Abstract
RNA uridylylation reactions catalyzed by terminal uridylyl transferases (TUTases) play critical roles in the formation of the mitochondrial transcriptome in trypanosomes. Two mitochondrial RNA editing TUTases have been described: RET1 catalyzes gRNA, rRNA and mRNA 3′-uridylylation and RET2 acts as a subunit of the RNA editing core complex, or the 20S editosome, to perform guided U-insertion mRNA editing. Although RET1 and RET2 carry out distinct functions and possess dissimilar enzymatic properties, their catalytic N-terminal (NTD) and base-recognition C-terminal (CTD) domains display a high degree of similarity while their middle domains are less conserved. Mitochondrial Editosome-like Complex Associated TUTase 1 (MEAT1), which interacts with an editosome-like assembly and is exclusively U-specific, nonetheless shows limited similarity with editing TUTases and lacks the middle domain. The crystal structures of apo and UTP-bound MEAT1 refined to 1.56 Å and 1.95 Å, respectively, reveal an unusual mechanism of UTP selection and domain organization previously unseen in TUTases. In addition to established invariant UTP-binding determinants, we have identified and verified by mutagenesis critical contributions of MEAT1-specific residues. Furthermore, MEAT1 possesses a novel bridging domain (BD), which extends from the CTD and makes hydrophobic contacts with the NTD, thereby creating a cavity adjacent to the UTP-binding site. Unlike the minimal TUT4 TUTase, MEAT1 shows no appreciable conformational change upon UTP binding and apparently does not require RNA substrate to select a cognate nucleoside triphosphate. Because MEAT1 is essential for viability of bloodstream and insect forms of Trypanosoma brucei, the unique organization of its active site renders this protein an attractive target for trypanocide development.
Keywords: Trypanosoma, mitochondria, RNA editing, TUTase
Introduction
Terminal RNA uridylyl transferases (TUTases) are functionally and structurally diverse enzymes that catalyze template-independent 3′-uridylylation of single-stranded RNAs and guide RNA-dependent internal U-insertions. Within the DNA polymerase β-type superfamily, TUTases are most closely related to non-canonical Trf4/5-type poly(A) polymerases (ncPAPs). It seems possible that the lack of transcriptional control for trypanosomal nuclear and mitochondrial genomes is partially compensated by a diversity of TUTases and ncPAPs.1 To that end, two nuclear ncPAPs (ncPAP1 and ncPAP2),2 two mitochondrial PAPs (KPAP1 and KPAP2),3, 4 two cytoplasmic (TUT3 and TUT4)5, 6 and three mitochondrial (RET1, RET2 and MEAT1) TUTases7–9 have been identified in Trypanosoma brucei. Presently, functional data are available only for mitochondrial TUTases.
Mitochondrial DNA of kinetoplastid protozoans represents a catenated network composed of few maxicircles and thousands of minicircles. Both strands of the maxicircles’ conserved region are transcribed as polycistronic RNAs that are then cleaved into ribosomal RNA and mRNA precursors. Uridylylation catalyzed by the founding member of the TUTase family, RNA editing TUTase 1 (RET1), takes place at the 3′-end of all known classes of mitochondrial RNAs. Ribosomal RNAs and guide RNAs receive ~20 nt-long U-tails while most mRNAs bear long (A/U) heteropolymers synthesized by concerted actions of RET1 and mitochondrial poly(A) polymerase KPAP1.10 RNA editing TUTase 2 (RET2) exists as a subunit of the RNA editing core complex (RECC), also referred to as the 20S editosome,11 and is responsible for the U-insertion mRNA editing activity.12, 13 A high degree of sequence similarity between the N-terminal catalytic (NTD) and C-terminal (CTD) domains in RET1 and RET2 indicates general conservation of these modules, but provides little cues for the differences in quaternary structure, UTP specificity, processivity and functional interactions.
Crystallographic studies of TbRET214 and cytosolic TbTUT46, 15 revealed a domain organization previously unseen among nucleotidyl transferases: the NTD and CTD share a large interface essentially creating a spherically-shaped bi-domain. The antiparallel β-sheet of the NTD and two helices in the CTD form a deep cleft which harbors three catalytic aspartates and a UTP-binding site. The distribution of critical active-center residues between the NTD and CTD revealed a clear functional distinction between these two domains. The highly conserved NTD bears three universal metal-binding carboxylates as well as residues interacting with the triphosphate moiety, a common feature of all NTPs. The specificity for UTP substrate comes from nucleotide base interactions with the CTD.6, 14 In RET2, an additional middle domain (MD) is inserted between two β-strands at the C-terminus of the NTD. Remarkably, in RET1 and RET2, the middle domains are inserted at a highly-conserved position while their primary structures are dissimilar. In RET2, the middle domain extends into the solvent while maintaining extensive interactions with the CTD; its positioning and surface charge are consistent with a potential role in RNA binding.14 Indeed, deletion of the middle domain in either RET116 or RET217 led to enzyme inactivation indicating its essential function.
Discovery of a third mitochondrial TUTase, Mitochondrial Editosome-like Complex Associated TUTase 1 (MEAT1), added more complexity to structure-function relationships among trypanosomal TUTases.18In vitro, similarly to RET2, this latest addition to the TUTase family is exclusively U-specific and is capable of both U-addition to single-stranded RNA and U-insertion into double-stranded RNA. RNAi knockdown of MEAT1 expression produced a cell growth-inhibitory phenotype, indicating that its in vivo function is essential for parasite viability, as is the case for RET1 and RET2. In the mitochondrial extract, MEAT1 associates with a protein complex resembling the RNA editing core complex. In this particle, MEAT1 effectively replaces the U-insertion subcomplex, which consists of MP81 structural protein, REL2 RNA ligase and RET2. Thus, RET2 and MEAT1 interact with similar multi-protein complexes in a mutually exclusive manner.18 At the protein sequence level, like TUT4, MEAT1 does not have a middle domain while the ~50 amino acid insertion within the CTD can be deduced by sequence alignments with other TUTases. Finally, several active-site residues, which are invariant among trypanosomal TUTases, are replaced in MEAT1 with either similar residues (position 181, S to N) or those resulting in altered charge (position 140, R to E) or polarity (position 183, Y to F).
In this work, we report crystallographic and mutational analyses of MEAT1 from Trypanosoma brucei (Genbank accession number ACT83521). X-ray structures of apo- and UTP-bound forms reveal a mechanism of UTP selection and domain organization that differs substantially from previously investigated TbRET214 and TbTUT4.6 In addition to establishing UTP-binding determinants, most of which are invariant among TUTases, we have identified, and verified by mutagenesis, additional critical contributions of MEAT1-specific residues. A MEAT1-specific domain, termed the bridging domain (BD), extends from the CTD and makes hydrophobic contacts with the NTD, thus creating a cavity adjacent to the UTP-binding site. Considering similar enzymatic properties of RET2 and MEAT1 TUTases, their unique domains are likely to be responsible for interactions with different protein partners and, therefore, association with distinct functional complexes.18
Results
Limited proteolysis and crystallization of apo and UTP-bound MEAT1
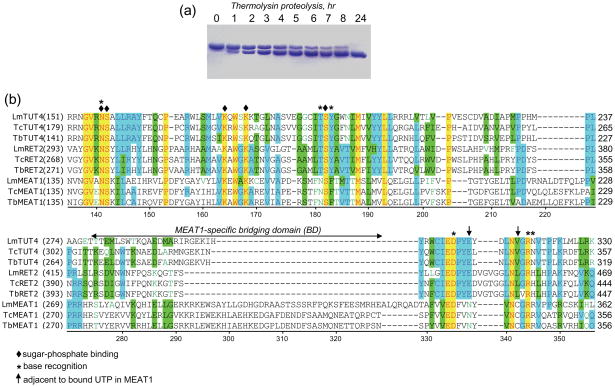
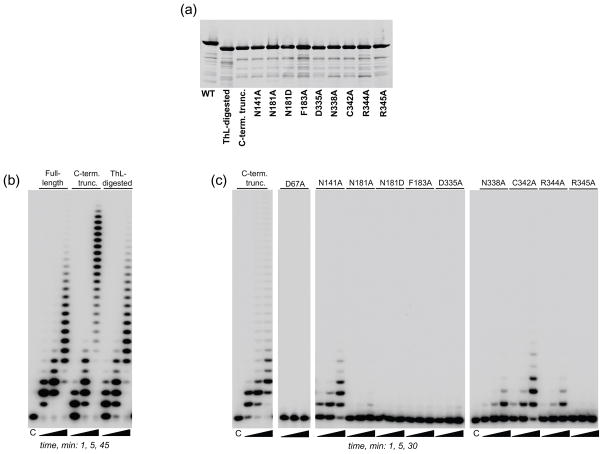
Initially, crystals were obtained for full-length C- and N-terminally 6His-tagged TbMEAT1 but despite numerous optimization trials the crystal quality and diffraction resolution were poor (7 Å and 3.2 Å, respectively). In an effort to obtain higher-quality crystals, limited proteolysis was employed. Digestion with thermolysin shortened MEAT1 by approximately 4 kDa, as monitored by SDS-PAGE, leading to an accumulation of a single fragment, which was resistant to further degradation (Fig. 1a). Proteolyzed TbMEAT1 was used without further purification to grow crystals in the presence or absence of UTP for data collection and structure solution. Diffraction data revealed electron density for 385 (44.3 kDa) out of 406 (46.8 kDa, excluding the affinity-tag) amino acids beginning with the N-terminal methionine. The remaining amino acids along with the C-terminal His-tag were removed by limited proteolysis with thermolysin.
Fig. 1.
Limited proteolysis of purified recombinant MEAT1. (a) Purified MEAT1 (0.5 mg/ml) was incubated at 4° C with thermolysin (2 μg/ml) in buffer containing 50 mM Tris-HCl, pH 8.0, 100 mM KCl and 50 mM CaCl2. Aliquots were removed at indicated periods of time, separated by SDS-PAGE and stained with Coomassie blue. (b) Partial multiple sequence alignment of C-terminal domains from T. brucei TUTases with known X-ray crystal structures (RET214, TUT46 and MEAT, this study) with orthologous proteins from other Kinetoplastida species. Lm: Leishmania major; Tc: Trypanosoma cruzi; Tb: Trypanosoma brucei. TbMEAT1 amino acids are numbered; UTP-binding residues are depicted by diamonds (sugar-phosphate) and asterisks (uracil base), and arrows indicate MEAT1 positions adjacent to bound UTP.
All crystals were of the space group P21 and, although the recombinant MEAT1 is a monomer in solution,18 contained two molecules in the asymmetric unit related by 2-fold non-crystallographic symmetry. The first crystal structure was obtained from selenomethionine-labeled protein in the presence of UTP and Mg2+ (TbMEAT1SeMet-UTP-Mg), followed by multiple anomalous dispersion (MAD) phasing. Apo and UTP-bound structures were then obtained by molecular replacement using TbMEAT1SeMet-UTP-Mg as the search model. The resolutions achieved for TbMEAT1-apo, TbMEAT1-UTP, and TbMEAT1SeMet-UTP-Mg were 1.56 Å, 1.95 Å, and 2.30 Å, respectively. The stereochemistry of each model was validated by PROCHECK19 and the quality of these structures and data is summarized in Table 1.
Table 1.
Crystal Data Collection and Refinement Statistics
| Crystal Data: UTP-Mg | Remote | Peak | Inflection |
|---|---|---|---|
| Wavelength (Å) | 0.97876 | 0.91837 | 0.97935 |
| Beamline* | 11-1 | 11-1 | 11-1 |
| Space Group | P21 | P21 | P21 |
| Cell Dimensions | a = 63.7 Å | a = 63.7 Å | a = 63.7 Å |
| b = 103.4 Å | b = 103.4 Å | b = 103.4 Å | |
| c = 63.7 Å | c = 63.8 Å | c = 63.8 Å | |
| β = 109.6° | β = 109.6° | β = 109.6° | |
| Resolution range (Å) | 50 – 2.30 | 50 – 2.30 | 50 – 2.30 |
| Total Reflections | 254,819 | 251,137 | 251,735 |
| Unique Reflections& | 67,571 | 67,371 | 67,988 |
| Redundancy | 3.8 (3.5) | 3.7 (3.4) | 3.7 (3.2) |
| Completeness (%) | 99.6 (98.7) | 99.1 (97.0) | 98.9 (96.3) |
| Mean I/σI | 21.7 (3.0) | 21.2 (2.9) | 20.3 (2.5) |
| Rsym (%) | 9.2 (48.0) | 9.3 (47.9) | 9.3 (51.7) |
| Crystal Data: | UTP | Apo |
|---|---|---|
| Wavelength (Å) | 0.97945 | 0.97945 |
| Beamline* | 11-1 | 11-1 |
| Space Group | P21 | P21 |
| Cell Dimensions | a = 62.9 Å | a = 63.2 Å |
| b = 102.0 Å | b = 102.0 Å | |
| c = 66.1 Å | c = 66.1Å | |
| β = 111.6° | β = 112.0° | |
| Resolution range (Å) | 99 – 1.95 | 99 – 1.56 |
| Total Reflections | 193,869 | 394,941 |
| Unique Reflections | 54,477 | 108,380 |
| Redundancy | 3.6 (3.0) | 3.6 (3.0) |
| Completeness (%) | 96.6 (84.0) | 98.2 (90.3) |
| Mean I/σI | 12.8 (2.2) | 25.2 (2.0) |
| Rsym (%) | 12.7 (49.1) | 6.8 (53.1) |
| Refinement: | UTP-Mg | UTP | Apo |
|---|---|---|---|
| Resolution range (Å) | 31.71 – 2.30 | 31.05 – 1.95 | 31.41 – 1.56 |
| Structure factors used | 30,731 | 51,550 | 102,739 |
| Rwork/Rfree† (%) | 19.8/24.8 | 20.4/24.6 | 16.2/20.3 |
| R.m.s.d. bonds(Å) | 0.014 | 0.013 | 0.013 |
| R.m.s.d. angles (°) | 1.5 | 1.4 | 1.4 |
| Number of Atoms/Avg. B value (Å2) | |||
| Protein | 6,182/29.1 | 6,261/17.4 | 6237/15.8‡ |
| Waters | 170/29.2 | 567/26.1 | 879/28.8‡ |
| UTP | 58/37.4 | 58/33.9 | |
| Mg | 2/32.3 | ||
| Ramachandran plot (favored/allowed/generously allowed, %)§ | 86.8/13.2/0 | 91.2/8.8/0 | 91.1/8.9/0 |
Structure of TbMEAT1
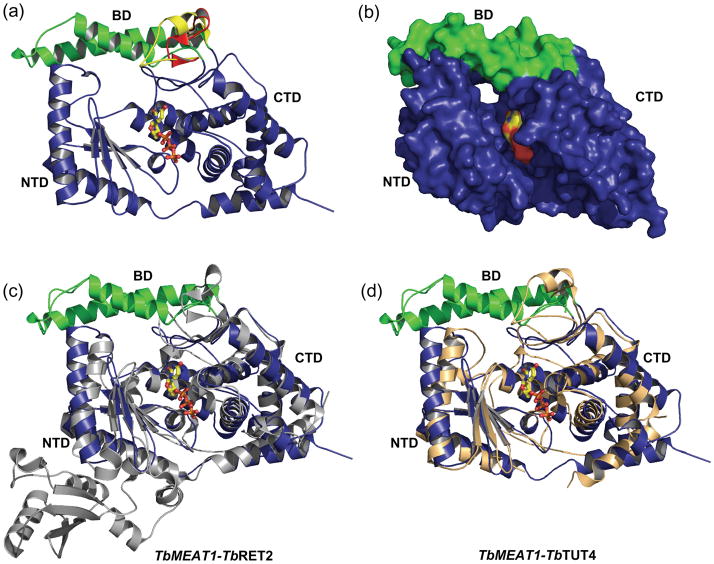
Notwithstanding its low sequence identity with TbRET214 (12%) and TbTUT46 (14%), TbMEAT1 adopts a similar bi-domain architecture which forms a deep cleft bearing both catalytic and nucleoside triphosphate-binding sites (Fig. 2). Global differences in the tertiary structure of UTP-bound TbMEAT1 account for Cα r.m.s.d. values of 2.2 Å and 2.3 Å with TbRET2 and TbTUT4, respectively, which are significantly greater than the 1.6 Å r.m.s.d. observed between TbRET2 and TbTUT4.6
Fig. 2.
Overall structure of TbMEAT1. (a) Ribbon representation illustrating the UTP bound in the deep cleft formed by the NTD and CTD. The unique two-α-helical bridging domain (green) and its differences from the shorter regions observed in TbRET2 (red) and TbTUT4 (yellow) are highlighted. (b) MEAT1-UTP surface representation. Structural superpositions of TbMEAT1 with (c) TbRET2 (gray) and (d) TbTUT4 (tan) depict the overall differences in these structures, which become increasingly evident away from the active site. Superpositions were generated with backbone α-carbons using the SSM algorithm41 in the program Coot.38
The N-terminal domain represents the DNA polymerase β-like (Pol β) catalytic fold containing a five-stranded anti-parallel β-sheet flanked by α-helices on either side. The twisted β-sheet provides carboxylate residues (D65 and D67) responsible for divalent metal coordination, a feature common to all members of the Pol β superfamily. The C-terminal domain, which resembles the ATP-cone domain20 and is otherwise conserved among TUTases, contains a MEAT1-specific insertion18 comprised of two α-helices (positions 274–329). These helices fold into a compact ~6.7 kDa bridging domain (BD) in a position that is occupied by unstructured loop regions in both TbRET2 and TbTUT4 (Fig. 2a). This loop region in TbRET2 and TbTUT4 overhangs the UTP-binding cleft and has little interaction with the remaining structure. In MEAT1, the bridging domain interacts with the NTD burying approximately 650 Å2 at the interface.21 A surface representation reveals a cavity that extends the UTP-binding cleft as a tunnel protruding from the backside of the molecule (Fig. 2b). The interface created by the BD consequently results in structural deviations in the opposing segment of the NTD relative to those regions in TbRET2 (Fig. 2c) and TbTUT4 (Fig. 2d).
Because MEAT1 interacts with an editosome-like complex via a binding partner that is distinct from MP81,18 a known structural protein responsible for RET2 docking into the core editing complex,13, 22 the BD may constitute a possible interface for MEAT1 interactions in vivo. The superimposition of apo and UTP-bound forms of TbTUT4 revealed significant conformational differences with an r.m.s.d. of 1.5 Å.6 On the contrary, apo and UTP-bound structures of TbMEAT1 are virtually identical (0.3 Å r.m.s.d.) which suggest a lack of structural rearrangement upon UTP binding. Higher relative rigidity of MEAT1 is likely caused by the additional BD-mediated interactions between the NTD and CTD.
Structural basis for UTP-binding specificity
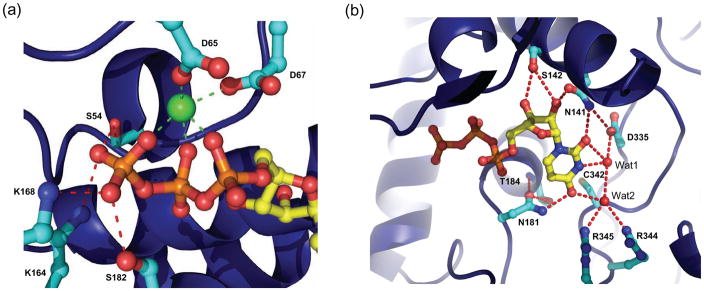
The mode of nucleoside’s sugar-phosphate binding appears conserved among TUTases with known crystal structures. A single Mg2+ (or water molecule in the absence of a metal ion) is tightly coordinated by one side of the triphosphate moiety as well as aspartate residues 65 and 67. In the case of TbMEAT1, the other side of the triphosphate is stabilized by side chains of amino acids S54, K164, K168, and S182, backbone NH groups of S54 and F183, and several ordered water molecules (Fig. 3a). The ribose 2′-hydroxyl group forms hydrogen bonds with the side chain carbonyl oxygen of N141 and the hydroxyl group of S142 (Fig. 3b).
Fig. 3.
TbMEAT1 active site. (a) Coordination of the catalytic Mg2+ ion (green) by the triphosphate of UTP and conserved aspartic acid residues. (b) The ribose sugar and uracil base moieties and their network of hydrogen bonds with neighboring active-site residues. Note MEAT1-specific N181, which donates a hydrogen bond to O4 of the uracil base. Two coordinated water molecules (Wat1 and Wat2) are included as red isolated spheres.
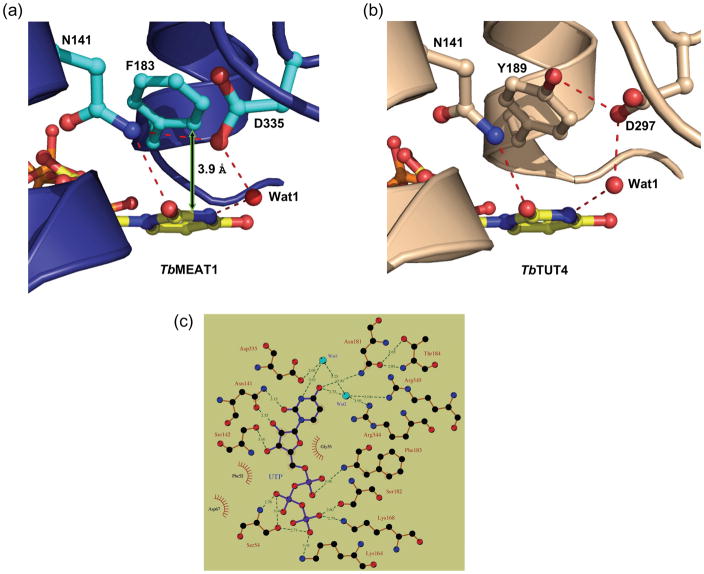
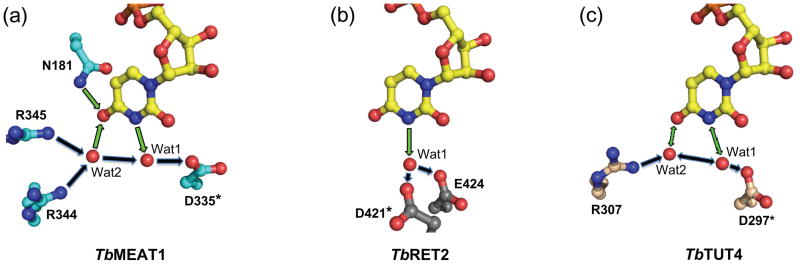
However, MEAT1’s exclusive uracil-base specificity18 is achieved both by contributions from amino acids that are conserved among TUTases and from MEAT1-specific residues. The side-chain amide nitrogen of N141 donates a hydrogen bond to the O2 carbonyl oxygen of the uracil base (Fig. 4a). However, this hydrogen bond is slightly longer than that observed in TbTUT4 and TbRET2, likely because of an additional hydrogen bond with D335. In TbRET2 (D421) and TbTUT4 (D297), this key aspartate accepts a hydrogen bond from the hydroxyl of a conserved tyrosine (Y319 in TbRET2, Y189 in TbTUT4) whose aromatic ring exhibits π–stacking interactions with the uracil base (Fig. 4b). In TbMEAT1, the base-stacking interaction is provided by a phenylalanine at position 183 (interplanar distance of ca. 3.9 Å), and the absence of the phenol hydroxyl allows interaction between N141 and D335 (Fig. 4a). In regard to other base-specific interactions, the water molecule that interacts with N3 of the uracil does not have the restricted directionality described for the respective water molecule in TbRET2 (Wat1, Fig. 5) which has been designated as TbRET2’s primary basis for U-specificity.14 Furthermore, glutamate 424 of TbRET2, which together with D421 facilitates the hydrogen-bonding pattern observed for Wat1, is replaced by an asparagine (N338) in TbMEAT1. This side-chain has no apparent involvement in nucleoside binding but may play a structural role. On the other hand, the invariant arginine in position 344, together with MEAT1-specific R345, coordinates a water molecule (Wat2), (B-factor: 27.1 Å2), which donates a hydrogen bond to O4 of the uracil base and also H-bonds with Wat1 (B-factor: 26.4 Å2) (Figs. 3b and 5).
Fig. 4.
Differences observed for (a) TbMEAT1 and (b) TbTUT4 in the conserved phenyl-containing residues involved in base-stacking interactions with bound UTP. In TbTUT4, the tyrosine side chain is locked down because of hydroxyl group H-bonding with invariant D297. In TbMEAT1, corresponding position is occupied by phenylalanine 183, which enables the formation of a hydrogen bond between N141 and D335. (c) Interactions of UTP, amino acid residues, and water molecules in the MEAT1 active site. 2D representation was generated using LIGPLOT.42
Fig. 5.
Comparison of the structural bases for achieving UTP specificity by TbMEAT1 (a), TbRET2 (b) and TbTUT4 (c) TUTases. Models were generated from PDB files 3HJ1, 2B51 and 2IKF, respectively. Hydrogen bonds involved in uracil base-specific interactions are depicted as green arrows. Invariant aspartic acid residues are indicated by asterisks.
Perhaps the most significant feature that confers U-specificity to TbMEAT1 by means alternative to TbRET2 and TbTUT4 is the contribution of N181 in a position universally occupied by serine in other trypanosomal TUTases. The side-chain amide nitrogen of N181 donates a hydrogen bond directly to O4 of the uracil base (Figs. 3b and 5). The side-chain carbonyl oxygen of N181 is locked by H-bonds from the backbone NH and side-chain hydroxyl of T184, thus making it energetically unfavorable for the side chain to adopt the opposite rotamer as would be preferred in the case of cytosine nucleotide base. The threonine in position 184 apparently co-varies with N181 in MEAT1 sequences from different trypanosomal species; in other TUTases the position corresponding to T184 is occupied by alanines or glycines, which possess small non-polar side chains incapable of H-bonding (Fig. 1b).
Binding of alternative substrates
MEAT118 and TUT46 TUTases show preferences for single-stranded RNA substrates with Us at their 3′-end while RET215 is more active on RNAs with terminal purines. Conversely, MEAT1 and RET2 are exclusively U-specific while TUT4 also incorporates CTP, although with lower efficiency, and residual levels of ATP and GTP. Co-crystallization and crystal soaking experiments with various NTPs and mono/dinucleotides serving as minimal RNA substrates demonstrated that, at physiological NTP concentrations, TbTUT4 binds all four nucleoside triphosphates. However, the coplanarity of purine bases relative to pyrimidines was suboptimal for both stacking interactions with the invariant aromatic residue (Y189) and the terminal base of the RNA substrate. These observations led to a model suggesting that specificity of U-addition by TUT4 is achieved via uracil-base binding in a specific orientation, which in turn creates a binding interface for the terminal RNA base.15 However, soaking RET2 crystals in non-UTP substrates revealed only a partial occupancy in the sugar-phosphate region, but virtually no density corresponding to nucleotide bases. RET2’s specificity, therefore, was explained based entirely on protein-UTP interactions.14
To distinguish which of these mechanisms is applicable to TbMEAT1, we solved structures of TbMEAT1 crystallized in the presence of ATP, GTP, and CTP. Similar to TbRET2, only partial occupancy was observed for each of these nucleoside triphosphates, with electron density peaking at the triphosphate moiety and diminishing toward the nucleotide bases (data not shown). Furthermore, unlike with TbTUT4, soaking of TbMEAT1-UTP crystals with high concentrations of UMP or UpU, as well as co-crystallization attempts of TbMEAT1 with UpU, revealed no electron density for UMP or UpU in either case. Nonetheless, the universal metal ion coordination by D65 and D67 suggests that RNA binding creates a pocket for the second metal, which in turn activates the 3′-OH as nucleophile. To conclude, it is likely that although the mechanisms of UTP recognition by TbMEAT1 and TbRET2 are only partially conserved, in both cases the uracil base specificity is achieved by direct or water-mediated contacts of the uracil base in the respective binding sites.
Mutational analysis of the UTP binding site
Because of considerable differences in the architecture of the uracil binding sites among TbMEAT1 and previously studied TUTases, we next determined the effects of point mutations introduced at positions that are invariant among TUTases as well as those occupied by MEAT1-specific residues. To ensure that the effects of these mutations were examined within the context of the same molecule used in crystallographic analysis, we genetically-engineered a C-terminally truncated protein to match a removal of 25 amino acids by thermolysin digestion (Fig. 6a). As demonstrated by comparison of U-addition profiles, neither limited proteolysis nor genetically-engineered C-terminal deletion had any discernable effects on enzymatic activity. A moderate lengthening of extension products for the truncated MEAT1 at longer incubation time was likely caused by increased protein stability (Fig. 6b).
Fig. 6.
Effects of catalytic-site mutations on U-addition activity. (a) Purified mutant proteins were separated on SDS gels and stained with Sypro Ruby. (b) U-addition patterns for full-length TbMEAT1, a genetically-engineered C-terminal truncated form and the thermolysin-resistant fragment. Reactions were performed in the presence of 300 nM MEAT1, 100 μM UTP and 1 μM RNA for indicated periods of time under conditions described in Materials and Methods. Products were separated on a 15% acrylamide/8M Urea gel. (c) U-addition activity of mutant proteins. Assays were carried out as in (b).
A triad of carboxyl residues (D65, D67 and D130, MEAT1 numbering), two of which are part of the signature sequence hG[G/S]x(9,13)Dh[D/E]h (x: any amino acid; h a hydrophobic residue) and the third positioned in close spatial proximity, represent a conserved functional assembly in TUTases and most other members of the Pol β superfamily. Predictably, substitution of D67, which is part of the catalytic metal-binding shell, led to a complete elimination of MEAT1 activity (Fig. 6c). Position N141 is universally conserved among TUTases as the asparagine’s side-chain carbonyl group is involved in recognition of the ribose ring while its side-chain amide group donates a hydrogen bond to O2 of the uracil base. The corresponding alanine substitution decreased catalytic efficiency of TbTUT4 by ~100-fold6 while the N141A mutation caused an approximately 10-fold drop in the kcat/Km value for MEAT1 (Fig. 6c and data not shown). The effects of mutating another invariant residue, R307 in TUT46 and R344 in MEAT1, which is engaged in water-mediated contact with O4 of the uracil moiety, were also very consistent among these two divergent TUTases: a decline in catalytic efficiency by ~20-fold (Fig. 6c). Finally, the key determinants of the uracil base binding,6 an aromatic ring at position 183 and aspartic acid in position 335, which provide π-stacking interactions and coordination of the water molecule (Wat1) receiving an H-bond from position 3, respectively, appear to be essential for MEAT1 activity: corresponding alanine substitutions inactivated the enzyme (Fig. 6c).
Next we analyzed amino acids that are divergent between MEAT1 and other TUTases (Fig. 1b) and are located within immediate proximity of the bound UTP (C342 and N338), as well as those that contact UTP directly or via coordinated water molecules (N181 and R345). The thiol group of C342 is positioned within 4 Å, or closer for some rotamers, of O4 of the uracil and, unlike hydrophobic amino acids in other TUTases, has a potential to form an H-bond with the base (Fig. 3b). The contribution of this interaction, however, appears to be insignificant as the C342A mutation had only a minor inhibitory effect. Unlike glutamate 424 in RET2, which is essential for coordination of Wat1 and uracil recognition (Fig. 5), in MEAT1 the corresponding asparagine residue in position 338 is tightly fixed by intra-protein hydrogen bonds and provides no apparent contribution to UTP binding. Therefore, the moderate inhibitory effect of alanine substitution manifested by a ~20-fold lower catalytic efficiency (Fig. 6c) was likely caused by structural permutations.
The asparagine in position 181, which is occupied by serine in other TUTases, and arginine in position 345, typically glutamine or histidine in other TUTases, clearly contribute the most critical uracil base contacts among MEAT1-specific amino acids tested in this study. The N181A mutation virtually eliminated TUTase activity and its effect could not be rescued by N181D substitution, indicating that the amide contribution by H-bonding to O4 is essential for UTP binding (Figs. 3b, 5a and 6c). In TUT4 and RET2, O4 receives a single H-bond and the disruption of this interaction by R307A mutation led only to moderate inhibition of TUT4 activity.6 Thus, the deleterious effect of the R345A mutation, which presumably eliminates Wat2-mediated H-bonding (Figs. 3b, 5a and 6c), further establishes O4 as the key element recognized by MEAT1. A direct interaction between R345 and O4 of the uracil base would be limited to a dipole-dipole interaction as the distance is too long for hydrogen bonding (3.6 Å).
Finally, testing of all mutant proteins for CTP-, ATP- and GTP-incorporation activity provided no indications that removal of UTP-specific positive binding determinates relaxes MEAT1 specificity (data not shown). To conclude, the atomic mechanism that ensures exclusive UTP binding by MEAT1 is a combination of hydrogen bonds and π-stacking interactions, which are likely universal among the TUTase enzyme family, as well as additional H-bonds provided by MEAT1-specific residues.
Discussion
Structural and functional diversity of RNA uridylyl transferases in trypanosomes provided an early recognition of 3′-uridylylation as a widespread post-transcriptional RNA modification with profound impact on eukaryotic RNA processing.1, 23 Recent reports demonstrate that TUTases participate in processes as diverse as U6 snRNA 3′-end processing,24 cell cycle-dependent regulation of histone mRNA stability in human cells,25 accelerated decay of let-7 miRNA precursor,26 3′-uridylylation of actin mRNAs in Schizosaccharomyces pombe27 and degradation of mRNA cleavage fragments in plants.28 Because three trypanosomal TUTases (RET1, RET2 and MEAT1) are essential for parasite viability and human Zcchc11 TUTase has implications in cancer and stem cell biology,29 the structures of TUTase active sites and mechanisms of UTP selection are of substantial interest from a pharmaceutical standpoint.
Crystallographic studies of trypanosomal RET214 and TUT46, 15 TUTases underscored a conserved bi-domain organization of the catalytic core and conservation of the key UTP-binding residues. However, significant differences were observed in the mechanisms of UTP selection. Whereas RET2, an exclusively U-specific enzyme active on double-stranded RNA, apparently relies entirely on protein-UTP contacts, the less specific TUT4 invokes active participation of the single-stranded RNA substrate to select a cognate nucleoside triphosphate.
Here we report crystallographic and mutational analyses of a recently identified third trypanosomal mitochondrial TUTase, MEAT1. Crystal structures were solved for the apo, UTP-, and UTP-Mg2+-bound forms at moderate to high resolution. In contrast to TUT4, these structures are virtually identical, indicating a lack of appreciable conformational changes upon UTP binding. The overall fold of the globular bi-domain formed by the NTD and CTD in MEAT1 is similar to TbRET2 and TbTUT4. In MEAT1, however, CTD contains an insertion of two α-helices which form a bridge extending across the top of the UTP-binding cleft and interacting with the N-terminal domain. This additional bridging domain likely confers greater stability to the entire fold but may also be involved in protein-protein interactions responsible for docking of MEAT1 into the editosome-like complex. The MEAT1-associated particle closely resembles the RNA editing core complex, also referred to as the 20S editosome, but lacks the RET2 TUTase. Although specific RNA substrates of MEAT1 and the MEAT1-associated complex remain to be firmly established, RNA interference studies demonstrated that MEAT1 expression is essential for viability of parasites in both bloodstream and procyclic forms.
The in vitro experiments demonstrate that TUTases differ substantially in their NTP selectivity: from U-specific (RET2, MEAT1) to moderate incorporation of CTP (RET1, TUT4), to being equally active in both U- and A-addition (Cid1 from S. pombe).30 Accordingly, TUTases and Trf4/5-type non-canonical poly(A) polymerases31 are not readily distinguishable at the protein sequence level. Furthermore, existing evidence suggests that nucleotide specificity of TUTases may be affected by interacting partners and/or the structures of RNA substrates in vivo.32, 33 Trypanosomal mitochondrial poly(A) polymerase KPAP1, for example, is highly similar to TUTases; most positions involved in UTP binding are also conserved in KPAP1.3 It is therefore not surprising that the active sites of TUTases, and perhaps of non-canonical PAPs, are quite spacious and that steric constraints do not appear to be discriminating factors between purine and pyrimidine bases. Instead, positive determinants (direct or water-mediated hydrogen bonding and π-stacking interactions) are exploited to select UTP even though the atomic details of uracil recognition vary among TUTases. TbRET2 employs unidirectional hydrogen bonding via a water molecule;14Tb TUT4 - prefers UTP over CTP with no obvious reason for selection of the former;6, 15 and TbMEAT1 relies on unidirectional hydrogen bonding via an active-site asparagine (Fig. 5). Whereas the promiscuity in nucleoside binding observed for TbTUT4 may explain its lower selectivity in U-addition reactions, the differences between TbRET2 and TbMEAT1 are rather intriguing, as both are exclusively U-specific, localized to the mitochondrion and associate with similar yet distinct complexes.
The mechanism for nucleoside incorporation involves nucleophilic attack by the 3′-OH of the terminal RNA residue on the α-phosphorus atom of the bound nucleoside triphosphate. The nucleophilic substitution reaction is catalyzed by the tightly coordinated Mg2+, which stabilizes the pyrophosphate leaving group. Adaptation of this mechanism for TUTases has been demonstrated by the crystal structure of TbTUT4 with bound UTP and UMP, in which UMP models the terminal nucleoside of an RNA substrate.15 In this example, a second Mg2+ catalyst, which was not observed in the TbTUT4-UTP binary complex, brings the nucleophile and electrophile into reacting proximity. This mechanism also implicates an active role for the RNA substrate in pyrimidine selectivity through subsequent base-stacking interactions between the terminal nucleoside base and the bound pyrimidine base. However, soaking of TbMEAT1-UTP co-crystals with UMP revealed no electron density for UMP. This may imply that MEAT1 requires a longer RNA substrate for productive binding or that UTP-protein contacts are indeed sufficient to confer the observed UTP selectivity. In the latter scenario, the coordination of the uracil’s O4 via additional hydrogen bonding from MEAT1-specific N181, an interaction not seen in RET2 or TUT4, appears to be a critical contribution as well as the stacking interaction and hydrogen bonding involving position N3 of the uracil base.
Furthering our understanding of the mechanisms by which TUTases edit RNA and regulate gene expression will enable exploration of these vital processes for trypanocide development. Specifically, atomic details of RET2 and MEAT1 active sites can be exploited for in silico docking to identify potential TUTase inhibitors.
Materials and Methods
Mutagenesis, expression and purification of recombinant MEAT1
C-terminally 6His-tagged TbMEAT1 was expressed in E. coli strain BL21(DE3) RIL Codon plus (Strategene) using M9 minimal media. The cells were grown at 37° C to an A600 of 0.7–0.8, reduced to a temperature below 18° C, and expression induced with 1 mM IPTG for 2 days at 10° C. The expression protocol for selenomethionine labeling was the same except for using the B834 (DE3) methionine auxotroph strain (Novagen) and supplementing the media with 60 mg/L of L-selenomethionine prior to induction and another 60 mg/L 24 hours after induction. Cell pellets were washed with phosphate-buffered saline, resuspended in 7.5 mL per gram of cell mass of lysis buffer containing 50 mM HEPES pH 8.0, 50 mM NaCl, 1 mg/ml of lysozyme, 1 tablet of Complete EDTA-free (Roche) protease inhibitors, and passed through a French Press at 1000 psi. The cellular extract was adjusted to 300 mM NaCl and 0.1% Triton X-100 and clarified by centrifugation at 40,000 g. The protein was purified by loading onto a 5 mL bed-volume of TALON (Clontech) affinity resin, washing with 10 column volumes of 50 mM HEPES, pH 8.0, 300 mM NaCl followed by 10 column volumes of a more stringent wash with the same buffer containing 10 mM imidazole. The protein was eluted from the column in 50 mM HEPES, pH 8.0, 300 mM NaCl, and 200 mM imidazole. Fractions from the Talon column were pooled, diluted 6-fold with 25 mM Tri-HCl, pH 8.0, 1 mM DTT, and loaded onto a 5 mL HiTrap Q HP anion exchange column (GE Healthcare). The column was washed with 10 column volumes of 25 mM Tris-HCl, pH 8.0, 1 mM DTT, 50 mM NaCl and eluted using the same buffer with a linear gradient of NaCl from 0.05–1 M over 20 column volumes. Fractions were analyzed by SDS-PAGE and stained with Sypro Ruby.
Point mutations were introduced by a PCR-based method using the QuikChange XL kit (Stratagene) and confirmed by DNA sequencing. Mutant proteins were isolated as above from two liters of bacterial culture.
Limited proteolysis and sample preparation
Purified fractions were diluted to achieve 100 mM concentration of NaCl and glycerol and CaCl2 were added to 5% and 50 mM, respectively. Thermolysin (Sigma) was added at a ratio of 1:250 (wt. protease: wt. substrate) and the reaction was carried out at 4° C for 24 hours. The cleaved protein was subsequently dialyzed against 2 L of 50 mM HEPES pH 8.0, 100 mM NaCl, 5% glycerol, 1 mM DTT, and 1 mM EDTA for 4 hours, and again in another 2 L of dialysis buffer for 16 hours. CHAPS was added to 0.3% (w/v), and the protein was concentrated to 10 mg/mL using a 10 kDa molecular weight cut-off Centricon centrifugal filter device (Amicon).
Crystallization and structure solution
The crystals were grown at 18° C at a protein concentration of 5 mg/mL and a 0–5-fold molar excess of UTP, in a crystallization buffer containing 0.2 M lithium acetate and 25% PEG 3350, using the hanging-drop vapor diffusion method. Crystals were soaked for 1–5 minutes in a similar crystallization buffer containing 30% PEG 3350, supplemented with 15% glycerol as cryoprotectant, and flash-cooled in liquid N2. Initial crystals of full-length C- and N-terminally His-tagged TbMEAT1 diffracted to 7 Å and 3.2 Å, respectively. Data for the proteolyzed TbMEAT1-UTP co-crystal were collected to 1.95 Å resolution. Numerous attempts of molecular replacement using various search models of TbTUT4 and TbRET2 were unsuccessful. Selenomethionine-labeled protein was then generated and crystallized under the same conditions; crystals were soaked in the same cryoprotectant solution with 10 mM MgCl2 (TbMEAT1SeMet-UTP-Mg). Data were collected at the Se peak, inflection and remote wavelengths, and the structure was solved by 3-wavelength multiple anomalous dispersion (MAD) to 2.30 Å resolution. The program SOLVE34 was used to locate the positions of the selenium atoms and to compute initial phases. Electron density was improved using RESOLVE35, 36 followed by further density modification in DM21. The structure factors and improved phases were utilized in Buccaneer37 for automated building of a virtually complete model. The apo structure (TbMEAT1-apo) was obtained in an unsuccessful attempt to co-crystallize the enzyme with UpU bound in the active site. Crystals of this type, which were grown in the presence of a 2-fold molar excess of UpU under the same crystallization conditions, exhibited a significant improvement in resolution (1.56 Å), yet devoid of electron density for the substrate in the active site. All data were processed in HKL2000 (18) and model building was carried out in Coot38. Refmac539 was employed for restrained refinement with TLS parameters, and NCS restraints were applied to the entire molecule. TbMEAT1-apo was refined using anisotropic B-factors, and hydrogens were generated for refinement but not included in the model. Due to non-isomorphism, the starting models of TbMEAT1-UTP and TbMEAT1-apo were generated by molecular replacement using the program PHASER40, and TbMEAT1SeMet-UTP-Mg as the search model. The test set was conserved for the TbMEAT1-UTP and TbMEAT1-apo structures. All Figures were generated using PyMOL (http://www.pymol.org).
Co-crystallization and crystal soaking with alternative substrates
Co-crystallization attempts of TbMEAT1 with ATP, GTP, and CTP, containing a 5-fold molar excess of NTPs, were carried out under the same conditions as with UTP. For attempts at UMP and UpU binding, TbMEAT1-UTP co-crystals were soaked in cryo-protecting solution containing either 10 mM UMP or 1 mM UpU, with soaking times ranging from 30 minutes to several hours. Data were collected, processed, and utilized to generate electron density maps in a similar manner as previously mentioned.
Enzymatic assays
Reactions were performed in buffer containing 50 mM Tris-HCl pH 8.0, 1 mM DTT, 10 mM KCl, 10 mM MgAc and 0.5 μM RNA of 5′-labeled RNA substrate (5′GCUAUGUCUGUCAACUUGUUUUUU-3′) at 27° C for 1–45 min. Reactions were started by adding recombinant proteins to 50–300 nM and stopped by adding 1.5 volumes of 10 mM EDTA, 95% formamide solution. Products were separated on a 15% acrylamide/8M urea gel. Initial velocities for U-addition were determined at UTP concentrations ranging from 0.5 to 150 μM and reaction times from 1 to 20 min. Quantity One software were used to calculate intensities of the input and product bands. Steady-state kinetic parameters were obtained using the enzyme kinetics module in the Sigma Plot package.
Protein Data Bank Accession Codes
The structure factors and atomic coordinates for TbMEAT1-UTP-Mg, TbMEAT1-UTP, and TbMEAT1-Apo have been deposited with the RCSB Protein Data Bank under accession codes 3HIY, 3HJ1, and 3HJ4, respectively.
Acknowledgments
We thank Elena Galitovskaya for technical assistance and members of Luecke and Aphasizhev laboratories for discussions. This work was supported by the UCI Center for Biomembrane Systems and NIH grants R01 AI078000 to H.L. and RO1 AI064653 to R.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim Biophys Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etheridge RD, Clemens DM, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol. 2009;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao CY, Read LK. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 2007;154:158–169. doi: 10.1016/j.molbiopara.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 8.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci U S A. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 14.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual Role of the RNA Substrate in Selectivity and Catalysis by Terminal Uridylyl Transferases. Proc Natl Acad Sci U S A. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J Biol Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 17.Ringpis GE, Aphasizheva I, Wang X, Huang L, Hatfield GW, Aphasizhev R. Mechanism of RNA Editing in Trypanosome Mitochondria: The Bimodal U-insertion Activity of the Core Complex. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.03.050. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 20.Aravind L, Wolf YI, Koonin EV. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J Mol Microbiol Biotechnol. 2000;2:191–194. [PubMed] [Google Scholar]
- 21.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Schnaufer A, Ernst NL, Palazzo SS, O’Rear J, Salavati R, Stuart K. Separate Insertion and Deletion Subcomplexes of the Trypanosoma brucei RNA Editing Complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 23.Aphasizhev R. RNA uridylyltransferases. Cell Mol Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 29.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc Natl Acad Sci U S A. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 32.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 33.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(a) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowtan K. The Buccaneer software for automated model building. 1 Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT 4. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 40.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions 4. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]