Abstract
Increasing evidence suggests that iron-sulfur proteins are the primary targets of NO (nitric oxide). Exposure of Escherichia coli cells to NO readily converts iron-sulfur proteins to the protein-bound DNICs (dinitrosyl iron complexes). While the protein-bound DNICs are stable in vitro under aerobic or anaerobic conditions, they are efficiently repaired in aerobically growing E. coli cells even without new protein synthesis. The cellular repair mechanism for the NO-modified iron-sulfur proteins remains largely elusive. Here we report that unlike aerobically growing E. coli cells, the starved E. coli cells fail to re-activate the NO-modified iron-sulfur proteins. Significantly, addition of L-cysteine, but not other related biological thiols, results in decomposition of the protein-bound DNICs in the starved E. coli cells and in the cell extracts under aerobic conditions. However, L-cysteine has little or no effect on the protein-bound DNICs in the starved E. coli cells and in vitro under anaerobic conditions, suggesting that oxygen is required for the L-cysteine-mediated decomposition of the protein-bound DNICs. Additional studies reveal that L-cysteine is able to exchange the DNIC with the protein-bound DNICs to form the L-cysteine-bound DNIC which is rapidly disrupted by oxygen, resulting in eventual decomposition of the protein-bound DNICs under aerobic conditions.
Keywords: nitric oxide, iron-sulfur clusters, dinitrosyl iron complex, L-cysteine, superoxide
Introduction
In activated macrophages and neutrophils, excessive amounts of NO (nitric oxide) are produced by inducible NO synthase to kill pathogenic bacteria and tumor cells [1]. Among the primary targets of NO cytotoxicity are the iron-containing redox centers hemes and iron-sulfur clusters in proteins [2–4]. Unlike reversible binding of NO to hemes in proteins [5, 6], iron-sulfur clusters are completely disrupted by NO forming the protein-bound DNICs (dinitrosyl iron complexes) [7–17]. Recent studies with synthetic compounds further demonstrated that NO is able to directly interact with iron-sulfur clusters to form the DNICs [18, 19]. The unique EPR (Electron Paramagnetic Resonance) signal at g = 2.04 of the protein-bound DNICs has been observed in bacterium Clostridium botulinum treated with NO produced from reduction of nitrite [20], in activated macrophages where NO was produced by inducible NO synthase [21], in tumor cells co-cultured with activated macrophages [22], in the post-operative-day-4 allografts [23], in the gastro-oesophageal junction where NO was generated luminally from nitrite [24], and in cultured mammalian cells treated directly with NO donors [25–27]. When Escherichia coli cells were exposed to pure NO gas at micromolar concentrations, a large number of iron-sulfur proteins were modified forming the protein-bound DNICs [16].
Purified protein-bound DNICs are stable under both aerobic and anaerobic conditions [13]. However, the protein-bound DNICs are efficiently repaired in aerobically growing E. coli cells even without new protein synthesis [12, 16]. Although a number of proteins including the iron-sulfur cluster assembly machinery [2, 14, 15, 28, 29] and the diiron protein YtfE [30] have been identified as crucial for re-activation of the NO-modified iron-sulfur proteins in E. coli cells, the underlying cellular mechanism has not been fully understood. In previous studies, we found that L-cysteine, but not proteins or other related biological thiols such as reduced glutathione and N-acetyl-L-cysteine, can efficiently decompose the protein-bound DNICs in vitro under aerobic conditions [13, 16]. Nevertheless, the physiological relevance of the L-cysteine-mediated decomposition of the protein-bound DNICs was not addressed. Here, we report that unlike aerobically growing E. coli cells, the carbon-starved E. coli cells in which the intracellular free amino acids including L-cysteine were largely depleted [31] fail to repair any NO-modified iron-sulfur proteins. Addition of L-cysteine, but not other related biological thiols or amino acids, to the starved E. coli cells restores the cellular activity for decomposing the protein-bound DNICs under aerobic conditions. However, L-cysteine has little or no effect on the protein-bound DNICs in the starved E. coli cells and in vitro under anaerobic conditions, suggesting that oxygen has an essential role for the L-cysteine-mediated decomposition of the protein-bound DNICs. Additional studies reveal that L-cysteine is able to extrude the DNIC from the protein-bound DNICs via the thiol ligand exchange to form the L-cysteine-bound DNIC which is rapidly disrupted by oxygen, resulting in eventual decomposition of the protein-bound DNICs. The physiological relevance of the L-cysteine/oxygen-mediated decomposition of the protein-bound DNICs will be discussed.
Materials and Methods
Gene cloning and protein preparation
The DNA fragment encoding an iron-sulfur enzyme aconitase B [32] was amplified from wild-type E. coli genomic DNA by PCR using two primers (AcnB-1, 5’-gaaccgccatggtagaagaatacc-3’; AcnB-2, 5’-tgactttttaaagcttagtctgga-3’). The PCR product was digested with NcoI and HindIII, and ligated to pBAD (Invitrogene co.) to produce pBACNB. Recombinant aconitase B was expressed in E. coli MC4100 by adding L-arabinose (0.02%) to LB (Luria-Bertani) medium and purified as described in [17]. The purity of purified aconitase B was over 95% judging from the SDS/PAGE followed with Coomassie staining. The aconitase activity was measured by transferring an aliquot of the E. coli cell extracts to a pre-incubated solution containing Tris (90 mM, pH 8.0) and D,L-isocitrate (20 mM) at 23°C as described in [17]. The reaction was monitored at 240 nm in a Beckman DU640 UV-Visible spectrometer using an extinction coefficient of 3.6 mM−1cm−1 [7].
NO exposure of E. coli cells
Overnight E. coli cells (MC4100) containing pBACNB plasmid were diluted 1:50 in LB medium and incubated at 37°C with aeration (250 rpm) for 3 hours, followed by induction with L-arabinose (0.02%) for additional 2 hours. Cells were then harvested, washed three times with the M9 minimal medium (without glucose), and re-incubated in the same minimal medium at 37°C with aeration for 3 hours. Chloramphenicol (34 µg/mL) was then added to E. coli cells to block new protein synthesis. Cells were subsequently purged with pure argon gas for 10 min before the NO exposure as described in [16]. NO gas (Air Co.) was first passed through a soda-lima column to remove NO2 and higher nitrogen oxides before being connected to Silastic tubing [16]. The length of Silastic tubing immersed in the cell culture was adjusted in such that about 100 nM NO per second was released under vigorous stirring to emulate the pathophysiological NO production in activated neutrophils [33, 34]. After NO exposure for 10 min, E. coli cells were re-purged with pure argon gas to remove residual NO and returned to different growth conditions. Because E. coli MC4100 has a reduced expression of transcription factor FNR [35], another “wild-type” E. coli strain GC4468 containing recombinant aconitase B was also used for the NO exposure experiments. The results using both MC4100 and GC4468 were essentially the same.
Preparation of the L-cysteine-bound DNIC
The L-cysteine-bound DNIC was prepared according to [36, 37] with slight modifications. Briefly, pre-degassed Fe(NH4)2(SO4)2 (100 µM) was injected into a pre-degassed vial containing L-cysteine (200 µM) and Tris (50 mM) (pH 8.0) anaerobically, followed by injection of a bolus of pure NO gas. Residual NO in the solution was removed by purging with pure argon gas.
Analyses of nitrite, nitrate, and ferrous iron
Nitrite in solutions was measured using the Griess reagents according to the manufacture’s instruction (Cayman Chemicals co.). Nitrate in solutions was measured after reduction to nitrite by means of cadmium reductor [38]. Ferrous iron in solutions was measured using an iron indicator ferroZine as described in [39].
EPR measurements
The X-band EPR spectra were recorded using a Bruker model ESR-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. Routine EPR conditions were: microwave frequency, 9.47 GHz; microwave power, 10.0 mW; modulation frequency, 100 kHz; modulation amplitude, 1.2 mT; temperature, 20 K; receive gain, 105.
Results
L-cysteine mediates decomposition of the protein-bound DNICs in the starved E. coli cells under aerobic conditions
Previous studies indicated that L-cysteine, but not other related biological thiols such as glutathione and N-acetyl-L-cysteine, can efficiently decompose the protein-bound DNICs in vitro under aerobic conditions [13, 16]. However, the physiological relevance of the L-cysteine-mediated decomposition of the protein-bound DNICs was not addressed. Here, we took advantage of the starvation approach under which the intracellular free amino acids in E. coli cells were largely depleted after 3 hours aerobic incubation in the M9 minimal medium without any carbon sources [31]. The starved E. coli cells (containing recombinant iron-sulfur enzyme aconitase B [32]) were subject to the NO exposure at a rate of 100 nM NO per second for 10 min using the Silastic tubing NO delivery system as described in [16]. Chloramphenicol was added to block new protein synthesis in the E. coli cells before the NO exposure. The NO-exposed E. coli cells were then re-incubated in the M9 minimal medium with or without glucose (0.2%) under aerobic conditions at 37°C for 60 min. The cell extracts were immediately prepared from the E. coli cells for the analysis of aconitase activity as described in [17].
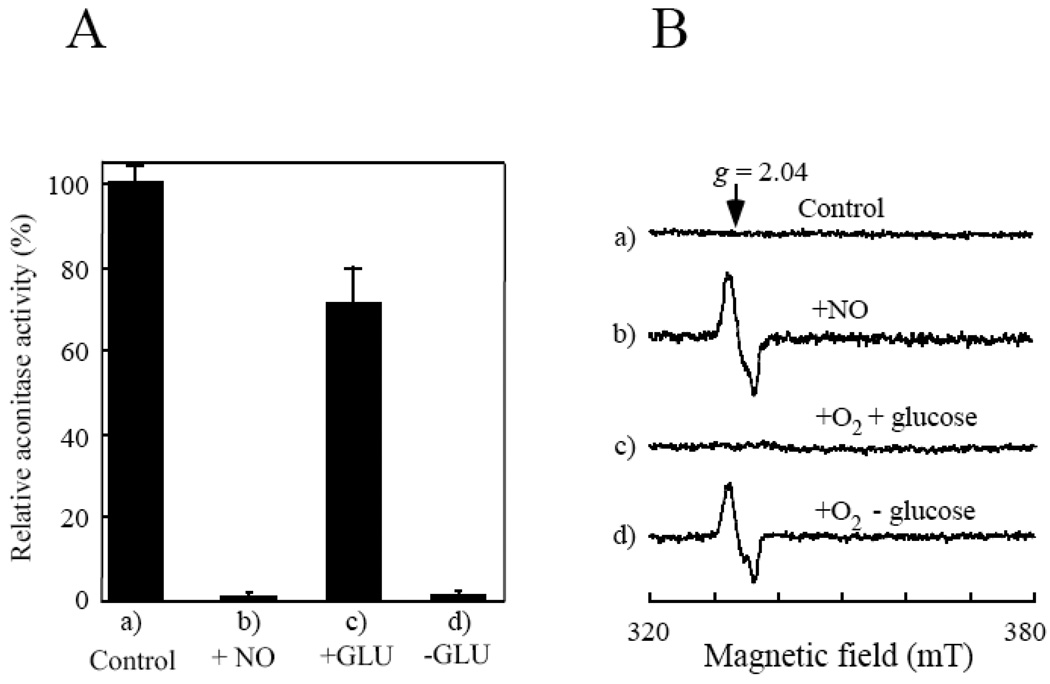
Figure 1A shows that the aconitase activity in the E. coli cells was abolished by the NO exposure and largely restored when the NO-exposed E. coli cells were re-incubated in the M9 minimal medium with glucose under aerobic conditions. However, when the NO-exposed E. coli cells were re-incubated in the M9 minimal medium without glucose under aerobic conditions, no aconitase activity was recovered after incubation (Figure 1A). The whole-cell EPR (Electron Paramagnetic Resonance) measurements further showed that unlike the aerobically growing E. coli cells, the starved E. coli cells failed to decompose the protein-bound DNICs (Figure 1B).
Figure 1. The starved E. coli cells fail to repair the NO-modified iron-sulfur proteins under aerobic conditions.
E. coli cells containing recombinant aconitase B (pBACNB) were washed three times with the M9 minimal medium (without glucose) and re-suspended in the same minimal medium to an OD at 600 nm of 4.0. After 3 hours starvation in the M9 minimal medium (without glucose) under aerobic conditions, chloramphenicol (34 µg/ml) was added to block new protein synthesis before the cells were exposed to NO using the Silastic tubing NO delivery system. A), relative aconitase activity in the cell extracts prepared from a), untreated starved E. coli cells; b), the NO-exposed starved E. coli cells; c), the NO-exposed starved E. coli cells after aerobic incubation in the M9 minimal medium with glucose (0.2%) at 37°C for 60 min; d), the NO-exposed starved E. coli cells after aerobic incubation in the M9 minimal medium without glucose at 37°C for 60 min. The results are averages from three independent experiments. B), the whole cell EPR spectra. The starved E. coli cells (spectrum a) were exposed to NO (spectrum b), followed by re-incubation in the M9 minimal medium with (spectrum c) or without (spectrum d) glucose (0.2%) at 37°C for 60 min under aerobic conditions.
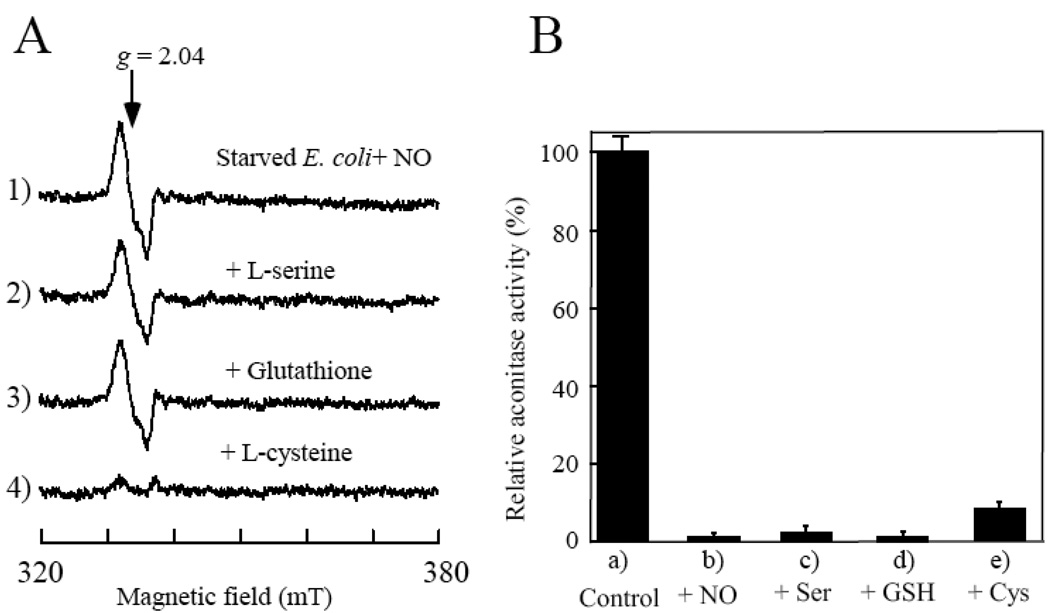
We then asked whether L-cysteine can promote the decomposition of the protein-bound DNICs in the starved E. coli cells under aerobic conditions. Because E. coli has at least two L-cysteine uptake transporters [40, 41], we assumed that at least some of L-cysteine supplemented in the minimal medium will be taken-up by the starved E. coli cells. Figure 2A shows that supplement with L-cysteine did indeed result in decomposition of the protein-bound DNICs in the starved E. coli cells under aerobic conditions. In contrast, addition of L-serine, or reduced glutathione had no effect on the protein-bound DNICs in the starved E. coli cells under aerobic conditions. Similarly, addition of N-acetyl-L-cysteine and L-cystine (oxidized L-cysteine) failed to decompose the protein-bound DNICs in the starved E. coli cells (data not shown). These results clearly suggested that L-cysteine has its unique activity in decomposing the protein-bound DNICs not only in vitro [13, 16] but also in the starved E. coli cells. Interestingly, supplement of L-cysteine only restored less than 10% of the total aconitase activity in the NO-exposed starved E. coli cells (Figure 2B), indicating that while L-cysteine can efficiently decompose the protein-bound DNICs in the starved E. coli cells under aerobic conditions, other cellular factors are required for a full re-activation of the NO-modified iron-sulfur proteins [14].
Figure 2. L-cysteine mediates decomposition of the protein-bound DNICs in the starved E. coli cells under aerobic conditions.
A), L-cysteine has a unique activity in decomposing the protein-bound DNICs in the starved E. coli cells under aerobic conditions. The NO-exposed starved E. coli cells (spectrum 1) were incubated in the M9 minimal medium (without glucose) supplemented with L-serine (1 mM) (spectrum 2), reduced glutathione (1 mM) (spectrum 3), or L-cysteine (1 mM) (spectrum 4) at 37°C for 60 min under aerobic conditions. B), relative aconitase activity in the cell extracts prepared from a), untreated starved E. coli cells; b), the NO-exposed starved E. coli cells; c), d), and e), the NO-exposed starved E. coli cells re-incubated aerobically at 37°C for 60 min in the M9 minimal medium supplemented with L-serine (Ser) (1 mM), reduced glutathione (GSH) (1 mM), or L-cysteine (Cys) (1 mM), respectively. The data are the averages from three independent experiments.
Oxygen is essential for the L-cysteine-mediated decomposition of the protein-bound DNICs
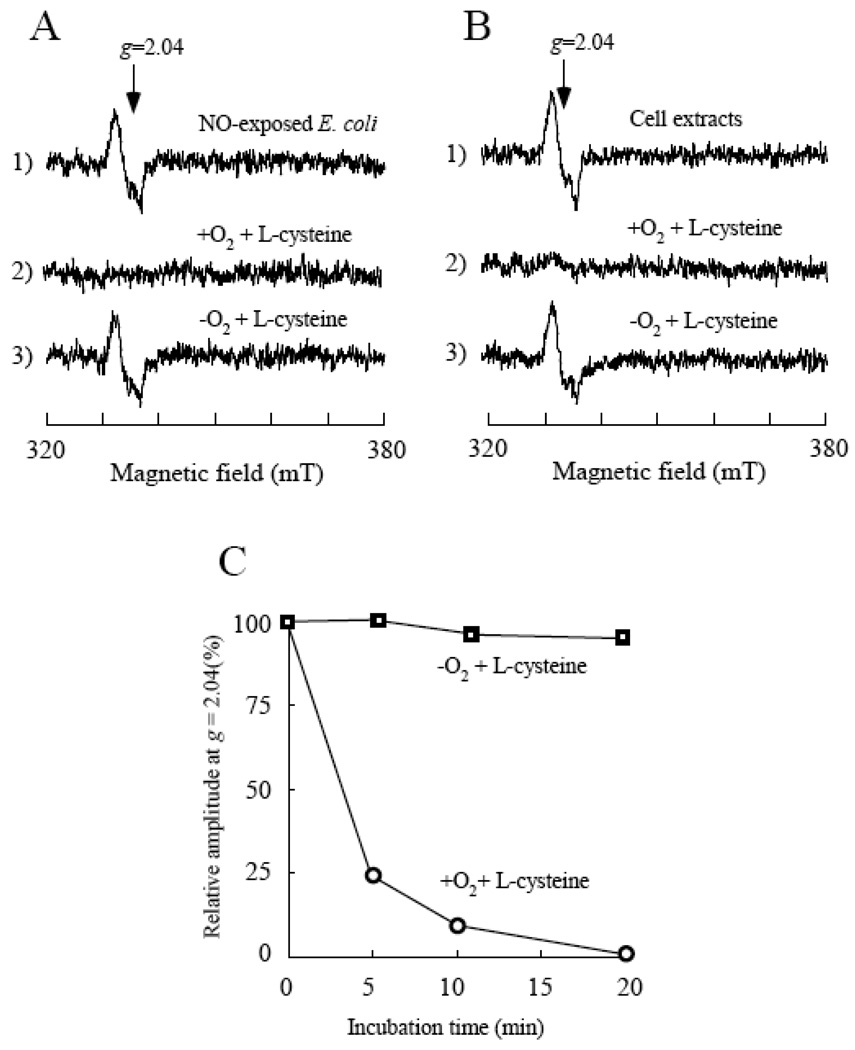
Because L-cysteine could be oxidized by oxygen in the presence of transition metals such as iron in solution [41, 42], we thought that L-cysteine would be more effective in decomposing the protein-bound DNICs in the starved E. coli cells under anaerobic conditions. To our surprise, addition of L-cysteine had little or no effect on the protein-bound DNICs in the starved E. coli cells under anaerobic conditions (Figure 3A). The requirement of oxygen for the L-cysteine-mediated decomposition of the protein-bound DNICs in the starved E. coli cells was not strainspecific, as the same results were observed when either E. coli MC4100 or GC4468 was used for the experiments (data not shown).
Figure 3. Oxygen is required for the L-cysteine-mediated decomposition of the protein-bound DNICs in the starved E. coli cells.
A), the NO-exposed starved E. coli cells (spectrum 1) were incubated at 37°C for 60 min in the M9 minimal medium (without glucose) supplemented with L-cysteine (1 mM) under aerobic (spectrum 2) or anaerobic (spectrum 3) conditions. B), the cell extracts prepared from the NO-exposed starved E. coli cells (spectrum 1) were incubated at 37°C for 30 min with L-cysteine (1 mM) under aerobic (spectrum 2) or anaerobic (spectrum 3) conditions. C), kinetics of the L-cysteine-mediated decomposition of the protein-bound DNICs in the cell extracts under aerobic and anaerobic conditions. The cell extracts prepared from the NO-exposed starved E. coli cells were incubated with L-cysteine (1 mM) at 37°C under aerobic (open circles) or anaerobic (open squares) conditions. The amplitudes of the EPR signal at g = 2.04 of the protein-bound DNICs were plotted as a function of incubation time.
One of the explanations would be that the starved E. coli cells may take up L-cysteine under aerobic conditions but not under anaerobic conditions. To overcome the cell’s membrane barriers, we prepared the cell extracts from the NO-exposed E. coli cells and incubated the cell extracts directly with L-cysteine under aerobic and anaerobic conditions. Figure 3B shows that L-cysteine had little or no effect on the protein-bound DNICs under anaerobic conditions. However, under aerobic conditions, L-cysteine efficiently decomposed the protein-bound DNICs in the cell extracts. Over 75% of the protein-bound DNICs in the cell extracts were decomposed by L-cysteine after 5 min incubation under aerobic conditions (Figure 3C). Thus, oxygen is required for the L-cysteine-mediated decomposition of the protein-bound DNICs in the starved E. coli cells and in the cell extracts.
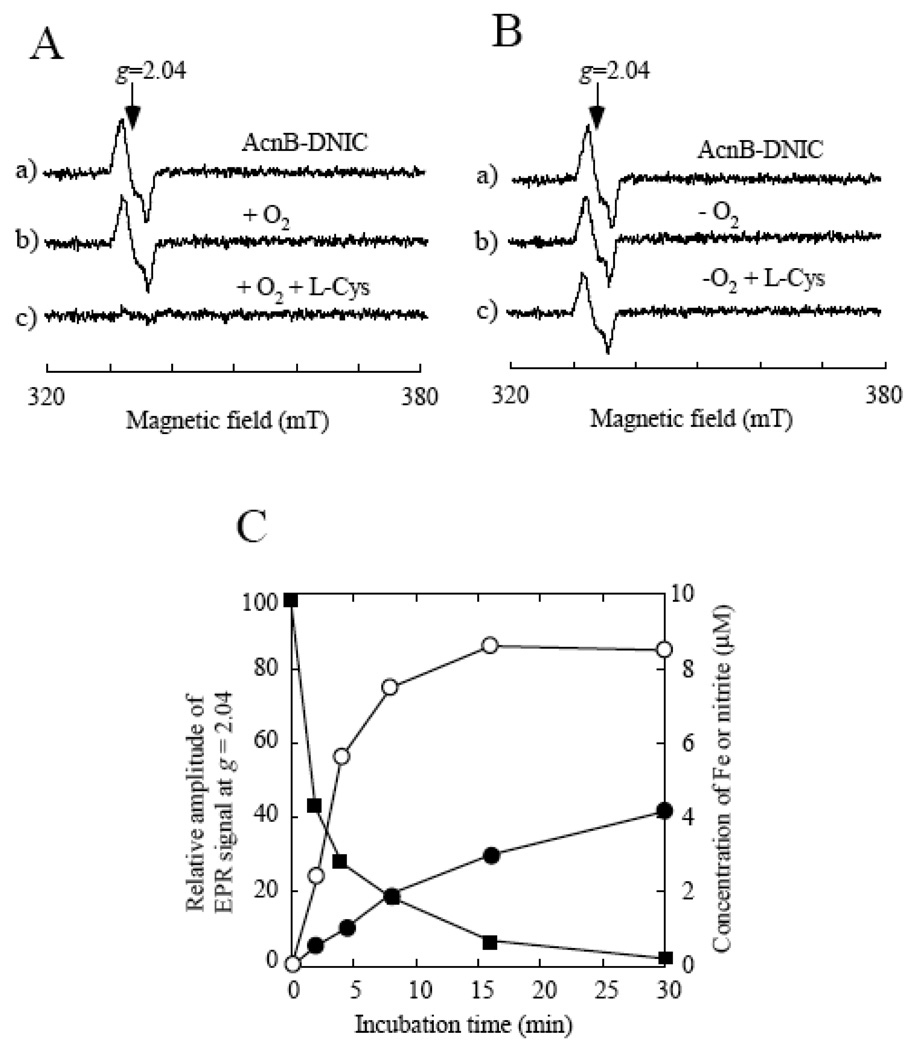
To rule out possible contributions of other cellular components to the L-cysteine-mediated decomposition of the protein-bound DNICs, we purified the aconitase B-bound DNIC from the NO-exposed E. coli cells and incubated the purified protein with L-cysteine under aerobic and anaerobic conditions. While L-cysteine quickly decomposed the aconitase B-bound DNIC under aerobic conditions (Figure 4A), L-cysteine had little or no effect on the aconitase B-bound DNIC under anaerobic conditions (Figure 4B). Parallel measurements of EPR signal at g = 2.04, nitrite, and ferrous iron in the incubation solution revealed that the L-cysteine-mediated decomposition of the aconitase B-bound DNIC was accompanied with release of nitrite and ferrous iron from the complex under aerobic conditions (Figure 4C). Nitrate and S-nitrosothiols were not detectable during the incubation process, indicating that nitrite is the major stable metabolite of the decomposition reaction. Taken together, the results indicated that oxygen and L-cysteine are necessary and sufficient for decomposition of the protein-bound DNICs.
Figure 4. Oxygen is required for the L-cysteine-mediated decomposition of the aconitase B-bound DNIC in vitro.
A), effect of L-cysteine on the aconitase B-bound DNIC under aerobic conditions. Purified aconitase B-bound DNIC (5 µM) (spectrum a) was incubated with buffer (spectrum b) or L-cysteine (1 mM) (spectrum c) under aerobic condition at 37°C for 30 min. B), effect of L-cysteine on the aconitase B-bound DNIC under anaerobic conditions. Purified aconitase B-bound DNIC (5 µM) (spectrum a) was incubated with buffer (spectrum b) or L-cysteine (1 mM) (spectrum c) under anaerobic condition at 37°C for 30 min. C), kinetics of the L-cysteine-mediated decomposition of the aconitase B-bound DNIC under aerobic conditions. Purified aconitase B-bound DNIC (5 µM) was incubated with L-cysteine (1 mM) at 37°C under aerobic conditions. At indicated time points, aliquot was taken for the measurements of the EPR signal at g = 2.04 (closed squares), “free” ferrous iron (closed circles), and nitrite (open circles) as described in the Materials and Methods. The data are representatives of three independent experiments.
Superoxide and hydrogen peroxide are not involved in the L-cysteine-mediated decomposition of the protein-bound DNICs
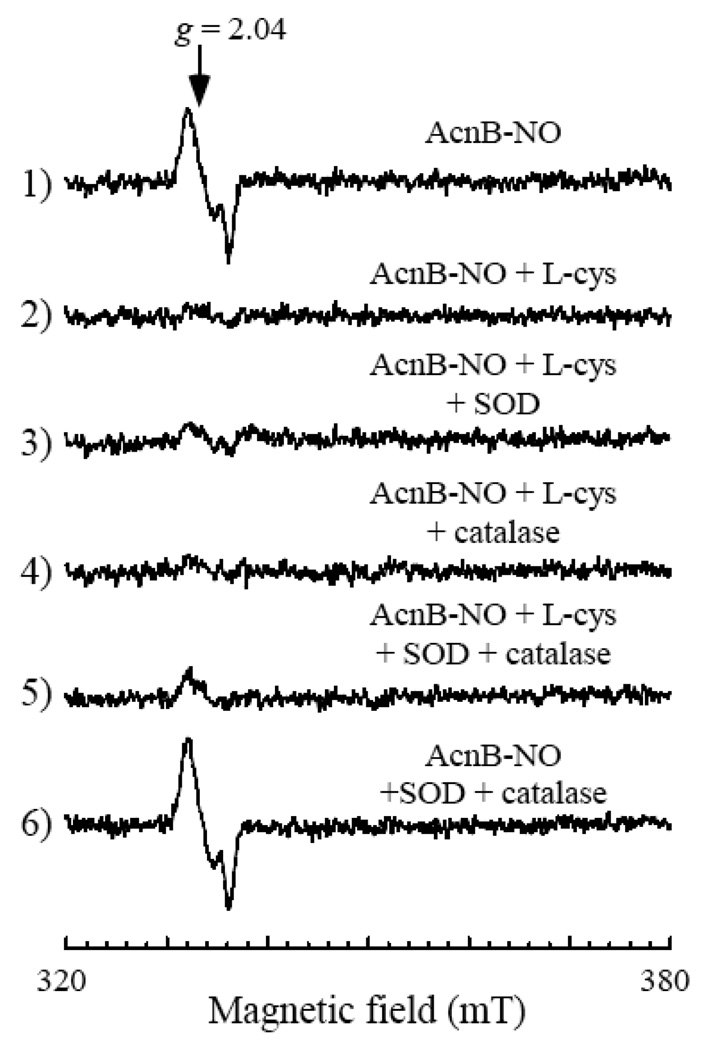
Vanin and colleagues recently reported that an excess of superoxide can partially disrupt the DNIC in bovine albumin in vitro [43]. To test whether L-cysteine decomposes the protein-bound DNICs by promoting the production of superoxide and/or hydrogen peroxide under aerobic conditions, we included excessive amounts of superoxide dismutase and/or bovine liver catalase (Sigma co.) in the incubation solution. Figure 5 shows that addition of superoxide dismutase and/or catalase failed to prevent the L-cysteine-mediated decomposition of the aconitase B-bound DNIC under aerobic conditions. Similar results were obtained when the cell extracts prepared from the NO-exposed E. coli cells were incubated with L-cysteine in the presence of superoxide dismutase and catalase under aerobic conditions (data not shown). Therefore, it is unlikely that superoxide and hydrogen peroxide are involved in the L-cysteine-mediated decomposition of the protein-bound DNICs under aerobic conditions.
Figure 5. Role of superoxide and hydrogen peroxide in the L-cysteine-mediated decomposition of the protein-bound DNICs.
Purified aconitase B-bound DNIC (5 µM) (spectrum 1) was incubated with L-cysteine (1 mM) (spectrum 2), or L-cysteine (1 mM) with superoxide dismutase (200 units/ml) (spectrum 3), L-cysteine (1 mM) with catalase (200 units/ml) (spectrum 4), or L-cysteine (1 mM) with superoxide dismutase (200 units/ml) and catalase (200 units/ml) (spectrum 5). Spectrum 6), purified aconitase B-bound DNIC (5 µM) was incubated with superoxide dismutase (200 units/ml) and catalase (200 units/ml) only. All samples were incubated at 37°C for 30 min under aerobic conditions.
L-cysteine is able to exchange the DNIC with the protein-bound DNICs
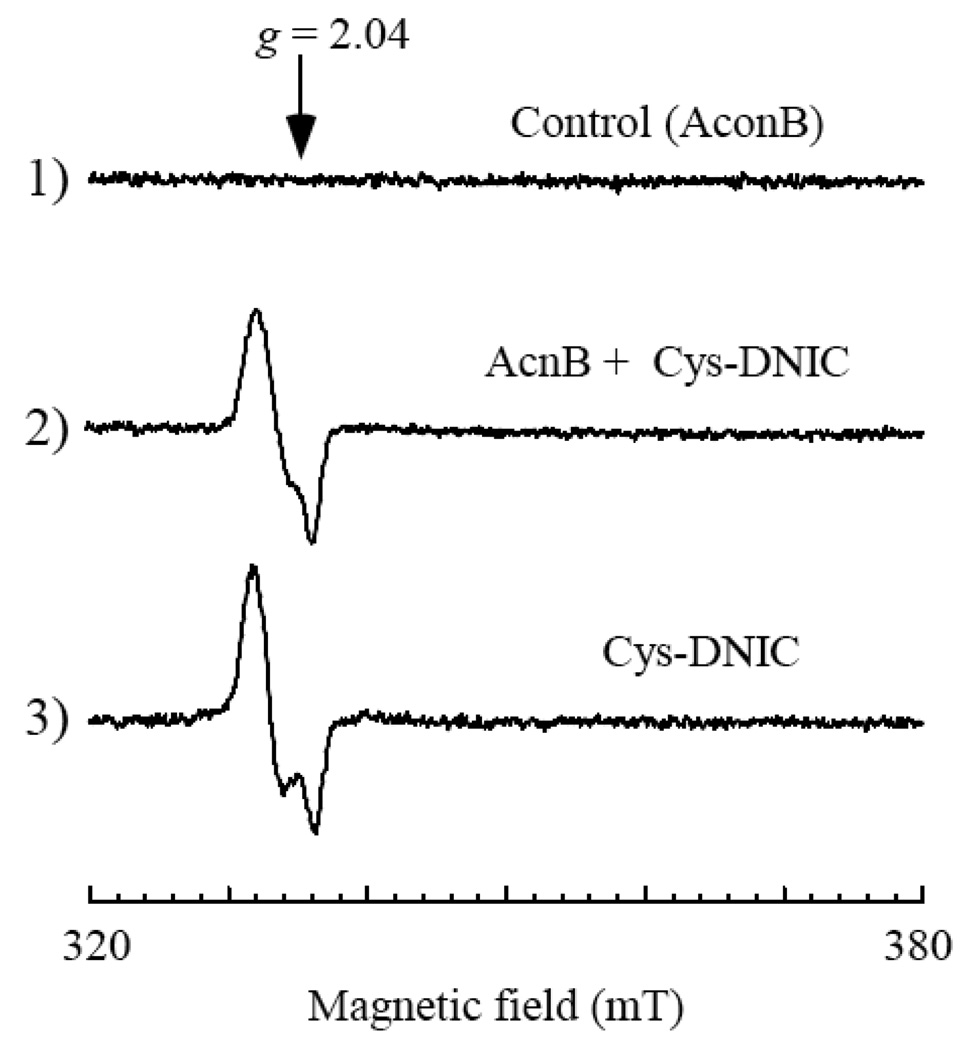
As an alternative model, we postulated that L-cysteine may extrude the DNIC from the protein-bound DNICs to form the L-cysteine-bound DNIC via the thiol ligand exchange. L-cysteine is able to form the DNIC with ferrous iron and NO in solution [44–46] and further to transfer the DNIC to proteins such as bovine serum albumin in vitro [36]. To determine whether the L-cysteine-bound DNIC can also transfer the DNIC to iron-sulfur proteins, we incubated the L-cysteine-bound DNIC with an equal concentration of purified E. coli aconitase B at 37°C under anaerobic conditions. Figure 6 shows that over 90% of the L-cysteine-bound DNIC was transferred to aconitase B to form the aconitase B-bound DNIC after 10 min anaerobic incubation. Furthermore, the transfer of the DNIC from the L-cysteine-bound DNIC to aconitase B did not require the iron-sulfur cluster in protein, as apo-aconitase B can equally accept the DNIC from the L-cysteine-bound DNIC under anaerobic conditions (data not shown).
Figure 6. The L-cysteine-bound DNIC can transfer the DNIC to aconitase B under anaerobic conditions.
Purified E. coli aconitase B (17 µM) was incubated with buffer (spectrum 1) or the L-cysteine-bound DNIC (20 µM) (spectrum 2) at 37°C for 10 min under anaerobic conditions, followed by re-purification of the protein. Spectrum 3) is the L-cysteine-bound DNIC (20 µM). The spectra are the representatives from three independent experiments.
The observed transfer of the DNIC from L-cysteine to aconitase B under anaerobic conditions suggested that if the protein-bound DNICs were incubated with an excess of L-cysteine, the DNICs in protein may be transferred back to L-cysteine to form the L-cysteine-bound DNIC. Indeed, when the aconitase B-bound DNIC was incubated with 500-fold excess of L-cysteine under anaerobic conditions, a small but reproducible amount (5–10%) of the aconitase B-bound DNIC was removed (data not shown). These results suggested that L-cysteine is able to exchange the DNIC with the protein-bound DNICs to form the L-cysteine-bound DNIC under anaerobic conditions.
The L-cysteine-bound DNIC is stable under anaerobic conditions and decomposed under aerobic conditions
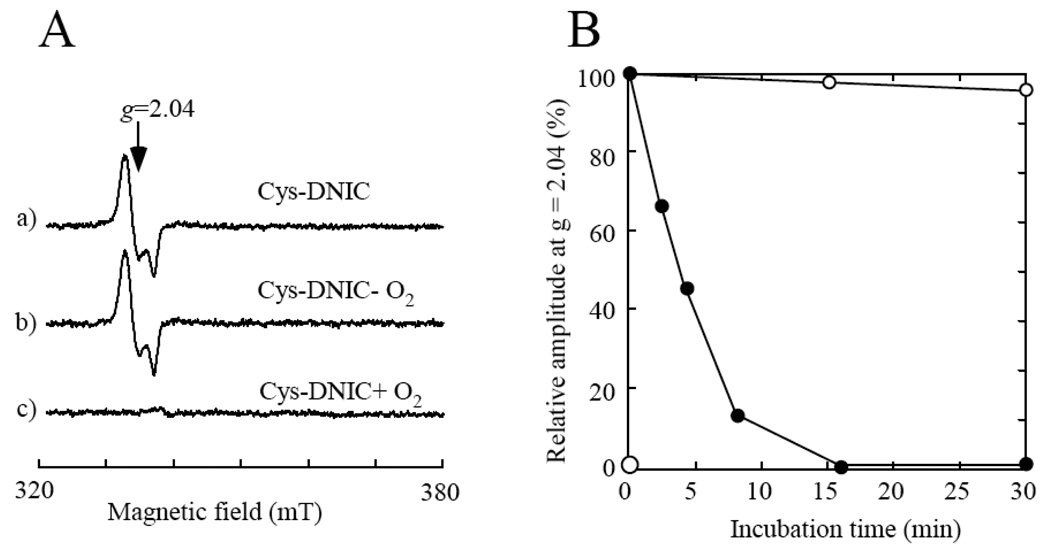
The proposed equilibrium between the protein-bound DNICs and the L-cysteine-bound DNIC would no longer be at equilibrium if the L-cysteine-bound DNIC is continuously decomposed by oxygen, resulting in eventual decomposition of the protein-bound DNICs. To test this idea, we incubated the freshly prepared L-cysteine-bound DNIC at 37°C under aerobic and anaerobic conditions. Figure 7A shows that the L-cysteine-bound DNIC was completely decomposed under aerobic conditions, but remained unchanged under anaerobic conditions. The half-life time of the L-cysteine-bound DNIC under aerobic conditions was about 5 min (Figure 7B), which is similar to that previously reported by Boese et al. [36].
Figure 7. Different stability of the L-cysteine-bound DNIC under aerobic and anaerobic conditions.
A), the EPR spectra of the L-cysteine-bound DNIC. The L-cysteine-bound DNIC (20 µM) (spectrum a) was incubated under anaerobic (spectrum b) or aerobic (spectrum c) conditions at 37°C for 20 min. B), decomposition kinetics of the L-cysteine-bound DNIC under aerobic (closed circles) or anaerobic (open circles) conditions. At indicated time points, aliquot was taken from the incubation solutions for the EPR measurements. The amplitudes of the EPR signal at g = 2.04 of the L-cysteine-bound DNIC were plotted as a function of incubation time.
Discussion
A complete repair of the NO-modified iron-sulfur proteins requires at least two steps: decomposition of the protein-bound DNICs and re-assembly of new iron-sulfur clusters in the proteins [13, 14]. Previous studies indicated that L-cysteine has its unique activity in decomposing the protein-bound DNICs in vitro under aerobic conditions [13, 16]. Here, we find that L-cysteine can also decompose the protein-bound DNICs in the starved E. coli cells under aerobic conditions (Figure 2). Other related biological thiols including reduced glutathione, N-acetyl-L-cysteine and L-cystine have little or no effects on the protein-bound DNICs in the starved E. coli cells, suggesting that L-cysteine is able to decompose the protein-bound DNICs not only in vitro [13, 16] but also in the starved E. coli cells. Under normal growth conditions, the intracellular L-cysteine concentration is estimated to be around 200 µM in E. coli cells [41, 42], which would be sufficient for decomposing the protein-bound DNICs formed in the cells by NO. However, upon the carbon starvation, the intracellular L-cysteine (and other free amino acids) is largely depleted [31], resulting in failure to decompose the protein-bound DNICs. Importantly, supplement of L-cysteine in the minimal medium is able to increase the intracellular L-cysteine content [41], thus restoring the cellular activity for decomposing the protein-bound DNICs in the starved E. coli cells under aerobic conditions (Figure 2A). Nevertheless, supplement of L-cysteine in the minimal medium only restores less than 10% of the activity of the iron-sulfur enzyme aconitase in the starved E. coli cells (Figure 2C), suggesting that additional cellular factors such as the iron-sulfur cluster assembly proteins [14] and the di-iron protein YtfE [30] could be required for a full re-activation of the NO-modified iron-sulfur proteins.
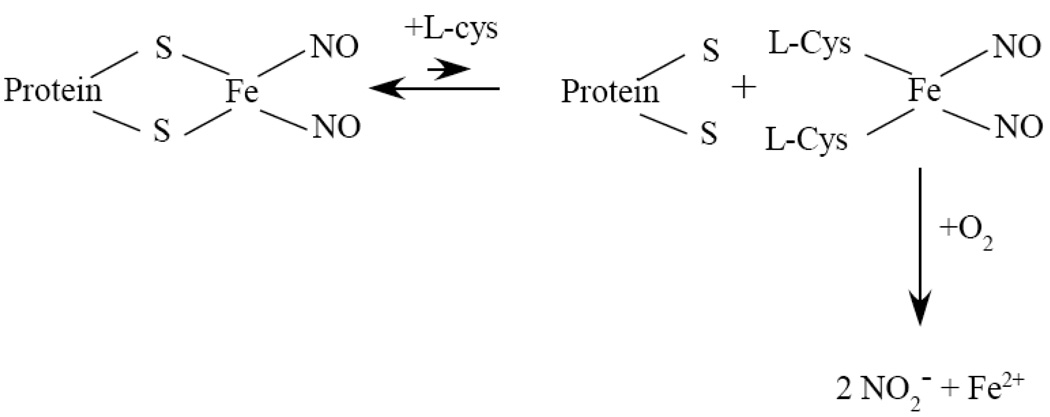
The salient finding reported in this study is that oxygen is required for the L-cysteine-mediated decomposition of the protein-bound DNICs in vitro and in vivo. While L-cysteine can efficiently decompose the protein-bound DNICs in the starved E. coli cells and in purified proteins under aerobic conditions, L-cysteine has very little or no effect on the protein-bound DNICs under anaerobic conditions (Figure 3 and Figure 4). At least two hypotheses could be considered for the mechanism of the L-cysteine/oxygen-mediated decomposition of the protein-bound DNICs. For the first hypothesis, L-cysteine may promote the production of reactive oxygen species which in turn disrupt the DNIC in proteins under aerobic conditions. Vanin’s group recently reported that an excess of superoxide can partially disrupt the DNIC in bovine albumin in vitro [43]. However, addition of excessive superoxide dismutase and catalase fails to prevent the L-cysteine-mediated decomposition of the protein-bound DNICs under aerobic conditions (Figure 5), indicating that superoxide and hydrogen peroxide are not directly involved in the L-cysteine/oxygen-mediated decomposition of the protein-bound DNICs. Alternatively, we propose that L-cysteine may extrude the DNIC from the protein-bound DNICs to form the L-cysteine-bound DNIC which is rapidly decomposed by oxygen (Figure 8). The results shown in Figure 6 and by others [36] clearly supported the idea that L-cysteine can exchange the DNIC with the protein-bound DNICs via the thiol ligands exchange. Unlike the protein-bound DNICs which are stable under aerobic and anaerobic conditions [13] (Figure 4), the L-cysteine-bound DNIC is rapidly disrupted by oxygen under aerobic conditions (Figure 7), resulting in eventual decomposition of the protein-bound DNICs. It should be pointed out that we still do not fully understand the chemistry for the observed stability of the L-cysteine-bound DNIC and the protein-bound DNICs under aerobic conditions. Perhaps, the rigid protein structure provides stable binding ligands for the DNICs, whereas the DNIC bound by “free” L-cysteine in solution is unstable and susceptible to oxidation and decomposition by oxygen. Evidently, additional experiments are required to further elucidate the molecular details of the L-cysteine/oxygen-mediated decomposition of the protein-bound DNICs. Nevertheless, the finding that oxygen is essential for the L-cysteine-mediated decomposition of the protein-bound DNICs provides a logical explanation for the puzzled observations that the NO-modified iron-sulfur proteins are efficiently repaired in the wild-type E. coli cells under aerobic conditions [2, 16] but not under anaerobic conditions [16].
Figure 8. A proposed model for the L-cysteine-mediated decomposition of the protein-bound DNICs.
L-cysteine extrudes the DNIC from the protein-bound DNICs to form the L-cysteine-bound DNIC via the thiol ligand exchange. Under anaerobic conditions, both the L-cysteine-bound DNIC and the protein-bound DNICs are stable, and no DNIC is decomposed. Under aerobic conditions, the L-cysteine-bound DNIC is rapidly disrupted by oxygen, resulting in eventual decomposition of the protein-bound DNICs.
The protein-bound DNICs have also been described as potential reservoirs for the protein S-nitrosation [27, 36, 46], a process involved in diverse signaling pathways [47–49]. Because the NO moieties in the DNICs have a partial positive charge [44], they may act as a natural source of NO+ for the protein S-nitrosation [27, 36]. In light of this study, if L-cysteine and oxygen are ample for decomposing the protein-bound DNICs, the protein-bound DNICs may have no chance to produce S-nitrosothiols in target proteins. However, if oxygen and/or L-cysteine are scarce, the protein-bound DNICs may effectively donate the NO moieties for S-nitrosation in proteins. The interplay between the L-cysteine/oxygen-mediated decomposition of the protein-bound DNICs and S-nitrosation of proteins remains to be further investigated.
Acknowledgements
This work was supported by the National Institutes of Health Grant CA107494 to H.D.
Abbreviations
- DNIC
dinitrosyl iron complex
- EPR
electron paramagnetic resonance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 2.Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiro S. Metalloregulatory proteins and nitric oxide signalling in bacteria. Biochem Soc Trans. 2008;36:1160–1164. doi: 10.1042/BST0361160. [DOI] [PubMed] [Google Scholar]
- 4.Jones-Carson J, Laughlin J, Hamad MA, Stewart AL, Voskuil MI, Vazquez-Torres A. Inactivation of [Fe-S] metalloproteins mediates nitric oxide-dependent killing of Burkholderia mallei. PLoS ONE. 2008;3:e1976. doi: 10.1371/journal.pone.0001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignarro LJ. Haem-dependent activation of guanylate cyclase and cyclic GMP formation by endogenous nitric oxide: a unique transduction mechanism for transcellular signaling. Pharmacol Toxicol. 1990;67:1–7. doi: 10.1111/j.1600-0773.1990.tb00772.x. [DOI] [PubMed] [Google Scholar]
- 6.Brookes PS, Kraus DW, Shiva S, Doeller JE, Barone MC, Patel RP, Lancaster JR, Jr, Darley-Usmar V. Control of mitochondrial respiration by NO*, effects of low oxygen and respiratory state. J Biol Chem. 2003;278:1603–1609. doi: 10.1074/jbc.M211784200. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy MC, Antholine WE, Beinert H. An EPR investigation of the products of the reaction of cytosolic and mitochondrial aconitases with nitric oxide. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 8.Drapier JC. Interplay between NO and [Fe-S] clusters: relevance to biological systems. Methods. 1997;11:319–329. doi: 10.1006/meth.1996.0426. [DOI] [PubMed] [Google Scholar]
- 9.Sellers VM, Johnson MK, Dailey HA. Function of the [2FE-2S] cluster in mammalian ferrochelatase: a possible role as a nitric oxide sensor. Biochemistry. 1996;35:2699–2704. doi: 10.1021/bi952631p. [DOI] [PubMed] [Google Scholar]
- 10.Foster MW, Cowan JA. Chemistry of Nitric Oxide with Protein-Bound Iron Sulfur Centers. Insights on Physiological Reactivity. J. Am. Chem. Soc. 1999;121:4093–4100. [Google Scholar]
- 11.Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. Embo J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron- sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers PA, Ding H. L-cysteine-mediated destabilization of dinitrosyl iron complexes in proteins. J Biol Chem. 2001;276:30980–30986. doi: 10.1074/jbc.M101037200. [DOI] [PubMed] [Google Scholar]
- 14.Yang W, Rogers PA, Ding H. Repair of nitric oxide modified ferredoxin [2Fe-2S] cluster by cysteine desulfurase (IscS) J Biol Chem. 2002;277:12868–12873. doi: 10.1074/jbc.M109485200. [DOI] [PubMed] [Google Scholar]
- 15.Rogers PA, Eide L, Klungland A, Ding H. Reversible inactivation of E. coli endonuclease III by nitric oxide via modification of its [4Fe-4S] cluster. DNA Repair. 2003;2:809–817. doi: 10.1016/s1568-7864(03)00065-x. [DOI] [PubMed] [Google Scholar]
- 16.Ren B, Zhang N, Yang J, Ding H. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol Microbiol. 2008;70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan X, Yang J, Ren B, Tan G, Ding H. Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem J. 2009;417:783–789. doi: 10.1042/BJ20081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrop TC, Tonzetich ZJ, Reisner E, Lippard SJ. Reactions of synthetic [2Fe-2S] and [4Fe-4S] clusters with nitric oxide and nitrosothiols. J Am Chem Soc. 2008;130:15602–15610. doi: 10.1021/ja8054996. [DOI] [PubMed] [Google Scholar]
- 19.Tonzetich ZJ, Do LH, Lippard SJ. Dinitrosyl iron complexes relevant to Rieske cluster nitrosylation. J Am Chem Soc. 2009;131:7964–7965. doi: 10.1021/ja9030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy D, Lancaster JR, Jr, Cornforth DP. Nitrite inhibition of Clostridium botulinum: electron spin resonance detection of iron-nitric oxide complexes. Science. 1983;221:769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]
- 21.Lancaster JR, Jr, Hibbs JB., Jr EPR demonstration of iron-nitrosyl complex formation by cytotoxic activated macrophages. Proc Natl Acad Sci U S A. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drapier JC, Pellat C, Henry Y. Generation of EPR-detectable nitrosyl-iron complexes in tumor target cells cocultured with activated macrophages. J Biol Chem. 1991;266:10162–10167. [PubMed] [Google Scholar]
- 23.Pieper GM, Halligan NL, Hilton G, Konorev EA, Felix CC, Roza AM, Adams MB, Griffith OW. Non-heme iron protein: a potential target of nitric oxide in acute cardiac allograft rejection. Proc Natl Acad Sci U S A. 2003;100:3125–3130. doi: 10.1073/pnas.0636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asanuma K, Iijima K, Ara N, Koike T, Yoshitake J, Ohara S, Shimosegawa T, Yoshimura T. Fe-S cluster proteins are intracellular targets for nitric oxide generated luminally at the gastro-oesophageal junction. Nitric Oxide. 2007;16:395–402. doi: 10.1016/j.niox.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Kim YM, Chung HT, Simmons RL, Billiar TR. Cellular non-heme iron content is a determinant of nitric oxide- mediated apoptosis, necrosis, and caspase inhibition. J Biol Chem. 2000;275:10954–10961. doi: 10.1074/jbc.275.15.10954. [DOI] [PubMed] [Google Scholar]
- 26.Toledo JC, Jr, Bosworth CA, Hennon SW, Mahtani HA, Bergonia HA, Lancaster JR., Jr Nitric oxide-induced conversion of cellular chelatable iron into macromolecule-bound paramagnetic dinitrosyliron complexes. J Biol Chem. 2008;283:28926–28933. doi: 10.1074/jbc.M707862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci U S A. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TM, Read RC, Green J, Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Justino MC, Almeida CC, Teixeira M, Saraiva LM. Escherichia coli Di-iron YtfE Protein Is Necessary for the Repair of Stress-damaged Iron-Sulfur Clusters. J Biol Chem. 2007;282:10352–10359. doi: 10.1074/jbc.M610656200. [DOI] [PubMed] [Google Scholar]
- 31.Reeve CA, Bockman AT, Matin A. Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984;157:758–763. doi: 10.1128/jb.157.3.758-763.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieglstein CF, Cerwinka WH, Laroux FS, Salter JW, Russell JM, Schuermann G, Grisham MB, Ross CR, Granger DN. Regulation of murine intestinal inflammation by reactive metabolites of oxygen and nitrogen: divergent roles of superoxide and nitric oxide. J. Exp. Med. 2001;194:1207–1218. doi: 10.1084/jem.194.9.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawers RG. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J Bacteriol. 2005;187:2609–2617. doi: 10.1128/JB.187.8.2609-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boese M, Mordvintcev PI, Vanin AF, Busse R, Mulsch A. S-nitrosation of serum albumin by dinitrosyl-iron complex. J Biol Chem. 1995;270:29244–29249. doi: 10.1074/jbc.270.49.29244. [DOI] [PubMed] [Google Scholar]
- 37.Vanin AF, Malenkova IV, Mordvintsev OI, Miul'sh A. Dinitrosyl complexes of iron with thiol-containing ligands and their reverse conversion into nitrosothiols. Biokhimiia. 1993;58:1094–1103. [PubMed] [Google Scholar]
- 38.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 39.Cowart RE, Singleton FL, Hind JS. A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal Biochem. 1993;211:151–155. doi: 10.1006/abio.1993.1246. [DOI] [PubMed] [Google Scholar]
- 40.Berger EA, Heppel LA. A binding protein involved in the transport of cystine and diaminopimelic acid in Escherichia coli. J Biol Chem. 1972;247:7684–7694. [PubMed] [Google Scholar]
- 41.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenberg JT, Demple B. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J Bacteriol. 1986;168:1026–1029. doi: 10.1128/jb.168.2.1026-1029.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shumaev KB, Gubkin AA, Serezhenkov VA, Lobysheva, Kosmachevskaya OV, Ruuge EK, Lankin VZ, Topunov AF, Vanin AF. Interaction of reactive oxygen and nitrogen species with albumin- and methemoglobin-bound dinitrosyl-iron complexes. Nitric Oxide. 2008;18:37–46. doi: 10.1016/j.niox.2007.09.085. [DOI] [PubMed] [Google Scholar]
- 44.Butler AR, Megson IL. Non-heme iron nitrosyls in biology. Chem Rev. 2002;102:1155–1166. doi: 10.1021/cr000076d. [DOI] [PubMed] [Google Scholar]
- 45.Richardson DR, Lok HC. The nitric oxide-iron interplay in mammalian cells: Transport and storage of dinitrosyl iron complexes. Biochim Biophys Acta. 2008;1780:638–651. doi: 10.1016/j.bbagen.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Vanin AF. Dinitrosyl iron complexes with thiolate ligands: physico-chemistry, biochemistry and physiology. Nitric Oxide. 2009;21:1–13. doi: 10.1016/j.niox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Bryan NS, Rassaf T, Maloney RE, Rodriguez CM, Saijo F, Rodriguez JR, Feelisch M. Cellular targets and mechanisms of nitros(yl)ation: an insight into their nature and kinetics in vivo. Proc Natl Acad Sci U S A. 2004;101:4308–4313. doi: 10.1073/pnas.0306706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Hogg N. S-Nitrosothiols: cellular formation and transport. Free Radic Biol Med. 2005;38:831–838. doi: 10.1016/j.freeradbiomed.2004.12.016. [DOI] [PubMed] [Google Scholar]