Abstract
Nucleotide excision repair (NER) is a major DNA repair pathway in eukaryotic cells. NER removes structurally diverse lesions such as pyrimidine dimers, arising upon UV irradiation or bulky chemical adducts, arising upon exposure to carcinogens and some chemotherapeutic drugs. NER defects lead to three genetic disorders that result in predisposition to cancers, accelerated aging, neurological and developmental defects. During NER, more than 30 polypeptides cooperate to recognize, incise, and excise a damaged oligonucleotide from the genomic DNA. Recent papers reveal an additional and unexpected role for the NER factors. In the absence of a genotoxic attack, the promoters of RNA polymerases I- and II-dependent genes recruit XPA, XPC, XPG, and XPF to initiate gene expression. A model that includes the growth arrest and DNA damage 45α protein (Gadd45α) and the NER factors, in order to maintain the promoter of active genes under a hypomethylated state, has been proposed but remains controversial. This paper focuses on the double life of the NER factors in DNA repair and transcription and describes the possible roles of these factors in the RNA synthesis process.
1. Introduction
A number of DNA repair pathways protect us from the deleterious effects of DNA damage. The importance of these mechanisms is highlighted by the existence of genetic disorders in which impaired DNA repair mechanisms predispose patients to cancer and early onset of aging. A major advance in our understanding of these DNA repair mechanisms has been to uncover the tangled connection existing between these systems and other fundamental cellular processes such as DNA replication and transcription. These cellular processes are not only highly connected with DNA repair pathways but they also share common factors with them. This complexity leads to new hypothesis about the cause of the phenotypes displayed by patients suffering from DNA repair disorders and may even force us to re-evaluate the place of the repair factors in cellular homeostasis.
2. The NER Pathway: The Fountain of Youth of Our Genome
We do not live forever young. We all have to experience aging, a functional decline coupled to an increased mortality risk from diseases such as cancer. The molecular origins of aging can be sought, at least in part, in an alteration of the expression of our genes that results from the physicochemical constitution of DNA, which does not guarantee life-long stability (for reviews see [1, 2]). Over time, DNA accumulates a tremendous diversity of lesions that, if unrepaired, lead to mutations that dysregulate the function of proteins. DNA lesions originate from environmental agents such as the ultraviolet (UV) component of sunlight, ionizing radiation, and numerous genotoxic chemicals, and also from the products of normal cellular metabolism. Aging is a relatively slow process for most of us, but unfortunately premature appearance of multiple symptoms of aging can be observed in a growing family of human syndromes [3, 4]. Among them, the xeroderma pigmentosum (XP), trichothyodystrophy (TTD), and Cockayne syndrome (CS) are remarkable as they all include two distinct phenotypes: either a 1000-fold elevated suninduced skin cancer risk, for XP patients, or a segmental progeria without an increase in cancer incidence, for CS and TTD [5]. These syndromes uncover what our lives would be if a “fountain of youth” was not protecting our genome day after day from endogenous and exogenous assaults. Indeed, the main molecular defect of the XP, CS, and TTD patients resides in a defect in the nucleotide excision repair (NER) pathway [6]. NER is an evolutionarily conserved DNA repair caretaker pathway involving about 30 proteins, ten of which (designated XPA to G; TTDA, CSA, and CSB) are differentially associated with XP, CS, or TTD disorders in an intricate network (Figure 1). NER is divided into two subpathways, which differentially remove damages from our genome depending on their location [7]. In the Global Genome NER (GGNER), the XPChHR23B complex recognizes damages. The DNA is then opened by the XPD and XPB helicasecontaining transcription/repair complex TFIIH together with XPA and RPA to generate the damaged single stranded DNA ready for incision by the specific endonucleases XPG and ERCC1-XPF. DNA gap filling is done by the replicative DNA polymerases δ and ε or the translesional polymerase k, in the presence of PCNA, RFC, and RPA [8] (Figure 2). In transcription-coupled NER (TC-NER), blockage of transcribing RNA Polymerase II (RNA-Pol II) on the damaged DNA template is thought to initiate the repair reaction in a process that requires, in addition to TFIIH, XPA, XPG, and ERCC1-XPF, the TCR-specific proteins CSB and CSA [9] (Figure 2). Although CSB is required to recruit NER factors to the stalled RNA-Pol II, CSA is coming later and is not needed for the formation of the TCR complex [10] (Figure 2).
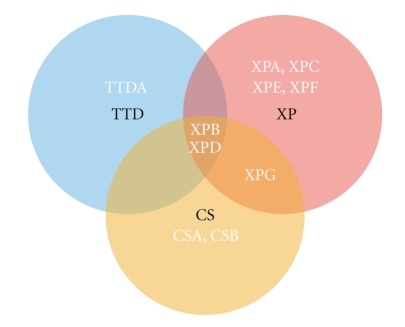
Figure 1.
Three disorders for nine genes. Mutations in ten genes are responsible for the xeroderma pigmentosum (XP), the trichothyodystrophy (TTD) or the Cockayne syndrome (CS). XPA, XPC, XPE and XPF are only involved in XP; CSA and CSB are only involved in CS; XPG is involved in pure XP or in an combined XP/CS syndrome; XPB and XPD are involved in TTD, XP, or in a combined XP/CS syndrome. TTDA is only involved in TTD.
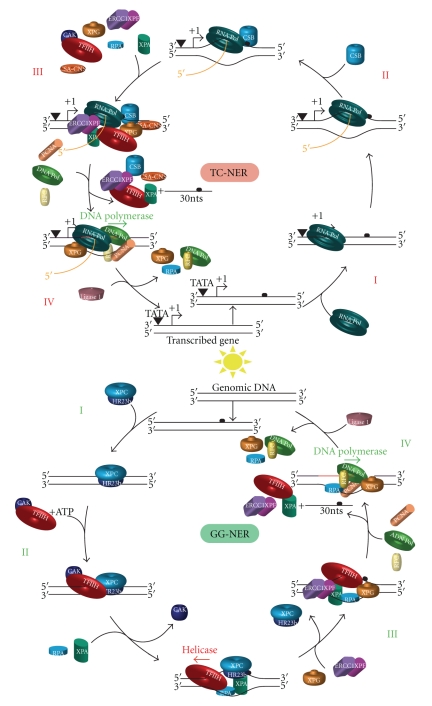
Figure 2.
The two subpathways of mammalian NER. Physical or chemical agents like UV, cis-platin, or benzopyrene can damage DNA and induce damage-mediated helix distortions anywhere in the genome (GG-NER in green, bottom panel) or on the transcribed strand of a gene (TC-NER in red, top panel). Bottom panel: (I) XPC-RAD23B recognizes and binds to DNA damage-mediated helix distortion to initiate GG-NER. (II) TFIIH is recruited in an ATP-dependent manner, followed by XPA and RPA, which verify the presence of the lesion. During this step, the CAK module of TFIIH is released from the preincision complex [29]. (III) Within the preincision complex, ERCC1-XPF and XPG structure-specific endonucleases incise the damaged strand on the 5′ and 3′ sides of the lesion, respectively. Following incision, NER factors are released from the DNA, except XPG and RPA that favour the recruitment of the replication machinery composed of PCNA, RFC, and the DNA Polymerases δ, ε, or κ (ref). (IV) Following replication of the gap, the DNA is sealed by the ligase 1 (or the ligase III-XRCC1 complex in nondividing cells). Top Panel: (I) TC-NER is triggered by DNA damage-mediated blockage of RNA-Pol II (Top panel). (II) CSB is then recruited to the stalled RNA-Pol II enzyme and triggers the recruitment of the NER factors TFIIH, XPA, RPA, ERCC1-XPF, and XPG together with the CSA-CNS complex (III). (IV) Following the excision of the damaged oligonucleotide, the same DNA replication machinery of the GG-NER subpathway fills the gap created by the incision/excision step.
Next to the basal NER machinery, additional factors modulate the efficiency of the NER reaction but are not required to incise a damaged oligonucleotide in vitro. The GG-NER damage recognition factor, XPC, forms in vivo a heterotrimeric complex involving one of the two human homologs of S.cerevisiae Rad23p (hHR23B) and centrin 2, a centrosomal protein [11]. The role of centrin 2 and hHR23B in NER has been elusive but they seem to increase the damage recognition capacity of XPC [12]. The XPE complex, mutated in XP-E patients, is another accessory NER factor composed of DDB1 and DDB2. The role of the XPE protein remains unclear, but it could participate in the recognition of lesions together with XPC [13]. Another NER factor, XPA-binding protein 2 (XAB2), was identified by virtue of its ability to interact with XPA [14]. XAB2 also associates with the TC-NER specific proteins CSA and CSB, as well as with RNA-Pol II, after UV irradiation and is specifically involved in the TC-NER subpathway [15]. Finally, the DDB2 and CSA polypeptides can be found integrated into nearly identical complexes containing cullin 4A, Roc1, and COP9 that seem to favor NER [16]. Although limited today, the list of proteins that modulate the NER reaction should increase in a near future and benefit from high through-put technologies. The study of these cofactors will constitute an important challenge, as the modulation of the efficiency of NER to eliminate DNA lesions may explain some cancer predispositions in healthy people. Moreover, identifying the complete set of proteins that participate in NER is a crucial aspect of cancer therapy since the resistance to chemotherapy treatment could partially rely on the capacities of the cell to eliminate drug-induced DNA lesions.
3. The NER Pathway in a Chromatin Context: Take Old Factors to Make Them New
New DNA repair players have also emerged from the study of NER in the chromatin context. Reorganisation of nucleosome structure following NER was observed over 30 years ago [17], and many studies demonstrate that chromatin acts as a barrier for the recognition of the lesions by NER factors [18]. Not surprisingly, chromatin remodelers identified in NER were already known to promote accessibility to the DNA for the transcription machinery. The ATP-dependent chromatin remodelling complexes SWI/SNF or ISW2 have been shown to act on UV-damaged nucleosomes and to stimulate repair in vitro [19]. In yeast, UV irradiation increases contacts between SWI/SNF and the homologs of XPC-hHR23B, and inactivation of SWI-SNF leads to a slow removal of CPD lesions [20]. Finally, the ATP-dependent chromatin assembly factor-1 (CAF-1) is required to restore the chromatin conformation after the removal of the lesions [21].
Apart from ATP-dependent nucleosome remodelling, many forms of histones modifications have been unveiled after UV irradiation. Histone acetylation was the first modification to be shown to play a role in NER. Treatment of nonreplicating human cells with the histone deacetylase inhibitor sodium butyrate enhances NER [22]. PCNA, the replicative protein involved in the DNA resynthesis step of NER, interacts with the p300 histone acetyltransferase following UV irradiation [23]. On the other hand, a complex containing the damaged DNA-binding protein DDB1, the CREB-binding protein CBP, and p300 has been isolated in vivo [24]. Another complex, TBP-free-TAFII complex (TFTC), directs histone H3 acetylation by hGCN5 after UV irradiation and facilitates access of DNA repair machinery to lesions within chromatin [25]. In addition to histone acetylation, UV damage also induces histone H2A monoubiquitination in the vicinity of DNA lesions [26]. Monoubiquitination of H2A depends on functional NER and occurs after incision [27].
Overall, these data show that histone modifications form part of the cellular response to UV damage and clearly play a role in chromatin remodelling during DNA repair. However, the exact nature of the modified histones and residues as well as the role of these modifications in the facilitation of DNA damage access or in the DNA damage response not clear. Much remains to be done to define a histone code in NER, comparable to that acquired in other fundamental cellular processes like transcription or double-strand break repair.
4. The Unveiled Side of the XP, CS, and TTD Syndromes
Although the UV sensitivity and/or cancer predisposition of XP, CS, and TTD patients can be explained by defects in NER, some other of their phenotypes (including neurological and developmental defects) are more difficult to rationalize. For instance, some group A patients show the most severe progressive neurological disorders while the XPA protein is only known for its role in the verification of the damages [5]. Thus, several studies have aimed to discover additional processes that may be disrupted in these pathologies and at a first glance have found evidence for transcription defect in TFIIH-, XPG-, and CSB-mutated cells.
TFIIH is a ten-subunit complex composed of a core (XPB, p62, p52, p44, p34, and TTDA) coupled to the Cdk-activating kinase complex (CAK) through the XPD subunit [28]. A recent study showed that CAK does not participate to NER and is released from the core TFIIH during the formation of the preincision complex following the recruitment of XPA [29] (see also Figure 2). As a component of TFIIH, CAK phosphorylates both the carboxyl terminal domain of RNA-Pol II and some nuclear receptors (NRs) including the retinoic acid receptors (RARα and γ) [30], the thyroid hormone receptor (TR) [31], and the peroxysome proliferator-activated receptor (PPAR) [32]. Phosphorylation of these NRs is required for the transactivation of specific genes. Cdk7 also activates the vitamin D receptor indirectly, by phosphorylating the Ets1 coactivator [33] (Table 1).
Table 1.
Repair/transcription factors and their functions.
| Factors | Role in NER | Role in transcription |
|---|---|---|
| TFIIH | Opens DNA around the damage. Favors 5′ incision by ERCC1-XPF. |
Opens DNA around the promoter. Phosphorylates the CTD of RNA-Pol II. Phosphorylates NR and coactivators. |
|
| ||
| XPG | Incises DNA in 3′ from the lesion | Involved in NR-dependent transcription. Stabilizes the interaction of CAK to the core TFIIH. |
|
| ||
| CSB | TC-NER-specific factor. Recruits NER factors to stalled RNA-Pol II. |
Chromatin-remodeling factor (SWI-SNF family). Reinitiates transcription after DNA damage removal. |
|
| ||
| XPC | Recognition of lesions |
Involved in NR-dependent transcription. Removal of 5meC Chromatin modification? |
|
| ||
| XPA | Verification of lesions |
Involved in NR-dependent transcription. Removal of 5meC Chromatin modification? |
|
| ||
| XPF | Incises DNA in 3′from the lesion |
Involved in NR-dependent transcription. Removal of 5meC, Chromatin modification? Incision? |
In bold, the new NER factors involved in transcription.
Patients with mutations in XPB and XPD display a transcriptional defect in specific genes, which may help clarifying the origin of their developmental or neurological problems. In TTD-XPD cells, mutations in XPD destabilize the CAK complex from TFIIH leading to defects in the phosphorylation of RAR, ER, and PPAR. In XP-B patients, two mutations in XPB (F99S and fs740) lead to the combined XP/CS defect with a very low level of residual NER activity [34]. However, only the fs740 mutation is cancer prone [35]. It was shown that this mutation specifically blocks transcription activation by the FUSE-Binding Protein (FBP), a regulator of c-myc expression, and inhibition by the FBP-Interacting Repressor (FIR) [36]. The fact that the regulation by FBP and FIR is impaired could directly affect proper regulation of c-myc expression and explain the development of malignancy in the corresponding patient. The XPB and XPD subunits of TFIIH are not the only NER polypeptides to be involved in transcription.
The first evidence for an involvement of XPG in transcription came from a study in yeast. RAD2, the S.cerevisiae counterpart of XPG, was shown to be required in promoting efficient RNA-Pol II transcription [37]. Later, it was demonstrated that mutations in human XPG, as found in XP-G/CS patient cells, prevent the association of XPG with TFIIH, resulting in the dissociation of the CAK and XPD from the core TFIIH [38]. This dissociation leads to an impair transactivation of the NR-dependent responsive genes.
The TCR-specific CSB protein belongs to the ATP-dependent SWI2/SNF2 family of chromatin remodeling proteins and has been shown to play a role in both remodeling the chromatin structure and disrupting protein–DNA interactions [39]. Besides its role in TCR, CSB is involved in the transcription recovery of housekeeping genes after UV irradiation [40]. CSB is specifically recruited to the promoters of these genes and helps in the recruitment of both the RNA-Pol II and the associated basal transcription factors, probably through its chromatin remodeling activity (Table 1).
Altogether, these data show that the transcription defect in XP/CS, CS, or TTD is subtle and more difficult to evaluate than the NER defect because this defect targets specific genes, under specific conditions, and probably in a cell-specific manner. However, the involvement of transcription dysregulation in aging and cancer makes these studies very important for the understanding of these diseases. Interestingly, a picture emerges from these studies, which shows that mutations in XP factors lead to a modification of the expression of specific genes by possibly two means; either through the accumulation of unrepaired lesions that will lead to mutations or through a direct involvement of repair factors in gene expression. However, a piece of the puzzle is missing. Even though a clear involvement of XPB, XPD, XPG, or CSB in transcription was documented, it has been more difficult to assign a transcriptional role to XPC, XPA, or ERCC1-XPF until the recent works discussed below.
5. Behind the Evidence: A Transcriptional Role for the NER Factors
Protein coding genes expression is the result of an acute process that starts at the promoter of a given gene and involves, in the addition to the RNA-Pol II and the basal transcription factors, a cocktail of proteins such as the NR, coactivators, mediator, and histone-modifying enzymes. A study from our group [41] shows that some NER factors are associated with the transcription machinery at the promoter of several activated NR-dependent genes. The recruitment occurs in a sequential order after the formation of the preinitiation complex (PIC) and induces XPC, CSB, XPA, and the XPG and ERCC1-XPF endonucleases. The transcriptional complex equipped with NER factors is formed in the absence of any exogenous genotoxic attack and is distinct from a repair complex, since it is specifically sensitive to transcription inhibitors and can be formed in the absence of the TCR specific-CSB protein (Figure 3). Following transcription initiation, NER factors escort the RNA-Pol II during the elongation step to form a complex that does not include XPC but requires CSB (Figure 3). These observations suggest a different function for the NER factors located at promoters in respect to those located at distal regions of the gene; while the latter may represent a pre-TCR complex ready to remove lesions on transcribed genes, the former may play an active role in transcription. In line with this hypothesis, patient cell lines mutated in XPC, XPA, or XPG show a dysregulation of the NR-dependent genes that results from a defect in the association of the NER factors with the transcription machinery. Although the corresponding XPC, XPA, XPG, and ERCC1-XPF repair factors are not essential for PIC formation, it remains that they optimize the efficiency of transcription.
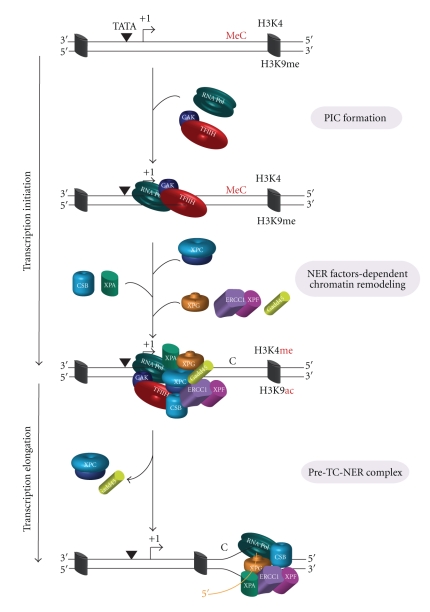
Figure 3.
NER factors and gene transcription. Upon gene activation, the formation of preinitiation complex (PIC) precedes the recruitment of XPC, which allows the sequential arrival of the other NER factors in the following order CSB, XPA/RPA, XPG/ERCC1-XPF, and Gadd45α. The association of NER factors and Gadd45α with the transcription machinery leads to a cascade of histone PTMs. Concomitantly, an active demethylation of 5′CpG islands occurs.
6. Insight into the Function of the NER Factor in Transcription
How do NER factors favor NR-dependent genes transcription? Several studies have reported a controversial role for Gadd45α in association with the endonuclease activity of XPG in transcription: the active demethylation of CpGs islands localized at proximal promoters [42–44]. Recent works support these findings and demonstrate that the recruitment of XPC, XPA, XPG, and ERCC1-XPF on the promoter of active RNA-Pol I- and II-dependent genes allows the association of Gadd45α to the PIC and induce the demethylation of promoters [41, 44]. Mutations in XPC, XPA and XPG found in XP patients dysregulate the corecruitment of the NER factors and Gadd45α to active promoters, thereby abolishing the active demethylation step and thus affecting transcription.
How can the NER factors demethylate DNA? Similar to a classical NER lesion, 5′-methylcytosine (meC) combined to the specific chromatin environment during transcription initiation could be recognized and eliminated by the NER machinery [45] (Figure 4(a)). Indeed, a previous study demonstrated a faster repair rates near the transcription initiation site linked to increased local concentrations of DNA repair factors associated with basal transcription factors [46]. The sequential recruitment of NER factors could help the incision and the replacement of meC with unmethylated nucleotides. Even if incision by XPG on the promoter of RNA Pol I-dependent genes has been reported [44], this hypothesis is highly controversial, and several groups propose other alternatives to explain the demethylation of meC. Recent studies have supported a model involving at least two steps [43, 47] (Figure 4(b)). The model predicts the conversion of meC to cytosine by the direct removal of the methyl group or by the hydrolytic deamination of meC to thymine further excised by a DNA repair enzyme. The first step concerns the deamination reaction and implies apolipoprotein B mRNA editing enzyme (APOBECS) proteins such as activation-induced cytidine deaminase (AID) and APOBEC1, which function in sequence specific context. Alternatively, it has also been suggested that enzymes called DNA methyltransferases (DNMTs) exhibit dual and opposite actions, not only to methylate CpG islands but also to deaminate them [48]. The second step is related to the action of a DNA glycosylase such as Mdb4 or TDG that remove thymine from G/T mispairs to generate abasic sites rapidly cleaved through the activity of apurinic/apyrimidinic endonuclease (APE) (Figure 4(b)) [43]. It was recently shown that Mdb4 is corecruited to active promoters with proteins from base-excision-repair (BER) process such as APE-1, DNA ligase I, or polymerase δ [48]. Even though the role of Gadd45α is controversial, it clearly increases the efficiency of the demethylation process.
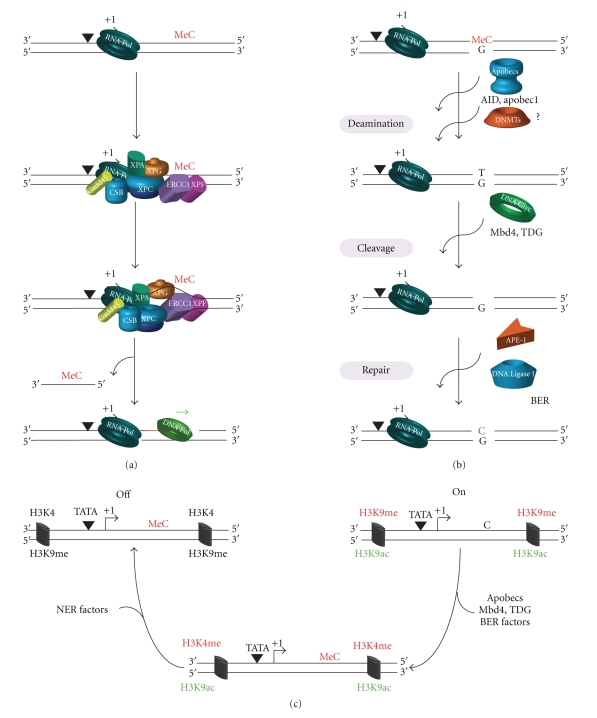
Figure 4.
Potential mechanisms of cytosine demethylation. (a) The NER machinery, recruited to the preinitiation complex, can recognize the 5-methyl cytosine (meC) as a NER-specific substrate, in the presence of Gadd45α, and eliminate it in a process closely related to the canonical NER process with the incision/excision of the oligonucleotide containing the meC. (b) Another pathway involves two steps; first, the deamination of 5-methylcytosine (meC) to thymine, which involves proteins from Apobec family such as AID or APOBEC1. A role in deamination has been also suggested for DNMTs proteins. Consequently, the impairment of the thymine with the guanine in the opposite strand induces the recruitment of DNA glycosylases such as Mbd4 or TDG that remove thymine through cleavage of the glycosidic bond. Following the action of DNA glycosylases, it remains an apyrimidinic site, which is cleaved by an AP endonuclease such as APE1 and repaired through the polymerase β and DNA ligases. NER factors and Gadd45α are involved in this mechanism but their roles are not determined. (c) We propose that NER factors control the epigenetic environment of the promoter favouring the demethylation of H3K9 and the methylation of H3K4. Following the action of the NER factors, Apobec proteins and BER factors demethylate the meC in a process similar to (b).
The results obtained recently by several groups lead us to propose another hypothesis that could account for the active demethylation of promoters during transcription and involves both the NER and the BER factors. Active DNA demethylation at promoters is intimately linked with histones posttranslational modifications (PTMs) [49]. Di/tri methylation of H3K4 (H3K4me) and di/trimethylation of H3K9 (H3K9me) correlate with active transcription and heterochromatin, respectively. In a repressed status, the methyltransferase G9a catalyzes the methylation of H3K9, which allows the binding of the heterochromatin protein 1 (HP1) to facilitate the local formation of heterochromatin. The G9a-containing complex also recruits the DNA methyltransferases DNMT3A and DNMT3B that catalyze the de novo methylation of DNA at promoters. Conversely, during active transcription and concomitantly to demethylation/acetylation of H3K9, methylation of H3K4 inhibits contacts between nucleosome and DNMT3 to facilitate active DNA demethylation [50]. NR-dependent altered transcription observed in XP-C, XP-A, or XP-G cells is accompanied by dysregulation of PTMs of histones. The hypermethylated status of these promoters in these cells is associated with absence of H3K4me and maintenance of the H3K9me marks. These observations imply that the promoter of NR-dependent genes persists in a heterochromatin environment, despite the formation of the RNA-Pol II machinery, thereby impairing optimal transcription. There is no evidence of a direct role of repair factors in the regulation of histones PTMs or histone-modifying enzymes; it can be hypothesised that NER factors function upstream of the BER factors to help to maintain a euchromatin environment characterised by a demethylation of H3K9 and a methylation of H3K4 (Figure 4(c)).
7. Conclusion
Almost twenty years after the discovery that the basal transcription factor TFIIH was also involved in NER [51], a new age arises from the discovery that basal NER factors are involved in activated transcription. The emergence of repair factors in transcription forces us to modify our approach for the understanding of the broad clinical features described for the so-called XP, TTD, and CS “repair syndromes”, but it also represents a breakthrough in gene expression studies. Indeed, the effects of DNA methylation variations on gene expression have been largely studied, but the mechanisms that promote active demethylation combined to histones modifications are just appearing with the finding that DNA repair factors may participate to this process.
Besides the 2D space organisation of a gene, one has also to consider the 3D space organisation of the nucleus. Transcription is deeply associated to genome organization; the location of a gene within the chromosome territories influences its ability to be reached by the suitable machinery [52]. Since the PTMs of histone and the methylated status of genomic DNA are connected to the dynamic topological regulation of chromatin, we have to consider that NER factors could contribute to transcription through a role in the nonrandom organization of the nucleus. Surprisingly, besides the DNA repair disorders, a second group of diseases that are characterized by accelerated aging comprises the Hutchinson-Gilford syndrome (or Progeria) that is due to a point mutation in Lamin A [53, 54]. This protein is a structural component of the nuclear matrix that plays a role in the 3D organization of the genome. It is then tempting to propose that changes in the nuclear architecture in these disorders participate in the modification of the transcription program and possibly to the impairment of the repair of some lesions, that altogether lead to accelerated aging and cancer.
Acknowledgments
The authors are more than grateful to Renier Velez Cruz, and Emmanuel Compe for the critical reading of this paper and for their suggestion. Frédéric Coin and Jean-Marc Egly are funded by the Institut National du Cancer (INCA-2008-041), the French National Research Agency (ANR-08-GENOPAT-042), and ERC advanced Grant (ERC-2008-ADG233077). Nicolas Le May is supported by the INSERM.
References
- 1.Hoeijmakers JHJ. DNA damage, aging, and cancer. The New England Journal of Medicine. 2009;361(15):1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 2.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends in Biochemical Sciences. 2010;35(6):333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher B, Garinis GA, Hoeijmakers JHJ. Age to survive: DNA damage and aging. Trends in Genetics. 2008;24(2):77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Kamenisch Y, Berneburg M. Progeroid syndromes and UV-induced oxidative DNA damage. Journal of Investigative Dermatology Symposium Proceedings. 2009;14(1):8–14. doi: 10.1038/jidsymp.2009.6. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85(11):1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nature Reviews Genetics. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21(58):8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 8.Ogi T, Limsirichaikul S, Overmeer RM, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Molecular Cell. 2010;37(5):714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Fousteri M, Mullenders LHF. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Research. 2008;18(1):73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 10.Spivak G, Cox RA, Hanawalt PC. New applications of the Comet assay: Comet-FISH and transcription-coupled DNA repair. Mutation Research. 2009;681(1):44–50. doi: 10.1016/j.mrrev.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki M, Masutani C, Takemura M, et al. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. Journal of Biological Chemistry. 2001;276(22):18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 12.Nishi R, Okuda Y, Watanabe E, et al. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Molecular and Cellular Biology. 2005;25(13):5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh T, Bennett D. The XPE gene of xeroderma pigmentosum, its product and biological roles. Advances in Experimental Medicine and Biology. 2008;637:57–64. doi: 10.1007/978-0-387-09599-8_7. [DOI] [PubMed] [Google Scholar]
- 14.Nakatsu Y, Asahina H, Citterio E, et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. Journal of Biological Chemistry. 2000;275(45):34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- 15.Kuraoka I, Ito S, Wada T, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. Journal of Biological Chemistry. 2008;283(2):940–950. doi: 10.1074/jbc.M706647200. [DOI] [PubMed] [Google Scholar]
- 16.Groisman R, Polanowska J, Kuraoka I, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113(3):357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 17.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA repair synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(9):4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Molecular and Cellular Biology. 2000;20(24):9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menoni H, Gasparutto D, Hamiche A, et al. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Molecular and Cellular Biology. 2007;27(17):5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nature Structural and Molecular Biology. 2006;13(10):902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 21.Gaillard P-HL, Martini EM-D, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86(6):887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 22.Smerdon MJ, Lan SY, Calza RE, Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. Journal of Biological Chemistry. 1982;257(22):13441–13447. [PubMed] [Google Scholar]
- 23.Hasan S, Hassa PO, Imhof R, Hottiger MO. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410(6826):387–391. doi: 10.1038/35066610. [DOI] [PubMed] [Google Scholar]
- 24.Datta A, Bagchi S, Nag A, et al. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutation Research. 2001;486(2):89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- 25.Brand M, Moggs JG, Oulad-Abdelghani M, et al. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. The EMBO Journal. 2001;20(12):3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergink S, Salomons FA, Hoogstraten D, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes and Development. 2006;20(10):1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends in Genetics. 2003;19(10):578–584. doi: 10.1016/j.tig.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Molecular Cell. 2008;31(1):9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly J-M. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARα . Cell. 2002;109(1):125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 31.Compe E, Malerba M, Soler L, Marescaux J, Borrelli E, Egly J-M. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nature Neuroscience. 2007;10(11):1414–1422. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 32.Compe E, Drané P, Laurent C, et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Molecular and Cellular Biology. 2005;25(14):6065–6076. doi: 10.1128/MCB.25.14.6065-6076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drané P, Compe E, Catez P, Chymkowitch P, Egly J-M. Selective regulation of vitamin D receptor-responsive genes by TFIIH. Molecular Cell. 2004;16(2):187–197. doi: 10.1016/j.molcel.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Vermeulen W, Scott RJ, Rodgers S, et al. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. American Journal of Human Genetics. 1994;54(2):191–200. [PMC free article] [PubMed] [Google Scholar]
- 35.Weeda G, van Ham RCA, Vermeulen W, Bootsma D, van der Eb AJ, Hoeijmakers JHJ. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell. 1990;62(4):777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Akoulitchev S, Weber A, et al. Defective interplay of activators and repressors with TFIIH in xeroderma pigmentosum. Cell. 2001;104(3):353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee S-K, Yu S-L, Prakash L, Prakash S. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription: implications for Cockayne syndrome. Cell. 2002;109(7):823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 38.Ito S, Kuraoka I, Chymkowitch P, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for cockayne syndrome in XP-G/CS patients. Molecular Cell. 2007;26(2):231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Citterio E, Van Den Boom V, Schnitzler G, et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Molecular and Cellular Biology. 2000;20(20):7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proietti-De-Santis L, Drané P, Egly J-M. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. The EMBO Journal. 2006;25(9):1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le May N, Mota-Fernandes D, Vélez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Molecular Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Barreto G, Schäfer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 43.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz K-M, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Molecular Cell. 2009;33(3):344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Ma DK, Guo JU, Ming G-L, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8(10):1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu Y, Tornaletti S, Pfeifer GP. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. The EMBO Journal. 1996;15(3):675–683. [PMC free article] [PubMed] [Google Scholar]
- 47.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Métivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 49.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 50.Ooi SKT, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaeffer L, Roy R, Humbert S, et al. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;259(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 52.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories—a functional nuclear landscape. Current Opinion in Cell Biology. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 53.De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300(5628):p. 2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 54.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423(6937):293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]