Abstract
Microtubule dynamics are dominated by events at microtubule plus ends as they switch between discrete phases of growth and shrinkage. Through their ability to generate force and direct polar cell transport, microtubules help to organise global cell shape and polarity. Conversely, because plus-end binding proteins render the dynamic instability of individual microtubules sensitive to the local intracellular environment, cyto-architecture also affects the overall distribution of microtubules. Despite the importance of plus-end regulation for understanding microtubule cytoskeletal organisation and dynamics, little is known about the signalling mechanisms that trigger changes in their behaviour in space and time. Here, we identify a microtubule-associated kinase, Drosophila Tao-1, as an important regulator of microtubule stability, plus-end dynamics and cell shape. Active Tao-1 kinase leads to the destabilisation of microtubules. Conversely, when Tao-1 function is compromised, rates of cortical-induced microtubule catastrophe are reduced and microtubules contacting the actin cortex continue to elongate, leading to the formation of long microtubule-based protrusions. These data reveal a role for Tao-1 in controlling the dynamic interplay between microtubule plus ends and the actin cortex in the regulation of cell form.
Keywords: Tao-1, Microtubule dynamics, +TIPs, Kinase, Actin cortex
Introduction
In animal cells, microtubule dynamics are dominated by events at the plus ends of individual microtubules, whose minus ends are anchored within γ-tubulin-rich microtubule-organising centres (Akhmanova and Steinmetz, 2008). Microtubules extend through the addition of tubulin-GTP monomers to their plus ends and, following a switch in state termed ‘catastrophe’ (Desai and Mitchison, 1997), are disassembled from the same ends (Akhmanova and Steinmetz, 2008). Microtubule behaviour is therefore dominated by environmentally induced changes in the rates of these transitions between discrete phases of growth and shrinkage (Desai and Mitchison, 1997), triggered for example by contact with the cell cortex, probably as a result of the loss of a protective plus-end β-tubulin-GTP cap (Akhmanova and Steinmetz, 2008). Significantly, this ability of individual growing microtubule tips to probe their local environment enables the population of microtubules as a whole to efficiently explore cellular space (Mitchison and Kirschner, 1985). Moreover, through their ability to direct polar cell transport and to generate forces through active polymerisation and depolymerisation, these same microtubules help to organise global cell architecture.
Establishing and maintaining a dynamic microtubule cytoskeleton that is sensitive to cell shape, mechanics and local polarity cues rests on the activities of a host of plus-end binding proteins (Akhmanova and Steinmetz, 2008). These form a visible comet at the growing tip of the microtubule, which results from the transient binding of a large number of proteins, probably through recognition of structural features within the growing microtubule GTP-tubulin cap (Akhmanova and Steinmetz, 2008). At the core of this plus-end binding complex is a protein called EB1 (Akhmanova and Steinmetz, 2008; Vaughan, 2005). EB1 is conserved from fungi to humans, binds directly to the plus ends of growing microtubules and is therefore able to track them in vitro (Bieling et al., 2007; Schuyler and Pellman, 2001). When bound to microtubules, EB1 recruits a host of other plus-end tracking proteins (Akhmanova and Steinmetz, 2008) including CLIP170 (Dixit et al., 2009; Goodson et al., 2003), CLASPs (Mimori-Kiyosue et al., 2005), RhoGEF2 (Rogers et al., 2004), ACF7/Kakapo/Shot (Kodama et al., 2003) and APC (Nathke et al., 1996). Together with EB1, these and other associated proteins dictate the behaviour and binding properties of the growing microtubule and regulate microtubule dynamic instability. As a result, in vivo, the loss of the tip-binding complex is correlated with catastrophe and microtubule depolymerisation (Akhmanova and Steinmetz, 2008). Interestingly, mechanical force can mimic the effect of these biochemical changes and induce catastrophe (Foethke et al., 2009; Janson et al., 2003), perhaps because it physically limits plus-end polymerisation, compromising the maintenance of the transient GTP-tubulin plus-end cap (Molodtsov et al., 2005).
Despite the importance of the regulation of microtubule plus ends for understanding microtubule cytoskeletal organisation and dynamics, little is known about the signalling mechanisms that trigger changes in their behaviour in space and time. Here, we report the identification of a conserved, microtubule-associated protein kinase Drosophila Tao-1 (Hutchison et al., 1998; Johne et al., 2008; Mitsopoulos et al., 2003; Zihni et al., 2007) as a protein whose ability to reduce microtubule stability is required for functional interactions between growing microtubule plus ends and the actin-rich cell cortex.
Results
Identification of Tao-1 as a regulator of cell shape
In a series of RNA inhibition (RNAi) screens in Drosophila cells in culture, we have identified a number of genes that share a common RNAi phenotype characterised by the loss of lamellipodia and the formation of multiple microtubule-based protrusions (Kiger et al., 2003; Kunda et al., 2003; Liu et al., 2009). The majority of these genes proved to encode known and novel regulators of the actin cytoskeleton, including Cdc42, Rac and components of the SCAR/WAVE and Arp2/3 complexes (Kunda et al., 2003; Rogers et al., 2003). Interestingly, however, our screens for dsRNAs that generate cells with microtubule-rich protrusions also identified several genes known to be associated with microtubule biology (supplementary material Table S1, left column). These included the kinesin-13 family member K1p10a (Mennella et al., 2005; Sharp et al., 2005), the microtubule-binding protein Shot [short stop/kakapo (Gregory and Brown, 1998; Kodama et al., 2003; Prokop et al., 1998; Roper et al., 2002)], dynein heavy chain Dhc64c (Rasmusson et al., 1994), the dynein light-chain binding protein SW (Song et al., 2007) and Tao-1 (Liu et al., 2009), a relatively poorly studied STE20 kinase family member implicated in the regulation of apoptosis, JNK (Zihni et al., 2007) and spindle-checkpoint signalling (Draviam et al., 2007). Because Tao-1 homologues had been described as binding to microtubules (Hutchison et al., 1998; Johne et al., 2008; Mitsopoulos et al., 2003; Zihni et al., 2007) and actin regulators (Johne et al., 2008), we chose to explore its function as a potential regulator of the actin and microtubule cytoskeleton and cell shape in more detail.
The genome of Drosophila melanogaster contains a single member of the conserved Tao protein kinase family, which has been reported to affect cell death in the Drosophila germline (Sato et al., 2007). The corresponding protein Tao-1 consists of an N-terminal kinase domain and two C-terminal coiled-coil domains. It also has a central region, which, according to the NCBI Conserved Domains tool, is predicted to contain a Smc domain (supplementary material Fig. S1A). A phylogenetic analysis of Tao kinase evolution (supplementary material Fig. S1B) suggests that Drosophila Tao-1 is related in a similar way to each of the three human Tao homologues.
RNAi-mediated depletion of Tao-1 induced the formation of microtubule-rich protrusions and bundled microtubule filaments, and exerted a strong effect on S2R+ cell shape (Fig. 1A,B). Using non-overlapping dsRNAs, we confirmed the specificity of this Tao-1-knockdown phenotype, which was associated with the loss of >95% of Tao-1 mRNA [measured relative to a control message (Rp49) by two-step RT-QPCR (Fig. 1C)]. To study the corresponding protein, we raised polyclonal antibodies against the C-terminus of Drosophila Tao-1 and against the putative active version of the kinase (Zihni et al., 2007) (phosphorylated on Ser180 in the T-loop within the kinase domain). Using these antibodies, we were able to identify a single protein band that ran at approximately 120 kDa in western blots of S2 cell lysates, a signal that was depleted 5 days after treatment with Tao-1 dsRNA (Fig. 2C). To further confirm the specificity of both Tao antibodies, we overexpressed a GFP-tagged version of full-length and truncated Tao-1-Δ423-900 in S2R+ cells. Using lysates from these cells, we were able to detect bands of protein running at approximately 150 kDa for the full-length, and 100 kDa for the truncated version, using our two anti-Tao antibodies and an anti-GFP antibody (Fig. 2D), as expected based upon the size of the GFP (30 kDa). We also used a brief treatment with lambda protein phosphatase to confirm that the antibody against phosphorylated Tao specifically recognised the phosphorylated form of the protein (Fig. 2D, top panel). These data show that Tao-1 is present as a single isoform in Drosophila cells in culture, and that a proportion of Tao-1 and its putative dominant-active form (Tao-1-Δ423-900, see below) is activated through phosphorylation of the T-loop during exponential growth.
Fig. 1.
Tao-1 RNAi phenotype in Drosophila S2R+ cells. Drosophila S2R+ cells were fixed and stained for microtubules using antibodies against α-tubulin 5 days after treatment with Tao-1 or control (lacZ) dsRNAs. (A) Following Tao-1 silencing, microtubules lose their normal organisation and form very long, microtubule-rich protrusions. Scale bar: 10 μm. (B) This phenotype was quantified in 100 randomly picked cells in triplicate. Spiky microtubule-rich cells were seen in an average of 90% of Tao-1 RNAi cells and in 3% of control lacZ RNAi cells. (C) Q-RT-PCR confirmed the knockdown of Tao-1 mRNA in S2R+ cells after a 5-day RNAi treatment. The ribosomal housekeeping gene Rp49 was used as an internal control. Values represent the means ± s.d. of triplicate Q-PCR values from duplicate samples.
Fig. 2.
Tao-1 colocalises with microtubules. Drosophila S2R+ cells were fixed and stained with microtubules (MT) as well as for polyclonal antibodies raised against Tao-1 (Tao) and phosphorylated Tao-1 (P-Tao), and examined by confocal microscopy. (A,B) Tao-1 is partially colocalised with microtubules, whereas the phosphorylated population manifests a more diffuse staining, though is enriched at the tips of a number of bundles. Scale bars: 5 μm. The specificity of the antibodies was confirmed by (C) disappearance of endogenous Tao-1 and phosphorylated Tao-1 signal (arrows) on dsRNA knockdown compared with a control protein (Sra1) on western blot, and (D) by the appearance of the appropriate signal upon overexpression of GFP-tagged Tao-1. The epitope for anti-phosphorylated Tao-1 was confirmed to be phospho-specific by lambda protein phosphatase treatment of lysates of overexpressed GFP-Tao-1 (D, upper panel). A GFP-tagged mutant of Tao-1 lacking the central domain (GFP-Δ423-900), which is a dominantly active form, was also phosphorylated.
When we used the anti-Tao antibody to immunostain fixed Drosophila S2R+ cells, endogenous Tao-1 was colocalised with α-tubulin along a subset of interphase microtubules (Fig. 2A,B). Similarly, during mitosis, this antibody stained the mitotic spindle (supplementary material Fig. S2D). By contrast, the antibody against phosphorylated Tao yielded a punctate staining pattern in interphase Drosophila cells (Fig. 2A-B; punctae were frequently seen closely associated with the tips of microtubule bundles) and was localised to peri-centrosomal material in mitotic cells, where the plus-tip binding protein EB1 has been shown to be concentrated (Rogers et al., 2002) (supplementary material Fig. S2D). As with the western blot analysis, the endogenous pool of Tao-1 was significantly depleted following Tao-1 RNAi (supplementary material Fig. S2A) and increased following overexpression of RFP-Tao-1 (supplementary material Fig. S2B), confirming that the signal corresponded to Tao-1. Inspection of cells by immunofluorescence in which ectopic Tao-1 was expressed at relatively low levels (e.g. GFP-Tao-1) revealed localisation of this fusion protein to a subset of microtubules in both interphase (supplementary material Fig. S2C) and mitotic (supplementary material Fig. S2E) cells. These localisation data are consistent with published data suggesting that Tao-1 homologues in human cells (Tao kinase 1-3) are associated with and act on microtubules (Johne et al., 2008; Mitsopoulos et al., 2003; Zihni et al., 2007).
Deconstructing Tao-1 protein structure and function
To carry out a structure-function analysis of Tao-1, we generated a number of expression constructs encoding mutant forms of the protein tagged with mRFP (Fig. 3A). S2R+ cells were transfected with these constructs and then fixed and stained to visualise both microtubules and the RFP-tagged protein. While the full-length RFP-tagged protein had a somewhat weak and variable effect on microtubule organisation and cell shape in this cell type (Fig. 3B, quantified in supplementary material Fig. S3A), the kinase-dead RFP-Tao-1 K56A protein, lacking a lysine residue at the catalytic core of the kinase domain, reproducibly induced the formation of microtubule-rich protrusions (Fig. 3C) similar to those seen in Tao-1 RNAi cells. A similar phenotype was also seen following the expression of a version of RFP-Tao-1 that lacked the entire kinase domain but which retained the central region and the C-terminal coiled-coil domain (RFP-Tao-1-Δ51-354, Fig. 3D). Tao-1 kinase activity is therefore critical for its function, as previously suggested (Raman et al., 2007; Sato et al., 2007). Because both these mutant forms of the protein retain the C-terminal coiled-coil domains, which in other proteins mediate oligomerisation, the dominant negative effect could be due to sequestration of endogenous Tao-1 by the form lacking kinase activity. The importance of the coiled-coil domains was confirmed when we examined the functional consequences of over-expressing an RFP-fusion construct deleted for the region corresponding to bases 900-1039. This truncated RFP-fusion protein did not influence interphase microtubule organisation and was found to localise to the nucleus (Fig. 3E), implying that the coiled-coil domains may help retain the protein in the cytoplasm under normal conditions. Finally, we explored the function of the central region of Tao-1. Its deletion, corresponding to bases 423-900 in the coding sequence, led to a profound and reproducible dismantling of the microtubule network. Moreover, cells expressing this construct remained small and rounded when replated (Fig. 3F), a phenotype that reflects a reduced ability to spread, as seen following a loss of microtubules (Picone et al., submitted manuscript).
Fig. 3.
Structure-function analysis of Tao-1. A structure-function analysis of Drosophila Tao-1 was carried out in Drosophila S2R+ cells to test the effects of mutating (K56A) or deleting the S-T protein kinase domain (Δ51-354), the central region containing homology to Smc (Δ423-900) and the C-terminal coiled-coil domain (Δ900-1039). (A) Schematic of the mutation and deletion constructs used. (B) RFP-Tao-1 has a variable, minor effect on microtubule organisation. Microtubules are shown in green, RFP-Tao-1 in red. Scale bars: 10 μm and 7.5 μm (bottom row). (C) Expression of the kinase point mutant RFP-Tao-1 K56A causes a spiky phenotype similar to that of Tao-1 RNAi, and localises with microtubules at the cell cortex and at the tip of microtubule bundles. (D) Expression of the kinase-domain deletion mutant Δ51-354 RFP-Tao-1 induces a phenotype similar to that seen following expression of RFP-Tao-1 K56A. (E) The coiled-coil deletion mutant, Δ900-1039 RFP-Tao-1, accumulates in the nucleus and does not induce a visible microtubule phenotype. (F) Expression of the central region deletion construct Δ423-900 RFP-Tao-1 causes drastic microtubule disassembly, which inhibits cell spreading.
These data suggest that the central region of Tao-1 negatively regulates the ability of the full-length protein to destabilise microtubules. Several other lines of evidence support the idea that ectopic Tao-1 compromises microtubule stability. For example, in a second Drosophila cell type, the non-adherent S2 line, overexpression of RFP-Tao-1 itself is sufficient to dismantle the microtubule network and to inhibit cell spreading (compare control and RFP-Tao-1 cells, supplementary material Fig. S3A,B). In addition, Tao-1 over-expression in these cells delayed the ability of the microtubule network to recover following the removal of a microtubule inhibitor, colcemid, from the medium (compare control and RFP-Tao-1 cells, supplementary material Fig. S3C). Taken together, the loss- and gain-of-function phenotypes suggest that Tao-1 functions to destabilise microtubules.
The role of microtubules in the generation of the Tao-1 phenotype
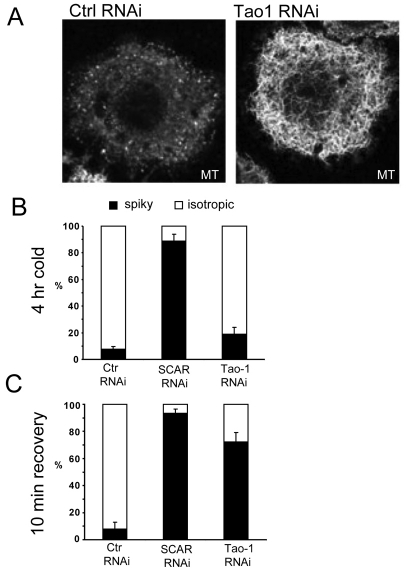
As mentioned above, Tao-1 was identified in screens by an RNAi phenotype similar to that seen when the actin cortex is compromised through loss of Arp2/3-dependent actin filament formation – namely, microtubule-based protrusions and a starfish-like cell shape (Kunda et al., 2003; Rogers et al., 2003) (Fig. 1). Following the loss of lamellipodial actin (as a result of RNAi or the addition of actin inhibitors), these microtubule-based protrusions are generated as growing microtubules push on and deform the weakened cell cortex (Kunda et al., 2003; Rogers et al., 2003). These observations imply that the normally isotropic cell shape is maintained by an active dialogue between actin filaments and microtubules. Thus, a similar phenotype could be generated by increased microtubular growth acting on a relatively normal cell cortex, as is seen in S2R+ cells treated with dsRNA to knock down Klp10A and Short stop/Kakapo (data not shown), where microtubules and microtubule-cortical interactions are likely to be perturbed (Sanchez-Soriano et al., 2010; Sharp et al., 2005; Applewhite et al., 2010). To determine whether microtubules have a direct role in the generation of the spiky shape that is characteristic of Tao-1 RNAi cells, we began by using a cold-shock regimen (Rogers et al., 2008) to test the effects of microtubule depolymerisation and recovery on cell shape 5 days after treatment with Tao-1 dsRNA. In this experiment, dsRNAs against lacZ and SCAR were used as controls.
The analysis revealed several important aspects of the Tao-1 RNAi phenotype. First, although both SCAR/WAVE and Tao-1 knockdown cells had long microtubule-rich protrusions before cold shock, after 2.5 hours in an ice-cold environment, microtubules were visible in Tao-1 RNAi cells (Fig. 4A shows a representative cell; note the loss of microtubule spikes), even though this treatment eliminated visible microtubules in the vast majority of control cells. This suggests that microtubules are unusually stable in cells lacking Tao-1. Nevertheless, after 4 hours on ice, tubulin polymer was completely disassembled in all control and RNAi cells (data not shown). Interestingly, under these conditions ~80% of Tao-1 RNAi cells resumed the isotropic shape characteristic of untreated S2R+ cells (Fig. 4B), whereas SCAR RNAi cells remained spiky. [A similar difference between SCAR and Tao-1 RNAi cells was also seen when microtubules were depolymerised using the drug nocodazole (data not shown).] However, microtubule-rich processes were rapidly re-established in Tao-1 RNAi cells following the return to room temperature (Fig. 4C). These data suggest that although the loss of lamellipodial actin appears to be the primary cause of the spiky SCAR RNAi phenotype (Kunda et al., 2003), microtubules have a crucial role in the generation of the spiky Tao-1 RNAi phenotype.
Fig. 4.
Tao-1 acts on microtubules. Microtubules were depolymerised with a cold-shock regimen in S2R+ cells that had been treated with Tao-1, SCAR or control dsRNA for 5 days, then fixed and stained for microtubules. (A) After 2.5 hours in an ice-cold environment, many long microtubules remain visible in Tao-1 RNAi cells but are completely eliminated in control cells. Cell shape was quantified in control, SCAR and Tao-1 RNAi cells after 4 hours on ice (B) and after a 10 minute recovery at room temperature (C). In each case, results show the average of 30 cells analysed in triplicate. Cells that failed to spread were not included in the analysis. Error bars represent s.d.
To examine the aetiology of Tao-1 versus SCAR RNAi phenotypes in more detail, we next performed a time-course dsRNA experiment to see how changes in microtubules and the actin cortex develop at 2, 3 and 4 days after treatment with dsRNA (supplementary material Fig. S4). Although the majority of SCAR and Tao-1 RNAi cells exhibited the expected microtubule-based protrusions after 4 days of RNAi, at 2 days and 3 days following dsRNA treatment, we observed a significant population of Tao-1 RNAi cells (20% and 30% respectively) with visibly bundled microtubules and a normal actin cytoskeleton (supplementary material Fig. S4A, arrow in Day 3 image). As expected for an intermediate state, the number of cells with this phenotype was reduced to 15% by day 4, when over 80% of Tao-1 RNAi cells had developed microtubule-based protrusions. During the same time-course in the corresponding control experiment, fewer than 5% of SCAR RNAi cells exhibited a normal actin cortex and a visibly disturbed microtubule cytoskeleton (see supplementary material Fig. S4B for the quantification). Again, this experiment supports the idea that although both Tao-1 and SCAR have a similar terminal RNAi phenotype, resulting from defects in the cross-talk between the actin and microtubule cytoskeletons, the two RNAi phenotypes arise in distinct ways: through a primary defect in the ability to nucleate lamellipodial actin in the case of SCAR, and through deregulation of microtubule dynamics in the case of Tao-1.
To further explore this potential role for Tao-1 in the regulation of microtubule dynamics and the dialogue between the actin cortex and the microtubule cytoskeleton, we used Drosophila EB1, a known plus-end microtubule-binding protein, tagged with GFP as a marker in live-cell experiments to image microtubules and changes in cell shape (Rogers et al., 2002) in control cells, in cells expressing the dominant-negative RFP-tagged K56A Tao-1 mutant and in Tao-1 RNAi cells (Fig. 5). In the control, the vast majority of microtubules slowed and underwent catastrophe soon after entering the lamellipodium (Fig. 5A). By contrast, microtubules were seen pushing at the cortex of cells overexpressing Tao-1 K56A or Tao-1 RNAi cells. Significantly, this unchecked local microtubule growth induced the formation of long microtubule-based protrusions over time (Fig. 5B,C). As expected based on the analysis of the SCAR RNAi cells, a similar phenotype was seen in cells with reduced cortical tension as a result of prior treatment with an actin poison, latrunculin B, which sequesters actin in the monomeric form (Fig. 5D).
Fig. 5.
Microtubules push on the cortex of cells depleted for active Tao-1, generating protrusions. (A-D) Stably expressed EB1-GFP visualised in control S2 cells (A), in cells overexpressing (OE) K56A RFP-Tao-1 (B), in Tao-1 RNAi cells (C) and in cells following treatment with latrunculin B (D). Times between frames are indicated in seconds. Scale bars: 2.5 μm. (E) The average (mean) lifetimes of GFP-EB1 comets pausing at the cell cortex before undergoing catastrophe determined by analysing frames taken every 2 seconds in movies of control, RFP-Tao-1- and K56A RFP Tao-1-expressing cells. Error bars represent s.d.
When we quantified the effects of overexpressing the Tao-1 dominant-negative K56A protein on microtubule catastrophe rates in cells expressing EB1-GFP (Fig. 5E), K56A was found to induce a threefold increase in the average lifetime of microtubule plus ends remaining paused at the cell cortex before catastrophe (30±5.6 seconds), compared with the control (10±1.6 seconds) and with cells overexpressing RFP-Tao-1 (8±1.4 seconds) (Fig. 5E). These data confirm a role for Tao-1 in the ability of growing microtubules to undergo catastrophe in response to contact with the actin-based cortex. If it is the case that Tao-1 acts primarily to destabilise microtubules, we reasoned that it should be possible to reduce the rate of formation of microtubule-induced protrusions in cells with a weakened actin cortex by expressing dominantly activated Tao-1. To test this, we examined the growth of individual microtubules in EB1-GFP S2 cells expressing control or RFP-Δ423-900 Tao-1 after the disruption of the actin cortex. As shown in supplementary material Fig. S5A (top row), growing microtubules rapidly induced the formation of protrusions in control cells following the addition of latrunculin B, but this effect was significantly reduced in cells overexpressing RFP-Δ423-900 Tao-1 protein (supplementary material Fig. S5A, bottom three rows).
To determine how Tao-1 might alter interactions between the plus ends of microtubules and the cell cortex, we tracked microtubule plus ends as they passed between central and peripheral regions of the cell, defined based on automatically thresholded background fluorescence levels (Fig. 6A). We then used EB1-GFP comets to measure the speed of individual growing microtubules within comparable regions of control cells, Tao-1 RNAi cells, and cells expressing RFP-Tao-1 and Tao-1 K56A (Fig. 6B-E). In both control cells and in cells overexpressing full-length Tao-1, microtubules were seen growing with a wide range of speeds (Fig. 6B). In both cases, the rate of microtubule polymerisation was dependent on the location of the microtubule plus end; specifically, EB1-GFP comets moved rapidly as if unimpeded when passing through the centre of these cells, and slowly upon entering the thin lamellipodial region of the cell (Fig. 6F). In striking contrast, in cells with reduced Tao-1 activity, growing microtubules formed a single kinetic population whose speeds were found to be independent of their position within the cell. This is shown most clearly in Fig. 6F, where the ratio of peripheral to internal velocity was found to be close to 1 for both Tao-1 RNAi cells and cells overexpressing the K56A-Tao-1 variant. Thus, plus ends appear blind to their position within the cell when Tao-1 function is compromised. When we examined unimpeded plus-end growth in cells immediately after recovery from drug-induced microtubule depolymerisation, the average growth rates of new plus-ends growing from the cell centre were found to be indistinguishable in control and Tao-1 K56A cells (data not shown). Thus, the slight reduction in the average velocity of EB1-GFP comets in the central part of cells lacking Tao-1 activity probably reflects a decrease in the available pool of tubulin monomer, which limits the speed of unimpeded growth of microtubule plus ends in these cells.
Fig. 6.
Tao-1-induced changes in growth rates of microtubule plus-ends vary with context. (A) Microtubule-plus-end growth rates were calculated from stills of S2 cells stably expressing EB1-GFP imaged every 2 seconds. Automatic thresholding identified distinct cell domains based on plus-end growth speeds, the cell edge (red) and cell centre (blue), to determine plus-end growth in different contexts. (B-E) The growth rates of individual microtubule plus ends at the edge (red, upper panels) or within the cell centre (blue, lower panels) determined using semi-automated image analysis. The average velocity of microtubule growth measured using EB1-GFP was quantified for individual microtubules in control and Tao-1 RNAi cells, and in cells expressing full-length RFP-Tao-1 or a K56A RFP-Tao-1 mutant. Data from individual samples was grouped in 20 nm/second bins. N denotes the number of individual microtubules analysed in each case. (F) The ratio of mean peripheral versus internal velocities calculated for each condition using the data from B-E (red versus blue). Varying plus-end growth, fast in the cell centre and reduced in lamellipodia, was evident in cells expressing Tao-1 but lost in cells with reduced Tao-1 activity.
Finally, in an attempt to identify proteins that function together with Tao-1 in the regulation of microtubule dynamics, we performed a small modifier RNAi screen in S2R+ cells (Kiger et al., 2003) for microtubule-associated proteins that genetically interact with Tao-1. We simultaneously silenced Tao-1 and several other microtubule regulators, and looked for dsRNAs that modified the spiky microtubule Tao-1 RNAi phenotype (supplementary material Table S1, right column). Of the genes tested, only EB1 RNAi was able to induce a partial but significant rescue of the Tao-1 phenotype (supplementary material Fig. S5B). This supports the idea that Tao-1 acts on the plus-end binding complex to regulate microtubule dynamics and the interactions between microtubule plus ends and the actin cortex.
Discussion
In this paper, we show that a conserved microtubule-associated kinase, Tao-1, regulates microtubule dynamics and is required to limit the growth of microtubule plus ends as they contact the actin-based cell cortex. The idea that Tao-1 acts on microtubules confirms the conclusions of previous work on the mammalian homologues of Tao-1. It should be noted however that one of its human homologues, Tao kinase 2, is reported to stabilise microtubules (Mitsopoulos et al., 2003), implying that the different homologues might have taken on divergent functions in human cells (Hutchison et al., 1998; Mitsopoulos et al., 2003).
Interestingly, in Drosophila cells in culture, the phenotypic effect of Tao-1 silencing mirrors the effects of a loss of lamellipodial actin, as seen for example following SCAR/WAVE RNAi. In this case, the actin cortex is weakened, allowing normally growing microtubules to generate microtubule-based protrusions as they polymerise and push on the plasma membrane (Kunda et al., 2003). By contrast, the spiky Tao-1 RNAi phenotype appears to develop in stages. First, cells appear to have a macroscopically normal actin cytoskeleton. The cell edge then becomes gradually distorted over time (supplementary material Figs S5 and S4) as a result of the unchecked growth of microtubules that fail to undergo catastrophe when reaching the cortex (Fig. 5). Although we cannot completely rule out an additional role for Tao-1 in the regulation of actin filaments in this process, as previously proposed (Johne et al., 2008), our analysis suggests that Tao-1 acts primarily on the microtubule cytoskeleton. The change in microtubule dynamics then alters the balance of forces acting to determine S2R+ cell shape, as the mechanically strong actin-rich cortex resists the protrusive forces generated by microtubule plus-end growth. A similar finely tuned balance has been proposed to underlie interactions between the actin cytoskeleton and microtubules in the regulation of axonal extension (Bradke and Dotti, 1999). Moreover, mechanical force has been shown to contribute directly to microtubule catastrophes and has been proposed to have a significant role in guiding microtubule organisation in yeast (Foethke et al., 2009; Janson et al., 2003). Force could therefore have a significant role in the Tao-1-dependent regulation of microtubule dynamics.
The ability of Tao-1 to regulate interactions between microtubule plus ends and the cell cortex could be mediated by a variety of molecular mechanisms. Because endogenous Tao-1 localises both to microtubules and at the cortex, as suggested by studies of the mammalian Tao-1 homologues (Johne et al., 2008; Mitsopoulos et al., 2003; Zihni et al., 2007), the kinase could act to bridge the interface between microtubules and actin filaments, as previously proposed for other microtubule-binding proteins such as ACF7/Kakapo/Shot (Gregory and Brown, 1998; Kodama et al., 2003; Prokop et al., 1998; Wu et al., 2008), CLIP170 (Akhmanova et al., 2001; Goode et al., 2000) and CLASPs (Lansbergen et al., 2006; Mimori-Kiyosue et al., 2005). In this context, we note that some algorithms identify a region within Tao-1 with homology to Smc proteins within its central regulatory domain, which was recently proposed to function as an actin-dependent ATPase in the context of ACF7 (Wu et al., 2008). In addition, the human Tao-1 homologue was recently reported to modulate the activity of Spred1 and TESK1 to affect actin-microtubule interactions (Johne et al., 2008). The role of Tao-1 in the ability of microtubules to sense and respond to these cortical forces is also reminiscent of the recent report that Drosophila Shot (a hit in our screen) acts in concert with cortical F-actin to check the growth of microtubules in neuronal growth cones (Sanchez-Soriano et al., 2010), suggesting that Tao-1 is part of a hitherto unappreciated class of proteins that can execute this important function of linking the two major cytoskeletal systems.
A detailed biochemical analysis will be required to reveal the molecules phosphorylated by Tao-1 to negatively regulate microtubule stability and dynamics. Nevertheless, our analysis shows that the kinase and C-terminal coiled-coil domains are required for this activity. Based on these findings, Tao-1 could function by phosphorylating one or more of the proteins on the plus ends of microtubules to mediate its effect on microtubule dynamics, as has been proposed for GSK-3 (Etienne-Manneville and Hall, 2003; Kumar et al., 2009; Wittmann and Waterman-Storer, 2005). In light of this, our preliminary results showing that EB1 and Tao-1 genetically interact are intriguing, but further experiments will be required to dissect the effect of Tao-1 activity on the plus-end complex.
In conclusion, this study identifies the Tao-1 kinase as an important component of the machinery required to make microtubule plus-end growth sensitive to local environmental cues. Significantly, this ability of Tao-1 to regulate the dialogue between the actin cortex and microtubules appears to be crucial for the ability of the actin-based cell cortex to alter microtubule dynamics, and hence for proper regulation of cell shape.
Materials and Methods
Bioinformatics analysis
Genius Pro 4.8 was used for multiple protein sequence alignment and the generation of the phylogenetic tree. DNAstar5.0 Megalign was used to obtain the consensus sequence. The conserved domain analysis was carried out based on NCBI protein domain prediction (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), SMART (http://smart.embl-heidelberg.de/) and the Coils server (www.ch.embnet.org/software/COILS_form.html).
Recombinant-DNA technology
Two pairs of primers were used to amplify the full-length Tao-1 sequence from a Drosophila embryo cDNA library (Brown and Kafatos, 1988). Primer pair LT-007 (5′-agatctccATGCCTTCGGCACGACC-3′) and LT-004 (5′-ACTGCATTGGAGATGGCCTGTTGC-3′) was used to amplify Tao-1 by PCR from the start codon to 1722 bp to generate Fragment 1. Primers LT-005 (5′-TCAGCCGCGCTATCTGACGAC-3′) and LT-008 (5′-aagcttAAATAGCGAACCAGCCGGAAAACTTG-3′) were used to amplify Tao-1 from 1556 bp to the stop codon, including an engineered HindIII restriction enzyme site, to form Fragment 2. Fragments 1 and 2 were ligated into the pGEM®-T-Easy Vector (Promega) as an intermediate subclone. BglII-ApaI and ApaI-EcoRI were then used to excise Fragment 1 and Fragment 2 from the pGEM-T vectors. Subsequently, the two fragments were simultaneously ligated with pENTR1A cut with BamH1 and EcoRI to generate a Drosophila Gateway® entry vector; Gateway recombination reagents were from Invitrogen. pENTR1A-FL-Tao-1 was sequenced after purification with the PureYield plasmid midiprep kit from Promega (Cat. no. A2492) to check for mutations. The K56A kinase domain point mutation, which is predicted to eliminate Tao-1 kinase activity, was generated using the QuikChange site-directed mutagenesis kit from Stratagene (Cat. no. 200519) and the primer pair K56A-f', 5′-GAGATCGTGGCCATCGCAAAGATGTCGTACACC-3′ and K56A-r', 5′-GGTGTACGACATCTTCGTGATGGCCACGATCTC-3′. The deletion mutants lacking the N-terminal protein kinase domain (Δ51-354), the central domain (Δ423-900) and the coiled-coil domains (Δ900-1039) were made based on the pENTR1A-FL-Tao so that they could be recombined into destination vectors with different fusion tags (GFP or RFP) using Gateway cloning. To generate the Δ51-354 deletion mutant, pENTR1-FL-Tao was digested with XcmI to release the segment from 153-1062 bp. The remaining part was then re-ligated to generate an in-frame mutant construct pENTR1A-Δ51-354. To generate the Δ900-1039 deletion mutant, the forward primer, NTaoΔ900-1039-f' (5′-ACAGCCGGTACCGCCTGGAGCCGTGTCCCGC-3′) and the reverse primer, TaoΔ900-1039-r' (5′-CGAAGCTTCTATTTGTAGCTCTGACTTTGGAAC-3′) were used to amplify the region 1260-2700 bp with terminal KpnI and HindIII sites. The forward primer-NTaoΔ423-900-f' containing a KpnI site (5′-GCGGTACCACTAGACGAAAGCCAAGTGATTGAGTGC-3′) and the reverse primer-TaoΔ546-900-r' (5′-CGAAGCTTCTATAGCGAACCAGCCGGAAAACTTG-3′) carrying a HindIII site were used to amplify the 420 bp segment to generate the pMT-GFP-TaoΔ423-900 mutant. PCR segments then were subcloned into the pGEM-T vector, digested with KpnI and HindIII, and the fragments ligated into the linearised pENTR1A-FL-Tao-1 vector digested respectively with KpnI and HindIII at 1266 bp and 3120 bp. Constructs were sequenced after ligation.
Quantitative real-time PCR
For QRT-PCR experiments, cytoplasmic RNA was harvested from both 5 day control RNAi and Tao-1 RNAi S2R+ cells using the RNeasy miniprep kit (Qiagen), following the manufacturer's guidelines. The SuperScript II Reverse Transcriptase kit (Invitrogen) was used to synthesise the first-strand cDNA according to the manufacturer's guidelines. E. coli RNase H was used to remove RNA complementary to the cDNA. Q-PCR was performed using the DyNAmo SYBR Green qPCR Kit With ROX Passive Reference Dye (New England Biolabs, UK) and an MX4000 real-time PCR machine (Stratagene, La Jolla, CA). SYBR green fluorescence was quantified using a serial dilution of template containing PCR products of known concentration. Relative abundance of transcript was normalised against control (rp49) RNA levels. Primer sequences (Eurogentec, Southampton, UK) for Tao-1 and rp49 were: Tao-1 f', AGTGCTGCAGGAAGAACACA; Tao-1 r', AAGGAGCTCCAGATACGCAA; Rp49 f', TACAGGCCCAAGATCGTGAA; Rp-49 r', ACGTTGTGCACCAGGAACTT.
Antibodies, enzymatic manipulation and western blotting
A rat polyclonal anti-peptide-antibody was generated against the C-terminal portion of Tao-1 (α-Tao-1): 967-981: C+FNQERAERLRMKHEK-CONH2, and a rabbit antibody against phosphorylated Tao (α-P-Tao-1) was generated against the sequence CONH2-RAQRATS[P]NVFAMC+3 (Eurogentec); both antibodies were affinity purified using standard methods. For western blotting, samples were either lysed directly in 2× Laemmli buffer and disrupted with a fine-gauge needle, or, for experiments involving phosphatase treatment, lysed in ice-cold RIPA buffer (50 mM HEPES, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS adjusted to pH 7.7) supplemented with protease inhibitor cocktail (Sigma, P2714), incubated and agitated for 10 minutes on ice, centrifuged to harvest protein-containing supernatants, then treated with lambda protein phosphatase (New England Biolabs, P0753S) using the manufacturer's protocol at 30°C for 1 hour. Corresponding controls were treated identically except that an equivalent amount of buffer was added instead of enzyme. After enzymatic treatment, samples were made up to 1× Laemmli buffer. All samples were heated at 95°C for 10 minutes before loading onto 8% SDS-PAGE gels. Protein was then transferred to Immobilon-P (Millipore) by wet western blotting. For non-phosphospecific antibodies, membranes were blocked with 5% non-fat dry milk in Tris-buffered saline with 0.05% Triton X-100 for 1 hour; for phosphorylated Tao antibody, membranes were instead blocked in 5% BSA in TBS-T (0.01% Triton X-100). All primary antibody incubations were carried out at room temperature for several hours with either 1% milk (non-phospho) or 1% BSA (phospho) [1:1000 for the P-Tao-1 antibody; 1:75 for the Tao-1 antibody; 1:1000 for GFP antibody (Molecular Probes A11122), 1:1500 for the loading control dSra1], and then washed five times with TBS-T (same respective Triton X-100 concentration as used for blocking). Membranes were then incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (DAKO, 1:1000) in TBS-T for 1 hour, and washed as before. Enhanced chemiluminescent reagent (Amersham/GE Healthcare) was used for detection on Hyperfilm EC (Amersham/GE Healthcare). When re-probing was required, membranes were stripped in 100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8, at 50°C for 30 minutes with agitation, then washed twice for 10 minutes in TBS-T. Membranes were tested with ECL to confirm absence of signal before re-washing and re-blocking as normal.
Drosophila cell culture, RNAi and cell biology methods
The Drosophila cell lines S2R+ and GFP-EB1 S2 used in this study were all cultured with Shields and Sang M3 insect medium (Sigma) with 10% heat-inactivated fetal bovine serum (JRH Biosciences) and 1% penicillin-streptomycin (Sigma). Drosophila RNAi experiments were carried out as previously described (Baum and Cherbas, 2008). Briefly, cells suspended in serum-free medium at a concentration of 2×106 cells/ml were mixed with dsRNA designed and prepared as described (Liu et al., 2009), to give a final concentration of 30 ng/μl, centrifuged briefly in their culture vessels, and then incubated at 24°C for 15 minutes before addition of complete medium. Cells were grown for 2-7 days at 24°C, depending on the assay. For microscopy, cells were replated onto FBS- or concanavalin A (ConA)-coated 35mm glass coverslips and allowed to spread before analysis. For modifier RNAi experiments, a large pool of cells received Tao-1 dsRNA first as a pre-treatment in 0.1 μg/μl dsRNA in serum-free medium for 15 minutes. These Tao-1 dsRNA pretreated cells were then seeded into a 384-well plate that already contained droplets consisting of 300 ng individual dsRNA targeted to a different gene per well. dsRNA targeting the lacZ gene was used as a negative control in all experiments.
When overexpression was required, Drosophila S2R+ and S2 cells were transiently transfected with Tao-1 expression constructs using FugeneHD transfection reagent (Roche). In each case, 1 μg purified plasmid was mixed with 3 μl of FugeneHD reagent in 50 μl of serum-free M3 medium and incubated at room temperature for 30 minutes. The plasmid and FugeneHD mixture were then added to 5×105 cells in serum-free medium in a well of a four-well dish. After 30 minutes, an additional 300 μl complete medium was then added and cells were incubated for a further 12 hours at 24°C before changing the medium. After 24-48 hours, cells were replated onto FCS-coated or ConA-coated 35 mm glass coverslips. Where necessary (EB1-GFP S2 cells), EB1-GFP expression was induced (via the MT promoter) the following day by addition of CuSO4 solution to a final concentration of 70 nM.
For immunostaining, S2 and or S2R+ cells were fixed with either −20°C methanol (to image microtubules) or 4% formaldehyde (to image actin and microtubules simultaneously). Antibodies used were diluted to concentrations ranging from 1 to 20 μg/ml [Tao-1 and P-Tao-1, DM1α-tubulin (Sigma) and anti-GFP (LabVision, Fremont, CA; MS-1315-PO)]. Secondary antibodies (conjugated with FITC, TRITC, Cy5 and TRITC; Jackson ImmunoResearch) were used at 1:1000. DAPI (Sigma) was used at a concentration of 5 μg/ml. Cells were mounted in FluorSave™ Reagent (Calbiochem). For the modifier and EB1 or Tao experiments, cells were replated on ConA-coated glass coverslips 5 days after RNAi; after spreading, cells were fixed for 15 minutes at −20°C in 100% methanol for optimal preservation of microtubules, then stained with tubulin antibody and imaged as above.
For cold-shock experiments, S2R+ cells were allowed to spread on ConA-coated 35mm optical quality glass-bottomed culture dishes (MatTek). Cells were then placed on a partially submerged aluminium block in a water-ice bath at 0°C for 2.5 hours or 4 hours in a 4°C cold room. Cells were then fixed immediately in ice-cold methanol or allowed to recover for 10 minutes at room temperature before being fixed. All images were captured using confocal microscopy.
To analyse the effect of microtubule-destabilising drugs on the behaviour of Tao-1, we used EB1-GFP S2 cells transiently overexpressing RFP-Tao-1. Cells were plated onto glass and visualised with confocal microscopy; EB1-GFP comets were visualised in cells expressing the RFP-Tao and compared with adjacent non-expressing cells as a control in the presence of 0.5 μM colcemid, and at 4 and 12 minutes after drug wash-out. To assess the effect of a lack of an actin cell cortex on the ability of microtubules to respond to Tao-1 activity, we imaged EB1-GFP S2 cells growing on glass-bottomed, ConA-coated dishes and expressing control or RFP-Δ423-900 Tao-1 immediately on addition of 0.5 μM latrunculin B, and at various time-points afterwards.
To analyse the effect of Tao-1 activity on EB1-marked comet behaviour, we first prepared the cells as follows. Both control and Tao-1 RNAi pMT-GFP-EB1 S2 cells (from DGRC) were treated with 3 μg of either lacZ dsRNA or Tao-1 dsRNA in four-well plastic-bottom dishes. For the RFP-K56A overexpressing cell movie, cells were transfected 48 hours before the experiment in the plastic-bottomed four-well dishes. CuSO4 was added as above, 24 hours before the experiment, to induce EB1-GFP expression. The cells were replated onto ConA-coated 35mm glass-bottomed dishes 5 days after RNAi. The cells were allowed to spread for 30 minutes before they were imaged using confocal microscopy; a 40× oil lens was used for taking 512×512-resolution live images at a frequency of one frame every 2-4 seconds for 60 time-points. Representative movies are available as supplementary material: supplementary material Movie 1 is the control, supplementary material Movie 2 is the K56A null mutant and supplementary material Movie 3 is the Tao-1 RNAi condition. Analysis of EB1-GFP comets was carried out in ImageJ on processed images as follows. Because the lamellipodia manifested lower GFP background fluorescence when compared with the central portion of the cell, thresholding was used to differentially identify the two regions, and pixels with grey values below and above this identified threshold were reassigned with values of 255 and 0 respectively. 231-313 EB3-GFP comets from 2-5 individual cells (depending on the conditions) were tracked by hand and recorded as ImageJ ROI files. Finally, localisation of the tracked EB1-ROI spots onto either cell edge or central regions were obtained by taking pixel values (0 or 255) from the cell binary movies at the EB1-ROI positions. The velocity of the EB1-GFP spots was calculated by dividing their displacement by the time interval between frames (2-4 seconds).
Supplementary Material
Acknowledgments
We would like to acknowledge the rest of the Baum lab, Jonathan Morris, Joe Howard and Marija Zanic for help and advice, and Andrew Vaughan and Guillaume Charras for help with microscopy. We thank Jeroen Dobbelaere for constructs, Alexis Gautreau for the anti-dSra1 antibody, and Nicholas Brown for the cDNA library. For funding, T.L. would like to thank the Ludwig Institute of Cancer Research, UCL, the Wellcome Trust and the AICR. J.R. thanks the Wellcome Trust, R.P. thanks UCL CoMPLEX, P.K. thanks Cancer Research UK, the Wellcome Trust and the European Union, and B.B. thanks the Royal Society, the EMBO YIP programme, UCL and Cancer Research UK. Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/16/2708/DC1
References
- Akhmanova A., Steinmetz M. O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309-322 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C. C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B. M., De Zeeuw C. I., Grosveld F., et al. (2001). Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923-935 [DOI] [PubMed] [Google Scholar]
- Applewhite D. A., Grode K. D., Keller D., Zadeh A., Slep K. C., Rogers S. L. (2010). The spectraplakin short stop is an actin-microtubule crosslinker that contributes to organization of the microtubule network. Mol. Biol. Cell. Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Cherbas L. (2008). Drosophila cell lines as model systems and as an experimental tool. Methods Mol. Biol. 420, 391-424 [DOI] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100-1105 [DOI] [PubMed] [Google Scholar]
- Bradke F., Dotti C. G. (1999). The role of local actin instability in axon formation. Science 283, 1931-1934 [DOI] [PubMed] [Google Scholar]
- Brown N. H., Kafatos F. C. (1988). Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203, 425-437 [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117 [DOI] [PubMed] [Google Scholar]
- Dixit R., Barnett B., Lazarus J. E., Tokito M., Goldman Y. E., Holzbaur E. L. (2009). Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. USA 106, 492-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V. M., Stegmeier F., Nalepa G., Sowa M. E., Chen J., Liang A., Hannon G. J., Sorger P. K., Harper J. W., Elledge S. J. (2007). A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 9, 556-564 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. (2003). Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature 421, 753-756 [DOI] [PubMed] [Google Scholar]
- Foethke D., Makushok T., Brunner D., Nedelec F. (2009). Force- and length-dependent catastrophe activities explain interphase microtubule organization in fission yeast. Mol. Syst. Biol. 5, 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Drubin D. G., Barnes G. (2000). Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 12, 63-71 [DOI] [PubMed] [Google Scholar]
- Goodson H. V., Skube S. B., Stalder R., Valetti C., Kreis T. E., Morrison E. E., Schroer T. A. (2003). CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil. Cytoskeleton 55, 156-173 [DOI] [PubMed] [Google Scholar]
- Gregory S. L., Brown N. H. (1998). kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J. Cell Biol. 143, 1271-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison M., Berman K. S., Cobb M. H. (1998). Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 273, 28625-28632 [DOI] [PubMed] [Google Scholar]
- Janson M. E., de Dood M. E., Dogterom M. (2003). Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161, 1029-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne C., Matenia D., Li X. Y., Timm T., Balusamy K., Mandelkow E. M. (2008). Spred1 and TESK1-two new interaction partners of the kinase MARKK/TAO1 that link the microtubule and actin cytoskeleton. Mol. Biol. Cell 19, 1391-1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A. A., Baum B., Jones S., Jones M. R., Coulson A., Echeverri C., Perrimon N. (2003). A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2, 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. (2003). ACF7: an essential integrator of microtubule dynamics. Cell 115, 343-354 [DOI] [PubMed] [Google Scholar]
- Kumar P., Lyle K. S., Gierke S., Matov A., Danuser G., Wittmann T. (2009). GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell Biol. 184, 895-908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. (2003). Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867-1875 [DOI] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A. B., Grosveld F., et al. (2006). CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell 11, 21-32 [DOI] [PubMed] [Google Scholar]
- Liu T., Sims D., Baum B. (2009). Parallel RNAi screens across different cell lines identify generic and cell type-specific regulators of actin organization and cell morphology. Genome Biol. 10, R26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. (2005). Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235-245 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., et al. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Kirschner M. W. (1985). Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J. Cell Biol. 101, 766-777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsopoulos C., Zihni C., Garg R., Ridley A. J., Morris J. D. (2003). The prostate-derived sterile 20-like kinase (PSK) regulates microtubule organization and stability. J. Biol. Chem. 278, 18085-18091 [DOI] [PubMed] [Google Scholar]
- Molodtsov M. I., Ermakova E. A., Shnol E. E., Grishchuk E. L., McIntosh J. R., Ataullakhanov F. I. (2005). A molecular-mechanical model of the microtubule. Biophys. J. 88, 3167-3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke I. S., Adams C. L., Polakis P., Sellin J. H., Nelson W. J. (1996). The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J. Cell Biol. 134, 165-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A., Uhler J., Roote J., Bate M. (1998). The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. J. Cell Biol. 143, 1283-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Earnest S., Zhang K., Zhao Y., Cobb M. H. (2007). TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 26, 2005-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson K., Serr M., Gepner J., Gibbons I., Hays T. S. (1994). A family of dynein genes in Drosophila melanogaster. Mol. Biol. Cell 5, 45-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Hacker U., Turck C., Vale R. D. (2004). Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14, 1827-1833 [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Peifer M., Rogers S. L. (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell 19, 3163-3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper K., Gregory S. L., Brown N. H. (2002). The ‘spectraplakins’: cytoskeletal giants with characteristics of both spectrin and plakin families. J. Cell Sci. 115, 4215-4225 [DOI] [PubMed] [Google Scholar]
- Sanchez-Soriano N., Goncalves-Pimentel C., Beaven R., Haessler U., Ofner-Ziegenfuss L., Ballestrem C., Prokop A. (2010). Drosophila growth cones: a genetically tractable platform for the analysis of axonal growth dynamics. Dev. Neurobiol. 70, 58-71 [DOI] [PubMed] [Google Scholar]
- Sato K., Hayashi Y., Ninomiya Y., Shigenobu S., Arita K., Mukai M., Kobayashi S. (2007). Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc. Natl. Acad. Sci. USA 104, 7455-7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S. C., Pellman D. (2001). Microtubule ‘plus-end-tracking proteins’: the end is just the beginning. Cell 105, 421-424 [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Mennella V., Buster D. W. (2005). KLP10A and KLP59C: the dynamic duo of microtubule depolymerization. Cell Cycle 4, 1482-1485 [DOI] [PubMed] [Google Scholar]
- Song Y., Benison G., Nyarko A., Hays T. S., Barbar E. (2007). Potential role for phosphorylation in differential regulation of the assembly of dynein light chains. J. Biol. Chem. 282, 17272-17279 [DOI] [PubMed] [Google Scholar]
- Vaughan K. T. (2005). TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 171, 197-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C. M. (2005). Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J. Cell Biol. 169, 929-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Kodama A., Fuchs E. (2008). ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell 135, 137-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni C., Mitsopoulos C., Tavares I. A., Baum B., Ridley A. J., Morris J. D. (2007). Prostate-derived sterile 20-like kinase 1-alpha induces apoptosis. JNK- and caspase-dependent nuclear localization is a requirement for membrane blebbing. J. Biol. Chem. 282, 6484-6493 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.