Abstract
Aurora kinases are central regulators of mitotic-spindle assembly, chromosome segregation and cytokinesis. Aurora B is a member of the chromosomal passenger complex (CPC) with crucial functions in regulation of the attachment of kinetochores to microtubules and in cytokinesis. We report here that Aurora B contains a conserved SUMO modification motif within its kinase domain. Aurora B can bind SUMO peptides in vitro when bound to the IN-box domain of its CPC partner INCENP. Mutation of Lys207 to arginine (Aurora BK207R) impairs the formation of conjugates of Aurora B and SUMO in vivo. Expression of the SUMO-null form of Aurora B results in abnormal chromosome segregation and cytokinesis failure and it is not able to rescue mitotic defects in Aurora-B-knockout cells. These defects are accompanied by increased levels of the CPC on chromosome arms and defective centromeric function, as detected by decreased phosphorylation of the Aurora-B substrate CENP-A. The Aurora-BK207R mutant does not display reduced kinase activity, suggesting that functional defects are probably a consequence of the altered localization, rather than decreased intrinsic kinase activity. These data suggest that SUMOylation of Aurora B modulates its function, possibly by mediating the extraction of CPC complexes from chromosome arms during prometaphase.
Keywords: Aurora kinases, Cell cycle, Chromosome segregation, Cancer target, SUMO modifications
Introduction
Aurora B (serine/threonine-protein kinase 12) is the enzymatic core of a multiprotein complex, the chromosomal passenger complex (CPC), which comprises other three non-enzymatic subunits, INCENP, Survivin and Borealin (Carmena and Earnshaw, 2003; Ruchaud et al., 2007). The CPC is an important upstream regulator of centromere-kinetochore function, and is responsible for the recruitment to the kinetochore and centromere of a number of proteins, including inner centromere-kinetochore proteins (Sgo1, Sgo2, MCAK), regulators of the microtubule-kinetochore interaction (such as Hec1 and Plk1, among others) and proteins involved in the spindle-assembly checkpoint (SAC; such as Mad2, BubR1 and Mps1) (Kelly and Funabiki, 2009; Ruchaud et al., 2007). Some kinetochore components, including Hec1, Dam1, MCAK and CENP-A, are Aurora-B substrates, suggesting a crucial role for the CPC in the destabilization of aberrant microtubule-to-kinetochore attachments and in the SAC-dependent delay of mitotic progression until these defects are corrected. Recent data suggests that substrate phosphorylation depends on the distance of the substrate from Aurora B at the inner centromere, and artificial recruitment of the CPC to the kinetochore prevents the stabilization of improper attachments and activates the SAC to delay the metaphase-to-anaphase transition (Kelly and Funabiki, 2009; Liu et al., 2009). During cytokinesis, Aurora B is localized to the mid-body where its local inactivation is crucial for completion of abscission (Guse et al., 2005; Steigemann et al., 2009).
Activity of Aurora B is regulated through several mechanisms, including controlled interactions with INCENP, Survivin and Borealin. Knockdown of any member of the complex delocalizes the others, disrupts mitotic progression and might destabilize one or more subunits (Jeyaprakash et al., 2007; Ruchaud et al., 2007). In addition, several post-translational modifications are known to be required for proper activity and control of the dynamic behavior of the CPC. Activation of Aurora B is triggered by autophosphorylation of the T-loop residue after association with its substrate, INCENP. Full activation involves a positive-feedback loop after Aurora-B phosphorylation of INCENP in the conserved IN-box region, possibly as a result of structural rearrangements (Bishop and Schumacher, 2002; Honda et al., 2003).
CPC function is also modulated by ubiquitin and ubiquitin-related modifiers. Aurora B is targeted by a Cul3-containing SCF ubiquitin ligase during early mitosis (Sumara et al., 2007). Interestingly, this modification is necessary to remove a fraction of Aurora B from mitotic chromosomes, allowing its accumulation on the central spindle during anaphase. In the absence of this SCF-Cul3 ubiquitin ligase, Aurora B spreads along chromosome arms and fails to dissociate from them during anaphase. Aurora B also requires APC/C-Cdh1 activity to efficiently translocate to the spindle mid-zone in anaphase. In APC/C-Cdh1-depleted cells Aurora B and the CPC prematurely accumulate in the equatorial cortex, resulting in a weak anaphase spindle (Floyd et al., 2008). Finally, after completion of cytokinesis, the remaining pool of Aurora B is targeted for degradation by APC/C-Cdh1 (Floyd et al., 2008; Garcia-Higuera et al., 2008). These post-translational modifications also affect other CPC components. For instance, Survivin is ubiquitylated during mitosis and its deubiquitylation is required for Survivin dynamics at centromeres (Vong et al., 2005).
Post-translational modifications by the covalent attachment of small ubiquitin-related modifiers (SUMOs) are important in several cellular processes, including the cell-division cycle (Geiss-Friedlander and Melchior, 2007; Hay, 2005; Ulrich, 2008). In vertebrates, at least three SUMO forms (SUMO1, SUMO2 and SUMO3) are covalently attached to a lysine residue in the target protein in three enzymatic steps analogous to the ubiquitylation cascade. SUMO conjugation requires a SUMO-activating E1 enzyme (SAE1 or SAE2), the SUMO-conjugating E2 enzyme UBC9 and, in some cases, additional E3 SUMO ligases (Hay, 2005). SUMO modification is reversible by the action of SUMO-specific isopeptidases that belong to the family of ubiquitin-like proteases (ULPs), also known as sentrin-specific proteases (SENPs) (Hay, 2007). Recent work has revealed the importance of SUMOylation for mitotic progression (Dasso, 2008; Watts, 2007) and has suggested a crucial role of SUMO modifications in kinetochore and centromere function (Dawlaty et al., 2008; Di Bacco et al., 2006; Mukhopadhyay et al., 2010; Zhang et al., 2008), including direct SUMOylation of the CPC component Borealin (Klein et al., 2009).
In this manuscript, we provide evidence for SUMOylation of Aurora B at Lys207. Mutation of this residue results in abnormal mitotic progression and the generation of multinucleated cells. In those mutant cells, Aurora B and INCENP display an abnormal localization at the chromosome arms during prometaphase and metaphase and defective CENP-A phosphorylation, indicating a role for Aurora-B SUMOylation in the proper localization and activity of the CPC during chromosome alignment.
Results
A conserved SUMOylation motif in Aurora-B proteins
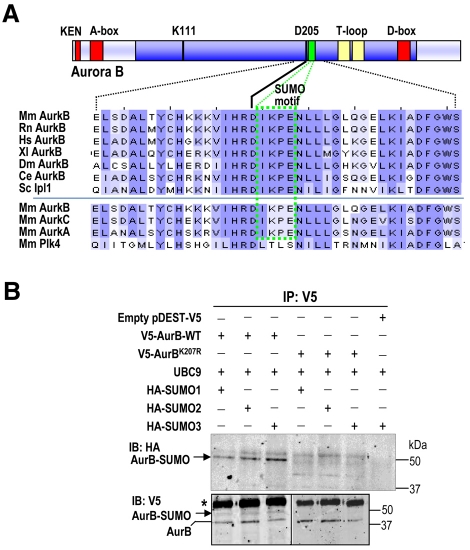
Aurora B contains a consensus motif for SUMO modification (Ψ-Lys-x-Glu, in which Ψ is a hydrophobic residue and x, any residue) within the kinase domain (Fig. 1A). This motif is readily identified by using different protein-domain-recognition algorithms (see the Materials and Methods). The SUMO motif is conserved among Aurora-B orthologues from yeast to human, and it is also present in the other mammalian members of the Aurora family, but not in other cell cycle kinases, including Polo-like kinases (Fig. 1A).
Fig. 1.
A SUMOylation motif in Aurora B. (A) Multiple sequence alignment of Aurora B proteins from different species using Clustal software. Mm, Mus musculus; Rn, Rattus norvegicus; Hs, Homo sapiens; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, single Aurora family member (Ipl1) from Saccharomyces cerevisiae. The SUMO motif (green box) is highly conserved in all these Aurora-B proteins, as well as in Aurora A and Aurora C, but it is not present in Polo-like kinase 4 (Plk4), a close mitotic relative of these mitotic kinases. The kinase domain is shown in dark blue, whereas the degradation motifs (KEN, A- and D-boxes) are represented as red boxes. Note that the SUMO motif is located one residue downstream of the aspartate (D205) that is crucial for ATP binding. Residue numbers correspond to the mouse Aurora-B protein. (B) In vivo SUMOylation of Aurora B at K207. HEK293 asynchronous cells were transiently transfected with vectors expressing V5-Aurora-B WT, Aurora BK207R, HA-SUMO1, -SUMO2 or -SUMO3 and the E2 SUMO ligase UBC9, as indicated. The empty vector pDEST-V5 was used as a negative control. V5-Aurora-B proteins were immunoprecipitated (IP) 48 hours after transfection using anti-V5 antibodies and immunoblotted (IB) with antibodies against the HA and V5 tags. Aurora-B-SUMO conjugates are found after detection of the HA and V5 tags in cells transfected with WT V5-Aurora-B but not in cells transfected with the V5-Aurora-BK207R mutant form. V5-Aurora-B immunoprecipitated in all lanes. Mouse IgGs are indicated with an asterisk.
To test the possible SUMOylation of this domain, we mutated the lysine (K207) predicted to accept the SUMO peptide into a charge-conservative arginine residue. The Aurora-BK207R mutation is predicted to impair covalent binding of SUMO with little effect on the overall kinase structure. Next, we expressed V5-epitope-tagged versions of wild-type (WT) Aurora-B and Aurora-BK207R mutant forms along with the E2 SUMO ligase UBC9 and HA-tagged versions of the three SUMO isoforms (HA-SUMO1 to HA-SUMO3; ~12 kDa) in HEK293 cells (Fig. 1B). The ectopically expressed proteins were immunoprecipitated using anti-V5 antibodies and detected with antibodies against the HA epitope. The predicted SUMO-Aurora-B conjugates (~50 kDa) were detected after expression of all three SUMO isoforms (Fig. 1B), suggesting that Aurora B can be SUMOylated by the three SUMO peptides in this assay. Importantly, the Aurora-B-SUMO conjugates were not observed in cells expressing the Aurora-BK207R isoform, indicating that K207 is required for covalent modification of Aurora B by SUMO residues.
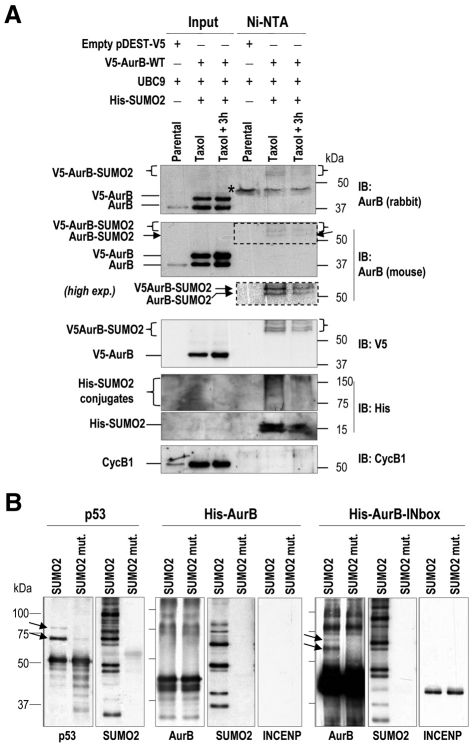
We next tested SUMOylation of endogenous Aurora B by taking advantage of HeLa cells stably expressing histidine-tagged SUMO2 proteins (~16 kDa). These cells were transfected with V5-Aurora-B-expressing vectors and SUMO2 conjugates were exposed to antibodies against V5 or two different antibodies against Aurora B. SUMO2 conjugates were affinity purified under denaturing conditions with Ni-NTA beads. As depicted in Fig. 2A, all these three antibodies detected a band of ~60 kDa, and possibly additional higher-molecular-weight bands in SUMO2 conjugates. This band was detected in taxol-treated cells and it was barely visible in asynchronously growing cells or in parental cells that do not overexpress SUMO2 (data not shown). An additional band of ~55 kDa that might correspond to the endogenous Aurora-B-(His)SUMO2 conjugates is detected by a monoclonal antibody against Aurora B. In fact, this band was also observed in similar assays in which the V5-Aurora-B vector was not used (data not shown), thus further suggesting modification of the endogenous protein.
Fig. 2.
In vivo and in vitro SUMOylation of Aurora B. (A) HeLa-SUMO2 or parental cells were transfected with UBC9 or V5-Aurora-B-expressing vectors as indicated. Cells were harvested after 18 hours in the presence of taxol or 3 hours after release from taxol. His-SUMO2 conjugates were recovered on Ni-NTA beads and analyzed by immunoblotting (IB). The indicated antibodies were used for immunoblot analysis on total protein lysates (input) or SUMO2 conjugates purified with Ni-NTA beads. The position of V5-Aurora-B-SUMO2 conjugates (~60 kDa and higher) is indicated by brackets. An additional band of ~55 kDa (predicted size for SUMO conjugates with endogenous Aurora B) is detected by the mouse monoclonal antibody against Aurora B. The asterisk indicates an unspecific band detected by the rabbit, but not the mouse, anti-Aurora-B antibody. (B) In vitro SUMOylation of Aurora B. Recombinant p53, Aurora B or Aurora B in complex with the IN-box of INCENP were incubated in the presence of SUMOylation machinery and SUMO2 or a SUMO2 non-conjugatable mutant. Proteins were immunodetected using specific antibodies against the indicated proteins. p53- or Aurora-B-SUMO2 conjugates are indicated by arrows.
We then used a reconstituted in vitro SUMO modification system to further test the molecular requirements for this SUMOylation. Recombinant Aurora B was incubated with SUMO E1-activating enzymes (SAE1/2), the E2-conjugating enzyme UBC9 and SUMO2 in the presence of ATP. A mutant SUMO2 molecule unable to conjugate to substrates was used as a control. As recently suggested (Klein et al., 2009), we did not detect obvious SUMOylation of Aurora B in this assay, whereas p53 was significantly SUMOylated by SUMO2, but not SUMO2 mutant peptides (Fig. 2B). Since Aurora B seems to be SUMOylated in vivo, we then asked whether additional CPC domains were required for this post-translational modification. Indeed, higher-molecular-weight Aurora B conjugates (Fig. 2B, arrows) were observed after incubation of Aurora B with the IN-box segment of INCENP, a domain that directly binds and activates Aurora B (Adams et al., 2000). INCENP-bound Aurora B was conjugated with SUMO2, but not with SUMO2 mutants, indicating the specificity of this signal. All together, these results suggest that Aurora-B K207 can be conjugated to SUMO residues and this modification requires binding of Aurora B to its activator INCENP.
Defective SUMOylation of Aurora B results in abnormal chromosome segregation and reduced cell viability
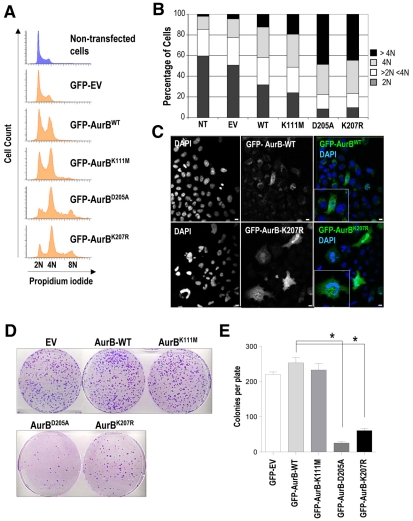
We transiently transfected HEK293 cells with GFP-tagged vectors for wild-type Aurora B, kinase-dead mutants (Aurora BK111M and Aurora BD205A) and the SUMO-null form (Aurora BK207R) to understand the relevance of SUMOylation of Aurora B in cell-cycle progression. Overexpression of wild-type Aurora B slightly increased the percentage of 4N and >4N cells, which is consistent with a mitotic delay and defects in completion of cytokinesis. These defects were more pronounced upon expression of the kinase-dead form Aurora BK111M (19.1% of >4N) and, at an even higher degree with Aurora BD205A (47.6% of >4N) (Fig. 3A,B). The phenotypic differences observed with the Aurora-BK111M and Aurora-BD205A mutants suggest distinct degrees of inhibition of the kinase activity in these mutants (see below). Interestingly, overexpression of the SUMO-null form (Aurora BK207R) induces polyploidy at the same level as the Aurora-BD205A mutant (44.1% in Aurora-BK207R mutants vs 47.6% in Aurora-BD205A mutants). Microscopy analysis indicated that >4N cells overexpressing the Aurora-BK207R mutant were mostly multinucleated and displayed severe nuclear abnormalities (Fig. 3C), which were similar to those of kinase-dead mutants (not shown). Stable expression of Aurora BD205A and Aurora BK207R in U2OS cells severely reduced the number of colonies, suggesting that mutations in these residues impair Aurora-B function in a way that is incompatible with cell proliferation (Fig. 3D,E).
Fig. 3.
Aurora BK207R induces polyploidy and defective cell viability. (A) DNA content of HEK293 cells transiently transfected with a GFP empty vector (EV) and GFP-tagged vectors for wild-type Aurora B and Aurora B mutants (Aurora BK111M, Aurora BD205A and Aurora BK207R) and fixed 72 hours after transfection. HEK293 GFP-positive cells were detected and used for cell-cycle analysis. Overexpression of a SUMO-null Aurora-B protein (Aurora BK207R) deregulates cell-cycle progression inducing polyploidy in the same way as Aurora-BD205A kinase-dead form. (B) Bar graph quantification of cellular DNA content, showing a dramatic increase of polyploidy in cells overexpressing Aurora BD205A and Aurora BK207R mutants. (C) Immunofluorescence (GFP, green; DAPI, blue) images revealing that cells overexpressing Aurora BK207R are mostly multinucleated and display nuclear aberrancies. Scale bars: 10 μm. (D) Colony-formation assay in U2OS cells stably expressing GFP empty vector (EV) and GFP-tagged vectors for Aurora B (wild-type and mutants). U2OS GFP-positive cells were sorted 48 hours after transfection, replated in the presence of 1 mg/ml G418, then fixed 12 days later and stained with Crystal Violet to determine colony number. 50,000 GFP-positive cells were sorted and plated in 10 cm plates. (E) Quantification of the number of macroscopic colonies per 10 cm plate showing a significant (*P<0.001) reduction in the number of colonies in cells expressing Aurora BD205A and Aurora BK207R.
Altered mitotic progression and cytokinesis in Aurora-BK207R mutants
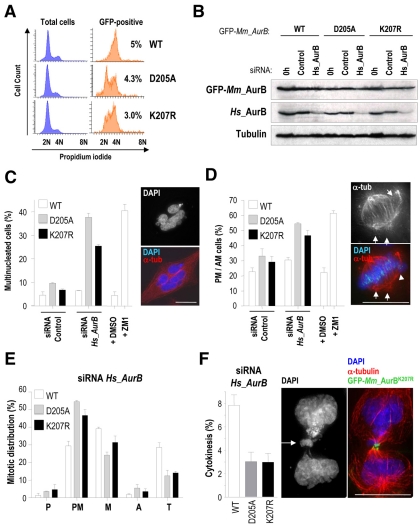
Deregulation of the function of Aurora B by chemical inhibition, small interfering RNA (siRNA) or overexpression of the kinase-dead form all lead to similar phenotypes, including chromosome misalignment and failure to complete cytokinesis, resulting in multinucleated cells (Ruchaud et al., 2007). To gain further insights into the cellular consequences of defective Aurora-B SUMOylation, we analyzed stable HeLa cell lines expressing the different mutant constructs. As reported above, proliferation of clones stably expressing GFP-Aurora-B fusion proteins was relatively normal, whereas expression of the kinase-dead and SUMO-null forms resulted in reduced viability. Accumulation of the GFP-Aurora-B fusion protein was mostly observed in G2-M cells (Fig. 4A) in agreement with its role at these cell-cycle stages. The Aurora-B mutants exhibited a slightly lower percentage of GFP-positive cells (4.3% and 3.0% in GFP-Aurora-BD205A and GFP-Aurora-BK207R, respectively, vs 5% in GFP-Aurora-BWT). Strikingly, the accumulation of these mutant proteins was not restricted to G2-M. This altered behaviour of the mutants might be explained by a higher stability of the mutant proteins or by the existence of aneuploid cells with a DNA content other than 4N.
Fig. 4.
Defective mitotic progression and cytokinesis in the presence of Aurora BK207R. HeLa cells stably expressing siRNA-resistant mouse Aurora B fused to GFP were nucleofected with human AURKB siRNA oligonucleotides or a control siRNA and analyzed 36 hours after nucleofection by immunoblotting and immunofluorescence. (A) Generation and validation of HeLa stable cell lines by cytometry cell-cycle analysis. The analysis of the total cell population is shown in blue histograms, whereas the analysis of the GFP-positive population is shown in orange histograms accompanied by the percentage of these cells over the whole population. At least three different stable clones were analyzed for each GFP-Aurora-B fusion protein. Depicted histograms are from representative stable clones. (B) Western blot analysis showing efficient depletion of human endogenous Aurora B and protein levels of siRNA-resistant mouse GFP-Aurora-B proteins in HeLa stable cell lines. Tubulin was used as loading control. (C) Quantification of the percentage of interphase multinucleated cells. Stable expression of GFP-Aurora-BD205A and Aurora-BK207R induces accumulation of multinucleated cells (see picture) upon depletion of endogenous Aurora B. Around 1000 interphase cells were counted per point. (D) Quantification of the percentage of mitotic cells in prometaphase (PM) or abnormal metaphases (AM) with misaligned chromosomes (arrowheads) and multiple poles (arrows) revealing a significant arrest at these stages in cells lacking the endogenous Aurora B and stably expressing GFP-AuroraBD205A and -AuroraBK207R. About 50-100 mitotic cells were counted per point. Chemical inhibition of Aurora-B kinase activity with ZM447439 (ZM1) was included in C and D as a positive control. (E) Mitotic distribution of stable cell lines after depletion of endogenous Aurora B. Expression of GFP-Aurora-BD205A and -Aurora-BK207R provokes an arrest at prometaphase and a reduction in cells at telophase. P, prophase; PM, prometaphase; M, metaphase; A, anaphase; T, telophase. (F) GFP-Aurora-BD205A and -Aurora-BK207R stable cell lines display a reduced number of cells undergoing cytokinesis. In these mutant cells, the remaining cytokinesis figures are usually abnormal with chromosomal bridges (arrow). α-tubulin (α-tub) is in red, GFP-Aurora-B in green and DAPI (DNA) in blue. Scale bars: 10 μm.
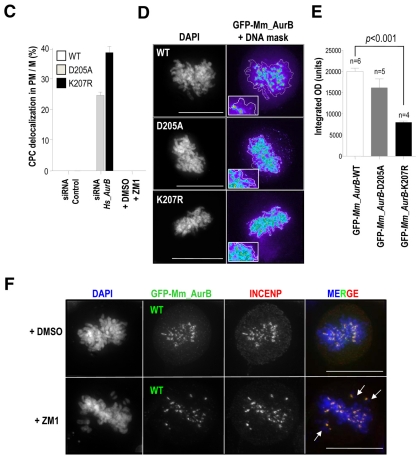
We then depleted endogenous Aurora B by siRNA in the stable clones expressing GFP-Aurora-B fusion proteins. The mouse Aurkb cDNA, encoding Aurora B, was used to generate these constructs and they were therefore resistant to siRNA treatment directed against the endogenous human transcript (Fig. 4B). The wild-type GFP-Aurora-B construct efficiently rescued the formation of multinucleated cells after depletion of endogenous Aurora B proteins and these rescued cells progressed normally through mitosis (Fig. 4C). However, the expression of Aurora-BD205A and Aurora-BK207R mutants failed to rescue the defects caused by knockdown of the endogenous protein. These cultures delayed in prometaphase with unaligned chromosomes, failure to execute cytokinesis and accumulation of multinucleated cells (Fig. 4C,D). Similar results were observed after chemical inhibition of Aurora-B kinase activity with ZM447439 (ZM1; Fig. 4C,D). Quantification of the distribution of mitotic phases in these stable cell lines after depletion of endogenous Aurora B confirmed that expression of GFP-Aurora-BD205A and GFP-Aurora-BK207R provokes a similar arrest at prometaphase and a reduction of cells in telophase (Fig. 4E). This was accompanied by a decrease in the percentage of cells in cytokinesis. Furthermore, those cells that were undergoing cytokinesis in cultures expressing Aurora BD205A and Aurora BK207R were mostly aberrant and frequently displayed chromosomal bridges between the two daughter cells (Fig. 4F).
Altered chromosomal dynamics of the CPC in Aurora-BK207R mutants
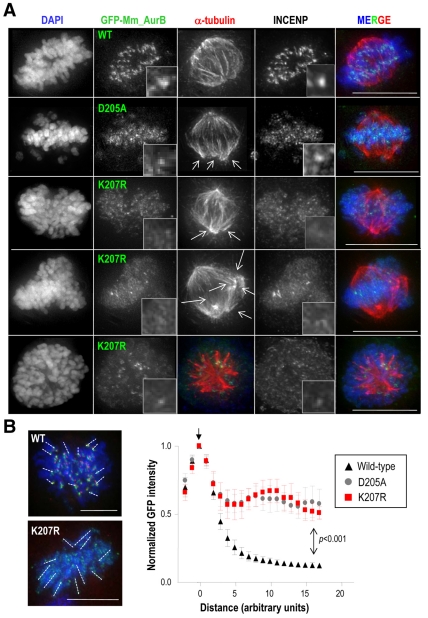
Endogenous Aurora B concentrates in the centromeric region of chromosomes during prometaphase, similarly to other CPC components such as INCENP. In the absence of endogenous Aurora B, both INCENP and exogenous wild-type GFP-Aurora-B localized properly to these centromeric structures, consistent with the functional rescue by this wild-type protein (Fig. 5A,B). Interestingly, the Aurora BK207R mutant (as well as the kinase-dead isoform) was dispersed along the entire chromosome arms in prometaphase (PM) and metaphase (M) cells. INCENP also failed to concentrate in inner centromeres, suggesting that there are CPC localization defects upon lack of Aurora B SUMOylation. Further analysis of the distribution of Aurora B using Metamorph algorithms confirmed that Aurora BD205A and Aurora BK207R were dispersed along the chromosome arms (Fig. 5C-E). Quantification of the integrated optical density (OD), a measure of the opacity of an object when exposed to light, showed that the chromosomal staining of the SUMO-null Aurora B mutant is significantly (P<0.001) less opaque than Aurora-BWT staining. This means that Aurora BK207R does not concentrate at any chromosomal region (Fig. 5E). These defects were reduced in the kinase-dead mutant and did not reach statistical significance in this assay. We also tested whether Aurora-B kinase activity was important for proper localization of Aurora B in the centromere, by treating GFP-Aurora-BWT cells with the ZM1 inhibitor. In agreement with previous reports (Ditchfield et al., 2003; Xu et al., 2009), chemical inhibition of Aurora B kinase activity did not perturb localization of Aurora B or INCENP to the centromere, indicating that an active kinase is not required for their centromeric localization (Fig. 5F).
Fig. 5.
The Aurora-B K207 residue is required for proper localization of CPC proteins at the centromere. (A) Immunofluorescence images of HeLa stable cell lines after depletion of endogenous Aurora B. Wild-type Aurora B colocalizes with INCENP at the centromeric regions. However, GFP-Aurora-BD205A and -Aurora-BK207R mutants display a disperse localization over the chromatin that also affects to INCENP in PM/M cells. Multipolar spindles are also present in these mutant cells (arrows). Scale bars: 10 μm. (B) Quantification of GFP-Aurora-B at chromosome arms from the centromere (Position 0; vertical arrow). Results are means ± s.e.m. Scale bars: 5 μm. (C) Quantification of the percentage of PM/M cells in which GFP-Aurora-B fusions and INCENP are delocalized. Delocalization of Aurora B is only observed in Aurora-BD205A and Aurora-BK207R mutants after depletion of the endogenous protein. At least 50 mitotic cells were counted per point. (D) Images pseudo-coloured with Metamorph software to visualize the distribution of GFP-Aurora-B signal in stable cell lines after depletion of endogenous Aurora B. Intensity of the signal is represented in a colour scale: (low) purple, blue, green, yellow, red (high). The contour of DNA (DAPI staining) is delineated. Scale bars: 10 μm. (E) Quantification of the integrated optical density (OD) of GFP-Aurora-B signal over the chromatin shows a significant reduction (P<0.0001) of GFP-Aurora-BK207R, but not -Aurora-BD205A, signal density. (F) Chemical inhibition of Aurora B with ZM1 does not delocalize the protein although it provokes chromosome misalignment (arrows). α-tubulin is in red in A, INCENP is in red in E, GFP-Aurora B is in green and DAPI (DNA) in blue. Scale bars: 10 μm.
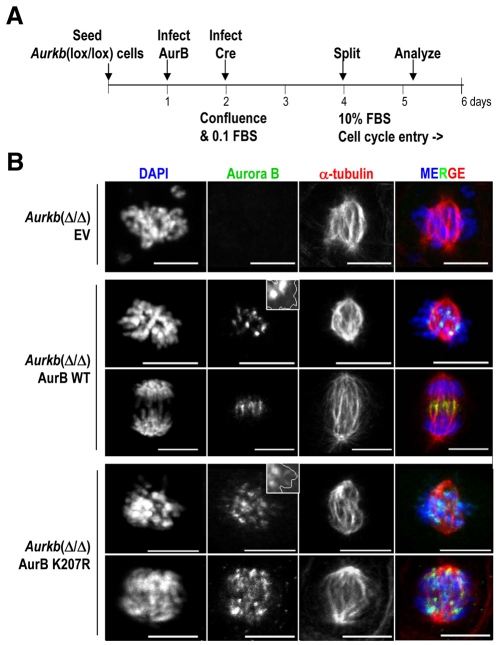
Since the presence of endogenous Aurora B might interfere with the localization of exogenous Aurora-BK207R mutants, we decided to further study these mutants in Aurora-B-knockout cells. These cells derive from Aurora-B conditional-knockout mice in which exons 2 to 6 are flanked by loxP sites [Aurkb(lox) allele; G.F.-M., I.P.C. and M.M., unpublished results]. Upon expression of the Cre recombinase (Fig. 6A), exons 2 to 6 are excised, resulting in a null allele [Aurkb(Δ)]. Aurkb(Δ/Δ) MEFs progress normally through prophase and prometaphase. However, whereas 65% of mitotic cells (n=40 of each genotype) were in metaphase, anaphase or telophase in Aurkb(lox/lox) cultures, these figures were reduced to 8% in Aurkb(Δ/Δ) cells (most of these anaphases and telophases showed immunoreactivity against Aurora B, probably as a consequence of inefficient gene deletion; data not shown). Aurkb(Δ/Δ) prometaphases displayed several misaligned chromosomes and abnormal spindles (Fig. 6B). These defects could be rescued by infection with retroviruses expressing wild-type Aurora B, and Aurkb(Δ/Δ); Aurora BWT cells displayed normal mitotic progression (about 60% mitotic cells were in metaphase, anaphase or telophase) and resulted in normal cytokinesis. In these Aurkb(Δ/Δ) MEFs, the exogenous Aurora B was properly located at centromeres and migrated to the midzone during anaphase (Fig. 6B). By contrast, Aurkb(Δ/Δ); Aurora BK207R cells displayed abnormal spindles and misaligned chromosomes and did not progress to anaphase (about 10% of mitotic figures in metaphase, anaphase or telophase). In these cells, the exogenous Aurora BK207R displayed an abnormal centromeric concentration and remained diffuse at chromosome arms (Fig. 6B).
Fig. 6.
Aurora BK207R does not rescue mitotic defects in Aurora-B-knockout MEFs. (A) Schematic representation of the protocol followed for rescue upon acute deletion of Aurora B in quiescent cells. Primary (passage 2) Aurkb(lox/lox) MEFs were seeded at 80% confluence and infected the following day with retroviruses expressing Aurora-BWT or Aurora-BK207R. Next day, cultures reach confluence and were cultured in 0.1% FBS and infected with adenoviruses expressing Cre recombinase (AdCre). Two days after the infection, cells were split in new plates at low confluence in the presence of 10% FBS to induce entry into the cell cycle. Mitotic cells were analyzed by immunofluorescence during the first round of division (28 hours after splitting). (B) Acute deletion of Aurora B in MEFs provokes chromosome misalignment and spindle defects that are rescued by exogenous, properly localized, wild-type Aurora B [Aurkb(Δ/Δ); Aurora BWT; green signal in merged images]. The exogenous Aurora BK207R, however, remains diffuse on chromosome arms and it does not rescue the Aurkb-null defects resulting in chromosome misalignment and impaired cytokinesis in Aurkb(Δ/Δ); Aurora BK207R cells. DNA (DAPI; blue) is delineated in the insets. α-tubulin is in red in merged images. Scale bars: 10 μm.
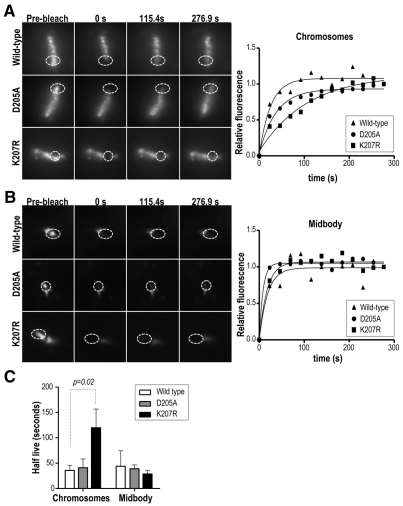
Fluorescence recovery after photobleaching (FRAP) was then used to compare the relative mobility of Aurora-B isoforms at metaphase chromosomes and the mid-body. Wild-type Aurora B showed a more dynamic behavior in the chromosomes located at the metaphase plate than the other mutant isoforms (Fig. 7). Although the recovery half-life (t1/2) was 35.6±10.2 seconds for wild-type Aurora B (n=7), the kinase-dead mutant Aurora BD205A displayed a higher value (40.8±17.4 seconds; n=4), as previously reported in similar assays (Murata-Hori and Wang, 2002). A significantly higher t1/2 value (119.9±36.8 seconds; n=4) was scored in the Aurora-BK207R mutant, suggesting defective dynamics in the SUMO mutant protein. A similar recovery pattern for all the Aurora B isoforms was observed at the mid-body [t1/2=44.00±30.5 seconds in wild-type Aurora B (n=4); t1/2=38.9±7.4 seconds in Aurora BD205A (n=3); t1/2=22.5±12.9 seconds in Aurora BK207R (n=4)] (Fig. 7B,C) indicating a specific chromosomal defect in the dynamics of Aurora B following mutation of the SUMOylation motif.
Fig. 7.
Defective dynamic localization of Aurora-B K207R at metaphase chromosomes. HeLa cells stably expressing GFP-tagged Aurora-B isoforms were used in a series of fluorescence recovery after photobleaching (FRAP) experiments. (A) Representative cases of recovery of wild-type Aurora B, Aurora BK207R and Aurora BD205A at the metaphase chromosomes. Delocalization of the Aurora-B mutant isoforms is not obvious as in Fig. 4 because these cells express the endogenous Aurora-B protein. (B) Representative images and quantification of similar FRAP assays at the mid-body. Graphs in both cases show the curves obtained using a regression analysis to integrate data from each stable clone. (C) The half-lives of GFP-Aurora-B at metaphase chromosomes and mid-bodies were compared using a t-test showing significant differences between the AurBK207R and the wild-type forms. Results are means ± s.e.m.
All together, these data indicate that Aurora BK207R is functionally defective and suggest that SUMOylation at K207 is crucial to regulate Aurora-B and INCENP dynamics at chromosomes.
The Aurora BK207R mutant displays no compromised kinase activity
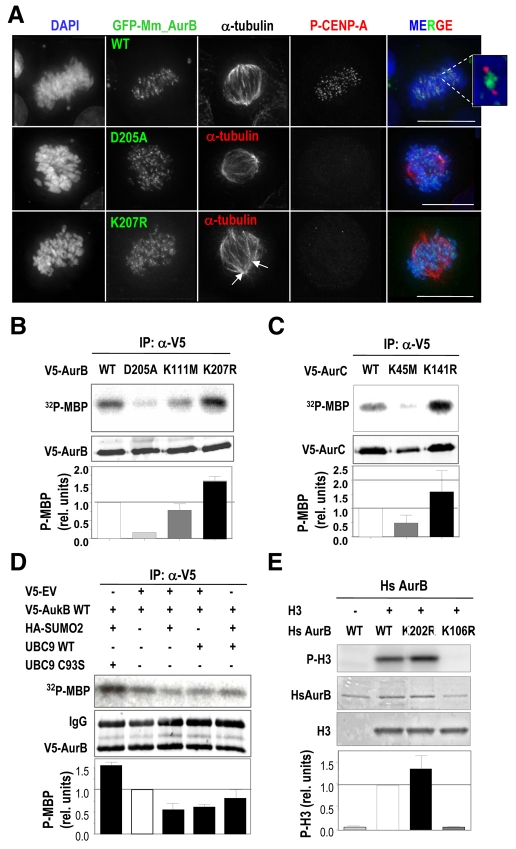
The differences in localization described above suggest that the molecular defects caused by the kinase-dead and SUMO-null mutations of Aurora B are not completely identical. We therefore tested the effects of these two mutations on kinase activity. Staining with phospho-specific antibodies against CENP-A, a known Aurora-B substrate, reveals that phosphorylation of CENP-A was significantly decreased in cells expressing either Aurora BD205A or Aurora BK207R (Fig. 8A). By contrast, GFP-Aurora-BWT, which was concentrated at centromeres, efficiently phosphorylated CENP-A, resulting in positive staining for phospho-CENP-A at kinetochores flanking the GFP-Aurora BWT signal (Fig. 8A). We next directly tested the kinase activity of V5-tagged Aurora-B proteins immunoprecipitated from transiently transfected HEK293 cells (Fig. 8B). Aurora BK111M and Aurora BD205A failed to efficiently phosphorylate the substrate MBP in vitro, although Aurora BK111M still displayed some kinase activity, in agreement with the previous phenotypic analysis of these kinase-dead mutants.
Fig. 8.
Centromeric mislocalization of Aurora-BK207R mutant prevents CENP-A phosphorylation without reducing its kinase activity. (A) Immunofluorescence images from HeLa stable cell lines treated with siRNA against endogenous Aurora B. GFP-Aurora-BD205A and -Aurora-BK207R mutants accumulate along chromosome arms and fail to phosphorylate CENP-A. α-tubulin staining is in red (only merged in the mutant forms), phosphorylated CENP-A, if present, is in red (only merged in the wild-type forms), GFP-Aurora-B in green and DAPI (DNA) in blue. The inset represents proper phosphorylation of CENP-A (red) by centromeric Aurora B (green). Scale bars: 10 μm. (B) In vitro kinase activity toward recombinant MBP of V5-tagged Aurora B proteins. HEK293 cells were transiently transfected with the indicated V5-Aurora-B expression vectors and the resulting exogenous expressed proteins were immunoprecipitated with anti-V5 antibody. The kinase activity of a SUMO-null (Aurora BK207R) Aurora B form is not reduced but partially increased. Aurora-B kinase-dead mutants K111M and D205A were included as negative controls of MBP phosphorylation. Quantification of [32P]MBP signal was calculated considering the total amount of V5-Aurora-B protein in each case. (C) Similar kinase assays using Aurora C constructs with the corresponding kinase-dead (Aurora CK45M) or SUMO-null (Aurora CK141R) mutations. (D) In vitro kinase assay in the presence of SUMO2, UBC9 or a dominant-negative UBC9 form (C93S mutant). Whereas the overexpression of SUMO2 and UBC9 results in decreased activity, the presence of UBC9 C93S results in increased Aurora-B-kinase activity in this assay. Data were normalized to the intensity of phosphorylated MBP in cells transfected only with wild-type Aurora B. (E) Phosphorylation of histone H3 (P-H3) by recombinant human wild-type (WT) Aurora B or the kinase-dead (K106R) and SUMO-null (K202R) mutants. Data were normalized to total levels of human Aurora B (HsAurB).
Strikingly, the kinase activity of the Aurora BK207R mutant was not reduced at all and indeed it was slightly increased when compared with the activity of the wild-type protein. Similar results were obtained when a similar SUMO-defective mutant (K141R) of the highly related kinase Aurora C was used (Fig. 8C). To further corroborate whether lack of SUMOylation modulates the intrinsic kinase activity of these proteins, we performed similar kinase assays in cells transfected with Aurora B, SUMO2 and wild-type or dominant-negative forms of UBC9. Interestingly, promoting Aurora-B SUMOylation by overexpressing SUMO2 and UBC9 resulted in a slight overall decrease in Aurora-B activity, whereas preventing SUMOylation by expressing a dominant-negative form of UBC9 led to increased Aurora B kinase activity (Fig. 8D). Finally, we also tested kinase activity in human recombinant proteins produced in baculovirus. As shown in Fig. 8E, the recombinant Aurora-BK202R mutant (corresponding to mouse K207) was also active and displayed a slight increase in kinase activity. Whether this increase represents differences in the intrinsic kinase properties of the K202,K207 mutants, or is a consequence of impaired SUMOylation of the wild-type molecule in mammalian or insect cells is not clear at present. In any case, these results indicate that the functional defects in the Aurora-BK207R mutant are not a consequence of decreased kinase activity, but rather suggest that SUMOylation modulates Aurora-B function by impairing its proper removal from chromosome arms.
Discussion
Global mapping of the SUMO network by protein-protein interactions and genetic-network analysis has linked SUMOylation to many different biological processes, including chromosome segregation and the cell cycle (Makhnevych et al., 2009). In recent years, the SUMO pathway has been implicated in several aspects of mitosis, including chromosome structure, cell-cycle progression, kinetochore function and cytokinesis (Dasso, 2008; Watts, 2007). In mammals, cells depleted of the SUMO E2-conjugating enzyme UBC9 show major deficiencies in chromosome condensation and segregation, along with severely altered nuclear morphology (Hayashi et al., 2002; Nacerddine et al., 2005), revealing the importance of this post-translational modification for mitotic progression. The CPC has been widely proposed to be regulated by SUMOylation. The yeast CPC component Bir1 (Survivin) is known to be modified by SUMO (Montpetit et al., 2006) and Sli15 (INCENP) is also a candidate for SUMO modifications in this organism (Wohlschlegel et al., 2004). Similarly, Borealin is regulated by binding to SUMO2 and SUMO3 in human cells (Klein et al., 2009).
In this work, we have identified a SUMO target site in mammalian Aurora B and analyzed the consequences of lack of SUMOylation in Aurora-B regulation and function. The SUMOylation site is highly conserved among Aurora B orthologs and in the other members of the Aurora family, Aurora A and Aurora C. Our data indicate that Aurora B can be SUMOylated in vivo by the three SUMO isoforms at Lys207 (K207). According to observations in Xenopus egg extracts and human cells, SUMO2 and SUMO3 are found at centromeres and chromatin during prometaphase and metaphase, whereas SUMO1 localizes to the mitotic spindle and the spindle mid-zone during these stages (Ayaydin and Dasso, 2004; Azuma and Dasso, 2002; Zhang et al., 2008). Indeed, Aurora B and SUMO2 are known to colocalize (Azuma et al., 2005) (our unpublished results), suggesting that this kinase is preferentially modified by SUMO2, and possibly SUMO3, in vivo.
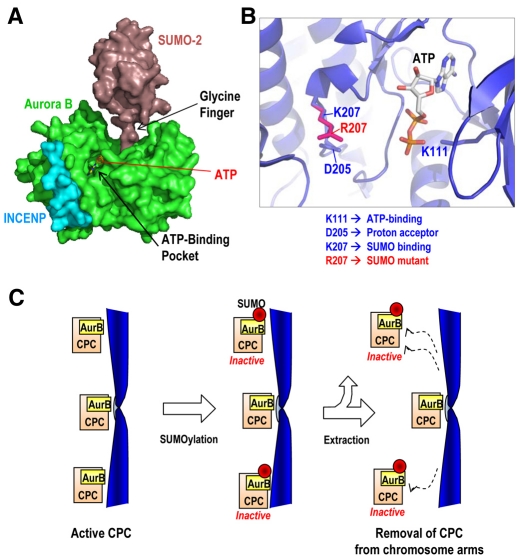
Recent work has reported that Aurora B is not significantly modified by SUMO1 in vitro and that it is not able to interact with E2 SUMO-conjugating enzyme UBC9 or SUMO isoforms in yeast two-hybrid assays (Klein et al., 2009). An intriguing possibility is that other CPC components might be necessary for interactions between Aurora B and the SUMOylation machinery. Indeed, Aurora B seems to be modified by SUMO when immunoprecipitated from cell extracts where the CPC integrity is maintained. In addition, SUMOylation of Aurora B is favoured in vitro by the presence of the IN-box domain of INCENP. Indeed, a computer simulation of the interaction between these proteins suggests that the presence of the IN-box domain of INCENP facilitates the proper energetic environment required for SUMOylation of Aurora B (Fig. 9A,B).
Fig. 9.
Modelling the Aurora-B-SUMO covalent interaction. (A) The crystal structures of human SUMO2 (Reverter and Lima, 2006) (PDB 2I03) and Xenopus Aurora B in complex with IN-box segment of INCENP (Sessa et al., 2005) (PDB 2BFY) were used to simulate the interaction between these molecules. SUMO2 might covalently bind the Aurora-B K207 residue (K218 in Xenopus crystal structure) through a finger composed of two glycine residues. SUMOylation is not predicted to affect binding of the Aurora-B partner and activator INCENP, but it might disturb the transfer of phosphate groups to its substrates. (B) Details of the ATP-binding pocket showing the localization of K111, D205 and K207 residues (K122, D216 and K218 in Xenopus, respectively) used in this study. The position of the ATP molecule was modelled after a superposition with the Aurora-A-TPX2 structure (PDB 1OL5). (C) A model for the role of SUMOylation of Aurora B in CPC function. SUMOylation of Aurora B might result in decreased kinase activity, probably as a consequence of interference of the SUMO residue with kinase function. In addition, the presence of the SUMO residue might trigger an unknown pathway that favours the removal of Aurora B and the CPC from chromatin. MT, microtubule.
How might SUMO modifications modulate Aurora-B function? SUMOylation frequently alters protein surfaces and thereby positively or negatively influences interactions with other macromolecules (Geiss-Friedlander and Melchior, 2007). Thus, SUMOylation can interfere with or promote protein-protein interactions and provoke conformational changes in the modified target. The consequences of those molecular changes might influence the localization, stability or activity of a target protein in vivo. As shown in this work, SUMOylation of Aurora B seems to regulate the dynamics of Aurora-B localization at the chromosomes. SUMOylation of Aurora B seems to affect a small fraction of the Aurora B pool, although it is not clear at present whether this is a consequence of the transient nature of SUMOylation or technical problems owing to strong demodifying activity in mammalian protein extracts (Geiss-Friedlander and Melchior, 2007; Pichler, 2008). Rescue experiments performed with the Aurora-B SUMO-deficient form (Aurora BK207R) demonstrates that both Aurora B and INCENP fail to concentrate in inner centromeres during prometaphase or metaphase, thus suggesting that lack of Aurora-B SUMOylation affects the localization of other CPC components. Other examples of mitotic proteins whose localization at kinetochores or centromeres is regulated by SUMO include the kinesin CENP-E (Zhang et al., 2008), the CENP-H/I/K kinetochore complex (Mukhopadhyay et al., 2010) and the GTPase-activating protein RanGAP1 (Joseph et al., 2002).
It has been recently confirmed that phosphorylation of Aurora-B substrates is influenced by the relative locations of the substrate and kinase at the inner centromere (Liu et al., 2009). This mechanism offers an efficient mechanism to selectively destabilize improper microtubule-kinetochore attachments (Kelly and Funabiki, 2009). As described in the present manuscript, the SUMO-deficient form of Aurora B, despite exhibiting normal kinase activity in vitro, fails to phosphorylate CENP-A in vivo. Thus, the lack of dynamic localization of SUMO-null Aurora B might impair its interaction with its substrates, resulting in failure to phosphorylate them in vivo but not in vitro.
SUMOylation of Aurora B might also directly modulate the intrinsic kinase activity. Aurora-B D205 is crucial for kinase function and it is only two residues upstream from the SUMO-binding site (K207). Indeed, both residues are in the same structural loop (Fig. 9B). Rescue experiments with a kinase-dead mutant (Aurora BD205A) result in phenotypes that are similar to those obtained with the SUMO-null (Aurora BK207R) mutant. This led us to initially hypothesize that the Aurora-BK207R mutant is kinase dead. However, this is not the case, as the kinase activity in vitro is, if anything, slightly increased in this mutant and in a similar mutant in Aurora C. Indeed, based on our kinase assays, the Aurora BK207R mutant might act as a delocalized, partially hyperactive mutant. A detailed examination of the predicted structure reveals that SUMO binding might block the entrance of ATP to the kinase and/or the transference of phosphate groups to a given substrate (Fig. 9A,B). Therefore, it is reasonable to think that SUMOylation might act as a transient and reversible means of inhibiting Aurora B activity that is crucial for CPC localization and dynamics, analogous to T14,Y15 phosphorylation of Cdk1, which also blocks ATP binding and transfer. In general, our data are consistent with a model where the non-SUMOylated, active CPC exerts its function at unattached or merotelic kinetochores. Upon SUMOylation of Aurora B, the kinase activity would be reduced and the SUMOylated complexes can be removed from chromatin by an unknown pathway that recognizes the SUMO residues (Fig. 9C). In the Aurora-BK207R mutant, the Aurora-B kinase might be hyperactive, as a result of the lack of SUMO residues, but the CPC is diffuse over the chromatin, resulting in deficient localized phosphorylation of its substrates.
Several lines of evidence link SUMO modifications with ubiquitin-mediated processes (Geoffroy and Hay, 2009). Recent studies have shown that ubiquitylation of Aurora B by the Cul3-Klhl9/Klhl13 E3-ligase complex during early mitosis might serve as a signal for its removal from chromosomes (Sumara et al., 2007). The AAA-ATPase p97 complex specifically interacts with ubiquitylated Aurora B and extracts it from chromosomes, leading to inactivation of the kinase on chromatin (Ramadan et al., 2007). In the absence of Cul3-Klhl9/Klhl13 activity, Aurora B is not ubiquitylated and accumulates on chromosomes (Sumara et al., 2007). The fission yeast Aurora kinase (Ark1p) interacts with two RING-finger proteins that possess SUMO-interacting motifs (SIMs), called Rfp1p and Rfp2p (Sun et al., 2007). These E3 ubiquitin ligases recognize SUMOylated proteins and heterodimerize with Slx8, another RING-finger protein, to form a functional ubiquitin ligase that is functionally similar to vertebrate RNF4 (Geoffroy and Hay, 2009; Sun et al., 2007; Uzunova et al., 2007). It is therefore possible that, in mammals, SUMOylation of Aurora B facilitates the formation of a functional Cul3-Klhl9/Klhl13 ligase complex during mitosis. Indeed, we have found three predicted SIMs in each coadaptor, Klhl9 and Klhl13, and one additional SIM in the Cul3 ligase that are conserved in human and mouse in all cases (data not shown). It is also possible that SUMOylation of Aurora B modulates its ubiquitylation by the APC/C-Cdh1. However, Aurora B does not accumulate on chromosomes upon RNA interference to knock down Cdh1 (Floyd et al., 2008), suggesting that these two pathways are not linked. Future experiments will be necessary to test and validate these hypotheses.
In summary, our data suggest that Aurora-B function is modulated by SUMOylation at K207. This modification is likely to interfere with Aurora-B kinase activity and it might participate in the extraction of the CPC from chromatin during prometaphase and the metaphase-to-anaphase transition. The effects of SUMOylation on the dynamic localization of Aurora B might be linked to ubiquitylation by the Cul3-Klhl9/Klhl13 ligase, because prevention of either SUMOylation or ubiquitylation by this complex results in a similar phenotype where the CPC is delocalized on chromosome arms. The regulation of Aurora B by SUMOylation might also have relevant implications, not only in preventing genomic instability, but also in tumorigenesis and cancer therapy (Girdler et al., 2006; Keen and Taylor, 2004; Mo and Moschos, 2005; Perez de Castro et al., 2007; Perez de Castro et al., 2008).
Materials and Methods
Sequence analysis and gene constructs
For protein-domain analysis, we first used the ELM (Eukaryotic Linear Motif, http://elm.eu.org) algorithm. To further confirm the presence of SUMO motifs we used the specific SUMO prediction software SUMOplot™ Analysis Program (http://www.abgent.com/tools/SUMOplot). Sequence alignments were obtained with the Clustal software (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Mouse Aurkb cDNA was amplified by PCR from the plasmid mAA0155 (Mammalian Gene Collection), cloned into pENTR-D-TOPO vector using TOPO technology (Invitrogen) and transferred to a destiny vector coexpressing GFP or V5 tag by a LR recombination reaction of the Gateway system (Invitrogen). Aurora-B mutants (Aurora BK111M, Aurora BD205A and Aurora BK207R) were prepared using the Quik-Change site-directed mutagenesis kit (Stratagene) and were verified by DNA sequencing. Plasmids expressing histidine- or HA-tagged SUMO1, SUMO2 and SUMO3 as well as wild-type UBC9 or the UBC9 C93S mutant were kindly provided by Ron Hay (Wellcome Trust Centre for Gene Regulation and Expression, University of Dundee, UK).
Cell culture, transfection and cell-cycle analysis
HEK293, U2OS and HeLa human cells were maintained in DMEM medium supplemented with 10% fetal bovine serum and antibiotics and were grown at 37°C in a humidified 5% CO2 atmosphere. The generation of conditional alleles at the mouse Aurkb locus will be described elsewhere. Mouse embryo fibroblasts were obtained from E14.5 Aurkb(lox/lox) embryos and cultured using routine protocols (Garcia-Higuera et al., 2008). Human Aurora B was silenced using validated siRNA oligos from Qiagen with the following sequence: 5′-AACGCGGCACTTCACAATTGA-3′. HEK293, U2OS and HeLa cells were transfected with 1 μg plasmid DNA using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. For silencing experiments, siRNA oligos were nucleofected in HeLa cells using Amaxa kit and the conditions provided by Amaxa I13 program. Cell-cycle distribution was determined by flow cytometry after DNA staining with propidium iodide (Sigma) and analyzed on a FACSCanto flow cytometer (Becton Dickinson). Data were processed using FACSDiva software (Becton Dickinson). For chemical inhibition of Aurora B, HeLa cells were treated with 2 μM ZM447439 (Tocris Bioscience). For acute deletion of Aurora B in MEFs, we followed the scheme represented in Fig. 5A. Adenoviruses expressing Cre recombinase (Ad5 CMV-Cre) were obtained from The University of Iowa (Iowa City, IA). Adenoviral infection was carried out during 2 days in a cell culture synchronized in G0 by serum deprivation and confluence. Rescue experiments were performed with retroviral vectors derived from pBabe-derivative plasmids expressing Aurora BWT and AuroraK207R.
HeLa cells stably expressing His-SUMO2 (kindly provided by Ron Hay) were transfected with His-SUMO2, UBC9 and V5-Aurora-WT and incubated for 24 hours. A parental asynchronous culture was used as a negative control. Taxol was added at a concentration of 100 nM, and cells were incubated for an additional 18 hours. To obtain a G1 cell population, taxol-arrested cells were released in fresh medium for 3 hours. Cell lysates and Ni-NTA pull-downs were performed as described (Muller et al., 2000; Tatham et al., 2009).
Expression and analysis of recombinant proteins
Recombinant baculovirus-expressing human His-Aurora B-WT, His-Aurora B-K106R and His-Aurora B-K202R were generated using the pFastBacHTA vector and the Bac-to-Bac Expression System (Invitrogen), as described previously (Gassmann et al., 2004). For in vitro SUMOylation assays, His-Aurora BWT or a complex of His-Aurora BWT and the IN-box fragment (human INCENP amino acids 821 to end; Millipore) were incubated with purified E1-activating enzyme (SAE1/2), E2-conjugating enzyme (UBC9) and either SUMO2 or mutant SUMO2 protein (harbouring a proprietary mutation that impedes SUMO conjugation) in the presence of ATP and following the manufacturer's instructions (Active Motif).
Immunofluorescence and FRAP analysis
For immunofluorescence, cells were fixed with 4% PFA and permeabilized with 0.15% Triton X-100. These cells were then blocked with 3% BSA and incubated with the primary antibodies against the following proteins: α-tubulin (Sigma), Aurora B (Abcam, BD Transduction), INCENP (Human GLPL serum, a gift from Manuel Valdivia, Hospital Puerta del Mar, Cadiz, Spain), Phospho-CENP-A (Ser7; Upstate Biotechnology). The matching secondary antibodies, with different Alexa Fluor dyes (488, 594, 647), are from Molecular Probes (Invitrogen), Images were obtained using a confocal ultraspectral microscope (Leica TCS-SP5) or an Olympus IX70 microscope controlled by Delta Vision SoftWorx (Applied Precision, Issequa, WA). Image stacks were deconvolved, quick-projected and saved as tiff images to be processed using Adobe Photoshop.
Fluorescence recovery after photobleaching (FRAP) was performed on HeLa stable cell lines expressing GFP-Aurora-B wild-type and mutants (D205A and K207R). All experiments were conducted using a FRAP-enabled DeltaVision. Images of fluorescence recovery of centromeres and mid-bodies were captured every 20-30 seconds. Intensity values were normalized using the following equation; It=(Xt–Y)/(Z–Y), where I is the normalized intensity at time t, X is the actual intensity at time t, Y is the intensity immediately following the photobleach (when t is equal to zero), and Z is the intensity at the final time point. This sets the initial post-bleach intensity (at t=0 seconds) to zero and the final intensity to 1 arbitrary units. Regression analysis was performed using the GraphPad Prism software by calculating the least squares fit using the standard first order logarithmic equation. The t1/2 value was the time at which the normalized intensity reaches 0.5 arbitrary units.
Protein extraction and analysis
For immunodetection in protein lysates, cells were washed twice with ice-cold PBS and lysed in RIPA lysis buffer (37 mM NaCl, 0.5% NP-40, 0.1% SDS, 1% Triton X-100, 20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 10% glycerol 1 mM PMSF) supplemented with protease and phosphatase inhibitory cocktails (Sigma). Additionally, for detecting SUMOylated proteins, an inhibitor of SUMO proteases, N-ethylmaleimide (NEM), was added at 10 mM final concentration. After 30 minutes on ice, samples were cleared by centrifugation. Proteins were separated on SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), probed using specific antibodies and detected using goat anti-rabbit or anti-mouse secondary antibodies (Dako) for ECL (Perkin Elmer) detection. After transfer of the protein lysates, we probed nitrocellulose membranes with primary antibodies against Aurora B (BD Transduction and Abcam), INCENP (Abcam), SUMO2/3 (MBL), Cyclin B1 (Santa Cruz Biotechnology), p53 (Active Motif), HA tag (Abcam), V5 tag (Invitrogen), His6 tag (Roche) and α-tubulin (Sigma).
Immunoprecipitation and in vitro kinase assays
Total protein lysates extracted from HEK293 cells transiently transfected with V5-tagged Aurora-B vectors were precleared with protein-G-agarose bead suspension for 1 hour (Amersham). Supernatants were first incubated for 2 hours at 4°C on a rotating wheel with mouse anti-V5 (Invitrogen) and later with 50 μl of the blocked bead suspension for an additional hour. Immunoprecipates were then washed three times in RIPA buffer and, in case of the kinase assay, one extra wash was performed with RIPA plus 0.5 M NaCl to remove possible bound unspecific kinases. One-fifth of the immunoprecipitates bound to the beads were used for V5 detection by western blot. For the kinase assay, four-fifths of the beads from the previous immunoprecipitations or recombinant proteins (Millipore) were washed in kinase assay buffer [10 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, 10 mM MgCl2, and 1 mM EGTA] supplemented with 0.5 mM DTT and phosphatase inhibitor cocktail (Calbiochem) and later incubated in 25 μl of 2× kinase buffer supplemented with 0.2 mM ATP, 2.5 μCi [γ-32P]ATP and 3 μg MBP (Sigma) or histone H3.1 (New England Biolabs) as an Aurora substrate. The reactions were incubated for 30 minutes at 30°C, stopped by addition of 5 μl of 5× Laemmli sample buffer, separated by 12.5 % SDS-PAGE and analyzed by autoradiography.
Acknowledgments
We thank Susana Temiño for technical assistance, Diego Megías for support in image processing and Ron Hay and Manuel Valdivia for reagents. G.F.M. was supported by a fellowship from the Comunidad de Madrid. I.P.C. was supported by a Ramón y Cajal contract. Work in the W.C.E. lab is funded by The Wellcome Trust. G.M. is funded by grants CSD2006-00023, and BFU2008-01344 from the Spanish Ministry of Science and Innovation (MICINN). The Cell Division and Cancer Group of the CNIO is funded by grants from the Association for International Cancer Research (AICR 08-0188), Fundación Mutua Madrileña Automovilista, Fundación Ramón Areces, MICINN (SAF2007-64571 to I.P.C.; and SAF2006-05186 and SAF2009-07973 to M.M.), the OncoCycle Programme (S-BIO-0283-2006) from the Comunidad de Madrid, and the OncoBIO Consolider-Ingenio 2010 Programme (CSD2007-00017) from the MICINN, Madrid. Deposited in PMC for release after 6 months.
References
- Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., Earnshaw W. C. (2000). INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10, 1075-1078 [DOI] [PubMed] [Google Scholar]
- Ayaydin F., Dasso M. (2004). Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15, 5208-5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Dasso M. (2002). A new clue at the nuclear pore: RanBP2 is an E3 enzyme for SUMO1. Dev. Cell 2, 130-131 [DOI] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Anan T., Dasso M. (2005). PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172-2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. D., Schumacher J. M. (2002). Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277, 27577-27580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Earnshaw W. C. (2003). The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842-854 [DOI] [PubMed] [Google Scholar]
- Dasso M. (2008). Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. (2008). Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133, 103-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A., Ouyang J., Lee H. Y., Catic A., Ploegh H., Gill G. (2006). The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 26, 4489-4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S., Pines J., Lindon C. (2008). APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr. Biol. 18, 1649-1658 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I., Manchado E., Dubus P., Canamero M., Mendez J., Moreno S., Malumbres M. (2008). Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10, 802-811 [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A. J., Ruchaud S., Hudson D. F., Honda R., Nigg E. A., Gerloff D. L., Earnshaw W. C. (2004). Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166, 179-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947-956 [DOI] [PubMed] [Google Scholar]
- Geoffroy M. C., Hay R. T. (2009). An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell. Biol. 10, 564-568 [DOI] [PubMed] [Google Scholar]
- Girdler F., Gascoigne K. E., Eyers P. A., Hartmuth S., Crafter C., Foote K. M., Keen N. J., Taylor S. S. (2006). Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 119, 3664-3675 [DOI] [PubMed] [Google Scholar]
- Guse A., Mishima M., Glotzer M. (2005). Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15, 778-786 [DOI] [PubMed] [Google Scholar]
- Hay R. T. (2005). SUMO: a history of modification. Mol. Cell 18, 1-12 [DOI] [PubMed] [Google Scholar]
- Hay R. T. (2007). SUMO-specific proteases: a twist in the tail. Trends Cell. Biol. 17, 370-376 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Seki M., Maeda D., Wang W., Kawabe Y., Seki T., Saitoh H., Fukagawa T., Yagi H., Enomoto T. (2002). Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 280, 212-221 [DOI] [PubMed] [Google Scholar]
- Honda R., Korner R., Nigg E. A. (2003). Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325-3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A., Conti E. (2007). Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131, 271-285 [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S. H., Karpova T. S., McNally J. G., Dasso M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N., Taylor S. (2004). Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 4, 927-936 [DOI] [PubMed] [Google Scholar]
- Kelly A. E., Funabiki H. (2009). Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21, 51-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U. R., Haindl M., Nigg E. A., Muller S. (2009). RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell 20, 410-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M. J., Lampson M. A., Lens S. M. (2009). Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350-1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., et al. (2009). Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33, 124-135 [DOI] [PubMed] [Google Scholar]
- Mo Y. Y., Moschos S. J. (2005). Targeting Ubc9 for cancer therapy. Expert Opin. Ther. Targets 9, 1203-1216 [DOI] [PubMed] [Google Scholar]
- Montpetit B., Hazbun T. R., Fields S., Hieter P. (2006). Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 174, 653-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Arnaoutov A., Dasso M. (2010). The SUMO protease SENP6 is essential for inner kinetochore assembly. J. Cell Biol. 188, 681-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Berger M., Lehembre F., Seeler J. S., Haupt Y., Dejean A. (2000). c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275, 13321-13329 [DOI] [PubMed] [Google Scholar]
- Murata-Hori M., Wang Y. L. (2002). Both midzone and astral microtubules are involved in the delivery of cytokinesis signals: insights from the mobility of aurora B. J. Cell Biol. 159, 45-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769-779 [DOI] [PubMed] [Google Scholar]
- Perez de Castro I., de Carcer G., Malumbres M. (2007). A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis 28, 899-912 [DOI] [PubMed] [Google Scholar]
- Perez de Castro I., de Carcer G., Montoya G., Malumbres M. (2008). Emerging cancer therapeutic opportunities by inhibiting mitotic kinases. Curr. Opin. Pharmacol. 8, 375-383 [DOI] [PubMed] [Google Scholar]
- Pichler A. (2008). Analysis of sumoylation. Methods Mol. Biol. 446, 131-138 [DOI] [PubMed] [Google Scholar]
- Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., Gotta M., Meyer H. H. (2007). Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258-1262 [DOI] [PubMed] [Google Scholar]
- Reverter D., Lima C. D. (2006). Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat. Struct. Mol. Biol. 13, 1060-1068 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. (2007). Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8, 798-812 [DOI] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. (2005). Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379-391 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M. H., Held M., Guizetti J., Maar S., Gerlich D. W. (2009). Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473-484 [DOI] [PubMed] [Google Scholar]
- Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., Ricci R., Peter M. (2007). A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12, 887-900 [DOI] [PubMed] [Google Scholar]
- Sun H., Leverson J. D., Hunter T. (2007). Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102-4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M. H., Rodriguez M. S., Xirodimas D. P., Hay R. T. (2009). Detection of protein SUMOylation in vivo. Nat. Protoc. 4, 1363-1371 [DOI] [PubMed] [Google Scholar]
- Ulrich H. D. (2008). The fast-growing business of SUMO chains. Mol. Cell 32, 301-305 [DOI] [PubMed] [Google Scholar]
- Uzunova K., Gottsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., et al. (2007). Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167-34175 [DOI] [PubMed] [Google Scholar]
- Vong Q. P., Cao K., Li H. Y., Iglesias P. A., Zheng Y. (2005). Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310, 1499-1504 [DOI] [PubMed] [Google Scholar]
- Watts F. Z. (2007). The role of SUMO in chromosome segregation. Chromosoma 116, 15-20 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd (2004). Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279, 45662-45668 [DOI] [PubMed] [Google Scholar]
- Xu Z., Ogawa H., Vagnarelli P., Bergmann J. H., Hudson D. F., Ruchaud S., Fukagawa T., Earnshaw W. C., Samejima K. (2009). INCENP-aurora B interactions modulate kinase activity and chromosome passenger complex localization. J. Cell Biol. 187, 637-653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. D., Goeres J., Zhang H., Yen T. J., Porter A. C., Matunis M. J. (2008). SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729-741 [DOI] [PMC free article] [PubMed] [Google Scholar]