusual DNA structures, mainly mismatch-containing DNA [197–200], abasic sites [201], and trinucleotide repeats [202]. However, only two members of this family, namely, B O Q 1 (for Bis-Ortho-Quinacridine, Figure 13) [203–205] and BisA (for Bis-Acridine, Figure 13), [206] have been already evaluated for their ability to interact with quadruplex-DNA.
BisA and more particularly B O Q 1 display promising G-quadruplex-binding properties. Both compounds are composed of two large polycyclic aromatic moieties, namely, acridine [207–209] or quinacridine [210–212], that enable efficient π-stacking interactions with nucleic bases in particular guanines [213]. These dimeric macrocycles were found to bind much better to quadruplexes than their planar monomeric acyclic counterparts [203–206]. This strong association has been demonstrated by numerous techniques, including FRET-melting assay, SPR, and equilibrium dialysis: B O Q 1 and BisA efficiently induce thermal stabilization of the human telomeric quadruplex (ΔT 1/2 = 28 and 15°C, resp.) displaying high affinity for this target (K a = 1.2 × 107 M−1 for B O Q 1 measured by SPR and 1.1 × 105 M−1 for BisA, estimated by dialysis) [203–206]. The difference between the two compounds clearly demonstrates the importance of the size of the aromatic unit in the binding interaction with the quadruplex structure.
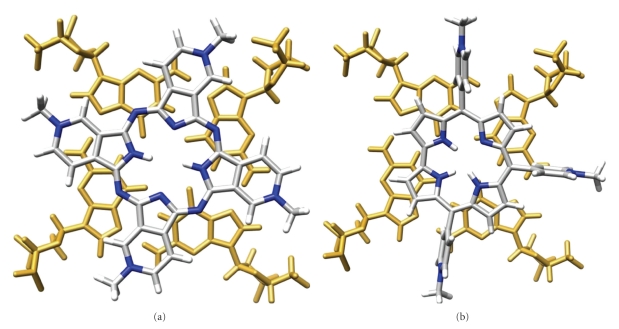
Most interestingly the two compounds exhibit a high selectivity for quadruplex over duplex DNA attributed to their particular topology. CBIs are clearly distinct from the planar rigid macrocycles mentioned precedently since they exist in a semiclosed conformation with the two aromatic units facing each other in more or less parallel planes (Figures 13 and 14) [39]. This particular nonplanar conformation impedes binding to duplex-DNA, both for steric reasons and because the length of the linkers does not enable bisintercalation between contiguous base pairs due to the neighbour-exclusion principle [214, 215]. In the case of B O Q 1, this particular topology was found both in the free state (Figures 14(a)–14(c)) and in the complex formed with the human telomeric quadruplex (Figure 14(d)) [205], meaning that there is little conformational change upon binding. This would suggest that the ligand is already preorganized to fit into the quadruplex target resulting in a gain in the binding entropy. In addition, the modelling studies performed both with the telomeric and with the c-myc quadruplex demonstrate that the semiclosed conformation of B O Q 1 enables interaction via a mixed binding mode, combining G-quartet and loop (Figure 14(d), c-myc not shown) [205]. In fact, careful examination of this complex strongly suggests the occurrence of electrostatic and H-bonding interactions between the linkers and the loop backbone (Figure 14(d)).
Figure 14.
Structure of B O Q 1 (a), side (b), and front views (c) of the lowest-energy conformation during the molecular dynamic simulation (see [39]) and modelled interaction between B O Q 1 and the human telomeric quadruplex ((d), see [205]); guanine residues appear in gold; the carbon, nitrogen, oxygen, and hydrogen atoms appear in grey, blue, red, and white, respectively.
The second category of polyammonium cyclophane-type macrocycles, namely, the neomycin-capped macrocycles was developed with the goal to validate the ditopic interactive design. To this aim three aromatic platforms (acridine, phenanthroline, and quinacridine) able to stack on a G-quartet were combined with a well-known loop-binding motif, that is, the aminoglycoside neomycin. The most active prototype of this series is the quinacridine derivative NCQ shown in Figure 13 [216, 217]. This ligand that has a high degree of molecular flexibility is a potent binder of the telomeric quadruplex as shown by the FRET melting assay (ΔT 1/2 = 15°C) whereas it elicits a poor affinity for duplex-DNA. Most importantly NCQ elicits a high degree of selectivity for looped quadruplex (i.e., intramolecular telomeric quadruplex), compared with the tetramolecular quadruplex lacking the loop motifs. The preference for intramolecular over tetramolecular quadruplexes confirms the potential of the multitopic approach and altogether this work proposes a new innovative design for achieving intra-quadruplex selectivity. Subsequent studies have since then fully supported this approach using various G-quadruplex recognition scaffolds, including a macrocyclic oxazole-based tripeptide [218] or a small-molecule ligand (acridine) [219] equipped with loop and/or groove recognition elements like carbohydrates [218] or peptides [219].
In conclusion, polyammonium cyclophane-type macrocycles have been only sparingly studied as G-quadruplex ligands. They are able to establish various types of interaction with their DNA target, thereby opening the innovative perspective of a selective interaction with quadruplex DNA as a function of the nature of the quadruplex structure itself. This intra-quadruplex selectivity, motivated by the putative presence of quadruplexes at various genomic localizations (vide supra), is one of the major issues that will have to be addressed in the near future. The results previously obtained with CBIs and presented herein, although limited to a few examples, are promising. On this solid basis, an extensive study of binding of CBIs to G-quadruplex has been undertaken which is further reported in the companion paper (“One Ring to Bind Them All”—Part II, by A. Granzhan et al.), a research article in the present issue.
5. Conclusion
Macrocyclic scaffolds are particularly attractive for designing selective G-quadruplex ligands essentially for the two following reasons: on one hand, they show a poor affinity for the “standard” B-DNA conformation, due to their sterically difficult intercalation between the base pairs of the double helix; on the other hand, in contrast, they fit nicely with the external G-quartets of quadruplexes that constitute accessible planar sites of large aromatic area. Although synthetic accesses to macrocycles are often difficult, this chemical class is nevertheless a fascinating tool that can be chemically engineered to generate new biological properties. The few macrocyclic families depicted herein have only begun to mine this incredible potential and we hope that the present article will be helpful to understand what the crucial guidelines are to control their efficiencies and selectivities for recognizing quadruplexes.
Acknowledgments
The authors would like to thank sincerely their coworkers for the synthetic work and stimulating scientific discussions related to G-quadruplex DNA and ligands; a particular acknowledgment will be dedicated to A. De Cian, M. Kaiser, and L. Lacroix. The authors are also grateful to all of the collaborators who contributed to the works and to all of their colleagues worldwide whose works are described herein. A special acknowledgment is dedicated to J. R. R. Tolkien for the title inspiration and to J. Martin for careful reading of the manuscript.
References
- 1.Watson JD, Crick FHC. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins MHF, Stokes AR, Wilson HR. Molecular structure of nucleic acids: molecular structure of deoxypentose nucleic acids. Nature. 1953;171(4356):738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 3.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171(4356):740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 4.Kwan ICM, Mo X, Wu G. Probing hydrogen bonding and ion-carbonyl interactions by solid-state 17O NMR spectroscopy: G-ribbon and G-quartet. Journal of the American Chemical Society. 2007;129(8):2398–2407. doi: 10.1021/ja067991m. [DOI] [PubMed] [Google Scholar]
- 5.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Research. 2006;34(19):5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Current Opinion in Structural Biology. 2006;16(3):288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Research. 2007;35(22):7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90(8):1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angewandte Chemie International Edition. 2004;43(6):668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 10.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes and Development. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 11.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS Journal. 2010;277(5):1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 12.De Cian A, Lacroix L, Douarre C, et al. Targeting telomeres and telomerase. Biochimie. 2008;90(1):131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Oganesian L, Karlseder J. Telomeric armor: the layers of end protection. Journal of Cell Science. 2009;122(22):4013–4025. doi: 10.1242/jcs.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nature Reviews Cancer. 2002;2(3):188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 15.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nature Reviews Drug Discovery. 2002;1(5):383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 16.Mergny J-L, Riou J-F, Mailliet P, Teulade-Fichou M-P, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Research. 2002;30(4):839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annual Review of Pharmacology and Toxicology. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 18.Neidle S, Thurston DE. Chemical approaches to the discovery and development of cancer therapies. Nature Reviews Cancer. 2005;5(4):285–296. doi: 10.1038/nrc1587. [DOI] [PubMed] [Google Scholar]
- 19.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. BioEssays. 2007;29(2):155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 20.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nature Structural and Molecular Biology. 2006;13(12):1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 21.Fry M. Tetraplex DNA and its interacting proteins. Frontiers in Bioscience. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 22.Kelland L. Targeting the limitless replicative potential of cancer: the telomerase/telomere pathway. Clinical Cancer Research. 2007;13(17):4960–4963. doi: 10.1158/1078-0432.CCR-07-0422. [DOI] [PubMed] [Google Scholar]
- 23.Pagano B, Giancola C. Energetics of quadruplex-drug recognition in anticancer therapy. Current Cancer Drug Targets. 2007;7(6):520–540. doi: 10.2174/156800907781662257. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Current Opinion in Chemical Biology. 2009;13(3):345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Cian A, Cristofari G, Reichenbach P, et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(44):17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90(8):1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene C-kit. Journal of the American Chemical Society. 2007;129(24):7484–7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Research. 2008;36(4):1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monchaud D, Teulade-Fichou M-P. A hitchhiker’s guide to G-quadruplex ligands. Organic and Biomolecular Chemistry. 2008;6(4):627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 30.Arola A, Vilar R. Stabilisation of G-quadruplex DNA by small molecules. Current Topics in Medicinal Chemistry. 2008;8(15):1405–1415. doi: 10.2174/156802608786141106. [DOI] [PubMed] [Google Scholar]
- 31.Franceschin M. G-quadruplex DNA structures and organic chemistry: more than one connection. European Journal of Organic Chemistry. 2009;(14):2225–2238. [Google Scholar]
- 32.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Current Opinion in Structural Biology. 2009;19(3):239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Shin-ya K, Wierzba K, Matsuo K, et al. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus . Journal of the American Chemical Society. 2001;123(6):1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 34.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. Journal of the American Chemical Society. 1999;121(15):3561–3570. [Google Scholar]
- 35.Shi D-F, Wheelhouse RT, Sun D, Hurley LH. Quadruplex-interactive agents as telomerase inhibitors: synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. Journal of Medicinal Chemistry. 2001;44(26):4509–4523. doi: 10.1021/jm010246u. [DOI] [PubMed] [Google Scholar]
- 36.Mergny J-L, Lacroix L. UV melting of G-quadruplexes. Current Protocols in Nucleic Acid Chemistry. 2009;(supplement 37):17.1.1–17.1.15. doi: 10.1002/0471142700.nc1701s37. [DOI] [PubMed] [Google Scholar]
- 37.De Cian A, Guittat L, Kaiser M, et al. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42(2):183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Fajkus J. Detection of telomerase activity by the TRAP assay and its variants and alternatives. Clinica Chimica Acta. 2006;371(1-2):25–31. doi: 10.1016/j.cca.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Sun D, Hurley LH. Binding of G-quadruplex-interactive agents to distinct G-quadruplexes induces different biological effects in MiaPaCa cells. Nucleosides, Nucleotides and Nucleic Acids. 2005;24(10–12):1801–1815. doi: 10.1080/15257770500267238. [DOI] [PubMed] [Google Scholar]
- 41.Sumi M, Tauchi T, Sashida G, et al. A G-quadruplex-interactive agent, telomestatin (SOT-095), induces telomere shortening with apoptosis and enhances chemosensitivity in acute myeloid leukemia. International Journal of Oncology. 2004;24(6):1481–1487. [PubMed] [Google Scholar]
- 42.Tauchi T, Shin-ya K, Sashida G, et al. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25(42):5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 43.Tahara H, Shin-ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25(13):1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 44.Gomez D, Wenner T, Brassart B, et al. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. Journal of Biological Chemistry. 2006;281(50):38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 45.Kim M-Y, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of Telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Research. 2003;63(12):3247–3256. [PubMed] [Google Scholar]
- 46.Gomez D, O'Donohue M-F, Wenner T, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Research. 2006;66(14):6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 47.Shammas MA, Shmookler Reis RJ, Li C, et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clinical Cancer Research. 2004;10(2):770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 48.Binz N, Shalaby T, Rivera P, Shin-ya K, Grotzer MA. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. European Journal of Cancer. 2005;41(18):2873–2881. doi: 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 49.Arnoult N, Shin-ya K, Londoño-Vallejo JA. Studying telomere replication by Q-CO-FISH: the effect of telomestatin, a potent G-quadruplex ligand. Cytogenetic and Genome Research. 2009;122(3-4):229–236. doi: 10.1159/000167808. [DOI] [PubMed] [Google Scholar]
- 50.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Research. 2005;33(18):6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. Journal of the American Chemical Society. 2009;131(31):10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seenisamy J, Bashyam S, Gokhale V, et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. Journal of the American Chemical Society. 2005;127(9):2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 53.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. Journal of the American Chemical Society. 2006;128(16):5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. Journal of the American Chemical Society. 2007;129(33):10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y, Shin-ya K, Brosh RM., Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Molecular and Cellular Biology. 2008;28(12):4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temime-Smaali N, Guittat L, Sidibe A, Shin-ya K, Trentesaux C, Riou J-F. The G-quadruplex ligand telomestatin impairs binding of topoisomerase IIIα to G-quadruplex-forming oligonucleotides and uncaps telomeres in ALT cells. PLoS ONE. 2009;4(9, article e6919) doi: 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M-Y, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. Journal of the American Chemical Society. 2002;124(10):2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 58.Agrawal S, Ojha RP, Maiti S. Energetics of the human tel-22 quadruplex—telomestatin interaction: a molecular dynamics study. Journal of Physical Chemistry B. 2008;112(22):6828–6836. doi: 10.1021/jp7102676. [DOI] [PubMed] [Google Scholar]
- 59.Rezler EM, Seenisamy J, Bashyam S, et al. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. Journal of the American Chemical Society. 2005;127(26):9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 60.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Research. 2008;36(12):4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deeley J, Pattenden G. Synthesis and establishment of stereochemistry of the unusual polyoxazole-thiazole based cyclopeptide YM-216391 isolated from Streptomyces nobilis. Chemical Communications. 2005;(6):797–799. doi: 10.1039/b416530f. [DOI] [PubMed] [Google Scholar]
- 62.Atkins JM, Vedejs E. A two-stage iterative process for the synthesis of poly-oxazoles. Organic Letters. 2005;7(15):3351–3354. doi: 10.1021/ol051244x. [DOI] [PubMed] [Google Scholar]
- 63.Marson CM, Saadi M. Synthesis of the penta-oxazole core of telomestatin in a convergent approach to poly-oxazole macrocycles. Organic and Biomolecular Chemistry. 2006;4(21):3892–3893. doi: 10.1039/b613791c. [DOI] [PubMed] [Google Scholar]
- 64.Chattopadhyay SK, Biswas S. Convergent synthesis of a 24-membered macrocyclic hexaoxazole derivative related to the novel telomerase inhibitor telomestatin. Tetrahedron Letters. 2006;47(45):7897–7900. [Google Scholar]
- 65.Hernandez D, Riego E, Francesch A, Cuevas C, Albericio F, Alvarez M. Preparation of penta-azole containing cyclopeptides: challenges in macrocyclization. Tetrahedron. 2007;63(39):9862–9870. [Google Scholar]
- 66.Doi T, Yoshida M, Shin-ya K, Takahashi T. Total synthesis of (R)-telomestatin. Organic Letters. 2006;8(18):4165–4167. doi: 10.1021/ol061793i. [DOI] [PubMed] [Google Scholar]
- 67.Minhas GS, Pilch DS, Kerrigan JE, LaVoie EJ, Rice JE. Synthesis and G-quadruplex stabilizing properties of a series of oxazole-containing macrocycles. Bioorganic and Medicinal Chemistry Letters. 2006;16(15):3891–3895. doi: 10.1016/j.bmcl.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 68.Barbieri CM, Srinivasan AR, Rzuczek SG, Rice JE, LaVoie EJ, Pilch DS. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Research. 2007;35(10):3272–3286. doi: 10.1093/nar/gkm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tera M, Sohtome Y, Ishizuka H, et al. Design and synthesis of telomestatin derivatives and their inhibitory activity of telomerase. Heterocycles. 2006;69(1):505–514. [Google Scholar]
- 70.Tsai Y-C, Qi H, Lin C-P, et al. A G-quadruplex stabilizer induces M-phase cell cycle arrest. Journal of Biological Chemistry. 2009;284(34):22535–22543. doi: 10.1074/jbc.M109.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satyanarayana M, Rzuczek SG, LaVoie EJ, et al. Ring-closing metathesis for the synthesis of a highly G-quadruplex selective macrocyclic hexaoxazole having enhanced cytotoxic potency. Bioorganic and Medicinal Chemistry Letters. 2008;18(13):3802–3804. doi: 10.1016/j.bmcl.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pilch DS, Barbieri CM, Rzuczek SG, LaVoie EJ, Rice JE. Targeting human telomeric G-quadruplex DNA with oxazole-containing macrocyclic compounds. Biochimie. 2008;90(8):1233–1249. doi: 10.1016/j.biochi.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rzuczek SG, Pilch DS, LaVoie EJ, Rice JE. Lysinyl macrocyclic hexaoxazoles: synthesis and selective G-quadruplex stabilizing properties. Bioorganic and Medicinal Chemistry Letters. 2008;18(3):913–917. doi: 10.1016/j.bmcl.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Calculator Plugins were used for pKa calculation, Marvin 5.0.1, 2008, ChemAxon, http://www.chemaxon.com/
- 75.Tera M, Ishizuka H, Takagi M, Suganuma M, Shin-ya K, Nagasawa K. Macrocyclic hexaoxazoles as sequence- and mode-selective G-quadruplex binders. Angewandte Chemie International Edition. 2008;47(30):5557–5560. doi: 10.1002/anie.200801235. [DOI] [PubMed] [Google Scholar]
- 76.Iida K, Tera M, Hirokawa T, Shin-ya K, Nagasawa K. G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer: greater selectivity than monomer. Chemical Communications. 2009;(42):6481–6483. doi: 10.1039/b910242f. [DOI] [PubMed] [Google Scholar]
- 77.Tera M, Iida K, Ishizuka H, et al. Synthesis of a potent G-quadruplex-binding macrocyclic heptaoxazole. ChemBioChem. 2009;10(3):431–435. doi: 10.1002/cbic.200800563. [DOI] [PubMed] [Google Scholar]
- 78.Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. Oxazole-based peptide macrocycles: a new class of G-quadruplex binding ligands. Journal of the American Chemical Society. 2006;128(42):13662–13663. doi: 10.1021/ja064713e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chakraborty TK, Arora A, Roy S, Kumar N, Maiti S. Furan based cyclic oligopeptides selectively target G-quadruplex. Journal of Medicinal Chemistry. 2007;50(23):5539–5542. doi: 10.1021/jm070619c. [DOI] [PubMed] [Google Scholar]
- 80.Shirude PS, Gillies ER, Ladame S, et al. Macrocyclic and helical oligoamides as a new class of G-quadruplex ligands. Journal of the American Chemical Society. 2007;129(39):11890–11891. doi: 10.1021/ja073775h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jena PV, Shirude PS, Okumus B, et al. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. Journal of the American Chemical Society. 2009;131(35):12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fiel RJ, Howard JC, Mark EH, Datta Gupta N. Interaction of DNA with a porphyrin ligand: evidence for intercalation. Nucleic Acids Research. 1979;6(9):3093–3118. doi: 10.1093/nar/6.9.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fiel RJ, Munson BR. Binding of meso-tetra (4-N-methylpyridyl) porphine to DNA. Nucleic Acids Research. 1980;8(12):2835–2842. doi: 10.1093/nar/8.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pasternack RF, Gibbs EJ, Villafranca JJ. Interactions of porphyrins with nucleic acids. Biochemistry. 1983;22(10):2406–2414. doi: 10.1021/bi00279a016. [DOI] [PubMed] [Google Scholar]
- 85.Izbicka E, Wheelhouse RT, Raymond E, et al. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Research. 1999;59(3):639–644. [PubMed] [Google Scholar]
- 86.De Cian A, Guittat L, Shin-ya K, Riou JF, Mergny JL. Affinity and selectivity of G4 ligands measured by FRET. Nucleic Acids Symposium Series. 2005;(49):235–236. doi: 10.1093/nass/49.1.235. [DOI] [PubMed] [Google Scholar]
- 87.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38(49):16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 88.Monchaud D, Allain C, Teulade-Fichou M-P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorganic and Medicinal Chemistry Letters. 2006;16(18):4842–4845. doi: 10.1016/j.bmcl.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 89.Granzhan A, Ihmels H, Jäger K. Diazonia- and tetraazoniapolycyclic cations as motif for quadruplex-DNA ligands. Chemical Communications. 2009;(10):1249–1251. doi: 10.1039/b812891j. [DOI] [PubMed] [Google Scholar]
- 90.White EW, Tanious F, Ismail MA, et al. Structure-specific recognition of quadruplex DNA by organic cations: influence of shape, substituents and charge. Biophysical Chemistry. 2007;126(1–3):140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 91.Wang P, Ren L, He H, Liang F, Zhou X, Tan Z. A phenol quaternary ammonium porphyrin as a potent telomerase inhibitor by selective interaction with quadruplex DNA. ChemBioChem. 2006;7(8):1155–1159. doi: 10.1002/cbic.200600036. [DOI] [PubMed] [Google Scholar]
- 92.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nature Reviews Cancer. 2009;9(12):849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 93.Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. Journal of Biological Chemistry. 2001;276(7):4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 94.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grand CL, Han H, Muñoz RM, et al. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Molecular Cancer Therapeutics. 2002;1(8):565–573. [PubMed] [Google Scholar]
- 96.Seenisamy J, Rezler EM, Powell TJ, et al. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. Journal of the American Chemical Society. 2004;126(28):8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 97.Grand CL, Powell TJ, Nagle RB, et al. Mutations in the G-quadruplex silencer element and their relationship to c-MYC overexpression, NM23 repression, and therapeutic rescue. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(16):6140–6145. doi: 10.1073/pnas.0400460101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Yang D, Hurley L. Structure of the biologically relevant g-quadruplex in the c-MYC promoter. Nucleosides, Nucleotides and Nucleic Acids. 2006;25(8):951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 99.Freyer MW, Buscaglia R, Kaplan K, Cashman D, Hurley LH, Lewis EA. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophysical Journal. 2007;92(6):2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cashman DJ, Buscaglia R, Freyer MW, Dettler J, Hurley LH, Lewis EA. Molecular modeling and biophysical analysis of the c-MYC NHE-III1 silencer element. Journal of Molecular Modeling. 2008;14(2):93–101. doi: 10.1007/s00894-007-0254-z. [DOI] [PubMed] [Google Scholar]
- 101.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44(49):16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 102.Sun D, Liu W-J, Guo K, et al. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Molecular Cancer Therapeutics. 2008;7(4):880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Research. 2008;36(14):4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Research. 2006;34(18):5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.del Toro M, Bucek P, Avino A, et al. Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie. 2009;91(7):894–902. doi: 10.1016/j.biochi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 106.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Research. 2009;37(9):2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Research. 2007;35(22):7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gunaratnam M, Swank S, Haider SM, et al. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. Journal of Medicinal Chemistry. 2009;52(12):3774–3783. doi: 10.1021/jm900424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mikami-Terao Y, Akiyama M, Yuza Y, et al. Antitumor activity of TMPyP4 interacting G-quadruplex in retinoblastoma cell lines. Experimental Eye Research. 2009;89(2):200–208. doi: 10.1016/j.exer.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 110.Mikami-Terao Y, Akiyama M, Yuza Y, Yanagisawa T, Yamada O, Yamada H. Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Letters. 2008;261(2):226–234. doi: 10.1016/j.canlet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 111.Verma A, Halder K, Halder R, et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. Journal of Medicinal Chemistry. 2008;51(18):5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 112.Wieland M, Hartig JS. Turning inhibitors into activators: a hammerhead ribozyme controlled by a guanine quadruplex. Angewandte Chemie International Edition. 2006;45(35):5875–5878. doi: 10.1002/anie.200600909. [DOI] [PubMed] [Google Scholar]
- 113.Xu Y, Hirao Y, Nishimura Y, Sugiyama H. I-motif and quadruplex-based device that can control a protein release or bind and release small molecule to influence biological processes. Bioorganic and Medicinal Chemistry. 2007;15(3):1275–1279. doi: 10.1016/j.bmc.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 114.Pomerantz AK, Moerner WE, Kool ET. Visualization of long human telomere mimics by single-molecule fluorescence imaging. Journal of Physical Chemistry B. 2008;112(42):13184–13187. doi: 10.1021/jp806696u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen Z, Zheng K-W, Hao Y-H, Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. Journal of the American Chemical Society. 2009;131(30):10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- 116.Weisman-Shomer P, Cohen E, Hershco I, et al. The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n . Nucleic Acids Research. 2003;31(14):3963–3970. doi: 10.1093/nar/gkg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joachimi A, Mayer G, Hartig JS. A new anticoagulant-antidote pair: control of thrombin activity by aptamers and porphyrins. Journal of the American Chemical Society. 2007;129(11):3036–3037. doi: 10.1021/ja0677822. [DOI] [PubMed] [Google Scholar]
- 118.Ofer N, Weisman-Shomer P, Shklover J, Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Research. 2009;37(8):2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wei C, Jia G, Yuan J, Feng Z, Li C. A spectroscopic study on the interactions of porphyrin with G-quadruplex DNAs. Biochemistry. 2006;45(21):6681–6691. doi: 10.1021/bi052356z. [DOI] [PubMed] [Google Scholar]
- 120.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nature Chemical Biology. 2005;1(3):167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46(9):2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 122.Cavallari M, Garbesi A, Di Felice R. Porphyrin intercalation in G4-DNA quadruplexes by molecular dynamics simulations. Journal of Physical Chemistry B. 2009;113(40):13152–13160. doi: 10.1021/jp9039226. [DOI] [PubMed] [Google Scholar]
- 123.Ishikawa Y, Fujii S. Molecular docking study of binding of TMPyP4 to a bimolecular human telomeric G-quadruplex. Nucleic Acids Symposium Series. 2008;52(1):173–179. doi: 10.1093/nass/nrn088. [DOI] [PubMed] [Google Scholar]
- 124.Jia G, Feng Z, Wei C, Zhou J, Wang X, Li C. Dynamic insight into the interaction between porphyrin and G-quadruplex DNAs: time-resolved fluorescence anisotropy study. Journal of Physical Chemistry B. 2009;113(50):16237–16245. doi: 10.1021/jp906060d. [DOI] [PubMed] [Google Scholar]
- 125.Wei C, Jia G, Zhou J, Han G, Li C. Evidence for the binding mode of porphyrins to G-quadruplex DNA. Physical Chemistry Chemical Physics. 2009;11(20):4025–4032. doi: 10.1039/b901027k. [DOI] [PubMed] [Google Scholar]
- 126.Wei C, Wang L, Jia G, Zhou J, Han G, Li C. The binding mode of porphyrins with cation side arms to (TG4T)4 G-quadruplex: spectroscopic evidence. Biophysical Chemistry. 2009;143(1-2):79–84. doi: 10.1016/j.bpc.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 127.Zhang H-J, Wang X-F, Wang P, Ai X-C, Zhang J-P. Spectroscopic study on the binding of a cationic porphyrin to DNA G-quadruplex under different K+ concentrations. Photochemical and Photobiological Sciences. 2008;7(8):948–955. doi: 10.1039/b809322a. [DOI] [PubMed] [Google Scholar]
- 128.Zhang H, Xiao X, Wang P, et al. Conformational conversion of DNA G-quadruplex induced by a cationic porphyrin. Spectrochimica Acta A. 2009;74(1):243–247. doi: 10.1016/j.saa.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 129.Lubitz I, Borovok N, Kotlyar A. Interaction of monomolecular G4-DNA nanowires with TMPyP: evidence for intercalation. Biochemistry. 2007;46(45):12925–12929. doi: 10.1021/bi701301u. [DOI] [PubMed] [Google Scholar]
- 130.Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. Journal of Physical Chemistry B. 2009;113(25):8784–8792. doi: 10.1021/jp809486g. [DOI] [PubMed] [Google Scholar]
- 131.Haq I, Trent JO, Chowdhry BZ, Jenkins TC. Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin. Journal of the American Chemical Society. 1999;121(9):1768–1779. [Google Scholar]
- 132.Martino L, Pagano B, Fotticchia I, Neidle S, Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. Journal of Physical Chemistry B. 2009;113(44):14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 133.Arora A, Maiti S. Effect of loop orientation on quadruplex—TMPyP4 interaction. Journal of Physical Chemistry B. 2008;112(27):8151–8159. doi: 10.1021/jp711608y. [DOI] [PubMed] [Google Scholar]
- 134.Li H, Liu Y, Lin S, Yuan G. Spectroscopy probing of the formation, recognition, and conversion of a G-quadruplex in the promoter region of the bcl-2 oncogene. Chemistry: A European Journal. 2009;15(10):2445–2452. doi: 10.1002/chem.200801922. [DOI] [PubMed] [Google Scholar]
- 135.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. Journal of Physical Chemistry B. 2009;113(9):2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.del Toro M, Gargallo R, Eritja R, Jaumot J. Study of the interaction between the G-quadruplex-forming thrombin-binding aptamer and the porphyrin 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)-21,23H-porphyrin tetratosylate. Analytical Biochemistry. 2008;379(1):8–15. doi: 10.1016/j.ab.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 137.Vialas C, Pratviel G, Meunier B. Oxidative damage generated by an oxo-metalloporphyrin onto the human telomeric sequence. Biochemistry. 2000;39(31):9514–9522. doi: 10.1021/bi000743x. [DOI] [PubMed] [Google Scholar]
- 138.Maraval A, Franco S, Vialas C, Pratviel G, Blasco MA, Meunier B. Porphyrin-aminoquinoline conjugates as telomerase inhibitors. Organic and Biomolecular Chemistry. 2003;1(6):921–927. doi: 10.1039/b211634k. [DOI] [PubMed] [Google Scholar]
- 139.Dixon IM, Lopez F, Estève J-P, et al. Porphyrin derivatives for telomere binding and telomerase inhibition. ChemBioChem. 2005;6(1):123–132. doi: 10.1002/cbic.200400113. [DOI] [PubMed] [Google Scholar]
- 140.Keating LR, Szalai VA. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry. 2004;43(50):15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 141.Evans SE, Mendez MA, Turner KB, et al. End-stacking of copper cationic porphyrins on parallel-stranded guanine quadruplexes. Journal of Biological Inorganic Chemistry. 2007;12(8):1235–1249. doi: 10.1007/s00775-007-0292-0. [DOI] [PubMed] [Google Scholar]
- 142.Pan J, Zhang S. Interaction between cationic zinc porphyrin and lead ion induced telomeric guanine quadruplexes: evidence for end-stacking. Journal of Biological Inorganic Chemistry. 2009;14(3):401–407. doi: 10.1007/s00775-008-0457-5. [DOI] [PubMed] [Google Scholar]
- 143.Dixon IM, Lopez F, Tejera AM, et al. A G-quadruplex ligand with 10000-fold selectivity over duplex DNA. Journal of the American Chemical Society. 2007;129(6):1502–1503. doi: 10.1021/ja065591t. [DOI] [PubMed] [Google Scholar]
- 144.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42(2):150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 145.Gonçalves DPN, Ladame S, Balasubramanian S, Sanders JKM. Synthesis and G-quadruplex binding studies of new 4-N-methylpyridinium porphyrins. Organic and Biomolecular Chemistry. 2006;4(17):3337–3342. doi: 10.1039/b608494j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McGuire R, Jr., McMillin DR. Steric effects direct the binding of porphyrins to tetramolecular quadruplex DNA. Chemical Communications. 2009;(47):7393–7395. doi: 10.1039/b919273e. [DOI] [PubMed] [Google Scholar]
- 147.Barry NPE, Abd Karim NH, Vilar R, Therrien B. Interactions of ruthenium coordination cubes with DNA. Dalton Transactions. 2009;(48):10717–10719. doi: 10.1039/b913642h. [DOI] [PubMed] [Google Scholar]
- 148.Kieltyka R, Englebienne P, Fakhoury J, Autexier C, Moitessier N, Sleiman HF. A platinum supramolecular square as an effective G-quadruplex binder and telomerase inhibitor. Journal of the American Chemical Society. 2008;130(31):10040–10041. doi: 10.1021/ja8014023. [DOI] [PubMed] [Google Scholar]
- 149.Lu T, Hurley LH. Synthesis of 5,10,15,20-tetra(N-methyl-6-quinolyl)-21,23-dithiaporphyrin chloride as cationic core-modified porphyrin. Chinese Chemical Letters. 2004;15(11):1261–1264. [Google Scholar]
- 150.Wang SR, Zhang D, Luo FL, et al. Some cationic porphyrins: synthesis, stabilization of G-qua druplexes, and down-regulation of c-myc in Hep G2 cells. Journal of Porphyrins and Phthalocyanines. 2009;13(8-9):865–875. [Google Scholar]
- 151.Murashima T, Sakiyama D, Miyoshi D, et al. Cationic porphyrin induced a telomeric DNA to G-quadruplex form in water. Bioinorganic Chemistry and Applications. 2008;2008:5 pages. doi: 10.1155/2008/294756. Article ID 294756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kang C-C, Chen C-T, Cho C-C, Lin Y-C, Chang C-C, Chang T-C. A dual selective antitumor agent and fluorescence probe: the binary BMVC-porphyrin photosensitizer. ChemMedChem. 2008;3(5):725–728. doi: 10.1002/cmdc.200700336. [DOI] [PubMed] [Google Scholar]
- 153.Fu B, Huang J, Ren L, et al. Cationic corrole derivatives: a new family of G-quadruplex inducing and stabilizing ligands. Chemical Communications. 2007;(31):3264–3266. doi: 10.1039/b704599a. [DOI] [PubMed] [Google Scholar]
- 154.Fu B, Zhang D, Weng X, et al. Cationic metal-corrole complexes: design, synthesis, and properties of guanine-quadruplex stabilizers. Chemistry: A European Journal. 2008;14(30):9431–9441. doi: 10.1002/chem.200800835. [DOI] [PubMed] [Google Scholar]
- 155.Gonçalves DPN, Rodriguez R, Balasubramanian S, Sanders JKM. Tetramethylpyridiniumporphyrazines—a new class of G-quadruplex inducing and stabilising ligands. Chemical Communications. 2006;(45):4685–4687. doi: 10.1039/b611731g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Henn A, Joachimi A, Gonçalves DP, et al. Inhibition of dicing of guanosine-rich shRNAs by quadruplex-binding compounds. ChemBioChem. 2008;9(16):2722–2729. doi: 10.1002/cbic.200800271. [DOI] [PubMed] [Google Scholar]
- 157.Ren L, Zhang A, Huang J, et al. Quaternary ammonium zinc phthalocyanine: inhibiting telomerase by stabilizing G quadruplexes and inducing G-quadruplex structure transition and formation. ChemBioChem. 2007;8(7):775–780. doi: 10.1002/cbic.200600554. [DOI] [PubMed] [Google Scholar]
- 158.Zhang L, Huang J, Ren L, et al. Synthesis and evaluation of cationic phthalocyanine derivatives as potential inhibitors of telomerase. Bioorganic and Medicinal Chemistry. 2008;16(1):303–312. doi: 10.1016/j.bmc.2007.09.037. [DOI] [PubMed] [Google Scholar]
- 159.Alzeer J, Vummidi BR, Roth PJC, Luedtke NW. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angewandte Chemie International Edition. 2009;48(49):9362–9365. doi: 10.1002/anie.200903685. [DOI] [PubMed] [Google Scholar]
- 160.Membrino A, Paramasivam M, Cogoi S, Alzeer J, Luedtke NW, Xodo LE. Cellular uptake and binding of guanidine-modified phthalocyanines to KRAS/HRAS G-quadruplexes. Chemical Communications. 2010;46(4):625–627. doi: 10.1039/b918964e. [DOI] [PubMed] [Google Scholar]
- 161.Li Y, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35(21):6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 162.Li Y, Sen D. A catalytic DNA for porphyrin metallation. Nature Structural Biology. 1996;3(9):743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 163.Travascio P, Li Y, Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chemistry and Biology. 1998;5(9):505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 164.Travascio P, Bennet AJ, Wang DY, Sen D. A ribozyme and a catalytic DNA with peroxidase activity: active sites versus cofactor-binding sites. Chemistry and Biology. 1999;6(11):779–787. doi: 10.1016/s1074-5521(99)80125-2. [DOI] [PubMed] [Google Scholar]
- 165.Travascio P, Witting PK, Mauk AG, Sen D. The peroxidase activity of a hemin-DNA oligonucleotide complex: free radical damage to specific guanine bases of the DNA. Journal of the American Chemical Society. 2001;123(7):1337–1348. doi: 10.1021/ja0023534. [DOI] [PubMed] [Google Scholar]
- 166.Xiao Y, Pavlov V, Gill R, Bourenko T, Willner I. Lighting up biochemiluminescence by the surface self-assembly of DNA—hemin complexes. ChemBioChem. 2004;5(3):374–379. doi: 10.1002/cbic.200300794. [DOI] [PubMed] [Google Scholar]
- 167.Pavlov V, Xiao Y, Gill R, Dishon A, Kotler M, Willner I. Amplified chemiluminescence surface detection of DNA and telomerase activity using catalytic nucleic acid labels. Analytical Chemistry. 2004;76(7):2152–2156. doi: 10.1021/ac035219l. [DOI] [PubMed] [Google Scholar]
- 168.Xiao Y, Pavlov V, Niazov T, Dishon A, Kotler M, Willner I. Catalytic beacons for the detection of DNA and telomerase activity. Journal of the American Chemical Society. 2004;126(24):7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 169.Yin B-C, Ye B-C, Tan W, Hui W, Xie C-C. An allosteric dual-DNAzyme unimolecular probe for colorimetric detection of copper(II) Journal of the American Chemical Society. 2009;131(41):14624–14625. doi: 10.1021/ja9062426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li B, Du Y, Li T, Dong S. Investigation of 3, 3′,5, 5′-tetramethylbenzidine as colorimetric substrate for a peroxidatic DNAzyme. Analytica Chimica Acta. 2009;651(2):234–240. doi: 10.1016/j.aca.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 171.Weizmann Y, Beissenhirtz MK, Cheglakov Z, Nowarski R, Kotler M, Willner I. A virus spotlighted by an autonomous DNA machine. Angewandte Chemie International Edition. 2006;45(44):7384–7388. doi: 10.1002/anie.200602754. [DOI] [PubMed] [Google Scholar]
- 172.Deng M, Zhang D, Zhou Y, Zhou X. Highly effective colorimetric and visual detection of nucleic acids using an asymmetrically split peroxidase DNAzyme. Journal of the American Chemical Society. 2008;130(39):13095–13102. doi: 10.1021/ja803507d. [DOI] [PubMed] [Google Scholar]
- 173.Weizmann Y, Cheglakov Z, Willner I. A fok l/DNA machine that duplicates its analyte gene sequence. Journal of the American Chemical Society. 2008;130(51):17224–17225. doi: 10.1021/ja806222e. [DOI] [PubMed] [Google Scholar]
- 174.Nakayama S, Sintim HO. Colorimetric split G-quadruplex probes for nucleic acid sensing: improving reconstituted DNAzyme’s catalytic efficiency via probe remodeling. Journal of the American Chemical Society. 2009;131(29):10320–10333. doi: 10.1021/ja902951b. [DOI] [PubMed] [Google Scholar]
- 175.Li T, Shi L, Wang E, Dong S. Multifunctional G-quadruplex aptamers and their application to protein detection. Chemistry: A European Journal. 2009;15(4):1036–1042. doi: 10.1002/chem.200801282. [DOI] [PubMed] [Google Scholar]
- 176.Li D, Shlyahovsky B, Elbaz J, Willner I. Amplified analysis of low-molecular-weight substrates or proteins by the self-assembly of DNAzyme-aptamer conjugates. Journal of the American Chemical Society. 2007;129(18):5804–5805. doi: 10.1021/ja070180d. [DOI] [PubMed] [Google Scholar]
- 177.Li T, Wang E, Dong S. G-quadruplex-based DNAzyme for facile colorimetric detection of thrombin. Chemical Communications. 2008;(31):3654–3656. doi: 10.1039/b805565c. [DOI] [PubMed] [Google Scholar]
- 178.Li T, Dong S, Wang E. Label-free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated G-quadruplex-based dnazymes. Analytical Chemistry. 2009;81(6):2144–2149. doi: 10.1021/ac900188y. [DOI] [PubMed] [Google Scholar]
- 179.Kong D-M, Wu J, Wang N, Yang W, Shen H-X. Peroxidase activity-structure relationship of the intermolecular four-stranded G-quadruplex-hemin complexes and their application in Hg2+ ion detection. Talanta. 2009;80(2):459–465. doi: 10.1016/j.talanta.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 180.Elbaz J, Shlyahovsky B, Willner I. A DNAzyme cascade for the amplified detection of Pb2+ ions or L-histidine. Chemical Communications. 2008;(13):1569–1571. doi: 10.1039/b716774a. [DOI] [PubMed] [Google Scholar]
- 181.Li T, Wang E, Dong S. G-quadruplex-based DNAzyme as a sensing platform for ultrasensitive colorimetric potassium detection. Chemical Communications. 2009;(5):580–582. doi: 10.1039/b815814b. [DOI] [PubMed] [Google Scholar]
- 182.Li T, Wang E, Dong S. Potassium-lead-switched G-quadruplexes: a new class of DNA logic gates. Journal of the American Chemical Society. 2009;131(42):15082–15083. doi: 10.1021/ja9051075. [DOI] [PubMed] [Google Scholar]
- 183.Shlyahovsky B, Li Y, Lioubashevski O, Elbaz J, Willner I. Logic gates and antisense DNA devices operating on a translator nucleic acid scaffold. ACS Nano. 2009;3(7):1831–1843. doi: 10.1021/nn900085x. [DOI] [PubMed] [Google Scholar]
- 184.Kong D-M, Wu J, Ma Y-E, Shen H-X. A new method for the study of G-quadruplex ligands. Analyst. 2008;133(9):1158–1160. doi: 10.1039/b804627a. [DOI] [PubMed] [Google Scholar]
- 185.Cheng X, Liu X, Bing T, Cao Z, Shangguan D. General peroxidase activity of G-quadruplex-hemin complexes and its application in ligand screening. Biochemistry. 2009;48(33):7817–7823. doi: 10.1021/bi9006786. [DOI] [PubMed] [Google Scholar]
- 186.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Research. 1998;26(16):3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Triplex and quadruplex DNA structures studied by electrospray mass spectrometry. Rapid Communications in Mass Spectrometry. 2002;16(18):1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 188.Paramasivan S, Bolton PH. Mix and measure fluorescence screening for selective quadruplex binders. Nucleic Acids Research. 2008;36(17, article e106) doi: 10.1093/nar/gkn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43(4):313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hershman SG, Chen Q, Lee JY, et al. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Research. 2008;36(1):144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Johnson JE, Smith JS, Kozak ML, Johnson FB. In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie. 2008;90(8):1250–1263. doi: 10.1016/j.biochi.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Research. 2005;33(9):2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Research. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Research. 2007;35(2):406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Smith JS, Johnson FB. Isolation of G-quadruplex DNA using NMM-sepharose affinity chromatography. In: Baumann P, editor. G-Quadruplex DNA: Methods and Protocols. chapter 13. Vol. 608. 2010. pp. 207–221. (Methods in Molecular Biology ). [DOI] [PubMed] [Google Scholar]
- 196.Teulade-Fichou M-P, Vigneron J-P. Interactions of macrocyclic compounds with nucleic acids. In: Demeunynck M, Bailly C, Wilson WD, editors. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes. chapter 11. Vol. 1. 2003. pp. 278–314. [Google Scholar]
- 197.David A, Bleimling N, Beuck C, Lehn J-M, Weinhold E, Teulade-Fichou M-P. DNA mismatch-specific base flipping by a Bisacridine macrocycle. ChemBioChem. 2003;4(12):1326–1331. doi: 10.1002/cbic.200300693. [DOI] [PubMed] [Google Scholar]
- 198.Bahr M, Gabelica V, Granzhan A, Teulade-Fichou M-P, Weinhold E. Selective recognition of pyrimidine-pyrimidine DNA mismatches by distance-constrained macrocyclic bis-intercalators. Nucleic Acids Research. 2008;36(15):5000–5012. doi: 10.1093/nar/gkn392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Granzhan A, Teulade-Fichou M-P. A fluorescent bisanthracene macrocycle discriminates between matched and mismatch-containing DNA. Chemistry: A European Journal. 2009;15(6):1314–1318. doi: 10.1002/chem.200802421. [DOI] [PubMed] [Google Scholar]
- 200.Granzhan A, Largy E, Saettel N, Teulade-Fichou M-P. Macrocyclic DNA-mismatch-binding ligands: structural determinants of selectivity. Chemistry: A European Journal. 2010;16(3):878–889. doi: 10.1002/chem.200901989. [DOI] [PubMed] [Google Scholar]
- 201.Jourdan M, Garcia J, Lhomme J, Teulade-Fichou M-P, Vigneron J-P, Lehn J-M. Threading bis-intercalation of a macrocyclic bisacridine at abasic sites in DNA: nuclear magnetic resonance and molecular modeling study. Biochemistry. 1999;38(43):14205–14213. doi: 10.1021/bi991111h. [DOI] [PubMed] [Google Scholar]
- 202.Amrane S, De Cian A, Rosu F, et al. Identification of trinucleotide repeat ligands with a FRET melting assay. ChemBioChem. 2008;9(8):1229–1234. doi: 10.1002/cbic.200800062. [DOI] [PubMed] [Google Scholar]
- 203.Teulade-Fichou M-P, Carrasco C, Guittat L, et al. Selective recognition of G-quadruplex telomeric DNA by a bis(quinacridine) macrocycle. Journal of the American Chemical Society. 2003;125(16):4732–4740. doi: 10.1021/ja021299j. [DOI] [PubMed] [Google Scholar]
- 204.Allain C, Monchaud D, Teulade-Fichou M-P. FRET templated by G-quadruplex DNA: a specific ternary interaction using an original pair of donor/acceptor partners. Journal of the American Chemical Society. 2006;128(36):11890–11893. doi: 10.1021/ja062193h. [DOI] [PubMed] [Google Scholar]
- 205.Gabelica V, Baker ES, Teulade-Fichou M-P, De Pauw E, Bowers MT. Stabilization and structure of telomeric and c-myc region intramolecular G-quadruplexes: the role of central cations and small planar ligands. Journal of the American Chemical Society. 2007;129(4):895–904. doi: 10.1021/ja065989p. [DOI] [PubMed] [Google Scholar]
- 206.Alberti P, Ren J, Teulade-Fichou MP, et al. Interaction of an acridine dimer with DNA quadruplex structures. Journal of Biomolecular Structure and Dynamics. 2001;19(3):505–513. doi: 10.1080/07391102.2001.10506758. [DOI] [PubMed] [Google Scholar]
- 207.Belmont P, Bosson J, Godet T, Tiano M. Acridine and acridone derivatives, anticancer properties and synthetic methods: where are we now? Anti-Cancer Agents in Medicinal Chemistry. 2007;7(2):139–169. doi: 10.2174/187152007780058669. [DOI] [PubMed] [Google Scholar]
- 208.Demeunynck M. Antitumour acridines. Expert Opinion on Therapeutic Patents. 2004;14(1):55–70. [Google Scholar]
- 209.Denny WA. Acridine derivatives as chemotherapeutic agents. Current Medicinal Chemistry. 2002;9(18):1655–1665. doi: 10.2174/0929867023369277. [DOI] [PubMed] [Google Scholar]
- 210.Mergny J-L, Lacroix L, Teulade-Fichou M-P, et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3062–3067. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hounsou C, Guittat L, Monchaud D, et al. G-quadruplex recognition by quinacridines: a SAR, NMR, and biological study. ChemMedChem. 2007;2(5):655–666. doi: 10.1002/cmdc.200600286. [DOI] [PubMed] [Google Scholar]
- 212.Monchaud D, Allain C, Bertrand H, et al. Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008;90(8):1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 213.Baudoin O, Gonnet F, Teulade-Fichou M-P, Vigneron J-P, Tabet J-C, Lehn J-M. Molecular recognition of nucleotide pairs by a cyclo-bis-intercaland-type receptor molecule: a spectrophotometric and electrospray mass spectrometry study. Chemistry: A European Journal. 1999;5(9):2762–2771. [Google Scholar]
- 214.Crothers DM. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- 215.McGhee JD, von Hippel PH. Theoretical aspects of DNA protein interactions: cooperative and non cooperative binding of large ligands to a one dimensional homogeneous lattice. Journal of Molecular Biology. 1974;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 216.Kaiser M, De Cian A, Sainlos M, Renner C, Mergny J-L, Teulade-Fichou M-P. Neomycin-capped aromatic platforms: quadruplex DNA recognition and telomerase inhibition. Organic and Biomolecular Chemistry. 2006;4(6):1049–1057. doi: 10.1039/b516378a. [DOI] [PubMed] [Google Scholar]
- 217.De Cian A, Grellier P, Mouray E, et al. Plasmodium telomeric sequences: structure, stability and quadruplex targeting by small compounds. ChemBioChem. 2008;9(16):2730–2739. doi: 10.1002/cbic.200800330. [DOI] [PubMed] [Google Scholar]
- 218.Bugaut A, Jantos K, Wietor J-L, Rodriguez R, Sanders JKM, Balasubramanian S. Exploring the differential recognition of DNA G-quadruplex targets by small molecules using dynamic combinatorial chemistry. Angewandte Chemie International Edition. 2008;47(14):2677–2680. doi: 10.1002/anie.200705589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Redman JE, Granadino-Roldan JM, Schouten JA, et al. Recognition and discrimination of DNA quadruplexes by acridine-peptide conjugates. Organic and Biomolecular Chemistry. 2009;7(1):76–84. doi: 10.1039/b814682a. [DOI] [PMC free article] [PubMed] [Google Scholar]