Abstract
Proteins containing the basic Helix-Loop-Helix (bHLH) domain function as transcription factors and play important roles during the development of various metazoans including insects, nematodes and vertebrates. Insect genomes contain more than 50 bHLH transcription factors, but the function of only a few of these proteins in regulation of female reproduction is known. Using RNA interference, we have tested knock-down in the expression of genes coding for bHLH transcription factors in newly emerged adult females to determine their function in regulation of female reproduction in the red flour beetle, Tribolium castaneum. Knock-down in the expression of genes coding for four bHLH transcription factors (TcSRC, TcSim1, TcAsh and TcDaughterless) has caused mortality in the female beetles. In addition, knocking-down the expression of 16 bHLH genes has affected oogenesis and knock-down in the expression of 13 genes has affected embryogenesis. Two genes TcSide1 and TcSpineless are required for both oogenesis and embryogenesis. Thus, the data reported here showed that 31 bHLH transcription factors are required for female survival, reproduction and embryogenesis.
Keywords: Red flour beetle, RNA interference, oogenesis, vitellogenin, embryogenesis
1. Introduction
The proteins that contain basic Helix-Loop-Helix (bHLH) domain function as transcription factors and play important roles in a wide range of developmental and physiological processes including neurogenesis, myogenesis, hematopoiesis, sex determination, reproduction and gut development (Massari and Murre, 2000). The bHLH domain consists of two regions: the basic region that binds DNA and the Helix-Loop-Helix region that is involved in dimerization (Ferre D’Amar et al., 1993; Murre et al., 1989; Kadesh, 1993). Most bHLH transcription factors bind to a consensus hexameric DNA sequence called the E-box (CANNTG) that is present in the promoter regions of their target genes (Massari and Murre, 2000). This E-box sequence is further classified into two groups: class A (CACCTG or CAGCTG) and class B (CACGTG or CATGTG) (Murre et al., 1989; Van Dorren et al., 1991; Dang et al., 1992).
Recent advances in bioinformatics, genomics and proteomics research have helped in identifying genes coding for bHLH transcription factors in various organisms, including vertebrates and invertebrates. A number of bHLH genes have been identified in Drosophila melanogaster (Moore et al., 2000), Bombyx mori (Wang et al., 2007) and Tribolium castaneum (Bitra et al., 2009). However, the functions of only a few of these bHLH transcription factors are known. The bHLH transcription factors belonging to Hairy- Enhancer of split (HES) family play important roles in embryogenesis. Two HES family genes, Hey1 and Hey2 have been identified in D. melanogaster and these genes were shown to play important roles during embryogenesis. These two genes are highly expressed during the development of the nervous system, the somites, the heart and the craniofacial region (Leimeister et al., 1999). Mutation in dMyc, a member of Myc/Upstream Transcription Factor (USF2) family in D. melanogaster had resulted in an abnormally small body size and female sterility (Gallant et al., 1996). Mutations in D. melanogaster Taiman, a member of the bHLH-PAS family and a nuclear receptor coactivator, caused defects in the migration of special follicle cells and border cells in the ovary (Bai et al., 2000).
The two major insect hormones, juvenile hormone (JH) and ecdysteroids regulate reproduction. Juvenile hormone regulates vitellogenin (Vg) gene expression in the fat body and in the ovarian follicle epithelium in insects such as locust and cockroach (Wyatt and Davey, 1996; Englemann, 1983; Belles, 2004). In dipteran insects, ecdysteroids play a major role in regulating reproduction. Vitellogenin synthesis in the mosquito fat body is stimulated by a blood meal and the removal of ovaries, a source of ecdysteroids, prior to blood feeding blocks Vg synthesis (Hagedorn and Fallon, 1973). Receptors and transcription factors involved in ecdysteroid signal transduction play key roles in regulation of reproduction in mosquito, Ae. aegypti (Raikhel et al., 2002). However, there is very little information available on the function of bHLH transcription factors in regulation of insect reproduction.
In a previous study, the T. castaneum genome sequence deposited into the National Center for Biotechnology Information (NCBI) and the Human Genome Sequencing Center (HGSC) databases (Tribolium Genome Sequencing Consortium, 2008) were searched and 53 bHLH genes were identified (Bitra et al., 2009). Phylogenetic analysis using Bayesian analysis has classified these 53 bHLH genes into ten families as defined by bHLH-Per, Arnt, Sim (PAS) domain, HES, Myc/USF2, Atonal, Mesp, Hand, p48, Shout and Achaete-scute (AS-C). Ten genes that did not fall into any of these families were placed under a miscellaneous group. These studies used RNA interference (RNAi) to perform functional analysis of all the 53 bHLH superfamily members and showed that knocking-down the expression of 18 members of this superfamily affected the growth or development during larval or pupal stages.
In the current study, RNAi analyses showed that knocking-down the expression of four bHLH genes resulted in the death of female beetles. These studies also showed that 16 bHLH transcription factors are required for oogenesis and 13 bHLH transcription factors are required for embryogenesis and two bHLH transcription factors, TcSide1 and TcSpineless, are required for both oogenesis and embryogenesis.
2. Materials and methods
2. 1. Rearing and staging
The strain GA-1 of T. castaneum was reared on organic wheat flour containing 10% yeast at 30° C under standard conditions (Parthasarathy et al., 2008). The pupae were sexed based on the structural differences of genital papillae according to Tribolium rearing protocol (http://bru.gmprc.ksu.edu/proj/tribolium/wrangle). Adults were staged soon after emergence; the adults with untanned cuticle (teneral adults) were designated as 0 h and staged thereafter. The staged insects were maintained under similar conditions as mentioned above.
2.2. Double - stranded RNA synthesis
Genomic DNA was extracted from T. castaneum adults using DNeasy Tissue Kit (Qiagen). Synthesis of dsRNA was done according to the procedure described in Bitra et al., 2009. Primers containing the bHLH gene-specific sequences and T7 polymerase promoter at the 5′ end of both the forward primer and reverse primer were used to amplify the 150–550 bp regions (Table 1 and see Bitra et al., 2009 for primer sequences of PAS and HES family members). DsRNA quality was checked by running an aliquot on an agarose gel. The concentration of dsRNA was determined using Nanodrop 2000 (NanoDrop products, Wilmington, DE).
Table 1.
Primers used to amplify bHLH gene fragments for dsRNA Synthesis
| Name of gene | Length of dsRNA | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| HLH106 | 423 | ACATTGAACAAGCCAGACGTGCAG | TTTAACCTTGGGCTCCTTCCCAGT |
| Bigmax | 200 | AGGGTACAAGCTGAGCAAAGCCA | AAATCCGTGTTTCGGTGTGTCCAG |
| AP-4 | 418 | TTAGTCAACATGAAAGCGGTGGCG | AGCCTTTGTTCCGGTTCAGGACTA |
| Max | 129 | ACAGTTCGTACCTTGGAAA | TCTGGGTTGAACAGACCCTT |
| Mnt | 471 | AAGAGCAGCAACAGCATCATCAGG | TCAACGACTGAATGCATCGCAACG |
| Myc | 473 | ATCGATGTGGTGTCCCTCGAGAAGAA | AACTTGGCCAAATGCGCCCTATAC |
| Flocculin like | 630 | AAGTGAAGAACCAGCCAGTACCGT | ACGTGGTTTGTGGGTTGGTGAAAG |
| USF2 | 330 | CGAAGAGCCACGCATAACGAAGTA | TACGTCAAGTCCGTTTCT |
| HLH3B | 437 | TACATTGGCGACAACTCCTCGGAA | AGCCCTTGCAACCTCTGAACGATA |
| Twist | 413 | ACCTGACCAACTCCACCGAGAAAT | CAGCTTCAGCGTCTGTATCTTGGA |
| Hand | 157 | TTTCACAGCAGAGCTGGGTTCGAT | CCCAATATCGCCCTTTGATGAGAC |
| TF21 | 255 | ACCTGGAGGACTCTGACGACGA | TGAGTGGATGTTTGTGGACGTGGT |
| Paraxis | 310 | TGTAAACACGGCCTTCTCGACACT | ATAATAGATGGCGGTGAGGTCGTTGG |
| Delilah2 | 428 | ACTACACCGACTCCAACAACAACC | ACTATGCAACGGCAACGATTCACC |
| Delilah1 | 426 | ATGGATTCGCTCTTGCTCTACACC | ATCCGATAATTGCAACGACTCGCC |
| Shout | 383 | CTGAAGAAGCGCATGTGTCTGAA | TCTGGTTGTGCGAATAGGACGGAA |
| Fer3 | 510 | TCAAATCGGAAAGCGACGACAAGC | TCCCATTCCTTGCCGTGATACTGT |
| Fer1 | 256 | CCAACAGCAAATTCAGCAGCGACA | TTTGGGTTCGTCCTTCTGGGTGTT |
| Fer2 | 189 | ACTCTGCATTCGACGAGCTGAGAA | CTCTATCGGCTTTGATTTCGC |
| Dimmed | 150 | TCGTTGGAAGTGTCGACTCCTTGG | CCTCCATGCTTTCTTCGCTTTCTTCC |
| Tap | 510 | TCAAATCGGAAAGCGACGACAAGC | TCCCATTCCTTGCCGTGATACTGT |
| NeuroD | 415 | TCAGATTTGAGTTCGGCGCCAC | TCCACGATTATTCTCCCAGTACC |
| Beta3 | 405 | AACATTCCCGAGCGTCCCAAA | TTCCGTTGTTGCCCTCACTGTT |
| HLH4C | 220 | TAAGTCGCGAGGAGCGAAGACGAA | CACGCATCTAAGACGTGGTTCA |
| Nautilus | 232 | TCCCGGCTTCTATCCCAGCTATTT | AGTGGCCGCTTTCCTCCTATCAAT |
| Cato | 333 | ACTCCGAGCACCAATATCCCAACT | TCAATTTGTGGTCAGCGTCCAAGC |
| Atonal | 300 | AGGCTTACGGCTACCTCTACATGCT | CCGTAATGTAGGTCTGGGCCATTT |
| Amos | 459 | AAACGACTACTACCACCATCACGG | CAATGTACGTCTGAGCCATCTGA |
| Scute | 209 | ACACACGAAGACAACAAGCGA | ATGTACTCGATGGCCTGTTGCAG |
| Asense | 462 | CAACAAGAAGCTGAGCAAAGT | TCCCACCACTGCATGACATCCATA |
| Ash | 469 | ATCGTCTCTGGAAGCATCACCCAA | ATTGGTATTGGTCCGGACTGT |
| Daughterless | 282 | TTCGGATGAATTCAACGACTCGCC | CCTAAAGCTTTGTTCACACCGTCG |
| Emc | 317 | TGCAGACAACCTCACCGGAATCAA | AGAGGATGGTGTTGGG |
| Mitf | 424 | ATTGTCCCGAACGCAGTTGAAGC | ACTAGGGCACGAAACTTGAAAGGG |
2.3. Microinjection of adult beetles
Adult beetles were anaesthetized using ether vapors for 10 min and then placed in a row on the double sticky tape with the dorsal surface of the beetle facing up. The fore wings (Elytra) were lifted upwards and the hind wings were carefully lifted using the forceps and stuck to the side on the tape. The bHLH gene dsRNA was diluted to a concentration of 5 μg/μl, and 0.2 μl of the dsRNA was injected into each adult on the dorsal side in the abdominal region. DsRNA prepared using 800 bp bacterial malE gene was used as a negative control. After injection, the beetles were allowed to recover for 2–3 h and then transferred to the diet and reared under standard conditions.
2.4. Effect of bHLH transcription factors on reproduction and embryogenesis
Three days after injection of dsRNA, injected adult females were mated with uninjected virgin males. The number of eggs laid by each pair and the number of offspring produced from the eggs laid were monitored at weekly intervals for 3 weeks.
2.5. Fixing and DAPI staining
The ovaries were dissected from the injected adult female beetles at 5 days after injection. The ovaries were fixed overnight in 4 % paraformaldehyde. Then they were washed twice with 1X phosphate buffered saline (PBS) for 5 min each and stained using DAPI for 5 minutes, and mounted on a slide for taking pictures.
2.6. Oocyte scoring
The maturation of the basal primary oocyte in each ovariole was scored based on a scale developed for Tenebrio molitor (Ullmann, 1973). The scale includes stage 1, the oocyte is formed; stage 2, the oocyte leaves germarium; stage 3, the oocyte increases in size; stage 4, the oocyte continues to increase in size; stage 5, the follicle cells organize; stage 6, the oocyte rapidly increases in size; stage 7, the yolk is deposited into the oocyte; stage 8, the chorion is formed; stage 9, the oocyte matures with fully formed chorion.
2.7. Quantitative real-time PCR (qRT-PCR)
Vitellogenin mRNA levels were determined by qRT-PCR using RNA isolated from control malE dsRNA or bHLH dsRNA injected adults at five days after injection, primers designed based on Vg2 gene sequence (Forward primer, 5′ AACGCACACGATTTCGACCAAGTG 3′ and reverse primer, 5′ ACGGCAGCATTAACTTGGTTGCTC 3′) and MyiQ single color real-time PCR detection system (Bio-Rad laboratories, Hercules, CA). qRT-PCR reactions were performed using a common program as follows: initial incubation of 95° C for 3 min was followed by 40 cycles of 95° C for 10s, 60° C for 20 s, 72°C for 30 s. Standard curves were obtained using a 10-fold serial dilution of pooled cDNA. Quantitative mRNA measurements were performed in triplicate and normalized to an internal control of T. castaneum ribosomal protein rp49 mRNA.
2.8. Embryo collection and fixing
Eggs were collected within 24 h of laying. The eggs were dechlorinated in 25% bleach for 2 min. Then, the embryos were rinsed well in deionized water. The embryos were transferred into a glass scintillation vial containing fixing solution (7.2 ml of heptane + 2.4 ml of PEMS + 1.2 ml of 10% paraformaldehyde). The embryos were fixed for 20 min on a shaking platform. Then, the embryos were devitellinized by adding 8 ml of methanol. The embryos were then collected and washed twice with methanol and used for staining. The procedure for staining embryos with DAPI is similar to the procedure followed for staining ovaries described above.
2.9. Imaging and documentation
For documenting fluorescent images, an Olympus 1×71 Inverted Research Microscope fitted with reflected fluorescence system was used. Control of the microscope, as well as image acquisition and exportation as TIFF files, was conducted using MegnaFire software version 1.5. Exposure settings that minimized oversaturated pixels in the final images were used.
3. Results
3.1. Four bHLH transcription factors are required for survival of female beetles
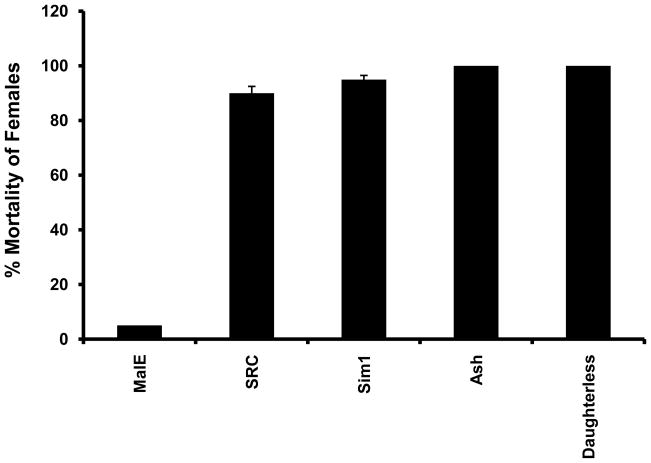
To determine the role of bHLH transcription factors in regulation of reproduction and embryogenesis, dsRNAs of 52 bHLH superfamily members (all bHLH transcription factors identified in T. castaneum except Met which is a subject of separate investigation) were injected into the female adults within 24 h after emergence and the survival of injected beetles were monitored over a three week period. Previous studies on bHLH transcription factor family members showed that RNAi works well to knock-down the expression of genes coding for all bHLH transcription factors by 50% or more (Bitra et al 2009). The dsRNAs for knocking-down the expression of the bHLH transcription factor superfamily members were designed based on the sequences in the variable regions of these genes; therefore these dsRNAs function specifically to knock-down the expression only target gene (Bitra et al. 2009). Knocking-down the expression of genes coding for four bHLH transcription factors caused mortality in female beetles. Knocking-down the expression of TcSRC or TcSim1 led to the death of female beetles within one week after injection (Table 2). Ninety and 95% of the female beetles injected with TcSRC and TcSim1 dsRNA respectively died within one week (Fig. 1). All TcDaughterless or TcAsh dsRNA injected female beetles also died within ten days after injection (Table 2, Fig. 1). In contrast, less than 5% of female beetles injected with control bacterial malE dsRNA died within ten days after injection (Fig. 1). These data suggest that four bHLH transcription factors (TcSRC, TcSim1, TcDaughterless and TcAsh) are required for survival of the adult female and may be involved in regulation of some vital physiological processes.
Table 2.
Effect of PAS and HES family genes silencing on the reproduction of female beetles
| Name of gene | No. of injected females | No. of dead females | No. of pairs matted | First Week | Second Week | Third Week | Total period | Total period (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | ||||
| MalE | 20 | 1 | 6 | 35 | 30 | 40 | 35 | 30 | 27 | 105 | 92 | 100 | 88 |
| Sim1 | 20 | 19 | 6 | - | - | - | - | - | - | - | - | - | - |
| Spineless | 20 | 4 | 5 | 18 | 7 | 15 | 5 | 14 | 7 | 47 | 19 | 45 | 40 |
| SRC | 20 | 18 | 5 | - | - | - | - | - | - | - | - | - | - |
| C ycle | 20 | 1 | 6 | 39 | 34 | 45 | 42 | 31 | 29 | 115 | 105 | 109 | 91 |
| Clock | 20 | 0 | 6 | 42 | 37 | 35 | 30 | 28 | 20 | 105 | 87 | 100 | 83 |
| Hypoxia | 20 | 4 | 5 | 15 | 10 | 16 | 10 | - | - | 31 | 20 | 28 | 65 |
| Dysfusion | 20 | 1 | 6 | 12 | 10 | 12 | 11 | 15 | 10 | 39 | 31 | 37 | 79 |
| Tango | 20 | 2 | 6 | - | - | - | - | - | - | - | - | - | - |
| Trachealess | 20 | 8 | 4 | - | - | - | - | - | - | - | - | - | - |
| Sim2 | 20 | 0 | 6 | 8 | 7 | 5 | 3 | 15 | 13 | 28 | 23 | 25 | 82 |
| MalE | 20 | 1 | 6 | 60 | 54 | 49 | 42 | 29 | 25 | 138 | 121 | 100 | 88 |
| Hey1 | 20 | 8 | 4 | 23 | 0 | 25 | 0 | 27 | 0 | 75 | 0 | 54 | 0 |
| Side1 | 20 | 1 | 6 | 30 | 0 | 13 | 5 | 20 | 19 | 63 | 24 | 46 | 38 |
| E(spl)2 | 20 | 1 | 6 | 25 | 2 | 25 | 14 | 24 | 22 | 74 | 38 | 53 | 51 |
| Hairy | 20 | 1 | 6 | 16 | 12 | 11 | 9 | 20 | 17 | 47 | 38 | 34 | 81 |
| Side2 | 20 | 1 | 6 | 39 | 34 | 70 | 64 | 31 | 29 | 140 | 127 | 101 | 91 |
| E(spl)2 | 20 | 1 | 6 | 21 | 0 | 23 | 0 | 26 | 0 | 70 | 0 | 51 | 0 |
| Deadpan | 20 | 1 | 6 | 0 | 0 | 35 | 30 | 27 | 25 | 62 | 55 | 45 | 89 |
| Hey2 | 20 | 8 | 4 | 21 | 0 | 25 | 0 | 26 | 0 | 72 | 0 | 52 | 0 |
Fig. 1.
Effect of Knock-down in the expression of bHLH superfamily genes on the survival of female beetles. TcSRC, TcSim1, TcAsh and TcDaughterless dsRNAs were injected into the female beetles within 24 h after adult emergence and the mortality was recorded at seven days after injection of dsRNA. Mean ± SE of three experiments are shown.
3.2. Function of bHLH transcription factors in reproduction
To determine the function of bHLH transcription factors in reproduction, dsRNAs of 48 bHLH superfamily members (excluding TcSRC, TcSim1, TcDaughterless and TcAsh whose knock-down caused 90–100% mortality) were injected into the female adults within 24 h after emergence and the dsRNA injected female beetles were mated with uninjected virgin male beetles. The number of eggs laid by each pair was recorded over a three week period. Knocking-down the expression of 16 bHLH genes (TcHypoxia, TcDysfusion, TcTango, TcTrachealess, TcSim2, TcHairy, TcDeadpan, TcSpineless, TcSide1, TcMyc, TcMax, TcBeta3, TcFer2, TcHLH106, TcAP-4, and TcEmc) has affected female reproduction. As a result, the female beetles injected with dsRNA of these genes laid fewer eggs when compared to the number of eggs laid by malE dsRNA injected control beetles (Tables 2& 3). Thus, Knocking-down the expression of 16 of the 48 bHLH genes tested affected female reproduction. Knock-down in the expression of TcTango, TcTrachealess and TcEmc caused the most severe effects. The females injected with dsRNAs of these genes and mated with uninjected male beetles laid no eggs over a three week period. Knocking-down the expression of TcMyc, TcMax, TcBeta3, TcFer2, TcHLH106, TcAP-4 caused more than 75% reduction in the number of eggs laid by the beetles when compared to the eggs laid by control beetles injected with malE dsRNA (Tables 2& 3). Knocking-down the expression of TcHypoxia, TcDysfusion, TcSim2, TcHairy, TcDeadpan, TcSpineless, TcSide1, and TcAP- 4 genes caused more than 50% reduction in the number of eggs laid by these beetles when compared to the eggs laid by control beetles injected with malE dsRNA (Tables 2& 3). These data suggest that these 16 bHLH transcription factors directly or indirectly regulate female reproduction.
Table 3.
Effect of My c/USF, Hand, Shout, p48, NeuroD/Neurogenin, Atonal, AS-C, and miscellaneous family genes silencing on the reproduction of female beetles
| Name of gene | No. of injected females | No. of dead females | No. of mating | First Week | Second Week | Third Week | Total period | Total period (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | Eggs laid | Offspring produced | ||||
| MalE | 20 | 1 | 6 | 45 | 42 | 50 | 45 | 40 | 32 | 135 | 119 | 100 | 88 |
| Myc | 20 | 2 | 6 | 8 | 6 | 3 | 1 | 6 | 3 | 17 | 10 | 13 | 59 |
| Max | 20 | 4 | 5 | 4 | 0 | 4 | 4 | 3 | 3 | 4 | 2 | 3 | 50 |
| Mnt | 20 | 4 | 5 | 25 | 21 | 28 | 17 | 30 | 21 | 83 | 59 | 61 | 71 |
| USF2 | 20 | 1 | 6 | 49 | 31 | 49 | 24 | 41 | 24 | 139 | 79 | 103 | 57 |
| Flocculin-like | 20 | 3 | 5 | 42 | 37 | 35 | 30 | 28 | 20 | 105 | 87 | 78 | 83 |
| Twist | 20 | 2 | 6 | 34 | 0 | 37 | 0 | 38 | 0 | 109 | 0 | 81 | 0 |
| Hand | 20 | 3 | 5 | 32 | 0 | 41 | 0 | 31 | 0 | 104 | 0 | 77 | 0 |
| TF21 | 20 | 5 | 5 | 28 | 21 | 34 | 25 | 23 | 16 | 85 | 62 | 63 | 73 |
| Delilah1 | 20 | 1 | 6 | 52 | 45 | 54 | 42 | 43 | 34 | 149 | 121 | 110 | 81 |
| Delilah2 | 20 | 4 | 5 | 58 | 0 | 42 | 0 | 40 | 0 | 140 | 0 | 104 | 0 |
| Shout | 20 | 2 | 6 | 42 | 22 | 52 | 35 | 54 | 40 | 148 | 97 | 110 | 66 |
| Fer1 | 20 | 1 | 6 | 25 | 5 | 48 | 15 | 51 | 15 | 124 | 35 | 92 | 28 |
| Fer2 | 20 | 0 | 6 | 5 | 2 | 8 | 4 | 9 | 5 | 22 | 11 | 21 | 50 |
| Fer3 | 20 | 4 | 5 | 35 | 0 | 52 | 0 | 42 | 0 | 129 | 0 | 96 | 0 |
| Scute | 20 | 1 | 6 | 45 | 24 | 54 | 27 | 51 | 32 | 150 | 83 | 111 | 55 |
| Asense | 20 | 2 | 6 | 42 | 21 | 39 | 25 | 41 | 21 | 122 | 67 | 90 | 55 |
| Ash | 20 | 20 | 6 | - | - | - | - | - | - | - | - | - | - |
| Dimmed | 20 | 4 | 5 | 33 | 0 | 29 | 0 | 41 | 0 | 103 | 0 | 76 | 0 |
| Tap | 20 | 1 | 6 | 29 | 0 | 50 | 2 | 45 | 0 | 124 | 2 | 92 | 2 |
| NeuroD | 20 | 4 | 5 | 29 | 20 | 35 | 25 | 44 | 30 | 108 | 75 | 80 | 70 |
| Beta3 | 20 | 1 | 6 | 9 | 2 | 5 | 4 | 4 | 0 | 18 | 6 | 13 | 33 |
| Cato | 20 | 1 | 6 | 32 | 0 | 40 | 0 | 51 | 0 | 140 | 0 | 104 | 0 |
| Atonal | 20 | 1 | 6 | 21 | 18 | 33 | 22 | 42 | 22 | 96 | 62 | 71 | 65 |
| Amos | 20 | 1 | 6 | 42 | 21 | 55 | 30 | 52 | 25 | 149 | 76 | 110 | 51 |
| HLH106 | 20 | 2 | 6 | 5 | 1 | 0 | 0 | 2 | 0 | 7 | 1 | 5 | 14 |
| Bigmax | 20 | 4 | 5 | 38 | 27 | 35 | 29 | 40 | 31 | 113 | 87 | 84 | 77 |
| AP-4 | 20 | 4 | 5 | 5 | 0 | 7 | 0 | 5 | 0 | 17 | 0 | 13 | 0 |
| HLH3B | 20 | 1 | 6 | 41 | 35 | 42 | 25 | 36 | 28 | 119 | 88 | 88 | 74 |
| Paraxis | 20 | 0 | 6 | 39 | 25 | 32 | 25 | 35 | 24 | 106 | 87 | 79 | 82 |
| HLH4C | 20 | 4 | 5 | 45 | 32 | 46 | 32 | 37 | 26 | 128 | 90 | 95 | 70 |
| Nautilus | 20 | 1 | 6 | 32 | 20 | 42 | 31 | 35 | 29 | 109 | 80 | 81 | 73 |
| Daughterless | 20 | 20 | 20 | - | - | - | - | - | - | - | - | - | - |
| Emc | 20 | 2 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mitf | 20 | 2 | 6 | 28 | 19 | 35 | 28 | 35 | 25 | 98 | 72 | 73 | 73 |
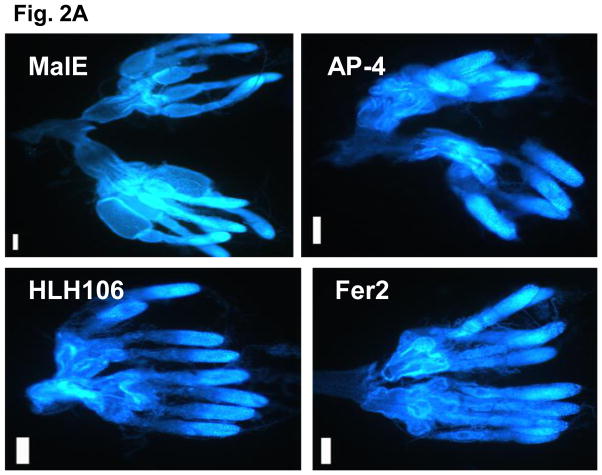
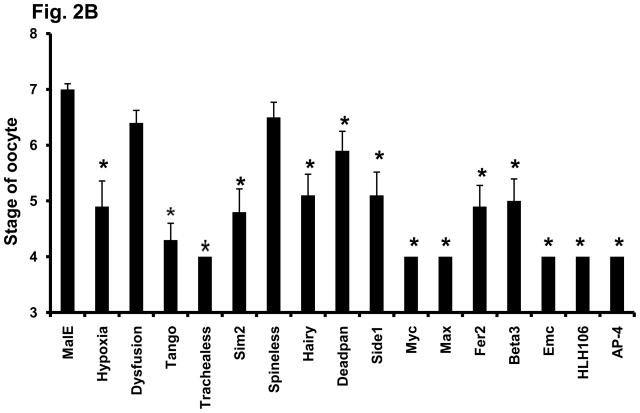
To determine whether any of these 16 bHLH transcription factors influence egg laying by affecting oocyte maturation, the ovaries from female beetles injected with dsRNA of these 16 bHLH genes were dissected on the 5th day after adult emergence and examined under a microscope. The oocytes were scored based on a scale of 1–9 as described in the Materials and Methods section. In the females injected with malE dsRNA, the primary oocyte matured by five days after adult emergence (Fig. 2A). Therefore, all the dsRNA injected females were dissected on 5th day after adult emergence. Knocking-down the expression of TcTrachealess, TcMyc, TcMax, TcEmc, TcHLH106 and TcAP-4 genes caused the most severe effect on oocyte maturation. The oocytes in ovaries dissected from the beetles injected with dsRNA of these genes were at stages between 3 and 4 (Figs. 2A&B). In contrast, the oocytes in ovaries dissected from control beetles injected with malE dsRNA were fully developed and scored at stage 7 (Figs. 2A&B). Knocking-down the expression of TcHypoxia, TcTango, TcSim2, TcHairy, TcSide 1, TcFer2 and TcBeta3 caused moderate effects on oocytes and therefore, the oocytes in ovaries dissected from beetles injected with dsRNA for these genes were scored between stages 4 and 5 (Fig. 2B). Knocking-down the expression of two bHLH genes, TcDysfusion and TcSpineless, did not cause significant effect on oocyte maturation as the oocytes in ovaries dissected from beetles injected with dsRNA for these genes were not significantly different from those in control insects. The oocyte development for these genes was scored between stages six and seven (Fig. 2B).
Fig. 2.
(A) Nuclear staining of female reproductive system of T. castaneum. Ovaries were dissected from the female beetles and stained by DAPI at 5 days after injection of malE, TcAP-4, TcHLH106 and TcFer2 dsRNA. Scale bar 100 μm.
Fig. 2. (B) Effect of Knock-down in the expression of bHLH superfamily genes on the development of oocytes in the female beetles. TcHypoxia, TcDysfusion, TcTango, TcTrachealess, TcSim2, TcHairy, TcDeadpan, TcSpineless, TcSide1, TcMyc, TcMax, TcFer2, TcBeta3, Tc Emc, TcHLH106 and TcAP-4 gene dsRNAs were injected into female beetles with in 24 h after emergence. Ovaries were dissected and scored at 5 days after injection of dsRNA. The ooctye stages 1–7 were determined as described in the Materials and Methods section. Mean score ± SE of ten individual beetles are shown. Paired T-test with unequal mean of variances was performed at P ≤ 0.05.
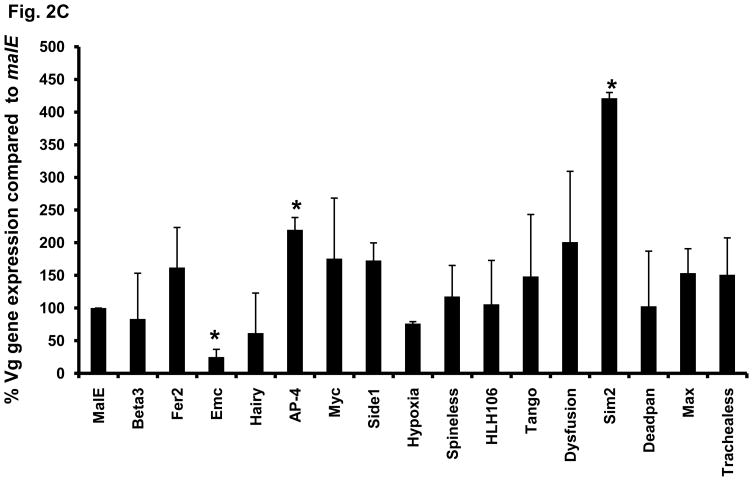
Fig. 2. (C) Effect of knock-down in the expression of bHLH superfamily genes on the Vg gene expression in the females injected with dsRNA. DsRNAs of MalE, TcBeta3, TcFer2, TcEmc, TcHairy, TcAP-4, TcMyc, TcSide1, TcHypoxia, TcSpineless, TcHLH106, TcTango, TcDysfusion, TcSim2, TcDeadpan, TcMax, TcTrachealess were injected into the females within 24h after adult emergence. At 5 days after injection, the RNAi females were used to quantify the Vg mRNA levels. Relative expression values for all the genes were calculated by keeping malE as 100%. Mean ± SE (n=3) are shown. Paired T-test with unequal mean of variances was performed at P ≤ 0.05.
To determine whether any of these 16 bHLH transcription factors influence egg laying by affecting the Vg synthesis, the Vg mRNA levels were quantified in female beetles injected with dsRNA of these genes at five days after injection. Only, three out of 16 genes tested caused significant effect on Vg synthesis. Injection of TcEmc dsRNA caused a decrease in Vg gene expression when compared to its expression in the control insects injected with malE dsRNA (Fig. 2C). In contrast, Injection of TcSim2 or TcAP-4 dsRNA caused an increase in Vg gene expression when compared to its expression in control insects injected with malE dsRNA (Fig. 2C). These data show that the majority of bHLH transcription factors effect female reproduction by regulating oogenesis.
3.4. Functions of bHLH transcription factors in embryogenesis
To determine the function of bHLH transcription factors in embryogenesis, 48 bHLH superfamily members (excluding TcSRC, TcSim1, TcDaughterless and TcAsh whose knockdown caused 90–100% mortality) dsRNAs were injected into the female adults within 24 h after emergence and the dsRNA injected female beetles were mated with uninjected virgin male beetles. The number of larvae hatched from the eggs laid was recorded over a three week period. Knocking-down the expression of 13 bHLH genes (TcSpineless, TcSide1, TcHey1, TcHey2, TcE(spl)2, TcTwist, TcHand, TcDelilah2, TcFer1, TcFer3, TcDimmed, TcTap, and TcCato) affected embryogenesis. As a result, fewer eggs laid by the females injected with dsRNA for these genes developed into larvae. Two bHLH genes TcSide1 and TcSpineless affected both the oogenesis and embryogenesis (Tables 2&3).
Knocking-down the expression of TcTango, TcTrachealess, TcEmc TcMyc, TcMax, TcFer2, TcBeta3, TcHLH106 and TcAP-4 caused severe reduction in eggs (less than 25 eggs over a three week period) laid by RNAi beetles (Tables 2&3) and therefore there were not enough eggs to study their effect on embryogenesis. The eggs laid by females injected with dsRNA for the rest of 39 bHLH genes and malE were monitored over a three week period and the number of larvae hatched from these eggs was recorded. Knocking-down the expression of 13 out of the 39 bHLH genes studied caused significant reduction in the percent eggs hatched when compared to the percent hatch observed in eggs laid by control beetles injected with malE dsRNA (Tables 2&3). Knocking-down the expression of TcHey1, TcHey2, TcE(spl)mgamma2, TcTwist, TcHand, TcDelilah 2, TcFer1, TcFer3, TcDimmed, TcTap and TcCato caused severe effects on the egg hatch and none of the eggs laid by the female beetles injected with dsRNA of these genes were able to develop into larvae (Tables 2&3). Knocking-down the expression of the other three bHLH genes, TcSpineless, TcSide1 and TcFer1 caused 60–72% reduction in percent egg hatch.
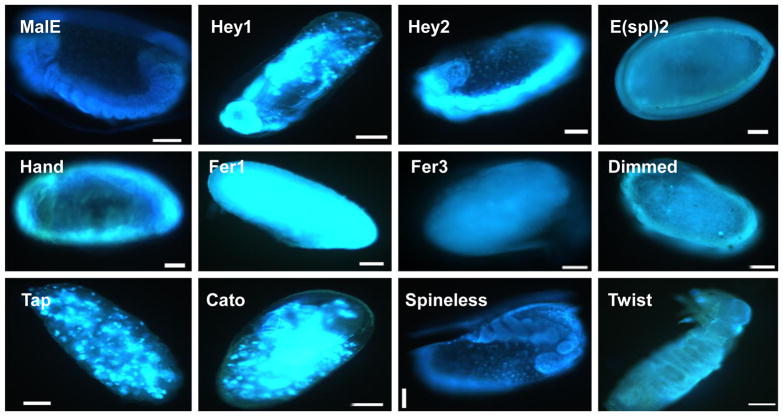
T. castaneum embryonic development is completed in 5–6 days after egg laying depending on the incubation temperature. To determine the stages at which the embryonic development was blocked due to the lack of bHLH transcription factors, the eggs were collected within 24 h of egg laying and on the fourth day after collection, then the embryos were fixed and stained using DAPI. Most of the eggs laid by control beetles injected with malE dsRNA successfully completed embryonic development (Fig. 3). The embryos in the eggs laid by TcHey1 dsRNA injected beetles were arrested during the mid-stage of germ band growth (Fig. 3). The embryos in the eggs laid by TcHey2 and TcSpineless dsRNA injected beetles were blocked near the stage of germ band growth completion (Fig. 3). The embryos in the eggs laid by TcE(spl)mgamma2 and TcDimmed dsRNA injected beetles were arrested at the beginning of the cellular blastoderm stage (Fig. 3). The embryos in the eggs laid by beetles injected with dsRNA of TcFer1 and TcFer3 belonging to the p48 family were arrested at the early blastoderm stage (Fig. 3). The embryos in the eggs laid by beetles injected with TcHand dsRNA were arrested at the formation of the germ band during the gastrulation stage. Most of the eggs laid by the females injected with TcTap and TcCato dsRNA were arrested at the beginning of germ band growth (Fig. 3). Embryogenesis was completed in the eggs laid by TcTwist or TcDelilah2 dsRNA injected beetles, but the larvae were unable to hatch (Fig. 3).
Fig. 3.
Nuclear staining of embryos of T. castaneum. Eggs were collected within 24 h after egg laying and stained after 5 days of egg laying to observe the stage of arrest of embryogenesis. Scale bar 100 μm.
4. Discussion
The major contribution of this study is the identification of 31 members of bHLH transcription factor superfamily that play key roles in reproduction, embryogenesis and survival of the adult female beetles of T. castaneum. The function of some of the bHLH transcription factors in reproduction and embryogenesis of Drosophila melanogaster has been previously reported. The D. melanogaster Spineless (Burgess and Duncan, 1990), Hypoxia (Smulders-Srinivasan and Lin, 2003), Sim2 (Mayer and Nusslein-Volhard, 1988), Hey1 and Hey2 (Leimeister et al., 1999), Myc (Gallant et al., 1996), Delilah (Armand et al., 1994), Hand (Han et al., 2006), Twist (Franch-Marro and Casanova, 2000), Beta3 (Kimble et al., 1989), Tap (Bush et al., 1996), Dimmed (Hewes et al., 2003), Cato (Goulding et al., 2000), Emc (Adam and Montell, 2004) play important roles in regulation of reproduction and embryogenesis. The T. castaneum homologues of these genes showed more or less similar effects described in D. melanogaster. This study identified six additional bHLH transcription factors, TcHLH106, TcAP-4 (miscellaneous group), TcMax (Myc/USF2 family), TcFer1, TcFer2 and TcFer3 (p48 family) that play key roles in reproduction and embryogenesis.
Four bHLH transcription factors, TcSim1, TcSRC, TcAsh and TcDaughterless, play key roles in the regulation of vital functions in adults and therefore are required for the survival of female beetles (Fig. 1). bHLH-PAS family transcription factor Sim was shown to play key roles in the specification of the ventral midline and the development of the nervous system in D. melanogaster (Thomas et al., 1988). Two Sim homologues have been identified in T. castaneum (Bitra et al., 2009) and they have been named as TcSim1 and TcSim2. In D. melanogaster only one Sim homolog has been identified. RNAi and misexpression studies in T. castaneum showed that TcAsh is necessary for neural precursor formation (Wheeler et al., 2003). As these two transcription factors, Sim1 and Ash, are known to be involved in the development of nervous system, knocking-down the expression of these two genes may have affected the nervous system function leading to the death of the beetles. TcSim2 affected the egg laying by female beetles when the expression of the gene coding for this transcription factor was knocked-down using RNAi methods. Previous studies have shown that DmSim is also required for normal development of the female germ line (Mayer and Nusslein-Volhard, 1988). The daughterless gene was shown to play an important role in sex determination and neurogenesis in D. melanogaster (Murre et al., 1989) and knocking-down the expression of this gene during the adult stage would have affected nervous system function, leading to the death of the adult beetles. Taken together these data suggest that the bHLH transcription factors, TcSim1, TcAsh and TcDaughterless, regulate the nervous system function and the presence of these transcription factors is critical for the survival of adult insects. TcSim1 knock-down did not have much effect on larval growth and development (Bitra et al., 2009), but the knock-down in the expression of TcAsh and TcDaughterless caused severe problems and death of most of the dsRNA injected larvae (Bitra et al., 2009).
TcSRC dsRNA injected female beetles showed 90% mortality within one week after injection and a few female beetles that lived showed immature ovaries and failed to lay eggs (data not shown). Previous studies that used SRC-2 knockout mice showed that this gene is required for female fertility and mammary morphogenesis (Mukherjee et al., 2007). Recently, we showed that knock-down in expression of TcSRC during the larval stage caused growth arrest by affecting the regulation of lipid metabolism (Bitra et al., 2009). Therefore, it is likely that TcSRC is required for lipid metabolism and other critical functions during the adult stages as well. This is another bHLH transcription factor with great potential for development of pest management methods.
Knocking-down the expression of TcHLH106, TcAP-4, TcMax and TcFer2 in the adult female beetles greatly reduced egg laying capacity of female beetles. The DmHLH106 gene was shown to play a key role in the fatty acid synthesis and is similar to the vertebrate sterol regulatory binding protein (SREBP) (Kunte et al., 2006). Knocking-down the expression of the TcHLH106 gene may have blocked the synthesis of sterol and fatty acids resulting in female sterility. Recent studies in our laboratory suggest that nutrition plays an important role in oocyte development and maturation (data not shown). The TcHLH106 transcription factor might have indirectly affected oocyte development and maturation by affecting the synthesis of nutrients required for oocyte development and maturation.
The dMyc mutant fruit flies are sterile as a result of defective oogenesis and the defects are observed in both germ cells and follicle cells (Gallant et al., 1996). TcMyc dsRNA injected females failed to lay eggs and the development of the oocyte was blocked at stage 4 (Fig. 2B). TcMax dsRNA injected females also showed similar phenotypes as TcMyc RNAi insects. Myc and Max proteins function as a network leading to alternative states of cell activation or quiescence (Amati and Land, 1994). TcAP-4 gene severely affected the egg laying of female beetles when its expression was knocked-down in female beetles (Fig. 2A) due to block in oocyte maturation (Fig. 2B). AP-4 gene has been shown to interact with distal region of insulin-like growth factor binding protein 2 (IGFBP-2) and may affect cell proliferation and differentiation via induction of IGFBP-2 synthesis in humans (Badinga et al., 1998). As AP-4 gene is shown to interact with IGFBP-2 and the T. castaneum genome contains IGFBP-2 homologue, knock-down in the expression of TcAP-4 gene would have affected the cell proliferation and differentiation, indirectly affecting the oocyte maturation.
The TcFer2 gene also affected egg laying ability of females when its expression was knocked-down. In D. melanogaster Fer1, Fer2 and Fer3 genes have been identified by Moore et al., 2000. These genes are similar to the pancreas transcription factor (Ptf) in mouse (Krapp et al., 1996). In the developing pancreas, the bHLH gene Ptf1a, promotes exocrine cell differentiation (Krapp et al., 1998) during vertebrate embryogenesis. The Hes1 gene belonging to the HES family represses Ptf1a expression by directly binding to the promoter of this gene in vertebrates. The Fer1 gene in D. melanogaster is expressed in the epidermis at the time of secretion of cuticle and shares a common function with the p48 in active exocrine cells of pancreas (Moore et al., 2000). DmFer2 shows a strong expression in the early embryo, whereas DmFer3 is highly expressed during the later stages of embryonic development (Moore et al., 2000).
Knock-down in the expression of TcFer1 and TcFer3 genes in the adult female beetles caused a severe effect on the embryogenesis in the eggs laid by the female beetles (Fig. 3). The p48 family genes play a key role in the secretion of enzymes involved in the digestion of food; as a result, knocking-down the expression of these genes would have affected the food metabolism, indirectly affecting the oogenesis. These genes are also highly expressed during the embryogenesis, as a result affecting the embryogenesis in RNAi beetles. HES family genes TcHey1 and TcHey2 showed a severe effect on the embryogenesis. Hey1 embryos stained with DAPI showed that most of the embryos were blocked at mid-stage of germ band growth and Hey2 embryos were blocked at completion of germband growth. These two genes in D. melanogaster were shown to play key roles during the later stages of embryonic development with high level of expression in the heart, craniofacial region and the nervous system (Leimeister et al., 1999). Based on the studies described above from D. melanogaster and our data, it is likely that most of the bHLH transcription factors identified in this study as those that are required for embryogenesis may affect embryonic development directly. However, the possibility of these gene products indirectly affecting embryonic development through their action during oogenesis could not be excluded. Further detailed examination of RNAi effects of each gene after injection of dsRNA into eggs need to be done to answer this question.
It is interesting that all the bHLH transcription factors (except TcSpineless) that are required for egg production are also required for oogensis. In contrast, only three of these transcription factors appear to regulate Vg gene expression. These data suggest that the major role of bHLH transcription factors in female reproduction is regulation of oogenesis. It is also interesting that only two bHLH transcription factors are required for both egg production and embryogenesis. The rest of the 11 bHLH transcription factors that are required for embryogenesis did not influence egg production when the expression of genes coding for these proteins were silenced by RNAi. These data suggest that most of these 11 bHLH transcription factors regulate embryogenesis.
In summary, 31 bHLH superfamily transcription factors that play key roles in female reproduction and survival of adult T. castaneum beetles were identified. The 31 bHLH transcription factors that affected adult survival and reproduction belong to bHLH-PAS (eight), HES (six), NeuroD/Neurogenin (three), p48 (three), Myc/USF2 (two), Hand (two), Shout (one), AS-C (one), Atonal (one), Miscellaneous (four) families. Out of the 11 members in bHLH-PAS family, eight of them affected adult survival or reproduction. Similarly, out of the eight members of the HES family, six affected embryogenesis. This study identified 31 out of 52 bHLH transcription factors tested as those that are required for female survival reproduction and embryogenesis. The rest of the 21 bHLH transcription factors that did not show phenotypes in our RNAi analyses may or may not regulate these processes. Since, we were not able to achieve 100% knock-down in the expression of genes coding for these 21 transcription factors, we cannot rule out the possibility that some of these transcription factors may regulate reproduction or embryogenesis. Further detailed studies on these 21 transcription factors will help in defining their function in regulation of reproduction and/or embryogenesis. This study provided the first over view on the function of 31 bHLH transcription factors in the female insects. Further detailed studies are required to determine the mechanism of action of these transcription factors in survival, reproduction and embryogenesis.
Acknowledgments
We dedicate this paper to Professor Judy Willis in gratitude for the guidance she provided throughout the career development of one of the authors (SRP). This work was supported by National Institutes of Health (GM070559-06) and National Science Foundation (IBN-0421856). This is contribution number 10-08-045 from the Kentucky Agricultural Experimental Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam JC, Montell DJ. A role for extra macrochaetae downstream of Notch in follicle cell differentiation. Development. 2004;131:5971–5980. doi: 10.1242/dev.01442. [DOI] [PubMed] [Google Scholar]
- Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Current Opinion in Genetics and Development. 1994;1:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Armand P, Knapp AC, Hirsch AJ, Wieschaus EF, Cole MD. A novel basic helix-loop-helix proteins expressed in muscle attachment sites of the Drosophila epidermis. Molecular and Cellular Biology. 1994;6:4145–4154. doi: 10.1128/mcb.14.6.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badinga L, Song S, Simmen RC, Simmen FA. A distal regulatory region of the insulin-like growth factor binding protein-2 (IGFBP-2) gene interacts with the basic helix-loop-helix transcription factor. Endocrine. 1998;8:281–289. doi: 10.1385/ENDO:8:3:281. [DOI] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a D. melanogaster protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Belles X. Vitellogenesis directed by juvenile hormone. In: Raikhel AS, Sappington TW, editors. Reproductive Biology of Invertebrates, Vol. 12, Part B: Progress in Vitellogenesis. Science Publishers; Enfield, USA/Plymouth, UK: 2004. pp. 157–198. [Google Scholar]
- Bitra K, Tan A, Dowling A, Palli SR. Functional characterization of PAS and HES family bHLH transcription factors during the metamorphosis of the red flour beetle, Tribolium castaneum. Gene. 2009;448:74–87. doi: 10.1016/j.gene.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess EA, Duncan I. Direct control of antennal identity by the spineless- aristapedia gene of Drosophila. Molecular and General Genetics. 1990;221:347–357. doi: 10.1007/BF00259398. [DOI] [PubMed] [Google Scholar]
- Bush A, Cole Y, Cole M. biparous: a novel bHLH gene expressed in neuronal and glial precursors in Drosophila. Developmental Biology. 1996;180:759–772. doi: 10.1006/dbio.1996.0344. [DOI] [PubMed] [Google Scholar]
- Dang CV, Dolde D, Gillison ML, Kato GJ. Discrimination between related DNA sites by a single amino acid residue of myc-related basic-helix-loop-helix proteins. Proceedings of the National Academy of Sciences. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englemann F. Vitellogenesis controlled by juvenile hormone. In: Downer RGH, Laufer H, editors. Endocrinology of Insects. Alan R. Liss; New York: 1983. pp. 259–270. [Google Scholar]
- Ferre-D’ Amar AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric B/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Franch-Marro X, Casanova J. The alternative migratory pathways of the Drosophila tracheal cells are associated with distinct subsets of mesodermal cells. Developmental Biology. 2000;227:80–90. doi: 10.1006/dbio.2000.9890. [DOI] [PubMed] [Google Scholar]
- Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- Goulding SE, White NM, Jarman AP. cato encodes a basic helix-loop-helix transcription factor implicated in the correct differentiation of Drosophila sense organs. Developmental Biology. 2000;221:120–131. doi: 10.1006/dbio.2000.9677. [DOI] [PubMed] [Google Scholar]
- Hagedorn HH, Fallon AM. Ovarian control of vitellogenin synthesis by the fat body in Aedes aegypti. Nature. 1973;244:103–105. doi: 10.1038/244103a0. [DOI] [PubMed] [Google Scholar]
- Han Z, Yi P, Li X, Olson EN. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development. 2006;133:1175–1182. doi: 10.1242/dev.02285. [DOI] [PubMed] [Google Scholar]
- Hewes RS, Park D, Gauthier SA, Schaefer AM, Taghert PH. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development. 2003;130:1771–1781. doi: 10.1242/dev.00404. [DOI] [PubMed] [Google Scholar]
- Kadesh T. Consequences of heterodimeric interactions among helix-loop-helix proteins. Cell Growth and Differentiation. 1993;4:49–55. [PubMed] [Google Scholar]
- Kimble M, Incardona J, Raff EC. A variant-tubulin isoform of Drosophila melanogaster (−3) is expressed primarily in tissues of mesodermal origin in embryos and pupae, and is utilized in populations of transient microtubules. Developmental Biology. 1989;131:415–429. doi: 10.1016/s0012-1606(89)80014-4. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO Journal. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knöfler M, Ledermann B, Bürki K, Berney C, Zoerkler N, Hagenbüchle O, Wellauer PK. The bHLH protain PTF-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes and Development. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in D. melanogaster larvae lacking SREBP. Cell Metabolism. 2006;6:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Enternbrink A, Klamt B, Gessier M. Hey genes: a novel subfamily of hairy and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mechanisms of Development. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Massari M, Murre C. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Molecular Cellular Biology. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Nusslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes and Development. 1988;2:1496–511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in D. melanogaster. Proceedings of the National Academy of Sciences USA. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Amato P, Allred DC, DeMayo FJ, Lydon JP. Steroid receptor coactivator 2 is required for female fertility and mammary morphogenesis: insights from the mouse, relevance to the human. Nuclear Receptor Signaling. 2007;5:e011. doi: 10.1621/nrs.05011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Mc Caw PS, Vaessin H, Caudy M, Jan LY, Cabrera CV, Buskin JN, Hauschka SD, Lasssar AB, Weintraub H. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Tan A, Bai H, Palli SR. Transcription factor broad suppres Precocious development of adult structures during larval-pupal metamorphosis in the red flour beetle, Tribolium castaneum. Mechanisms of Development. 2008;125:299–313. doi: 10.1016/j.mod.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel AS, Kokoza VA, Zhu J, Martin D, Wang SF, Li C, Sun G, Ahmed A, Dittmer N, Attardo G. Molecular biology of mosquito vitellogenesis: from basic studies to genetic engineering of antipathogen immunity. Insect Biochemistry and Molecular Bioliology. 2002;32:1275–1286. doi: 10.1016/s0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- Smulders-Srinivasan TK, Lin H. Screens for piwi suppressors in Drosophila identify dosage-dependent regulators of germline stem cell division. Genetics. 2003;165:1971–1991. doi: 10.1093/genetics/165.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;7190:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Ullmann SL. Oogenesis in Tenebrio molitor: Histological and autoradio-graphical observations on pupal and adult ovaries. J Embryol Exp Morph. 1973;30:179–217. [PubMed] [Google Scholar]
- Van Dorren M, Ellis HM, Posakony JW. The Drosophila Extramacrochaete Protein antagonizes sequence-specific DNA binding by Daughterless/Achaete-Scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen K, Yao Q, Wang W. The basic helix-loop-helix transcription factor family in Bombyx mori. Development Genes and Evolution. 2007;217:715–723. doi: 10.1007/s00427-007-0184-x. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Carrico ML, Wilson BA, Brown SJ, Skeath JB. The expression and function of the achaete-scute genes in Tribolium castaneum reveals conservation and variation in neural pattern formation and cell fate specification. Development. 2003;30:4373–4381. doi: 10.1242/dev.00646. [DOI] [PubMed] [Google Scholar]
- Wyatt GR, Davey KG. Cellular and molecular actions of juvenile hormone. II. Roles of juvenile hormone in adult insects. Advances in Insect Physiology. 1996;26:1–155. [Google Scholar]