Abstract
DNA adducts of carcinogens derived from tobacco smoke and cooked meat were identified, by liquid chromatography-electrospray ionization/multi-stage tandem mass spectrometry (LC-ESI/MS/MSn), in saliva samples from 37 human volunteers on unrestricted diets. The N-(deoxyguanosin-8-yl) (dG-C8) adducts of the heterocyclic aromatic amines 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP); 2-amino-9H-pyrido[2,3-b]indole (AαC); 2-amino-3,8-dimethylmidazo[4,5-f]quinoxaline (MeIQx); and the aromatic amine, 4-aminobiphenyl (4-ABP) were characterized and quantified, by LC-ESI/MS/MSn, employing consecutive reaction monitoring at the MS3 scan stage mode with a linear quadrupole ion trap (LIT) mass spectrometer (MS). DNA adducts of PhIP were found most frequently: dG-C8-PhIP was detected in saliva samples from 13 of 29 ever-smokers and in saliva samples from 2 of 8 never-smokers. dG-C8-AαC and dG-C8-MeIQx were identified solely in saliva samples of 3 current smokers, and dG-C8-4-ABP was detected in saliva from 2 current-smokers. The levels of these different adducts ranged from 1 to 9 adducts per 108 DNA bases. These findings demonstrate that PhIP is a significant DNA-damaging agent in humans. Saliva appears to be a promising biological fluid in which to assay DNA adducts of tobacco and dietary carcinogens, by selective LIT MS techniques.
Introduction
The covalent modification of DNA by chemical mutagens is recognized as the initiating step in chemical carcinogenesis (1). DNA adduct measurement is an important endpoint, both for cross-species extrapolation of the biologically effective dose and for the human risk assessment of exposure to chemical carcinogens (2,3). Identification and quantification of chemical-specific adducts in the target tissue are the most relevant findings for risk assessment (3). However, the opportunity to measure carcinogen DNA adducts in human tissues is often precluded by the unavailability of biopsy samples. Thus, accessible biological fluids, such as blood (4), urine (5), exfoliated bladder epithelial cells in urine (6), or exfoliated mammary epithelial cells in milk of lactating women (7,8), have served as surrogate matrices in which to assess exposure to chemicals, or their metabolites, or formation of DNA adducts. The identification of DNA adducts clearly demonstrates exposure to the biologically active metabolite; however, the adduct must correlate with cancer risk, if it is considered valid as a biomarker of health risk (9).
The oral cavity is the portal of entry for carcinogens that are ingested in the diet or inhaled through smoking. Epithelial buccal cells and leukocytes are the principal mammalian cells present in saliva (10,11). Cells of the oral cavity have been employed to measure biomarkers of genetic damage, molecular, and cellular changes following exposure to tobacco smoke or arsenic, as well as to assess the efficacy of chemopreventive agents (11-17). The expression of mRNA of several cytochrome P450 (P450) enzymes, including P450s 1A1 and 1A2, which bioactivate carcinogens, has been detected in buccal cells (18,19). When placed in primary culture, buccal cells were shown to bioactivate aflatoxin B1 (18), benzo[a]pyrene (B[a]P) (20), and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (21), to metabolites that bound to DNA or protein. More recently, the P450 1A2 protein was detected in human salivary glands, by immunohistochemical methods (22). P450 1A2 is a principal enzyme involved in the bioactivation of heterocyclic aromatic amines (HAAs) and the aromatic amine 4-aminobiphenyl (4-ABP) (23,24).
Both 32P-postlabeling (25-27) and immunohistochemical techniques (28,29) have been employed to screen for DNA adducts in cells of the oral cavity. Several of the studies reported differences in total DNA adduct levels between smokers and non-smokers; however, the complexity of the adduct profiles and the inability to identify specific DNA adducts precluded any interpretation on the principal DNA damaging agents and their significance in the risk of development of oral cancer or cancers of other organs (25-27,30). It is noteworthy that high levels of DNA adducts were detected in the oral cavity of nonsmokers: many of these lesions could be attributed to genotoxicants in the diet.
Electrospray ionization (ESI) is a soft-ionization technique employed to detect non-volatile and thermally labile compounds (31). The on-line coupling of liquid chromatography with ESI-tandem mass spectrometry can be used to provide structural information on DNA adducts, and the incorporation of stable, isotopically labeled internal standards allows precise and accurate quantification of DNA adducts (32,33). While the levels of sensitivity of recent vintage MS instruments are sufficient to detect carcinogen DNA adducts at trace levels in human tissues, there is still a need to establish DNA adduct biomarkers in surrogate tissues or fluids that can be obtained non-invasively, to permit assessment of DNA damage posed by chemical carcinogens.
We recently employed a linear quadrupole ion trap mass spectrometer (LIT MS) to screen for DNA adducts of environmental, dietary, and endogenous genotoxicants, by data-dependent constant neutral loss scanning, followed by triple-stage mass spectrometry from oral cells of smokers (34). Cyclic DNA adducts of acrolein, a highly reactive α,β-unsaturated aldehyde that is formed endogenously through lipid peroxidation, and that also arises in cigarette smoke (35), were identified. However, DNA adducts of other tobacco-related or meat-derived carcinogens, that include benzo[a]pyrene (B[a]P), 4-ABP, 2-AαC, PhIP, and MeIQx (34), were not detected. In this present study, we have screened for DNA adducts of the above tobacco and meat-associated carcinogens in saliva samples of subjects, employing liquid chromatography-electrospray ionization/multi-stage tandem mass spectrometry (LC-ESI/MS/MSn) at the MS3 scan stage with the LIT MS. DNA adducts of all of these carcinogens, except for B[a]P, were identified in a number of human saliva samples. Human saliva appears to be a highly promising biological fluid with which we can monitor exposure to various carcinogens, through detection of their DNA adducts.
Materials and Methods
Caution: AαC, 4-ABP, B[a]P, MeIQx, PhIP, and their derivatives are carcinogens, and they should only be handled in a well-ventilated fume hood with the appropriate protective clothing.
Chemicals. MeIQx, 3-[2H3C]-MeIQx, and PhIP were purchased from Toronto Research Chemicals (Toronto, ON, Canada). 2-Nitro-9H-pyrido[4,5-b]indole was a kind gift from Dr. D. Miller (National Center for Toxicological Research,(Jefferson, AR). (±)-r-7,t-8-Dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene ((±) (anti)B[a]PDE)) was purchased from the NCI Chemical Carcinogen Reference Standards Repository, Midwest Research Institute (Kansas City, MO). 4-Nitrobiphenyl was purchased from Aldrich (Milwaukee, WI). Calf thymus (CT) DNA, deoxyguanosine (dG), DNase I (Type IV, bovine pancreas), alkaline phosphatase (from E. coli), and nuclease P1 (from Penicillium citrinum) were purchased from Sigma (St. Louis, MO). [13C10]-dG was purchased from Cambridge Isotopes (Andover, MA). Phosphodiesterase I (from Crotalus adamanteus venom) was from GE Healthcare (Piscataway, NJ). All solvents used were high-purity B & J Brand from Honeywell Burdick and Jackson (Muskegon, MI). ACS reagent-grade formic acid (88%) was purchased from J.T. Baker (Phillipsburg, NJ). HyperSep™ filter SpinTips C18 (20 mg) were from Thermo Scientific (Palm Beach, FL). Oragene-DNA saliva kits were from Genotek Inc. (Ontario, Canada).
Preparation of the DNA adducts standards
N-(Deoxyguanosin-8-yl)-PhIP (dG-C8-PhIP) (36,37), N-(deoxyguanosin-8-yl)-MeIQx (dG-C8-MeIQx) (38,39), and N-(deoxyguanosin-8-yl)-AαC (dG-C8-AαC) (40) were prepared by reaction of their N-acetoxy-HAA derivatives with dG or [13C10]-dG (5 mg) in 100 mM potassium phosphate buffer (pH 8.0). N-(deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-4-ABP) was prepared by reaction of N-hydroxy-4-ABP with pyruvonitrile, followed by reaction with dG or [13C10]-dG (41). For the case of dG-C8-MeIQx, the internal standard was prepared by reaction of the N-acetoxy derivative of 3-[2H3C]-MeIQx with dG. The (±)-anti-B[a]PDE-derived dG adducts, 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene (dG-N2-B[a]P) were prepared by reaction of (±)-anti-B[a]PDE with dG as described (42). The adducts were purified by solid phase extraction (SPE), followed by HPLC purification (38,39), and were isolated as a mixture of unresolved isomers. The isotopic purity of the [13C10]-dG labeled internal standards exceeded 99%, whereas the isotopic purity of the dG-C8-[2H3C]-MeIQx adduct was 96.5%.
Tritium-labeled PhIP-modified, tritium-labeled 4-ABP-modified and tritium-labeled B[a]P-modified calf thymus DNA
[3H]-PhIP-modified CT DNA was prepared as described (36). The extent of [3H]-PhIP modification was estimated at 1 adduct per 106 unmodified DNA bases. [3H]-4-ABP-modified CT DNA (62 adducts per 108 unmodified DNA bases) (43), and [3H]-B[a]P-modified CT DNA (111 adducts per 108 unmodified DNA bases) (44) were provided by Dr. F. Beland (National Center for Toxicological Research, Jefferson, AR).
Isolation of DNA from saliva
Saliva (∼4 mL) was collected in Oragene-DNA kits, and stored at -20 or -80 °C prior to isolation of DNA. The DNA was isolated following the manufacturer's instructions. However, further purification of the DNA was required for the efficient enzymatic hydrolysis of DNA to its mononucleosides (unpublished observations). The purified salivary DNA was further processed by proteinase K treatment, and then treated with RNase A and RNase T1, followed by extraction with phenol and chloroform (45). The DNA was precipitated in 9:1 C2H5OH: 3M sodium acetate buffer (pH 6.0), followed by washing of the DNA filament with C2H5OH:H2O (7:3). The amount of DNA recovered from saliva samples ranged from 9 to 51 μg.
Enzyme Digestion and Solid Phase Extraction (SPE) of DNA adducts
Isotopically labeled internal standards were added to the DNA solution prior to enzyme digestion, at a level of 10 adducts per 108 bases. The enzymatic digestion conditions used for the hydrolysis of DNA (9 – 51 μg) in 5 mM Bis-Tris-HCl buffer (pH 7.1, 50 μL) employed DNAse I for 1.5 h, followed by incubation with nuclease P1 for 3 h, and then treatment with alkaline phosphatase and phosphodiesterase for 18 h (37). The DNA digest solution was diluted with high-purity water (200 μL; Burdick and Jackson), and the adducts were purified by SPE, using HyperSep™ filter SpinTips. The DNA digest extracts were applied to a SpinTip, which was placed on a vacuum manifold and prewashed with CH3OH containing 0.1% HCO2H (0.5 mL), followed by 10% CH3OH in 0.1% HCO2H (0.5 mL). The SpinTip was then washed with 10% CH3OH in 0.1% HCO2H (2 × 0.25 mL), to remove non-modified 2′-deoxynucleosides. Then, the desired adducts were eluted with CH3OH containing 0.1% HCO2H (0.3 mL) into silylated glass insert capillary LC vials (Microliter Analytical Supplies, Suwanee, GA). Samples were evaporated to dryness by vacuum centrifugation and then reconstituted in 1:1 DMSO/H2O (20 μL).
LC/MS Parameters
Chromatography was performed with an Agilent 1100 Series capillary LC system (Agilent Technologies, Palo Alto, CA) equipped with an Aquasil C18 column (0.32 × 250 mm) from Thermo Fisher (Bellafonte, PA). Samples (6 μL) were injected, and analytes were separated with a gradient. The solvent conditions were held at 100% A (solvent composition: 0.01% HCO2H and 10% CH3CN) for 2 min, followed by a linear gradient to 100% B (solvent composition: 95% CH3CN containing 0.01% HCO2H) over 30 min at a flow rate of 6 μL/min. The MS instrumentation was an LTQ MS (ThermoElectron, San Jose, CA), and Xcalibur version 2.07 software was used for data manipulations. Analyses were conducted in the positive ionization mode and employed an Advance nanospray source from Michrom Bioresource Inc. (Auburn, CA). Representative optimized instrument tuning parameters were as follows: capillary temperature 220 °C; source spray voltage 1.5 kV; source current 2.8 μA; no sheath gas, sweep gas or auxiliary gas was employed; capillary voltage 32 V; tube lens voltage 110 V; and in-source fragmentation 10 V.
We employed the LIT MS in the tandem MS/MS scan mode to monitor the loss of deoxyribose from the protonated molecules of the adducts ([M + H - 116]+), followed by the consecutive reaction monitoring can mode at the MS3 scan stage, to characterize the product ions of the aglycone adducts [BH2]+. The top two or three most abundant ions produced at the MS3 scan stage were used for quantitative measurements. The ions monitored in MS > MS2 > MS3 scan modes were as follows: dG-C8-PhIP (m/z 490.1 > 374.1 > 250.2, 329.2, 357.2); [13C10]-dG-C8-PhIP (m/z 500.1 > 379.1 > 251.2, 333.3, 362.2); dG-C8-MeIQx (m/z 479.1 > 363.1 > 239.2, 318.3, 346.3); dG-C8-[2H3C]-C8-MeIQx (m/z 482.1 > 366.1 > 242.2, 321.2, 349.3); dG-AαC (m/z 449.1 > 333.1 > 288.2, 316.3); [13C10]-dG-AαC (m/z 459.1 > 338.1 > 292.3, 321.2); dG-C8-4-ABP (m/z 435.1 > 319.1 > 208.2, 274.2, 302.2); [13C10]-dG-C8-4-ABP (m/z 445.1 > 324.1 > 210.2, 278.2, 307.2), dG-N2-B[a]P (m/z 570.1 > 454.1 > 257.2, 285.2, 303.1); and [13C10]-dG-N2-B[a]P (m/z 580.1 > 459.1 > 257.2, 285.2, 303.1).
For DNA adducts of HAAs and 4-ABP, the normalized collision energies were set at 32 and 40, and the isolation widths were set at 3.0 and 1.0 Da, respectively, for the MS2 and MS3 scan modes. The activation Q was set at 0.35 and the activation time was 5 ms, for both scan modes. For dG-N2-B[a]P, the isolation widths were set at 5.0 Da for both MS2 and MS3 scan modes. The activation Q was set for 0.35 and the activation times were 20 ms for MS2 and 5 ms for MS3. Helium was used as the collision damping gas in the ion trap and was set at a pressure of 1 mTorr. One μscan was used for data acquisition. The automatic gain control settings were full MS target 30,000 and MSn target 10,000, and the maximum injection time was 50 ms. With these MS parameters, about 12 scans were acquired for each adduct and its corresponding internal standard.
Subjects
Both men and women were participants in this study. Some subjects were residents of New York City or the immediate vicinity; other subjects resided in Albany, New York. The subjects were either current-smokers, former-smokers, or never-smokers. All the subjects were on unrestricted diets. This study was approved by the Institutional Review Boards at the Wadsworth Center and the Albert Einstein College of Medicine.
Accuracy and Performance of the Analytical Method
Calibration curves were constructed with unlabeled DNA adducts added to 15 μg of DNA digest (45 nmol of deoxynucleoside), which was assayed by LC-ESI/MS/MS3, following SPE. The range in the level of spiking, reported as adducts per 108 DNA bases (and average amount of adduct in pg) per 15 μg of DNA was: 0 (0 pg), 0.3 (0.06 pg), 0.6 (0.12 pg), 1.0 (0.21 pg), 3.0 (0.62 pg), 6.0 (1.2 pg) and 10 (2.1 pg). The internal standards were added at a level equivalent to 10 adducts per 108 bases for dG-C8-MeIQx and dG-C8-PhIP (2.2 pg); 7 adducts per 108 bases for dG-C8-ABP (1.4 pg); and 13 adducts per 108 bases (2.7 pg) for dG-C8-AαC. The calibration curves were done in triplicate for each calibrant level, and the data were fitted to a straight line (area of response of the adduct/internal standard versus the amount of adduct/internal standard) using ordinary least-squares with equal weightings. The coefficient of determination (r2) values of the slopes exceeded 0.98 (Supporting Information, Figure S-1). The dissociation efficiency of the dG-N2-B[a]P adduct was poor, and the recovery of total ion counts, when going from MS2 to MS3 stage scan mode with the LIT MS was very low in comparison to the other adducts investigated (unpublished observations). The poor sensitivity prevented the acquisition of a complete calibration curve for the dG-N2-B[a]P adduct. Therefore, the estimates of B[a]P adducts in DNA were based upon peak area and assumed that the response of the adduct was equal to the response of the internal standard, with injection of the equivalent of 10 adducts per 108 bases (2.6 pg of [13C10]-dG-N2-B[a]P).
The accuracy of the method and the limit of quantification (LOQ) values of the DNA adducts were determined with DNA isolated from saliva from a non-smoker, that contained non-detectable levels of DNA adducts (<3 adducts per 109 DNA bases). CT-DNA modified with PhIP, 4-ABP, and B[a]P with defined levels of dG-C8-PhIP (36), dG-C8-ABP (43), and dG-N2-B[a]P (44) was diluted with salivary DNA from the volunteer, to achieve a level of carcinogen DNA modification of 0, 1, 3, or 10 adducts per 108 DNA bases in 50 μg of DNA.
Results
Method Development and Validation
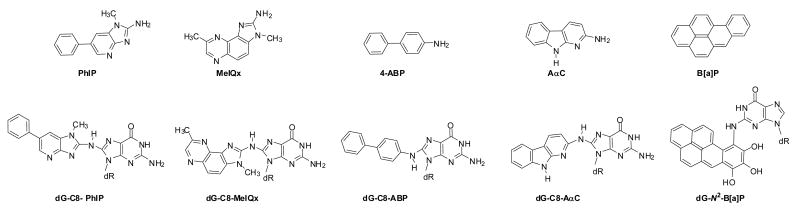
PhIP, 4-ABP, AαC, MeIQx, and B[a]P were selected for screening, because these carcinogens arise in tobacco smoke (35,46-48) and/or are formed in cooked meats (49). These compounds undergo metabolic activation by cytochrome P450 enzymes to produce electrophiles that react with DNA (32,44). The structures of these carcinogens and their DNA adducts are shown in Figure 1. We employed the LIT MS in tandem MS/MS to monitor the loss of deoxyribose from the protonated molecules of the adducts ([M+H-116]+), followed by the consecutive reaction monitoring scan mode at the MS3 stage, to characterize and measure product ions of the aglycone adducts [BH2]+.
Figure 1.
Chemical structures of tobacco and meat carcinogens and their DNA adducts monitored in human saliva.
The estimates and LOQ values of the DNA adducts were determined with CT-DNA modified with [3H]-PhIP, [3H]-ABP and [3H]-B[a]P, with known levels of dG-C8-PhIP (36), dG-C8-4-ABP (43), and dG-N2-B[a]P (44) and diluted with salivary DNA from a volunteer who harbored non-detectable levels of adducts (<3 adducts per 109 bases). The level of carcinogen DNA modification was established at 0, 1, 3, or 10 adducts per 108 DNA bases in 50 μg of DNA. The spiking of salivary DNA with carcinogen-modified CT-DNA enabled us to determine the efficiency of enzyme hydrolysis, the accuracy of the method, and the recovery of DNA adducts. The estimates of DNA adducts are presented in Table 1. The estimates of dG-C8-PhIP were within 10% of the target value for all levels of spiking. The level of dG-C8-ABP was underestimated by about 30% at all levels of spiking, whereas the estimate of the dG-N2-B[a]P was ∼60% greater than the target value. The target value of dG-C8-PhIP in [3H]-PhIP-modified CT-DNA was determined by liquid scintillation counting of dG-C8-[3H]-PhIP, isolated by HPLC, after enzymatic digestion of the DNA (36); the target values of dG-C8-4-ABP and dG-N2-B[a]P were based upon the levels of adducts determined by triple stage quadrupole tandem mass spectrometry (TSQ/MS/MS), in an independent laboratory (43,44). We previously estimated, by TSQ/MS/MS (50), the amount of dG-N2-B[a]P adduct in this B[a]P-modified CT-DNA at a level of 15 ± 2.1 adducts per 108 DNA bases (mean ± SD, n = 3): the target value was 10 adducts per 108 bases. The response of the dG-N2-B[a]P adduct obtained by LIT MS at the MS3 scan stage was about 10-fold weaker than the responses observed for the other DNA adducts investigated. The inter-laboratory estimates of the levels of these DNA adducts are quite comparable, if we consider that the assays employed different sets of internal standards, different enzymes and DNA digestion conditions, and different MS instruments for analyses. Our data reveal that potential constituents co-purified with the human salivary DNA samples do not impair the hydrolysis of the DNA or the subsequent analysis of DNA adducts.
Table 1. Estimates of DNA Adducts in Human Salivary DNA Mixed with Carcinogen-Modified CT-DNA.
| Target Adduct Level per 108 DNA bases | dG-N2-B[a]P | dG-C8-PhIP | dG-C8-4-ABP | Independent measurements |
|---|---|---|---|---|
| 1.0 | N.A.a | 1.0 ± 0.1 | 0.8 ± 0.3 | 5 |
| 3.0 | N.A. | 2.9 ± 1.1 | 1.9 ± 0.4 | 6 |
| 10.0 | 15.7± 1.0 | 11.7 ± 1.2 | 6.0 ± 0.8 | 3 |
Not assayed (N.A.), the LOQ value for dG-N2-B[a]P was 5 adducts per 108 DNA bases
The reconstructed ion chromatograms of the LC-ESI/MS/MS3 traces of salivary DNA adducts from the volunteer, with and without addition of CT-DNA modified with PhIP and 4-ABP, diluted to a level of 1 adduct per 108 bases, are shown in Figure 2. Neither adduct is detected in the unspiked DNA sample (Figure 2A); however, both adducts are readily measured in the salivary DNA sample spiked with the CT-DNA modified with PhIP and 4-ABP (Figure 2B). The LC-ESI/MS/MS3 traces of dG-C8-MeIQx and dG-C8-AαC adducts spiked at a level of 1 adduct per 108 bases, in the same salivary DNA sample prior to enzyme digestion and SPE, were also readily measured (unpublished observations). On the basis of the level of the signal of the DNA adducts to the background signal (51), the LOQ values of all the adducts, except for dG-N2-B[a]P, are ∼5 – 10 adducts per 109 DNA bases, and the LOQ value of dG-N2-B[a]P was ∼50 adducts per 109 bases, when 15 μg of DNA are injected on the column.
Figure 2.
Reconstructed ion chromatograms of the LC-ESI/MS/MS3 traces of DNA adducts present in a saliva of a never-smoker with (A) non-detectable DNA adducts, and (B) LC-ESI/MS/MS3 traces of DNA adducts in saliva of the same subject, after spiking with PhIP- and 4-ABP-carcinogen-modified CT-DNA, at a level of 1 adduct per 108 bases. The dG-C8 adducts of MeIQx and AαC, and their internal standards, were also monitored. The peak observed for dG-C8-MeIQx (tR 20.5 min) is attributed to the isotopic impurity of the dG-C8-[2H3C]-MeIQx internal standard (96.5% purity), which is contaminated with the unlabeled adduct at a level of 3.5%. The retention time (tR) and area are reported.
Analysis of Carcinogen DNA Adducts in Human Saliva
Salivary DNA samples were obtained from 37 volunteers (Table 2): 19 subjects were male and 18 were female. The ages ranged from 32 to 84 years. The subjects were categorized as current-smokers, former-smokers, or never-smokers, on the basis of self-report. Some demographics of the volunteers and the estimates of DNA adducts measured are reported in Table 2. Representative reconstructed ion chromatograms of the LC-ESI/MS/MS3 traces of carcinogen DNA adducts found in saliva from two current-smokers are depicted in Figures 3A and 3B. The dG-C8 adducts of PhIP, MeIQx, AαC, and 4-ABP were identified in both saliva samples.
Table 2. Levels of Carcinogen DNA Adducts in Human Saliva.
| Adducts per 108 DNA Bases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subject | Gender | Racea | Age | Self-reported smoking historyb | Cig/day | DNA assayed (μg)c | dG-C8-PhIP | dG-C8-MeIQx | dG-C8-AαC | dG-C8-4-ABP |
| 27 | F | H | 51 | never | 0 | 51 | 2.7 | N.D.d | N.D. | N.D. |
| 11 | M | H | 57 | current | 30 | 47 | N.D. | N.D. | N.D. | N.D. |
| 23 | M | W | Unknown | current | 10 | 44 | 8.5 | N.D. | N.D. | N.D. |
| 19 | F | W | 63 | current | 20-40 | 42 | 4.5 | 1.5 | N.D. | N.D. |
| 37 | M | W | 53 | never | 0 | 36 | N.D. | N.D. | N.D. | N.D. |
| 26 | M | W | 34 | current | 20 | 35 | 7.8 | N.D. | N.D. | N.D. |
| 17 | F | B | 48 | current | 10 | 34 | 2.0 | N.D. | N.D. | N.D. |
| 33 | F | W | 43 | never | 0 | 33 | N.D. | N.D. | N.D. | N.D. |
| 36 | F | W | 49 | never | 0 | 33 | 1.1 | N.D. | N.D. | N.D. |
| 28 | M | W | 53 | never | 0 | 29 | N.D. | N.D. | N.D. | N.D. |
| 3 | F | H | 75 | 20 yr | 5 | 29 | N.D. | N.D. | N.D. | N.D. |
| 21 | M | W | 75 | current | 20-40 | 27 | 1.2 | N.D. | N.D. | N.D. |
| 1 | M | H | 84 | 20 yr | 3 | 27 | N.D. | N.D. | N.D. | N.D. |
| 9 | M | B | 68 | 5 yr | 14 | 27 | N.D. | N.D. | N.D. | N.D. |
| 34 | F | W | 55 | never | 0 | 26 | N.D. | N.D. | N.D. | N.D. |
| 25 | M | W | Unknown | current | 20 | 26 | 7.7 | N.D. | N.D. | N.D. |
| 20 | F | W | 63 | current | 20-40 | 26 | 6.5 | 5.1 | 9.2 | 5.8 |
| 30 | F | H | 44 | never | 0 | 26 | N.D. | N.D. | N.D. | N.D. |
| 24 | M | W | Unknown | current | 20 | 26 | 2.4 | N.D. | N.D. | N.D. |
| 14 | F | H | 69 | 30 yr | 20 | 24 | N.D. | N.D. | N.D. | N.D. |
| 15 | F | H | 48 | current | 3 | 24 | 9.7 | 3.5 | 5.4 | 5.5 |
| 12 | F | W | 63 | current | 40 | 21 | N.D. | N.D. | N.D. | N.D. |
| 29 | F | W | 74 | never | 0 | 21 | N.D. | N.D. | N.D. | N.D. |
| 13 | F | B | 48 | 7 yr | 30 | 20 | N.D. | N.D. | N.D. | N.D. |
| 35 | F | W | 43 | 10 yr | 10 | 20 | 2.8 | N.D. | N.D. | N.D. |
| 8 | M | H | 78 | 20 yr | 40 | 20 | N.D. | N.D. | N.D. | N.D. |
| 16 | M | W | 62 | 19 yr | 40-60 | 19 | 2.0 | N.D. | N.D. | N.D. |
| 32 | M | H | 47 | <20 yr | 1 time only | 18 | N.D. | N.D. | N.D. | N.D. |
| 31 | F | W | 87 | <20 yr | 1 | 18 | N.D. | N.D. | N.D. | N.D. |
| 22 | M | B | Unknown | 17 yr | 10 | 17 | 1.8 | N.D. | N.D. | N.D. |
| 10 | M | B | 65 | current | 10 | 16 | N.D. | N.D. | N.D. | N.D. |
| 5 | F | B | 58 | current | 10-20 | 14 | N.D. | N.D. | N.D. | N.D. |
| 2 | M | H | 80 | current | 3 | 13 | N.D. | N.D. | N.D. | N.D. |
| 7 | M | H | 45 | current | 20 | 13 | N.D. | N.D. | N.D. | N.D. |
| 6 | M | B | 70 | < 3 months | 10 | 12 | N.D. | N.D. | N.D. | N.D. |
| 18 | F | B | 50 | 3 yr | 20-40 | 12 | 1.8 | N.D. | N.D. | N.D. |
| 4 | M | H | 50 | < 1 yr | 20 | 9 | N.D. | N.D. | N.D. | N.D. |
W = White, B = Black, H = Hispanic
Reported time since quitting
Samples with the highest DNA content were assayed first, in batches of 5 or 6 samples.
N.D. = not detected, < 5 adducts per 109 DNA bases.
Figure 3.
Reconstructed ion chromatograms of the LC-ESI/MS/MS3 traces of DNA adducts present in saliva samples from two current-smokers. All DNA adducts were present at levels above the LOQ.
The MS3 scan stage mode was employed both for quantification of the adducts and for characterization of the product ion spectra of the aglycone adducts [BH2]+. The product ion spectra of the salivary DNA adducts and the respective internal standards, are shown in Figure 4. The MS3 product ion spectra provide rich structural information about these adducts and corroborate their identities. The proposed pathways of mass fragmentation of the DNA adducts have been previously reported (32,34,37,52): the product ion spectra of the salivary DNA adducts were in excellent agreement with the spectra of the synthetic adducts.
Figure 4.
MS3 product ion spectra of DNA adducts identified in saliva DNA samples of current-smokers and MS3 product ion spectra of the corresponding internal standards (after background subtraction), from upper to lowest panel: dG-C8-4-ABP; [13C10]-dG-C8-4-ABP; dG-C8-AαC; [13C10]-dG-C8-AαC; dG-C8-MeIQx; dG-C8-[2H3C]-MeIQx; dG-C8-PhIP; and [13C10]-dG-C8-PhIP.
The dG-C8-PhIP adduct was detected most frequently; it was found in 15 subjects, at levels that ranged from 1 to 9 adducts per 108 DNA bases. dG-C8-AαC and dG-C8-MeIQx were detected in saliva samples of 3 current-smokers, followed by dG-C8-4-ABP, which was identified in saliva samples from 2 current-smokers.
Pilot exposure analyses
Subjects were sorted into two groups based on the concentration of PhIP adducts (not detected versus detected). Logistic regression models were used for testing association between PhIP adducts and other variables. Because of the limited sample size, covariates including age, race, gender and lung cancer status were first tested. After univariate logistic analyses covariates, including gender and lung cancer status, were not included because they are not likely to be confounder (p>0.25). The multivariate logistic model examined parameters, including age, race, tobacco smoking status [never, former (quit x one year), current] and cumulative dose (pack years), quit years (recent <10 years, or greater), as well as dietary factors such as meat, grilled meat, and fruit and vegetable intake and amount, along with alcohol intake and quantity. No parameters were clearly significant predictors of the most commonly detected DNA adduct (PhIP), although the multivariate model suggested some borderline relationships between PhIP adduct detectability and smoking (current p=0.062, former p=0.060), and current alcohol intake (p=0.078).
Discussion
Saliva is a rich source of DNA (53), and is easily obtained, and routinely used for genetic analyses (45) and evaluation of DNA damage (17). The oral cavity is directly exposed to carcinogens present in tobacco smoke and in the diet. Over the past 20 years, cells of the oral cavity have been screened by 32P-postlabeling or immunohistochemical methods (25-29), for DNA adducts. A plethora of lesions have been detected; however, the identities of these presumed DNA adducts have never been confirmed by MS techniques. The objective of our study was to determine whether DNA adducts derived from several prototypical carcinogens present in tobacco smoke or cooked meat could be detected in saliva of subjects on unrestricted diets, by application of selective LIT MS scanning techniques.
Our findings demonstrate that LC-ESI/MS/MS3 with the LIT MS can be used to screen for carcinogen-DNA adducts in human saliva. An important advantage of the LIT MS over triple stage quadrupole mass spectrometer (TSQ MS) instruments, which have been more commonly used for the quantification of DNA adducts (32,33), is the ability of the LIT MS to acquire MS3 product ion spectra. We have used this multi-stage tandem scanning technique for structural characterization and identification of aglycone adducts [BH2]+ of several different carcinogen DNA adducts, in tissues of experimental animals and humans (34,37,54). We also employed LIT MS at the MS3 scan stage for quantitative measurements of dG-C8-PhIP (37). The intra-day and inter-day precision values (co-efficient of variation %) were <10% at the LOQ: this level of performance is close to the precision achieved by TSQ MS instruments (37). However, the slow duty cycle of the sequence of scanning events of the LTQ MS, which is termed the μscan, and which includes the time periods for the initial pre-scan event, the ion injection, isolation, activation and mass analysis, restricts the number of scans acquired. The paucity of scans can lead to an insufficient number of data points acquired for each peak and result in imprecise measurements, when multiple DNA adducts are measured in a single scan segment. The injection time of the LIT MS, the most time-consuming event of the μscan, can be reduced, so as to shorten the duty cycle and augment the number of scans acquired across the peaks. However, the sensitivity of response can decrease concomitantly, if the duration of the injection time is too brief. A too-brief injection time can adversely affect the quality and reproducibility of the product ion spectra, and the quantitative estimates become less precise. Indeed, we have observed a reduction in the precision of adduct measurements (Table 1), when the number of DNA adducts and internal standards assayed in the same scan segment is increased from 1 to 4 or 5 adducts, even though ∼12 scans were acquired for each adduct and its internal standard. In contrast to the extensive spectral data acquired by LIT MS at the MS3 scan stage for analyte characterization, the criteria used for DNA adduct identification using TSQ MS instruments are generally limited to a monitoring of the loss of deoxyribose ([M+H]+ → [M+H-116]+) in the selected reaction monitoring (SRM) mode combined, with the characteristic tR of the adduct (32,33). Product ion spectra can be obtained with the TSQ MS in the tandem MS/MS mode (39,55); however, the slow duty cycle of the TSQ MS in the full-scan mode, typically of the order of 0.1% (depending upon the monitored m/z range) results in a drastic reduction in sensitivity (56). Hence, the product ion spectra of DNA adducts are not routinely acquired in biomonitoring studies with TSQ MS instruments. However, the duty cycle of the TSQ MS is extremely rapid in the SRM mode, and the high level of sensitivity of the SRM mode allows accurate and precise quantitation, when multiple analytes are measured simultaneously (56). Thus, both LIT MS and TSQ MS instruments have important applications in the biomonitoring of DNA adducts.
In the present study, we have identified dG-C8-PhIP adduct in saliva of about 45% of the ever-smokers. DNA adducts of AαC, MeIQx, and 4-ABP were also detected, but at lower frequency. Firm conclusions about the source(s) of exposure to PhIP and other dietary carcinogens cannot be made as the concentrations of HAAs in cooked meats can vary over a 100-fold range (49,57): we did not detect a relationship between PhIP adduct formation and self-reported frequent grilled meat or total meat intake. The approximation of tobacco usage through self-reported smoking history is also likely to lead to uncertainty in the estimate of carcinogen exposure, but there may be a relationship among PhIP adduct formation, tobacco exposure and alcohol intake, albeit of borderline statistical significance in this pilot study. Moreover, both well-done meat consumption (58) and tobacco usage (59) induce the levels of expression of P4501A2, which bioactivate HAAs and 4-ABP (60) and can lead to elevated levels of DNA adducts.
The diet is considered to be the major source of exposure to PhIP. The amount of PhIP formed in cooked meats can range from less than 1 part-per-billion (ppb) up to 500 ppb (49), depending upon the type of meat, the cooking temperature and the method of cooking (57,61). AαC and MeIQx also form in cooked meats, and generally occur at <10 ppb levels. To our knowledge, 4-ABP has not been reported in cooked meats. PhIP, AαC, and 4-ABP also arise in mainstream tobacco smoke. The levels of PhIP have been reported to range from 11 to 23 ng/cig (47), whereas AαC can arise at levels as high as 258 ng/cig (48). The levels of 4-ABP occur at 0.1 to 4.3 ng/cig (46,62,63). There are no reports of MeIQx formation in tobacco smoke (47). On the basis of studies with model systems, creatinine, a constituent of muscle, is thought to be an essential precursor for the formation of PhIP (64). However, PhIP has been detected in incineration ash, and in airborne and diesel-exhaust particles (65), in addition to tobacco smoke (47). These findings suggest that PhIP is a ubiquitous environmental contaminant. The mechanisms of PhIP formation during combustion remain to be determined.
The high frequency of detection of dG-C8-PhIP in salivary DNA is noteworthy. PhIP has also been detected with high frequency in hair samples of omnivores (66-68), and a high percentage of mammary gland (69) and prostate gland biopsy (70) samples have tested positive for putative PhIP-DNA adducts, by immunohistochemical techniques. The presumed dG-C8-PhIP adduct was also detected, by 32P-postlabeling, in about 50% of the exfoliated mammary epithelial cells sampled from milk of lactating mothers (8). These data demonstrate that PhIP can damage DNA in multiple tissues of humans.
The bioactivation of PhIP, other HAAs, and 4-ABP is catalyzed by several enzymes present in different organs of the body. The liver is the most metabolically active organ in the biotransformation of HAAs and 4-ABP to genotoxicants. The carcinogenic N-hydroxy metabolites form principally by action of P450 1A2 (23,60) in the liver, and can reach the oral cavity through systemic circulation (71), followed by phase II activation in cells of the oral cavity. However, P450s 1A1, 1A2, or 1B1, that are expressed in the buccal cells or salivary glands of the oral cavity (18), can also directly bioactivate PhIP and the other procarcinogens noted above (72,73). Moreover, saliva contains peroxidases (74), which can catalyze the bioactivation of all these compounds (75).
The vast majority of DNA isolated from saliva with the Oragene kit is of human origin, with a median bacterial content of 11.8% (http://www.dnagenotek.com/pdf_files/PDWP002_BacterialContent.pdf). Epithelial buccal cells and leukocytes are the two principal mammalian cell types found in saliva (10,11). Like other epithelial cell types, buccal cells constantly and rapidly generate. For healthy oral epithelia, the time frame from new cell production to exfoliation of the buccal cell from the mucosal surface is estimated to be between 5 and 12 days (76). The leukocytes, which originate mainly from the gingival crevice (77), and then migrate into the oral cavity, are predominantly short-lived neutrophils and other granulocytes. Given the short lifespans of both buccal and leukocyte cell types, we believe that the DNA adducts present in saliva are likely to occur from recent exposures to carcinogens. Studies will be required to determine whether adducts are formed in both cell-types or whether they preferentially form in one type.
In summary, human saliva appears to be a promising fluid in which we can monitor DNA adducts of tobacco and meat-associated carcinogens, through the use of selective LIT MS methods. The high percentage of samples that are positive for dG-C8-PhIP is striking. Future studies that examine kinetics of PhIP-DNA adduct formation in cells of the oral cavity of humans exposed to defined amounts of PhIP, combined with studies that can unravel the myriad of plausible enzymes that contribute to PhIP adduct formation in oral cells will be required before this biomarker can be exploited in human population studies.
Supplementary Material
Acknowledgments
This work was funded by R01 CA122320 (R.J.T.) from the National Cancer Institute, by R21 ES014438 (E.E.B., S.D.S, A.K.G., R.J.T.) from the National Institute of Environmental Health Sciences, and by grant number 2007/58 funded by the World Cancer Research Fund International (E.E.B., R.J.T., and F.F.K.).
Abbreviations
- 4-ABP
4-aminobiphenyl
- MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- AαC
2-amino-9H-pyrido[2,3-b]indole
- B[a]P
benzo[a]pyrene
- (±) (anti)B[a]PDE)
(±)-r-7,t-8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- CT-DNA
calf thymus DNA
- dG
deoxyguanosine
- ESI
electrospray ionization
- HAA
heterocyclic aromatic amine
- LC-ESI/MS/MSn
liquid chromatography-electrospray ionization/multi-stage tandem mass spectrometry
- LIT MS
linear quadrupole ion trap mass spectrometer
- LOQ
limit of quantification
- MS
mass spectrometer
- ppb
part-per-billion
- SPE
solid phase extraction
- SRM
selected reaction monitoring
- TSQ MS
triple stage quadrupole mass spectrometer
- TSQ/MS/MS
triple stage quadrupole/tandem mass spectrometry
- dG-C8-4-ABP
N-(deoxyguanosin-8-yl)-4-ABP
- dG-C8-MeIQx
N-(deoxyguanosin-8-yl)-MeIQx
- dG-C8-AαC
N-(deoxyguanosin-8-yl)-AαC
- dG-C8-PhIP
N-(deoxyguanosin-8-yl)-PhIP
- dG-N2-B[a]P
10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene
Footnotes
Supporting Information Paragraph: Calibration curves of DNA adducts (Figure S-1) are shown in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Miller EC. Some current perspectives on chemical carcinogenesis in humans and experimental animals: Presidential address. Cancer Res. 1978;38:1479–1496. [PubMed] [Google Scholar]
- 2.Himmelstein MW, Boogaard PJ, Cadet J, Farmer PB, Kim JH, Martin EA, Persaud R, Shuker DE. Creating context for the use of DNA adduct data in cancer risk assessment: II. Overview of methods of identification and quantitation of DNA damage. Crit Rev Toxicol. 2009;39:679–694. doi: 10.1080/10408440903164163. [DOI] [PubMed] [Google Scholar]
- 3.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Creating context for the use of DNA adduct data in cancer risk assessment: I. Data organization. Crit Rev Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 4.Phillips DH. DNA adducts as markers of exposure and risk. Mutat Res. 2005;577:284–292. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Hecht SS. Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis. 2002;23:907–922. doi: 10.1093/carcin/23.6.907. [DOI] [PubMed] [Google Scholar]
- 6.Talaska G, Schamer M, Skipper P, Tannenbaum S, Caporaso N, Unruh L, Kadlubar FF, Bartsch H, Malaveille C, Vineis P. Detection of carcinogen-DNA adducts in exfoliated urothelial cells of cigarette smokers: association with smoking, hemoglobin adducts, and urinary mutagenicity. Cancer Epidemiol Biomarkers Prev. 1991;1:61–66. [PubMed] [Google Scholar]
- 7.Thompson PA, DeMarini DM, Kadlubar FF, McClure GY, Brooks LR, Green BL, Fares MY, Stone A, Josephy PD, Ambrosone CB. Evidence for the presence of mutagenic arylamines in human breast milk and DNA adducts in exfoliated breast ductal epithelial cells. Environ Mol Mutagen. 2002;39:134–142. doi: 10.1002/em.10067. [DOI] [PubMed] [Google Scholar]
- 8.Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Carcinogen-DNA adducts in human breast epithelial cells. Environ Mol Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- 9.Rundle A. Carcinogen-DNA adducts as a biomarker for cancer risk. Mutat Res. 2006;600:23–36. doi: 10.1016/j.mrfmmm.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Klinkhamer JM. Human oral leukocytes. Journal of the American Society of Periodontists. 1963;1:109–117. [Google Scholar]
- 11.Osswald K, Mittas A, Glei M, Pool-Zobel BL. New revival of an old biomarker: characterisation of buccal cells and determination of genetic damage in the isolated fraction of viable leucocytes. Mutat Res. 2003;544:321–329. doi: 10.1016/j.mrrev.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Prasad MP, Mukundan MA, Krishnaswamy K. Micronuclei and carcinogen DNA adducts as intermediate end points in nutrient intervention trial of precancerous lesions in the oral cavity. Eur J Cancer B Oral Oncol. 1995;31B:155–159. doi: 10.1016/0964-1955(95)00013-8. [DOI] [PubMed] [Google Scholar]
- 13.Rojas E, Valverde M, Sordo M, Ostrosky-Wegman P. DNA damage in exfoliated buccal cells of smokers assessed by the single cell gel electrophoresis assay. Mutat Res. 1996;370:115–120. doi: 10.1016/0165-1218(96)00062-6. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz JL, Muscat JE, Baker V, Larios E, Stephenson GD, Guo W, Xie T, Gu X, Chung FL. Oral cytology assessment by flow cytometry of DNA adducts, aneuploidy, proliferation and apoptosis shows differences between smokers and non-smokers. Oral Oncol. 2003;39:842–854. doi: 10.1016/s1368-8375(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz JL, Baker V, Larios E, Chung FL. Molecular and cellular effects of green tea on oral cells of smokers: A pilot study. Mol Nutr Food Res. 2004;49:43–51. doi: 10.1002/mnfr.200400031. [DOI] [PubMed] [Google Scholar]
- 16.Feng Z, Xia Y, Tian D, Wu K, Schmitt M, Kwok RK, Mumford JL. DNA damage in buccal epithelial cells from individuals chronically exposed to arsenic via drinking water in Inner Mongolia, China. Anticancer Res. 2001;21:51–57. [PubMed] [Google Scholar]
- 17.Proia NK, Paszkiewicz GM, Nasca MA, Franke GE, Pauly JL. Smoking and smokeless tobacco-associated human buccal cell mutations and their association with oral cancer--a review. Cancer Epidemiol Biomarkers Prev. 2006;15:1061–1077. doi: 10.1158/1055-9965.EPI-05-0983. [DOI] [PubMed] [Google Scholar]
- 18.Vondracek M, Xi Z, Larsson P, Baker V, Mace K, Pfeifer A, Tjalve H, Donato MT, Gomez-Lechon MJ, Grafstrom RC. Cytochrome P450 expression and related metabolism in human buccal mucosa. Carcinogenesis. 2001;22:481–488. doi: 10.1093/carcin/22.3.481. [DOI] [PubMed] [Google Scholar]
- 19.Spivack SD, Hurteau GJ, Jain R, Kumar SV, Aldous KM, Gierthy JF, Kaminsky LS. Gene-environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004;64:6805–6813. doi: 10.1158/0008-5472.CAN-04-1771. [DOI] [PubMed] [Google Scholar]
- 20.Autrup H, Seremet T, Arenholt D, Dragsted L, Jepsen A. Metabolism of benzo[a]pyrene by cultured rat and human buccal mucosa cells. Carcinogenesis. 1985;6:1761–1765. doi: 10.1093/carcin/6.12.1761. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Sundqvist K, Belinsky SA, Castonguay A, Tjalve H, Grafstrom RC. Metabolism and macromolecular interaction of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in cultured explants and epithelial cells of human buccal mucosa. Carcinogenesis. 1993;14:2383–2388. doi: 10.1093/carcin/14.11.2383. [DOI] [PubMed] [Google Scholar]
- 22.Kragelund C, Hansen C, Torpet LA, Nauntofte B, Brosen K, Pedersen AM, Buchwald C, Therkildsen MH, Reibel J. Expression of two drug-metabolizing cytochrome P450-enzymes in human salivary glands. Oral Dis. 2008;14:533–540. doi: 10.1111/j.1601-0825.2007.01415.x. [DOI] [PubMed] [Google Scholar]
- 23.Butler MA, Iwasaki M, Guengerich FP, Kadlubar FF. Human cytochrome P-450PA (P450IA2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc Natl Acad Sci USA. 1989;86:7696–7700. doi: 10.1073/pnas.86.20.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem Res Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 25.Stone JG, Jones NJ, McGregor AD, Waters R. Development of a human biomonitoring assay using buccal mucosa: comparison of smoking-related DNA adducts in mucosa versus biopsies. Cancer Res. 1995;55:1267–1270. [PubMed] [Google Scholar]
- 26.Foiles PG, Miglietta LM, Quart AM, Quart E, Kabat GC, Hecht SS. Evaluation of 32P-postlabeling analysis of DNA from exfoliated oral mucosa cells as a means of monitoring exposure of the oral cavity to genotoxic agents. Carcinogenesis. 1989;10:1429–1434. doi: 10.1093/carcin/10.8.1429. [DOI] [PubMed] [Google Scholar]
- 27.Chacko M, Gupta RC. Evaluation of DNA damage in the oral mucosa of tobacco users and non-users by 32P-adduct assay. Carcinogenesis. 1988;9:2309–2313. doi: 10.1093/carcin/9.12.2309. [DOI] [PubMed] [Google Scholar]
- 28.Romano G, Mancini R, Fedele P, Curigliano G, Flamini G, Giovagnoli MR, Malara N, Boninsegna A, Vecchione A, Santella RM, Cittadini A. Immunohistochemical analysis of 4-aminobiphenyl-DNA adducts in oral mucosal cells of smokers and nonsmokers. Anticancer Res. 1997;17:2827–2830. [PubMed] [Google Scholar]
- 29.Besarati NA, Van Straaten HW, Godschalk RW, Van Zandwijk N, Balm AJ, Kleinjans JC, Van Schooten FJ. Immunoperoxidase detection of polycyclic aromatic hydrocarbon-DNA adducts in mouth floor and buccal mucosa cells of smokers and nonsmokers. Environ Mol Mutagen. 2000;36:127–133. doi: 10.1002/1098-2280(2000)36:2<127::aid-em7>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 30.Santella RM. Immunological methods for detection of carcinogen-DNA damage in humans. Cancer Epidemiol Biomarkers Prev. 1999;8:733–739. [PubMed] [Google Scholar]
- 31.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 32.Turesky RJ, Vouros P. Formation and analysis of heterocyclic aromatic amine-DNA adducts in vitro and in vivo. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:155–166. doi: 10.1016/j.jchromb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 33.Singh R, Farmer PB. Liquid chromatography-electrospray ionization-mass spectrometry: the future of DNA adduct detection. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 34.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 36.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr, Kadlubar FF. Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem Res Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 37.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2′-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turesky RJ, Rossi SC, Welti DH, Lay OJ, Jr, Kadlubar FF. Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem Res Toxicol. 1992;5:479–490. doi: 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- 39.Paehler A, Richoz J, Soglia J, Vouros P, Turesky RJ. Analysis and quantification of DNA adducts of 2-amino-3,8- dimethylimidazo[4,5-f]quinoxaline in liver of rats by liquid chromatography/electrospray tandem mass spectrometry. Chem Res Toxicol. 2002;15:551–561. doi: 10.1021/tx010178e. [DOI] [PubMed] [Google Scholar]
- 40.Frederiksen H, Frandsen H, Pfau W. Syntheses of DNA-adducts of two heterocyclic amines, 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeA{alpha}C) and 2-amino-9H-pyrido[2,3-b]indole (A{alpha}C) and identification of DNA-adducts in organs from rats dosed with MeA{alpha}C. Carcinogenesis. 2004;25:1525–1533. doi: 10.1093/carcin/bgh156. [DOI] [PubMed] [Google Scholar]
- 41.Jones CR, Sabbioni G. Identification of DNA adducts using HPLC/MS/MS following in vitro and in vivo experiments with arylamines and nitroarenes. Chem Res Toxicol. 2003;16:1251–1263. doi: 10.1021/tx020064i. [DOI] [PubMed] [Google Scholar]
- 42.Ruan Q, Kim HY, Jiang H, Penning TM, Harvey RG, Blair IA. Quantification of benzo[a]pyrene diol epoxide DNA-adducts by stable isotope dilution liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1369–1380. doi: 10.1002/rcm.2457. [DOI] [PubMed] [Google Scholar]
- 43.Beland FA, Doerge DR, Churchwell MI, Poirier MC, Schoket B, Marques MM. Synthesis, characterization, and quantitation of a 4-aminobiphenyl-DNA adduct standard. Chem Res Toxicol. 1999;12:68–77. doi: 10.1021/tx980172y. [DOI] [PubMed] [Google Scholar]
- 44.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. High-performance liquid chromatography electrospray ionization tandem mass spectrometry for the detection and quantitation of benzo[a]pyrene-DNA adducts. Chem Res Toxicol. 2005;18:1306–1315. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 45.Lum A, Le ML. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–724. [PubMed] [Google Scholar]
- 46.Patrianakos C, Hoffmann D. Chemical studies on tobacco smoke LXIV. On the analysis of aromatic amines in cigarette smoke. J Assoc Off Anal Chem. 1979;3:150–154. [Google Scholar]
- 47.Manabe S, Tohyama K, Wada O, Aramaki T. Detection of a carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, in cigarette smoke condensate. Carcinogenesis. 1991;12:1945–1947. doi: 10.1093/carcin/12.10.1945. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida D, Matsumoto T. Amino-alpha-carbolines as mutagenic agents in cigarette smoke condensate. Cancer Lett. 1980;10:141–149. doi: 10.1016/0304-3835(80)90037-3. [DOI] [PubMed] [Google Scholar]
- 49.Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K. Contents in Foods, Beverages and Tobacco. In: Nagao M, Sugimura T, editors. Food Borne Carcinogens Heterocyclic Amines. John Wiley & Sons Ltd.; Chichester, England: 2000. pp. 31–71. [Google Scholar]
- 50.Spink BC, Bennett JA, Pentecost BT, Lostritto N, Englert NA, Benn GK, Goodenough AK, Turesky RJ, Spink DC. Long-term estrogen exposure promotes carcinogen bioactivation, induces persistent changes in gene expression, and enhances the tumorigenicity of MCF-7 human breast cancer cells. Toxicol Appl Pharmacol. 2009;240:355–366. doi: 10.1016/j.taap.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacDougall D, Amore FJ, Cox GV, Crosby DG, Estes FL, Freeman DH, Gibbs WE, Gordon GE, Keith LH, Lal J, Langner RR, McClelland NI, Phillips WF, Pojasek RB, Sievers RE. Guidelines for data acquistion and data quality evaluation in environmental chemistry. Anal Chem. 1980;52:2242–2249. [Google Scholar]
- 52.Turesky RJ, Bendaly J, Yasa I, Doll MA, Hein DW. The impact of NAT2 acetylator genotype on mutagenesis and DNA adducts from 2-amino-9H-pyrido[2,3-b]indole. Chem Res Toxicol. 2009;22:726–733. doi: 10.1021/tx800473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh DJ, Corey AC, Cotton RW, Forman L, Herrin GL, Jr, Word CJ, Garner DD. Isolation of deoxyribonucleic acid (DNA) from saliva and forensic science samples containing saliva. J Forensic Sci. 1992;37:387–395. [PubMed] [Google Scholar]
- 54.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rindgen D, Turesky RJ, Vouros P. Determination of in vitro formed DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chem Res Toxicol. 1995;8:1005–1013. doi: 10.1021/tx00050a003. [DOI] [PubMed] [Google Scholar]
- 56.Volmer DA, Sleno L. Mass analyzers, Part I: An overview over several designs and their applications. Spectroscopy. 2005;20:20–26. [Google Scholar]
- 57.Ni W, McNaughton L, LeMaster DM, Sinha R, Turesky RJ. Quantitation of 13 heterocyclic aromatic amines in cooked beef, pork, and chicken by liquid chromatography-electrospray ionization/tandem mass spectrometry. J Agric Food Chem. 2008;56:68–78. doi: 10.1021/jf072461a. [DOI] [PubMed] [Google Scholar]
- 58.Sinha R, Rothman N, Brown ED, Mark SD, Hoover RN, Caporaso NE, Levander OA, Knize MG, Lang NP, Kadlubar FF. Pan-fried meat containing high levels of heterocyclic aromatic amines but low levels of polycyclic aromatic hydrocarbons induces cytochrome P4501A2 activity in humans. Cancer Res. 1994;54:6154–6159. [PubMed] [Google Scholar]
- 59.Vistisen K, Poulsen HE, Loft S. Foreign compound metabolism capacity in man measured from metabolites of dietary caffeine. Carcinogenesis. 1992;13:1561–1568. doi: 10.1093/carcin/13.9.1561. [DOI] [PubMed] [Google Scholar]
- 60.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 61.Sinha R, Rothman N, Brown ED, Salmon CP, Knize MG, Swanson CS, Rossi SC, Mark SD, Levander OA, Felton JS. High concentrations of the carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) occur in chicken but are dependent on the cooking method. Cancer Res. 1995;55:4516–4519. [PubMed] [Google Scholar]
- 62.Luceri F, Pieraccini G, Moneti G, Dolara P. Primary aromatic amines from side-stream cigarette smoke are common contaminants of indoor air. Toxicol Ind Health. 1993;9:405–413. doi: 10.1177/074823379300900302. [DOI] [PubMed] [Google Scholar]
- 63.Grimmer G, Schneider D, Naujack KW, Dettbarn G, Jacob J. Intercept-reactant method for the determination of aromatic amines in mainstream tobacco smoke. Polycylic Aromat Compd. 1996;9:85–92. [Google Scholar]
- 64.Jagerstad M, Skog K, Grivas S, Olsson K. Formation of heterocyclic amines using model systems. Mutat Res. 1991;259:219–233. doi: 10.1016/0165-1218(91)90119-7. [DOI] [PubMed] [Google Scholar]
- 65.Manabe S, Kurihara N, Wada O, Izumikawa S, Asakuno K, Morita M. Detection of a carcinogen, 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine, in airborne particles and diesel-exhaust particles. Environ Pollut. 1993;80:281–286. doi: 10.1016/0269-7491(93)90049-t. [DOI] [PubMed] [Google Scholar]
- 66.Alexander J, Reistad R, Hegstad S, Frandsen H, Ingebrigtsen K, Paulsen JE, Becher G. Biomarkers of exposure to heterocyclic amines: approaches to improve the exposure assessment. Food Chem Toxicol. 2002;40:1131–1137. doi: 10.1016/s0278-6915(02)00053-4. [DOI] [PubMed] [Google Scholar]
- 67.Kobayashi M, Hanaoka T, Hashimoto H, Tsugane S. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) level in human hair as biomarkers for dietary grilled/stir-fried meat and fish intake. Mutat Res. 2005;588:136–142. doi: 10.1016/j.mrgentox.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 68.Bessette EE, Yasa I, Dunbar D, Wilkens LR, Marchand LL, Turesky RJ. Biomonitoring of carcinogenic heterocyclic aromatic amines in hair: A validation study. Chem Res Toxicol. 2009;22:1454–1463. doi: 10.1021/tx900155f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]
- 70.Tang D, Liu JJ, Rundle A, Neslund-Dudas C, Savera AT, Bock CH, Nock NL, Yang JJ, Rybicki BA. Grilled meat consumption and PhIP-DNA adducts in prostate carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2007;16:803–808. doi: 10.1158/1055-9965.EPI-06-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaderlik KR, Minchin RF, Mulder GJ, Ilett KF, Daugaard-Jenson M, Teitel CH, Kadlubar FF. Metabolic activation pathway for the formation of DNA adducts of the carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in rat extrahepatic tissues. Carcinogenesis. 1994;15:1703–1709. doi: 10.1093/carcin/15.8.1703. [DOI] [PubMed] [Google Scholar]
- 72.Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- 73.Crofts FG, Strickland PT, Hayes CL, Sutter TR. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) by human cytochrome P4501B1. Carcinogenesis. 1997;18:1793–1798. doi: 10.1093/carcin/18.9.1793. [DOI] [PubMed] [Google Scholar]
- 74.Ihalin R, Loimaranta V, Tenovuo J. Origin, structure, and biological activities of peroxidases in human saliva. Arch Biochem Biophys. 2006;445:261–268. doi: 10.1016/j.abb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Gorlewska-Roberts KM, Teitel CH, Lay JO, Jr, Roberts DW, Kadlubar FF. Lactoperoxidase-catalyzed activation of carcinogenic aromatic and heterocyclic amines. Chem Res Toxicol. 2004;17:1659–1666. doi: 10.1021/tx049787n. [DOI] [PubMed] [Google Scholar]
- 76.Squier CA, JOhnson NW, Hopps RM. Human Oral Mucosa: Development, Structure and Function. Blackwell; Oxford, UK: 1976. pp. 3–42. [Google Scholar]
- 77.Ashkenazi M, Dennison DK. A new method for isolation of salivary neutrophils and determination of their functional activity. J Dent Res. 1989;68:1256–1261. doi: 10.1177/00220345890680080901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.