Abstract
LINE-1 (L1) elements are retrotransposons that insert extra copies of themselves throughout the genome using a “copy and paste” mechanism. L1s have contributed ~20% to total human genome content and are able to influence chromosome integrity and gene expression upon reinsertion. Recent studies show that L1 elements are active and “jumping” during neuronal differentiation. New somatic L1 insertions may generate “genomic plasticity” in neurons by causing variation in genomic DNA sequences and by altering the transcriptome of individual cells. Thus, L1-induced variation may affect neuronal plasticity and behavior. Here, we discuss potential consequences of L1-induced neuronal diversity and propose that a mechanism generating diversity in the brain could broaden the spectrum of behavioral phenotypes that can originate from any single genome.
Introduction
When Barbara McClintock discovered transposons in 1948, she observed color variegation in maize kernels and leaves. Through cytological and genetic studies, she came to the conclusion that mobile DNA led to somatic mosaicism and phenotypic diversity in the affected tissues [1]. McClintock realized that, in addition to causing chromosome breakage and acting as insertional mutagens, transposons might also act as “controlling elements” and exert regulatory control over genes in their proximity. While the notion that DNA could be mobile was accepted, the idea of control was not [2].
McClintock favored the view that mobile DNA might play an important role in gene regulation, but conventional wisdom argued that transposons were “selfish” parasitic sequences that invaded host genomes and did more harm than good. In recent years, the view that not all transpositional events are detrimental has gained acceptance. Host genomes have evolved mechanisms to harness the unique properties of transposable elements to their own benefit. Transposed elements (elements that have been rendered incapable of transposition through mutation), and retroelements (elements that transpose via an RNA intermediate) in particular, have played important roles in mammalian genome evolution and in the generation of new human-specific genes [3–5]: transposed elements support genome integrity as part of centromeres and telomeres, impact the transcriptome and contribute to tissue-specific gene expression [6–8].
Recent evidence from our laboratory and others suggests that, within an individual, neuronal genomes are genetically diverse and that brains are somatic mosaics [9, 10]. Neuronal genetic diversity results from aneuploidy (whole chromosome gains and losses) [11], genomic copy number variations (CNVs) [12], and actively “jumping” transposable elements, termed long interspersed repeated sequences (LINE-1 or L1 elements) (Figure 1A) [9, 13].
Figure 1. Background on L1 retrotransposition.
A| Composition of L1 and dependent sequences. L1 elements belong to the long interspersed element (LINEs) class of repeat sequences and are the only active class of retroelements in the human genome. Full-length, functional L1s are ~6kb long and are autonomous because they encode proteins necessary for their retrotransposition. L1 elements contain a polymerase II promoter in their 5′UTR (blue band, with arrow), followed by two open reading frames (ORF1 and ORF2) and a short 3′-UTR that ends in a poly(A) tail. The poly(A) tail is preceded by a polyadenylation signal (pA). ORF1 encodes a nucleic acid chaperone [75]; ORF2 encodes an endonuclease and reverse transcriptase [76]. Both of the element-encoded proteins are essential for retrotransposition.
Another class of repeats in mammalian genomes are short interspersed elements (SINEs). SINEs are sequences that do not encode for their own proteins necessary for transposition but can be mobilized by L1- encoded proteins and are therefore non-autonomous. SINEs that can be transactivated by L1 are Alu and SVA elements. SVA elements are ~3kb long, do not contain a transcriptional promoter sequence and consist of several components (SINE-R, VNTR, Alu), including a hexanucleotide repeat sequence at their 5′ end (CCCTCTn) and a poly(A) tail at their 3′ end; (VNTR, variable number of tandem repeats). Alu elements are short ~300bp long sequences that contain two polymerase III promoter regions (blue bands, with arrows) and consist of two related monomers: left monomer (LM) and right monomer (RM). Both monomers are separated by an A-rich linker region (An). Alu elements usually end in a long poly(A) tract. A hallmark of all novel L1-mediated retrotransposon insertions is that they are flanked by target site duplications (TSDs, black arrows) of varying lengths.
Processed pseudogenes are generated from spliced mRNAs that have hijacked the L1 machinery, were reverse transcribed, and inserted into the genome. They consist of intron-less exonic sequences and typically do not include a promoter sequence. If inserted near a regulatory region they can become expressed and exert regulatory functions [77, 78].
B| L1 retrotransposition cycle. L1 transcription is repressed by methylation (Me) of the CpG island in its promoter. Once transcribed, L1 mRNA (blue line) is exported from the nucleus and translated in the cytoplasm, where it assembles with its own encoded proteins, which show a strong cis-preference and forms ribonucleoprotein complexes (RNPs). RNP complexes are re-imported into the nucleus, probably due to nuclear envelope breakdown during mitosis. L1 RNA is reinserted in the genome by a proposed mechanism referred to as target site primed reverse transcription (TPRT), whereby the endonuclease makes a single-stranded nick in the host DNA (consensus sequence: 5′-TTTT/AA-3′) [27] and the reverse transcriptase uses the nicked DNA to prime reverse transcription from the 3′ end of the L1 RNA. Frequently, reverse transcription fails to proceed to the 5′ end, resulting in truncated de novo insertions that are unable to jump again. L1 mRNA and proteins can be sequestered into stress granules (SGs) [79]. SGs are discrete cytoplasmic aggregates that are induced by a range of stress conditions, including hypoxia, heat shock and oxidative stress. Two possible functions of SGs in the context of L1 biogenesis have been discussed: L1-RNPs could be either targeted for degradation or return from SGs to the polysome-associated cytoplasmic pool of translated mRNAs [80], which upon re-entry into the nucleus would lead to new L1 insertions in the genomic DNA. In addition to L1, Alus, SVAs or cellular mRNAs can be “hijacked” by L1 proteins, thus facilitating their retrotransposition into the genome and possibly generating additional variation.
C| L1 regulation during neurogenesis. L1 transcription is repressed in neural stem cells (NSCs) by a complex comprised of the transcription factor SOX2 and histone deacetylase 1 (HDAC1). SOX2 binds to a bipartite sequence motif that also contains a LEF binding site. Derepression of the SOX2/HDAC1 complex occurs during the transition from NSCs to neural progenitor cells (NPCs); subsequently L1 expression is induced by Wnt signaling: WNT3A activates β–catenin, which then accumulates in the nucleus where it forms an activating complex with TCF/LEF. The TCF/LEF complex can now bind to the LEF binding site that has been vacated by SOX2 leading to the transcriptional activation (bent black arrow) and jumping (curved red arrows) of L1 elements.
Here, we discuss the intriguing hypothesis that L1-mediated retrotransposition provides a means for generating neuronal DNA sequence diversity [see Box 1] that could have an impact on the transcriptome and ultimately neuronal phenotypes. We propose that events related to L1 transposition are sources of genomic diversity within an individual mammalian brain that parallel conventional mechanisms generating genetic diversity in natural populations. This mechanism could account for the range of individual differences in behavior observed in isogenic animals that are presumed to be genetically identical, and it could underlie phenotypic discordance in monozygotic twins. In addition to germline mutations, the propensity to generate variability within individuals would provide another source of variation that could affect fitness and thereby be susceptible to natural selection.
Box 1. Genetic diversity among neurons is not a new idea.
A striking attribute of nervous systems is diversity. Neuronal diversity is seen in the remarkable array of morphological types and in the vast number of neuronal connections. For instance, it is estimated that the mouse brain contains ~75 million neurons, and the human brain has ~85-100 billion neurons [64, 65]. The total number of synapses in the human neocortex is estimated to be 0.15 quadrillion (1015) and a typical neuron is predicted to have ~5,000–200,000 synapses [65].
One hypothesis to explain how the vast extent of neuronal diversity in the human brain can be encoded by only ~30,000 genes was derived from analogy to the immune system [66], where extraordinary cellular diversity is the direct result of somatic DNA rearrangement. Evidence to support the hypothesis that a similar mechanism may occur during neurogenesis came from the observation that one component of the immune system recombinase (RAG-1) is expressed in cortical and hippocampal NPCs during mouse neurodevelopment [67]; however, the use of transgenic mice with rearrangement reporter genes failed to detect recombinase activity in the brain [68, 69].
Almost a decade later, the observation that nonhomologous end-joining (NHEJ) DNA repair proteins were required for NPC survival [70] and the description of candidate loci for rearrangement in neurons renewed the hypothesis that V(D)J-like recombination might occur during neurogenesis. Other mechanisms, such as stochastic promoter choice in the case of olfactory receptors [71] and trans-splicing of protocadherin transcripts [72], are now known to mediate diversity from these loci; yet a role for NHEJ and associated DNA damage signaling in generating genetic diversity in neurons cannot be ruled out [73, 74].
Somatic L1 retrotransposition occurs during neurogenesis and leads to somatic mosaicism
L1 elements are retrotransposons that are the most widespread class of transposons in mammals and constitute almost ~20% of mammalian genomic DNA content [14]. Whereas most L1 sequences in the genome are retrotransposition incompetent, ~150 full-length L1 elements are present in the human genome and ~3000 in the mouse genome [15, 16].
Retrotransposition by L1 occurs via a copy-and-paste mechanism (Figure 1B), in contrast to the cut-and-paste mechanism of DNA transposons. Most novel retrotransposition events result in truncated L1 copies that are no longer capable of jumping. As a consequence of this mechanism, retrotransposition leads to an increase in genomic DNA content. When L1 retrotransposition occurs in individual cells or cell lineages during development, it results in distinct genomes, here referred to as somatic mosaicism.
L1 elements are mobilized early in development during the formation of the central nervous system (CNS) and later during adult neurogenesis. Since this mobilization process appears to occur frequently and independently in individual cells, the result is a substantial number of newly transposed L1 elements in differentiated neurons. This neuron-to-neuron variation in genomic DNA content has been observed in both rodents and humans [9, 13, 17].
The idea that L1 retroelements may be involved in neural development first came from microarray transcriptome analyses that showed upregulation of L1 expression during differentiation of rat hippocampal neural stem cells (NSCs) [13]. Further experiments in cell culture corroborated the microarray results [18]. In this approach, a fluorescently-tagged L1 element was introduced into NSCs, and fluorescence was only observed after the L1 element had successfully retrotransposed into a new genomic location. These experiments revealed that L1 retrotransposed in NSCs at low but detectable frequency, but a strong increase of L1 retrotransposition was observed during a 48h time period after the onset of neuronal differentiation, consistent with an increase of L1 promoter activity (Figure 1C) [9, 13]. Furthermore, a fluorescently-tagged L1 element has been demonstrated to retrotranspose in the mouse brain in vivo [13, 17].
L1 activation is likely regulated through the canonical Wnt (wingless and int homolog)-signaling pathway [19]. The current model proceeds as follows: When L1 elements are inactive, SRY (sex determining region Y)-box2 (SOX2), a transcription factor that is essential to maintain self-renewal of undifferentiated embryonic stem cells (ESCs), binds to sequences in the promoter within the 5’ untranslated region (5’-UTR) where it acts as a repressor. After Wnt-induced activation of a signaling cascade, SOX2-mediated repression of L1 elements is relieved and L1 expression is induced concomitant with expression of the neurogenic transcription factor NeuroD1. This model suggests regulation of retrotransposition during neuronal cell fate specification (Figure 1C).
Expanding on these results, our laboratory [9] demonstrated that neural precursor cells (NPCs) isolated from human fetal brain or derived from human ESCs also supported retrotransposition of a transgenic human L1 element in vitro. In addition, an increase in endogenous L1 copy number in adult human brains compared to other somatic tissues could be demonstrated from the same individuals. Variability occurred not only between different brain regions in one individual but also between different individuals. The process of somatic L1 retrotransposition in developing neurons occurred with surprisingly high frequency: We estimate ~80–800 new insertions per cell in some brain regions, based on these quantitative assays [9].
These results were remarkable for two reasons: firstly, L1 retrotransposition in NPCs challenges the dogma that all neurons are genetically stable entities; secondly, it was believed that any L1 activity would be confined to the germline and L1 elements were not expected to be mobile in somatic tissues such as the brain. We believe that this L1-induced genomic variability could be a mechanism that contributes to neuronal diversity, thereby affecting neural plasticity, cognition and ultimately behavior. On the other hand, this mechanism may have negative implications for neuronal genomes, such as increased disease risk.
“Controlling elements” - how can L1 activity impact neuronal genomes?
A major implication of somatic mosaicism is that a subset of neurons would have distinct genomes, leading to unique transcriptomes. How L1 retrotransposition leads to such changes may be inferred from consequences of L1 germline insertions. Multiple genetic mechanisms whereby L1 elements impact host genomes, such as insertions, deletions and ectopic recombination, have been described (reviewed in [3, 20]).
New L1 insertions likely affect somatic genomes by modulating RNA abundance through changing the expression of messenger RNA (mRNA) and/or non-coding RNA (ncRNA). Transcript levels can increase or decrease through premature polyadenylation, aberrant splicing, formation of alternative transcripts or premature termination (Figure 2). Gene expression levels are modulated differently depending on where, and in which orientation, an L1 element inserts into the gene. For example, L1 elements inserted into an intron in the antisense orientation had almost no effect on transcript levels, whereas sense insertions generally caused a substantial decrease in transcript abundance [21, 22].
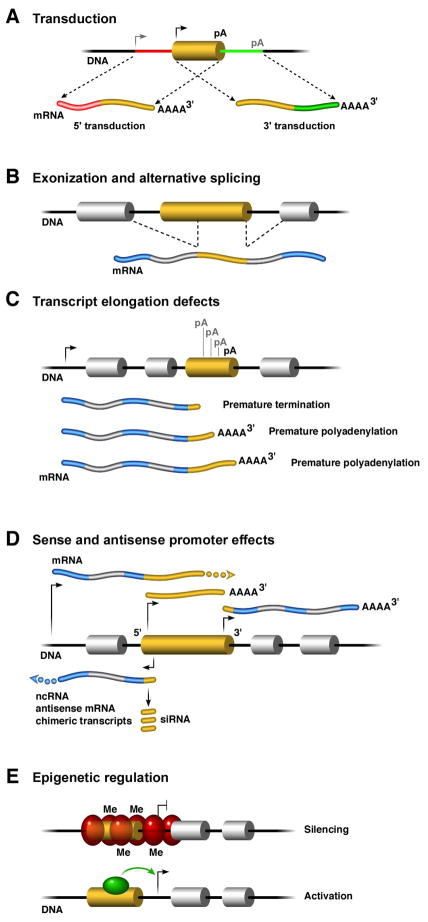
Figure 2. How L1 insertion may affect the transcriptome.
A| In a process termed transduction, L1 elements (golden cylinders) carry sequences flanking the element from their original source location to a new location; this can occur on the 5′ and 3′ end: transcription that initiates upstream from the L1 element results in mRNA that contains additional sequence 5′ of the L1 element (red line), whereas poly-adenylation sites that are located further downstream of the L1 element can lead to extension of L1 mRNA beyond the element’s 3′ end (green line). This process can cause exon shuffling or lead to insertion of regulatory sequences such as transcription factor binding sites at a new genomic location.
B| Mammalian L1 elements (golden cylinders) contain functional splice sites in both sense and antisense (AS) orientations; thus, when integrated into introns, L1 elements can induce aberrant splicing resulting in alternative mRNA transcripts.
C| L1 elements contain numerous internal polyadenylation [(p(A)] signals in both sense and antisense orientation that can lead to alternative mRNA transcripts or premature termination, thus affecting mRNA splicing and stability.
D| Transcript levels of genes can be changed through regulatory sequences that are contained within L1 elements. Human L1 elements contain one sense and one antisense promoter within their 5′ UTR and another promoter in the 3′ UTR (bent arrows). The antisense promoter in the 5′ and 3′ UTRs can initiate ectopic transcription of sequences flanking L1. Antisense transcripts can lead to production of non-coding RNA (ncRNA) or chimeric transcripts. Further, antisense RNA could reduce mRNA levels through the formation of double-stranded RNA (dsRNA), triggering the protein kinase R (PKR) degradation pathway [81], or leading to siRNA that induces RISC-dependent silencing [82]. In addition, ncRNAs could modulate epigenetic marks in cis and aid in the establishment of boundary elements.
E| An L1 element that has inserted upstream of another gene can become methylated (Me), which may induce formation of heterochromatin (red ovals) that can spread into flanking promoter sequences, effectively silencing transcription. Conversely, activation of an inserted L1 element could lead to ectopic activation of a nearby gene by facilitating binding of transcription factors (green oval).
Human and mouse L1 elements contain both a sense and antisense RNA polymerase II (pol II)-dependent promoter in their 5′UTR, as well as a newly discovered polII promoter in their untranslated 3′UTR [7]. Bidirectional transcription from the sense and antisense promoter in the 5′UTR can produce chimeric transcripts, non-coding RNA, antisense mRNA or double stranded RNA (dsRNA), which can affect gene expression in distinct ways (Figure 2) [23]. The promoter in the 3′UTR is in the sense orientation, reading “outwards” into the flanking genomic sequence (Figure 2) [7]. When genome-wide transcript abundance was measured, the majority of transcripts that derived from L1 sequence initiated at this promoter in a wide variety of somatic tissues, including brain [7]. Since the majority of new somatic L1 insertions are truncated near their 5′ end, it is likely that many newly inserted L1 sequences in neurons are not of full length but still contain the 3′ UTR promoter and may thereby initiate transcription of downstream sequences. Consequently, an alternative promoter derived from a novel L1 insertion could increase transcription of a nearby coding gene.
L1 elements are silenced through epigenetic mechanisms, such as DNA methylation and histone modification, and are activated when those repressive marks are relieved [24]. Ectopic activation of previously silenced retrotransposons can cause expression changes at neighboring genes by altering the timing or tissue specificity of gene expression, which can result in substantial somatic variability [25]; conversely, retrotransposons are targets for DNA methylation, which induces a repressive chromatin structure that can spread to coding sequences in their proximity, effectively silencing them (Figure 2).
We speculate that any of the mechanisms described can lead to transcript diversity in differentiated neurons that manifest themselves in individual neuronal proteomic variability. Indeed, variability of L1 expression has been observed not only between different individuals but also between different tissues in one individual, including the brain [7]. In this way, de novo inserted L1 elements can have considerable effects on the transcriptome in differentiated neurons, and hence act as “controlling elements” in McClintock’s sense.
Are there preferred target sites of L1 insertions during neuronal differentiation?
The extent of L1-mediated neuron-to-neuron variability depends on the conservation of L1 insertion sites within the genome; therefore, the question arises whether there are preferred target sites of L1 insertions during neuronal differentiation or whether integration occurs randomly?
Pre-existing L1 elements that are static in the human genome are significantly underrepresented in regions surrounding genes, and enriched in A+T rich regions. Interestingly, L1 elements that are in the sense orientation with respect to the nearest gene were significantly underrepresented. This bias was attributed to selective pressure to eliminate elements that disrupted gene transcription [26].
From analyses in mammalian cell culture it became clear that de novo insertions of engineered L1 elements occur preferentially within a small A+T-rich sequence window that contains the loosely conserved endonuclease recognition sequence [27, 28, 29]. New L1 insertions did occur within genes, preferably within introns and up- and downstream of coding sequences [28, 29]. One study came to the conclusion that no integration site preference for any specific genomic niche exists [30], while another study found de novo L1 insertions occurred in L1-poor, Alu element-rich and G+C neutral genomic regions [31]. Thus, despite the preference of the L1 endonuclease for A+T-rich sequences, this does not lead to an insertion bias over large genomic regions [31].
During embryonic or adult neurogenesis, L1 distribution patterns may be restricted by the context of chromatin structure that exists at specific timepoints during neuronal differentiation (Figure 1C). Insertions into open chromatin would be facilitated, whereas insertions into condensed heterochromatin would be unlikely. One possibility could be that de novo L1 insertions target a subset of genes causing similar transcriptional consequences; consequently, neurons would have similarly altered genomes, despite extensive retrotransposition. Another possibility could be that L1 insertions indiscriminately land in accessible chromatin, thus affecting many genes and causing a wide variety of transcriptional changes. As shown below, both scenarios are not necessarily mutually exclusive. Understanding if preferred genomic insertion sites exist for L1 is critical considering the amount of variability that can be caused by L1 retrotransposition in NPCs.
A hint that L1 element insertions may not be entirely random in NPCs came from an experiment in which a fluorescently-tagged L1 element was introduced into rat and human NPCs [9, 13]. Only a limited number of novel insertion events were cloned and sequenced, but the majority of the newly transposed elements landed in neuronally expressed genes. In one example, an insertion occurred upstream of the postsynaptic density 93 (Psd-93) gene in an antisense orientation. As a consequence, gene expression of Psd-93 was affected, resulting in a phenotypic change in the neuronal differentiation potential. In another example, two distinct insertions were found in the 5′ UTR of the Slc6a6 gene, which encodes a neurotransmitter transporter; two independent insertions within so close proximity would be exceedingly unlikely to occur randomly.
Chromatin modifications that occur during lineage commitment from ESCs to NPCs have been analyzed, revealing characteristic changes in active and repressive histone marks that correspond well with gene expression [32]. Two types of promoters were identified: CpG-rich promoters, generally associated with ubiquitously expressed ‘housekeeping’ genes; and CpG-poor promoters, generally associated with highly lineage-specific genes. In NPCs a large number (~8,000) of ‘housekeeping’ genes with CpG-rich promoters were associated with active chromatin marks and expressed, compared to the upregulation of only a small number (~200) of NPC-specific genes with CpG-poor promoters. Since L1 endonuclease recognizes an A+T-rich sequence motif, it is tempting to speculate that de novo L1 insertions could be preferentially targeted into CpG-poor promoters of NPC-specific genes; however, insertions into A+T-rich introns of housekeeping genes are equally likely.
It has been suggested that L1 mRNA could be retained in the cytoplasm in ribonucleoprotein complexes (RNPs) during germline transmission [33]; likewise, it remains a possibility that L1 persists in the same form in NPCs. This way, L1 mRNA could be passed on to daughter cells, retrotransposing during 2–3 cell divisions before cell cycle exit and before neuronal differentiation is complete (Figure 1C); thus, new insertions could be even more specifically targeted close to neuron-specific genes that are expressed at this stage.
Since massive cell death occurs during neurodevelopment [34–36], de novo L1 insertions in the mature brain probably represent only a subset of all L1 retrotranspositions that occurred during neurogenesis. Considering the fact that many essential housekeeping genes are expressed in NPCs, it is likely that some de novo insertions could have deleterious consequences that induce apoptosis.
Next-generation deep sequencing technologies will soon enable the discovery of the extent and exact locations of new L1 insertion sites in brain compared to other somatic tissues of the same individual. As many new L1 insertion sites are catalogued, it will become clear if preferred insertion sites exist or if L1 insertions are purely random. As these analyses extend to the single cell level, the relationship between new L1 insertion sites and the extent and character of neural mosaics will be revealed.
Does L1 retrotransposition cause “intangible variance” in the brain?
Despite exhaustive efforts to control genotype and environment, a wide range of phenotypes has been observed in isogenic mouse strains, in traits such as fear avoidance, stress response and learning [37]. Such variabilities usually take the form of a normally distributed, bell-shaped curve, indicating that the underlying causes are random. This phenomenon has been termed “intangible variance”, suggesting that there are intrinsic mechanisms that generate random variability besides the genetic blueprint within each individual [38]. The same effect, albeit smaller, can be observed in phenotypically discordant monozygotic twins. Mechanisms such as CNVs [12], aneuploidy [10], RNA editing [39] or epigenetic effects [25] could be involved in generating this random variation, all of which can have stochastic effects on genome structure and the transcriptome, and thus influence quantitative traits.
Could L1 retrotransposition play a role in generating this “intangible variance” in a population? Considering that many novel L1 insertions may occur in any given neuron during development and continue to occur during adult neurogenesis, this is an appealing proposition. As described above, novel L1 element insertions could have vast influences on neuronal genomes and transcriptomes. Further, L1 retrotransposition, with limited conservation of insertion sites, could fulfill the requirement of a stochastic mechanism that generates random variability (Figure 3). In addition, the L1-mediated effects need not be large in order to influence phenotypes. Recent studies have demonstrated altered rodent behavior resulting from the modulation of the firing pattern of a single neuron [40, 41]. Even if only a fraction of new L1 insertions have an impact on neuronal transcriptomes and thereby change neuronal properties subtly, effects on neural plasticity and behavior could occur. Thus, L1-mediated retrotransposition may be a mechanism that alters neuronal function in individuals, thereby broadening the spectrum of behavioral phenotypes that can originate from any single genome (Figure 3).
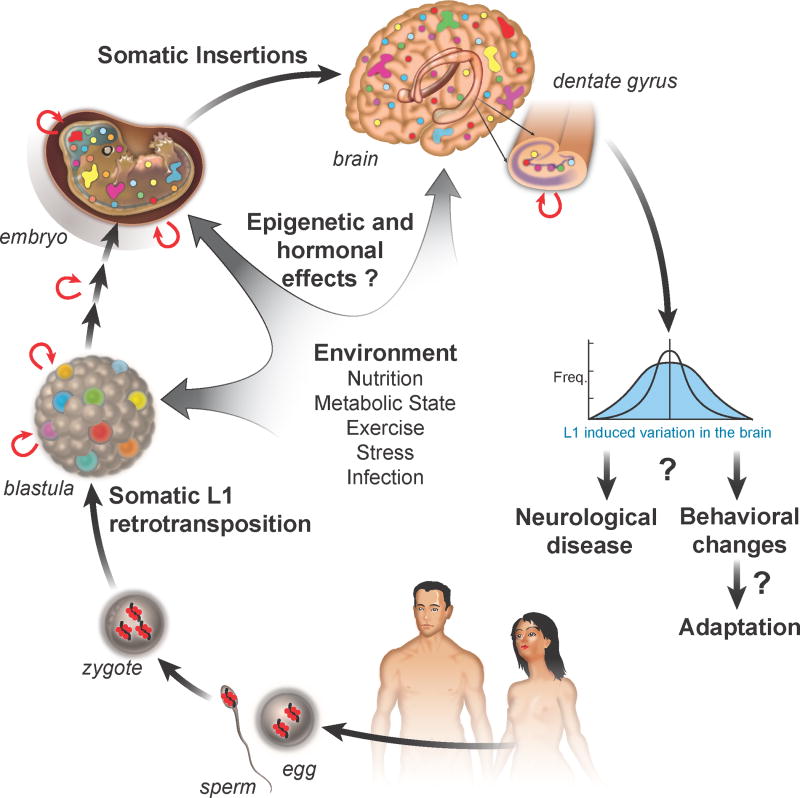
Figure 3. Putative implications of L1-mediated somatic mosaicism in the brain.
In a reversal of the commonly held belief that retrotransposition occurs primarily in the germline [83], it became clear that L1 elements are expressed in many somatic tissues, including the brain [7, 13, 84]. Recent evidence shows that L1 retrotransposition (curved red arrows) does not occur in the parental germline but in the soma during early embryonic development (colored dots), resulting in individuals that are genetically mosaic with respect to L1 composition [33]. It has been suggested, however, that L1 RNA may be transcribed in the parental germline and carried over in both male and female germ cells in the form of RNPs (black line with red dots) and integrated into the genome at the preimplantation stage [33] (colored spots); however, these events are probably rare, since retrotransposons are effectively silenced in the germline through a small RNA induced mechanism [78, 85]. Somatic L1 retrotranspositon events that occur during embryogenesis would result in clonal sectors of cells (colored patches) that carry the same insertion event. The size of clonal sectors depends on the developmental stage when the insertion occurred and the number of subsequent cell divisions. L1 insertion events that happen during embryonic brain development will be found in different brain regions (colored patches and dots), whereas events that happen during adult neurogenesis will be restricted to specific areas, such as the dentate gyrus (insert). According to our hypothesis, L1-induced mosaicism could increase variability in the brain (blue curve), which could have implications for behavioral phenotypes. The environment could influence regulation of somatic L1 retrotransposition in the brain and this influence could be mediated by epigenetic or hormonal mechanisms. Depending on its impact on the brain and the consequences, L1-induced somatic variability could either increase the risk for neurological disease or induce behavioral changes that could help the organism to better adapt to changing environments.
Can the environment influence L1-mediated neural plasticity?
McClintock proposed that activation of transposable elements is the genome’s response to environmental shock [42]. Since her original discovery, ample evidence has accumulated proving that environmental stress factors can have effects on the activity of transposons ranging from bacteria to higher eukaryotes: Regarding retroelements in mammals, physiological cell stressors and viral infection transcriptionally activated Alu elements [43–45] and genotoxic agents and carcinogens increased L1 retrotransposition in cell culture [46–48]. Oxidative stress or heavy metals were also implicated in increasing L1 retrotransposition activity in vivo and in cell culture, but these effects likely act on the post-transcriptional level influencing L1-mediated DNA repair and not L1 transcription [49, 50]. To date, no direct link between physiological cell stressors and enhanced L1 retrotransposition has been demonstrated in vivo.
Transposed elements can confer stress inducibility to genes in their proximity [51] or protect those genes against stress via a small RNA-mediated mechanism [52]; therefore, it has been speculated that, at least in plants, a burst of transposon activity aids in rapidly generating genetic diversity in changing environments. It is tempting to consider parallels to mammalian brain development, where environmental influences may impact L1 mobility in newly born neurons during embryonic development or adult neurogenesis [53, 54]. (Figure 3). For instance, stress, infections, nutrition or other environmental factors that affect the mother may increase or decrease L1 retrotransposition in the developing embryo. Cytokines and stress hormone levels can alter the in utero environment and can have direct influences on neurogenesis and brain development [55, 56]. In adult mice, exercise has been shown to enhance L1 retrotransposition two-fold [17], although whether this effect is due to an increase in newborn neurons or through metabolic changes due to the exercise itself is not yet clear.
Two pieces of evidence that link L1 activity with environmental stresses are the discoveries that L1 mRNA and proteins can accumulate in “stress granules” (SGs) (Figure 1C) and that L1 expression may be regulated hormonally. The sequestration of L1 RNA in SGs would provide a cellular “reservoir” that could be quickly activated in response to environmental stimuli [57]. In this way, environmental stressors could lead indirectly to increased L1 retrotransposition, which would affect neuronal plasticity. Regarding hormone-responsiveness, there is evidence that androgenic steroids and steroid-like compounds can induce L1 activity [58]. In addition, L1 expression has been observed in mouse steroidogenic tissues such as gonadal cell types and placenta [59]. The possibility that L1 activity may be induced in this manner is intriguing, considering evidence that implicates steroid hormones in embryonic brain development, stress responses, and behavior [60].
Although direct evidence of the induction of L1 jumping in the brain after exposing to stress or steroid hormones is currently lacking; we suggest that a gene regulatory network could exist that coordinates L1 expression in response to environmental factors, including stress and steroidal hormones. How could environmental cues be relayed back to L1 retrotransposition? A mechanism would have to include sensing of the environmental signal and its transduction to the genome to induce L1 activity. A testable working hypothesis could be based on the observation that a key component of the stress response is the increased secretion of adrenal steroids that can bind to specific receptors [61], inducing a signaling response that could lead to transcriptional activation of L1.
How could somatic variability in the brain be of evolutionary advantage?
One possibility for why L1 retrotransposition in neurons may have been retained during evolution is because it may have been metabolically too expensive to prevent it; consequently, genomic neuronal diversity was merely tolerated and evolved neutrally. However, a mechanism that creates somatic variability during neurogenesis and permits greater adaptability to changing environments would not only be subject to natural selection but also potentially favored by it [62]. According to the concept of Weismann’s barrier, the germ line is protected from somatic DNA changes during an individual’s lifetime [63]. This theory further negates the Lamarckian notion that adaptive somatic mutations in an individual could be passed on to their offspring; yet, if the mechanism that creates this variability has an impact on fitness, it will be subject to natural selection. Hence, we have to assume that the underpinnings of a mechanism generating somatic diversity in the brain are genetic. Thus, selection would act on maintaining this genetic mechanism and not on the somatic insertions themselves. This assumption can be tested by analyzing mutants that either show enhanced or decreased L1 retrotransposition or by finding ways to artificially alter the level of somatic L1 transposition in the brain. Since somatic mosaicism is observed in neurons and seems to be developmentally regulated, we favor the hypothesis that neural mosaics help to diversify the range of behavioral phenotypes that are possible from a given genome.
Conclusions and future directions
The discovery that somatic retrotransposition of L1 retroelements occurs in neural stem cells in the brain overturned the commonly held view that transposable elements in mammals are dormant genomic parasites that only “jump” in cells contributing to the germline to ensure their survival. Surprisingly, new somatic L1 insertions seem to occur frequently, probably in many (if not most) neurons, during embryonic neuronal differentiation as well as during adult neurogenesis. This leads to neuron-to-neuron variation in genomic DNA content, which may alter individual neuronal transcriptomes. Diversifying an individual neuronal ‘readout’ through retrotransposition could have implications for synaptic plasticity, leading to altered neuronal circuitry and behavior. Further, this event may be influenced by environmental factors, providing a potential genetic mechanism for generating neuronal diversity in response to changing environments.
A first step towards answering the question of a possible functional role of L1 induced genomic variation will be to comprehensively catalogue and characterize new insertion sites within the mammalian brain, ultimately at the single cell level. Even with the advent of ultra-high throughput sequencing methods this is still a costly and challenging endeavor. What we can already conclude is that an unexpected source of variation exists within mammalian brains that generates diversity within any individual. In unraveling the implications of L1 induced “genomic plasticity” in the brain we are just beginning to truly understand McClintock’s notion of a ‘dynamic genome’.
Acknowledgments
We thank Mary Lynn Gage for editorial comments, Stefan Aigner for helpful discussions and anonymous reviewers for their suggestions.
FUNDING
This work was supported by the The G. Harold and Leila Y. Mathers Foundation and NIMH Grant MH088485.
Footnotes
DISCLOSURE STATEMENT
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Comfort NC. “The real point is control”: the reception of Barbara McClintock’s controlling elements. J Hist Biol. 1999;32(1):133–62. doi: 10.1023/a:1004468625863. [DOI] [PubMed] [Google Scholar]
- 3.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gotea V, Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22(5):260–7. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Xing J, et al. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci U S A. 2006;103(47):17608–13. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chueh AC, et al. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5(1):e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41(5):563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 8.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006(1):1–16. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25(9):2176–80. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci U S A. 2001;98(23):13361–6. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82(3):763–71. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 14.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Goodier JL, et al. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11(10):1677–85. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100(9):5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muotri AR, et al. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19(10):1002–7. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prak ET, et al. Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci U S A. 2003;100(4):1832–7. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwabara T, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12(9):1097–105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135(1):23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429(6989):268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15(13):2146–56. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 23.Nigumann P, et al. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79(5):628–34. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 24.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–85. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 25.Morgan HD, et al. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 26.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12(10):1483–95. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37(51):18081–93. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 28.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–38. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–25. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 30.Berry C, et al. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2(11):e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasior SL, et al. Characterization of pre-insertion loci of de novo L1 insertions. Gene. 2007;390(1–2):190–8. doi: 10.1016/j.gene.2006.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23(11):1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaschke AJ, Staley K, Chun J. Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex. Development. 1996;122(4):1165–74. doi: 10.1242/dev.122.4.1165. [DOI] [PubMed] [Google Scholar]
- 35.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 36.McConnell MJ, MacMillan HR, Chun J. Mathematical modeling supports substantial mouse neural progenitor cell death. Neural Dev. 2009;4:28. doi: 10.1186/1749-8104-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakovcevski M, Schachner M, Morellini F. Individual variability in the stress response of C57BL/6J male mice correlates with trait anxiety. Genes Brain Behav. 2008;7(2):235–43. doi: 10.1111/j.1601-183X.2007.00345.x. [DOI] [PubMed] [Google Scholar]
- 38.Gartner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim. 1990;24(1):71–7. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 39.Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31(5):227–33. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Houweling AR, Brecht M. Behavioural report of single neuron stimulation in somatosensory cortex. Nature. 2008;451(7174):65–8. doi: 10.1038/nature06447. [DOI] [PubMed] [Google Scholar]
- 41.Li CY, Poo MM, Dan Y. Burst spiking of a single cortical neuron modifies global brain state. Science. 2009;324(5927):643–6. doi: 10.1126/science.1169957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 43.Li T, et al. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239(2):367–72. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 44.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276(1–2):135–41. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 45.Liu WM, et al. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23(10):1758–65. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee G, et al. Ultraviolet-induced transformation of keratinocytes: possible involvement of long interspersed element-1 reverse transcriptase. Photodermatol Photoimmunol Photomed. 2005;21(1):32–9. doi: 10.1111/j.1600-0781.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- 47.Farkash EA, et al. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34(4):1196–204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stribinskis V, Ramos KS. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 2006;66(5):2616–20. doi: 10.1158/0008-5472.CAN-05-3478. [DOI] [PubMed] [Google Scholar]
- 49.Rockwood LD, Felix K, Janz S. Elevated presence of retrotransposons at sites of DNA double strand break repair in mouse models of metabolic oxidative stress and MYC-induced lymphoma. Mutat Res. 2004;548(1–2):117–25. doi: 10.1016/j.mrfmmm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 50.El-Sawy M, et al. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J Mol Biol. 2005;354(2):246–57. doi: 10.1016/j.jmb.2005.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461(7267):1130–4. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 52.Hilbricht T, et al. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytol. 2008;179(3):877–87. doi: 10.1111/j.1469-8137.2008.02480.x. [DOI] [PubMed] [Google Scholar]
- 53.van Praag H, et al. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25(38):8680–5. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coe CL, et al. Prenatal stress diminishes neurogenesis in the dentate gyrus of juvenile rhesus monkeys. Biol Psychiatry. 2003;54(10):1025–34. doi: 10.1016/s0006-3223(03)00698-x. [DOI] [PubMed] [Google Scholar]
- 55.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64(1):61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Carpentier PA, Palmer TD. Immune influence on adult neural stem cell regulation and function. Neuron. 2009;64(1):79–92. doi: 10.1016/j.neuron.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32(4):683–96. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 58.Morales JF, Snow ET, Murnane JP. Environmental factors affecting transcription of the human L1 retrotransposon. I. Steroid hormone-like agents. Mutagenesis. 2002;17(3):193–200. doi: 10.1093/mutage/17.3.193. [DOI] [PubMed] [Google Scholar]
- 59.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci U S A. 1995;92(5):1520–4. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuloaga DG, et al. The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Horm Behav. 2008;53(5):613–26. doi: 10.1016/j.yhbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prager EM, Johnson LR. Stress at the synapse: signal transduction mechanisms of adrenal steroids at neuronal membranes. Sci Signal. 2009;2(86):re5. doi: 10.1126/scisignal.286re5. [DOI] [PubMed] [Google Scholar]
- 62.Mattick JS. Has evolution learnt how to learn? EMBO Rep. 2009;10(7):665. doi: 10.1038/embor.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weismann A. The Germ-Plasm. In: Robbins R, editor. ATheory of Heredity. Charles Scribner’s Sons Electronic Scholarly Publishing; 1893. http://www.esp.org/books/weismann/germ-plasm/facsimile/ [Google Scholar]
- 64.Williams RW. Mapping genes that modulate mouse brain development: a quantitative genetic approach. Results Probl Cell Differ. 2000;30:21–49. doi: 10.1007/978-3-540-48002-0_2. [DOI] [PubMed] [Google Scholar]
- 65.Pakkenberg B, et al. Aging and the human neocortex. Exp Gerontol. 2003;38(1–2):95–9. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- 66.Dreyer WJ, Gray WR, Hood L. Cold Spring Harb Symp Quant Biol. Cold Spring Harbor: 1967. The Genetic, Molecular, and Cellular Basis of Antibody Formation: Some Facts and a Unifying Hypothesis. [Google Scholar]
- 67.Chun JJ, et al. The recombination activating gene-1 (RAG-1) transcript is present in the murine central nervous system. Cell. 1991;64(1):189–200. doi: 10.1016/0092-8674(91)90220-s. [DOI] [PubMed] [Google Scholar]
- 68.Abeliovich A, et al. On somatic recombination in the central nervous system of transgenic mice. Science. 1992;257(5068):404–10. doi: 10.1126/science.1631561. [DOI] [PubMed] [Google Scholar]
- 69.Schatz DG, Chun JJ. V(D)J recombination and the transgenic brain blues. New Biol. 1992;4(3):188–96. [PubMed] [Google Scholar]
- 70.Ferguson DO, et al. The interplay between nonhomologous end-joining and cell cycle checkpoint factors in development, genomic stability, and tumorigenesis. Cold Spring Harb Symp Quant Biol. 2000;65:395–403. doi: 10.1101/sqb.2000.65.395. [DOI] [PubMed] [Google Scholar]
- 71.Serizawa S, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302(5653):2088–94. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 72.Tasic B, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10(1):21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 73.McConnell MJ, et al. Failed clearance of aneuploid embryonic neural progenitor cells leads to excess aneuploidy in the Atm-deficient but not the Trp53-deficient adult cerebral cortex. J Neurosci. 2004;24(37):8090–6. doi: 10.1523/JNEUROSCI.2263-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iourov IY, et al. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxiatelangiectasia brain. Hum Mol Genet. 2009;18(14):2656–69. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 75.Martin SL. The ORF1 Protein Encoded by LINE-1: Structure and Function During L1 Retrotransposition. J Biomed Biotechnol. 2006;2006(1):45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Q, et al. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 77.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 79.Goodier JL, et al. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodier JL, et al. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27(18):6469–83. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heinicke LA, et al. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390(2):319–38. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13(9):763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 83.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32(4):655–60. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 84.Rangwala SH, Zhang L, Kazazian HH., Jr Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10(9):R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]