Abstract
The protease a disintegrin and metalloprotease (ADAM) 17 cleaves tumor necrosis factor (TNF), L-selectin, and epidermal growth factor receptor (EGF-R) ligands from the plasma membrane. ADAM17 is expressed in most tissues and is up-regulated during inflammation and cancer. ADAM17-deficient mice are not viable. Conditional ADAM17 knockout models demonstrated proinflammatory activities of ADAM17 in septic shock via shedding of TNF. We used a novel gene targeting strategy to generate mice with dramatically reduced ADAM17 levels in all tissues. The resulting mice called ADAM17ex/ex were viable, showed compromised shedding of ADAM17 substrates from the cell surface, and developed eye, heart, and skin defects as a consequence of impaired EGF-R signaling caused by failure of shedding of EGF-R ligands. Unexpectedly, although the intestine of unchallenged homozygous ADAM17ex/ex mice was normal, ADAM17ex/ex mice showed substantially increased susceptibility to inflammation in dextran sulfate sodium colitis. This was a result of impaired shedding of EGF-R ligands resulting in failure to phosphorylate STAT3 via the EGF-R and, consequently, in defective regeneration of epithelial cells and breakdown of the intestinal barrier. Besides regulating the systemic availability of the proinflammatory cytokine TNF, our results demonstrate that ADAM17 is needed for vital regenerative activities during the immune response. Thus, our mouse model will help investigate ADAM17 as a potential drug target.
Many membrane proteins are cleaved at the plasma membrane to release soluble ectodomains, which may exert different biological activities (Murphy, 2008). Membrane-bound growth factors and cytokines become systemically available upon shedding. Soluble receptors for growth factors and cytokines can be antagonists of the cognate cytokines, as is the case for IL-1 and TNF (Müllberg et al., 2000). Alternatively, soluble receptors can be agonistic, i.e., together with their respective ligands they stimulate cells, which are otherwise unresponsive to the cytokine, as demonstrated for IL-6 trans-signaling (Rose-John et al., 2006). A disintegrin and metalloprotease (ADAM) 17 is an important sheddase involved in the proteolysis of membrane proteins such as TNF, IL-6R, L-selectin, and ligands of the epidermal growth factor receptor (EGF-R; Peschon et al., 1998). ADAM17-deficient mice are not viable. Interestingly, these mice were reminiscent of mice lacking TGF-α (Peschon et al., 1998). Recently, conditional ADAM17 knockout animals have been generated (Horiuchi et al., 2007, 2009). Although these studies confirmed that ADAM17 is the major endotoxin-stimulated TNF sheddase in myeloid cells in vivo and that ADAM17 is involved in the control of physiological bone remodeling, they did not clarify the role of ADAM17 in complex settings such as inflammation and cancer. Studies of ADAM17 have been complicated by the fact that it is not clear whether the protease and its substrates need to be expressed on the same cell or whether shedding in trans (i.e., the protease is expressed on one cell, the substrate on a different cell) is possible (Janes et al., 2005). Therefore, the choice of cre-transgenic mice for tissue-specific deletion of the ADAM17 gene in conditional mice is ambiguous.
In this paper, we developed a novel strategy to generate mice with barely detectable levels of ADAM17 in all tissues. The strategy is based on the generation of a new exon within the ADAM17 gene, which starts with an in-frame translational stop codon and which was flanked by splice donor/acceptor sites, which slightly deviated from the canonical consensus sequence. This strategy has been named exon-induced translational stop (EXITS). Homozygous mice used the new exon for ∼95% of the ADAM17 mRNAs, resulting in a dramatic loss of ADAM17 protein in all cell types. Nevertheless, homozygous ADAM17ex/ex mice were viable and developed eye, hair, and skin defects reminiscent of mice lacking TGF-α. Although the intestine of the homozygous ADAM17ex/ex mice showed no overt abnormalities, the animals displayed dramatically increased susceptibility to intestinal inflammation induced by dextran sulfate sodium (DSS) as a consequence of impaired EGF-R–dependent regeneration caused by failure of shedding of EGF-R ligands. Results show that during inflammation, ADAM17 is not only involved in shedding the proinflammatory cytokine TNF but also in the regulation of regenerative responses. Thus, our mouse model will help investigate ADAM17 as a potential drug target in TNF- and/or EGF-R–dependent pathologies in inflammation and cancer.
RESULTS AND DISCUSSION
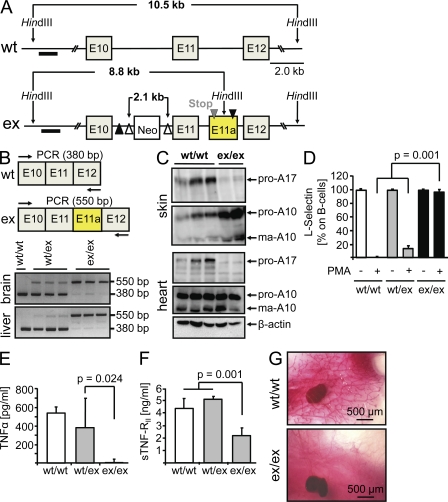
To construct a targeting vector for the ADAM17 gene, we inserted a loxP sequence followed by a noncanonical donor splice site downstream of a cryptic acceptor splice site within intron11. This manipulation generated a new exon between exons 11 and 12, which started with an in-frame translational stop codon (Fig. 1 A). Importantly, mice homozygous for the ADAM17 ex allele (Fig, 1 A; Fig. S1 A) were viable. The usage of the new exon between exons 11 and 12 of the mouse ADAM17 gene was tested by RT-PCR as outlined in Fig. 1 B. Whereas in WT animals only a band of 380 bp was detected, heterozygous ADAM17WT/ex mice showed an additional band of 550 bp corresponding to the insertion of the new exon sequence. In homozygous ADAM17ex/ex mice, ∼95% of the ADAM17 mRNAs contained the new exon. Consequently, almost no ADAM17 protein was detected in all tissues analyzed (Fig. 1 C; Fig. S1 B). These results indicated that we had generated viable mice with barely detectable ADAM17 protein levels in all tissues. ADAM17ex/ex mice were born at a frequency clearly below the expected Mendelian distribution pattern (Table S1). Homozygous ADAM17ex/ex mice showed abnormalities of the eyes, skin, and heart, but not in the brain, body segmentation, or vascular development, indicating that ADAM17 is not crucial for developmental Notch or Notch ligand function in vivo (Fig. S2).
Figure 1.
Targeted generation of ADAM17ex/ex mice. (A) Scheme for targeting the ADAM17 gene. Open arrowheads denote FLP recombinase sites, and LoxP sites are indicated by closed arrowheads. The altered allele contains an artificial exon starting with an in-frame translational stop codon placed downstream of exon 11. The artificial exon (E11a, yellow) containing the stop codon is flanked by noncanonical splice donor and acceptor sites (see Materials and methods). The black bar indicates the hybridization probe used for Southern blotting. (B) mRNA from liver and brain was analyzed by RT-PCR. (C) ADAM17 and ADAM10 Western blots of skin and heart tissues. (D and E) Shedding of L-selectin from B cells (D) and TNF from spleen cells (E) was analyzed by FACS and ELISA, respectively. (F) Serum levels of sTNF-RII were measured by ELISA. Data in D–F are shown as mean values ± SD. (G) Formation of milk ducts in mice at the age of 12 wk. Representative pictures from four animals are shown. Bars, 500 µm.
Generation of soluble L-selectin and TNF was completely abrogated in ADAM17ex/ex mice and levels of sTNF-RII were significantly reduced, indicating that shedding of ADAM17 substrates was severely impaired (Fig. 1, D–F). ADAM17 has been implicated in the cleavage of membrane-bound ligands of the EGF-R (Sternlicht et al., 2005). Soluble, but not membrane-bound, amphiregulin (AR) has been shown to be sufficient and necessary for the formation of milk ducts in developing female animals (Sternlicht et al., 2005). As shown in Fig. 1 G, in contrast to WT mice, milk duct development was reduced in ADAM17ex/ex mice at 12 wk (Fig. 1 G, right), indicating that AR-induced milk duct formation required ADAM17-mediated cleavage of this EGF-R ligand.
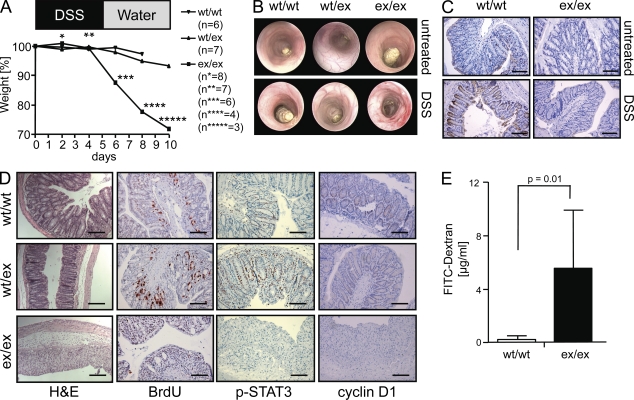
Blockade of TNF has been shown to be beneficial for the treatment of patients with Crohn’s disease (van Dullemen et al., 1995). Therefore, it was of interest to examine such a condition in ADAM17ex/ex animals. When WT and heterozygous ADAM17WT/ex mice were treated with a mild regimen (2%) of DSS, they showed mild weight loss (Fig. 2 A) and minimal signs of inflammation upon endoscopy (Fig. 2 B). In contrast, homozygous ADAM17ex/ex animals exhibited severe weight loss and five of eight mice died. Colonoscopy revealed severe inflammation and ulcerations in these animals (Fig. 2 B) and an increase in endoscopic index of colitis severity (Fig. S3). ADAM17 protein was expressed in the crypts of the intestine of WT mice and was up-regulated upon DSS challenge but could not be detected in ADAM17ex/ex animals (Fig. 2 C). In ADAM17ex/ex animals, the crypt structure was completely lost after day 10 (Fig. 2 D). Interestingly, a similar damage was seen in mice with targeted disruption of STAT3 in epithelial cells of the intestine. These mice, upon treatment with azoxymethane/DSS, showed destruction of the colon architecture (Bollrath et al., 2009; Grivennikov et al., 2009; Pickert et al., 2009). Stimulation of the EGF-R was shown to lead to activation and phosphorylation of STAT3 (Itoh et al., 2006). We therefore hypothesized that the regenerative response of colonic epithelial cells of ADAM17ex/ex mice was compromised.
Figure 2.
ADAM17ex/ex mice are highly susceptible to DSS-induced colitis. (A) Body weight after the onset of treatment with DSS. Numbers of mice per group used are indicated. The experiment was performed three times. (B) Colonoscopy of untreated (top) and treated (bottom) mice. Representative pictures are shown. (C) DSS-treated (4 d) and untreated WT and ADAM17ex/ex mice were immunostained with anti-ADAM17 (magnification, 100×). Bars, 100 µm. Representative macroscopic pictures of four mice per group are shown. (D) Hematoxylin and eosin (H&E), BrdU, anti–phosho-STAT3, and anti-cyclin D1 staining of colons from mice challenged for 10 d with DSS. Bars: (H&E) 200 µM; (BrdU, p-STAT3, and cyclin D1) 100 µm. Representative microscopic images of three experiments (H&E and BrdU), two experiments (pSTAT3), and one experiment (cyclin D1) of ADAM17ex/ex, ADAM17WT/ex, and WT controls (five mice per group) are shown. (E) DSS colitis was induced and plasma FITC-dextran concentrations in ADAM17WT/WT (n = 7) and ADAM17ex/ex (n = 7) mice 4 h after FITC-dextran administration by oral gavage (60 mg/100 g of body weight) are shown. Data are shown as mean values ± SD. The experiment was performed twice with similar results.
DSS is toxic to gut epithelial cells of the basal crypts and affects the integrity of the mucosal barrier (Wirtz et al., 2007). Colon sections of WT and ADAM17WT/ex mice exhibited proliferating nuclei throughout the crypts of the gut (Fig. 2 D). No such staining was seen in ADAM17ex/ex mice. In WT and ADAM17WT/ex mice, but not in ADAM17ex/ex mice, phosphorylated STAT3 and cyclin D1 were readily detectable (Fig. 2 D). These results indicated that in ADAM17ex/ex mice the proliferative response of intestinal epithelial cells to DSS was blocked, presumably as a result of the inability to activate EGF-R–mediated STAT3 phosphorylation (Itoh et al., 2006). Importantly, upon DSS challenge, the intestinal barrier became highly permeable for FITC-dextran in ADAM17ex/ex mice (Fig. 2 E).
Surprisingly, in unchallenged ADAM17ex/ex mice the colon mucosa was regularly arranged (Fig. 3 A) and unaltered proliferation in colon tissue sections was detected (Fig. 3 B). Gene expression analysis in the intestine of unchallenged mice revealed significant up-regulation of genes involved in immunity and defense as well as of genes playing a role in interferon-mediated immunity (Fig. 3 C and Fig. S4). These findings indicated that loss of ADAM17 activity, although it did not result in an overt inflammatory phenotype, led to a significant inflammatory gene expression signature, possibly resulting in sensitization toward inflammatory stimuli.
Figure 3.
Intestinal architecture, epithelial proliferation, and gene expression in unchallenged ADAM17ex/ex mice. (A) H&E staining of paraffin-embedded colon tissue from unchallenged ADAM17ex/ex mice. Bar, 100 µM. Representative microscopic pictures of eight ADAM17ex/ex mice are shown. (B) ADAM17ex/ex (n = 3) and WT (n = 3) mice were injected intraperitoneally with BrdU and stained for BrdU in the distal colon. The arrowheads indicate BrdU-positive nuclei. Bars, 100 µm. The experiment was performed three times with three animals per group. (C) Pathway analysis of microarray data from colon tissue of WT (n = 3) and ADAM17ex/ex (n = 3) mice. Regulated genes, which are significantly enriched or depleted during biological processes, are displayed. The bar represents the −log(p) of the probability of the enrichment or depletion occurring by chance. Processes that are associated with down-regulated transcripts are shown on the left, and those associated with up-regulated transcripts are shown on the right. The numbers in the bars represent the number of observed regulated transcripts in the category.
In DSS-treated animals we detected a strong increase in myeloperoxidase activity in ADAM17ex/ex animals (Fig. S5 A), which is considered a marker for activated neutrophils (Breckwoldt et al., 2008). In colon organ cultures from DSS-treated animals the inflammatory chemokines KC and MCP-1 were elevated in ADAM17ex/ex mice. Interestingly, the antiinflammatory cytokine IL-10 and the IL-6–related cytokine IL-11 were also up-regulated in these animals (Fig. S5, B–E). Such an up-regulation of IL-10 and IL-11 was also seen in DSS-challenged mice with a deletion of STAT3 in intestinal epithelial cells (Bollrath et al., 2009). Consequently, an increase of CD68-positive mononuclear cells and CD3-positive T cells in intestinal tissue sections of DSS-treated ADAM17ex/ex animals was detected (Fig. S5, F and G). The cytokines TNF, IL-21, IL-12, and IL-17 were not significantly up-regulated (unpublished data).
The inflammatory response seen in homozygous ADAM17ex/ex mice was not the result of an intrinsic activation of the immune system in these animals, as demonstrated in a T cell transfer colitis model where T cells from WT or homozygous ADAM17ex/ex mice were transferred into rag−/− animals (Powrie et al., 1994; Atreya et al., 2000). Importantly, no significant difference in colitis development was detected when comparing animals transplanted with WT or ADAM17ex/ex T cells (unpublished data).
We hypothesized that the failure of intestinal epithelial cells to proliferate was caused by the fact that ADAM17ex/ex mice could not cleave and thereby systemically activate ligands of the EGF-R (Berasain et al., 2009). As shown in Fig. 4 A, after DSS challenge, higher levels of cell surface–expressed TGF-α were detected in ADAM17ex/ex mice as compared with WT animals. We therefore asked whether injection of recombinant TGF-α would restore proliferation of intestinal epithelial cells. As shown in Fig. 4 B, treatment of ADAM17ex/ex mice with this EGF-R ligand induced intestinal epithelial cell proliferation to a degree, which clearly surpassed the proliferation seen in WT animals. Concomitantly, phosphorylation of STAT3 was induced in TGF-α–treated mice. Interestingly, WT and ADAM17ex/ex mice upon daily injection of EGF-R ligand lost less weight than PBS-injected animals (Fig. 4 C). These results demonstrated that in ADAM17ex/ex mice signaling downstream of the EGF-R was intact and that ectopic stimulation of this pathway compensated for the loss of proliferation caused by the deficiency of ADAM17.
Figure 4.
Cellular accumulation of TGF-α and compensatory proliferation induced by recombinant TGF-α. (A) Colonic tissue sections from unchallenged and challenged WT and ADAM17ex/ex mice (4 d, 2% DSS) were stained for TGF-α. Representative microscopic pictures of three mice per group are shown. Bars, 100 µm. (B) Colitis was induced in WT (n = 5) and ADAM17ex/ex (n = 9) mice with 2% DSS for 10 d. ADAM17ex/ex mice were injected daily with recombinant TGF-α (15 µg/mouse; n = 5) or with PBS (n = 4). Representative microscopic images of one experiment with five WT mice, four ADAM17ex/ex mice treated with PBS, and five ADAM17ex/ex mice treated with TGF-α are shown. Bars, 100 µm. The experiment was performed twice with similar results. (C) Colitis was induced in WT and ADAM17ex/ex mice with 2% DSS and the animals were injected daily with the EGF-R ligand EGF (15 µg/mouse; WT, n = 3; ADAM17ex/ex mice, n = 8) or with PBS, (WT, n = 8; ADAM17ex/ex mice, n = 9) and body weight was recorded. Data were pooled from two independent experiments.
Although the cre/loxP technology (Kühn et al., 1995) has been helpful in the analysis of gene defects in animals when total deletion of the gene of interest was lethal, there are limitations to this method. In cases when the expression profile and the activity of the gene of interest is complex or when it is not known which cell type is responsible for the synthesis and secretion of a soluble protein, it can be difficult to choose the appropriate transgenic cre mouse. In the case of ADAM17, it is not clear whether the protease exclusively cleaves its targets in cis or whether shedding in trans is possible (Janes et al., 2005). Furthermore, many functions of ADAM17 remained unknown as a result of embryonic lethality caused by ADAM17 gene deletion (Peschon et al., 1998). The phenotypes described in this paper are based on the development of a novel knockin targeting strategy, leading to an almost complete absence of functional ADAM17 protein as the result of a premature translational stop codon encoded by the artificially introduced exon. Our approach can be adapted to other genes as we showed in the case of the human CNTF gene (unpublished data) and will therefore be generally applicable.
Our experiments showed that for all substrates tested, shedding in ADAM17ex/ex animals was severely compromised. Physiological proliferation in the intestine of ADAM17ex/ex mice was normal, indicating that ADAM17 activation was not needed. In contrast, formation of milk ducts in 12-wk-old mice was abrogated as a result of the lack of AR processing. During the regenerative response i.e., after DSS treatment, the STAT3-mediated proliferative response of the intestinal epithelial cells was completely absent in ADAM17ex/ex mice. This finding is consistent with a study showing that mice lacking TGF-α have an increased susceptibility to DSS-induced colitis (Egger et al., 1997) and that TGF-α–overexpressing mice were less susceptible to DSS-induced colitis (Egger et al., 1998). The EGF-R can be stimulated by its cognate ligands in a paracrine and in a systemic fashion (Rodland et al., 2008). Thus, ADAM17ex/ex mice will be a good model to study the differences between EGF-R signaling via membrane-bound and soluble ligands.
The activation mechanisms of ADAM17 are not fully understood. In contrast to matrix metalloproteases, which are activated by the removal of the prodomain, ADAM17 additionally needs to be activated by intra- or extracellular signaling (Murphy, 2008). Recently, we showed that induction of apoptosis in neutrophils led to selective activation of ADAM17 and subsequent shedding of the IL-6R (Chalaris et al., 2007) and that this mechanism might represent a gauge to measure the extent of damage encountered by neutrophilic cells. In view of the pleiotropic activities governed by ADAM17, it will be interesting to study how the activity of this protease is regulated under stress conditions.
The intestine of unchallenged ADAM17ex/ex mice appeared normal but failed to respond adequately to the tissue damage by DSS, although we detected an up-regulation of inflammatory genes. These results show that ADAM17 plays a role in the coordination of reactions toward stress caused by wounding. The intestinal epithelium acts as an intrinsic barrier against microbial invaders. Our data show that lack of regeneration and failure to maintain the barrier function of the intestine act dominantly over the reduced activity of the immune system as a result of impaired shedding of, for example, TNF, caused by diminished ADAM17 activity. As a consequence, ADAM17ex/ex mice were more susceptible in a model of inflammatory bowel disease.
Because ADAM17 is the main sheddase for TNF and several ligands of the EGF-R (Peschon et al., 1998), this protease has been considered to be a target of therapeutic drugs for the treatment of inflammatory diseases (Moss et al., 2008) and cancer (Kenny, 2007). In view of the role ADAM17 played in the coordination of the regeneration response in the gut, it now becomes clear that inhibition of ADAM17 might have severe side effects, such as lack of tissue regeneration. In contrast, many tumors largely depend on the activity of the EGF-R, which can be efficiently blocked by inhibition of ADAM17. In this respect, our mouse model is well suited to evaluate the consequences of ADAM17 deficiency in vivo. It is noteworthy that our strategy is based on the presence of some residual ADAM17 activity, which is also likely to be present upon pharmacological inhibition of this protease.
In summary, we presented the novel EXITS method for gene targeting, which will be applicable to other genes. Using this technique, viable mice with greatly diminished ADAM17 activity in all tissues were generated. Models of inflammatory bowel disease showed that these mice were highly susceptible to intestinal damage by DSS and point to a function of ADAM17 in the coordination of pro- and antiinflammatory activities in response to body stress and damage.
MATERIALS AND METHODS
Reagents and antibodies.
Reagents were purchased from Carl Roth unless otherwise specified. Restriction enzymes, molecular weight markers, Taq-polymerase, and reverse transcription were obtained from Fermentas. Cell culture media, trypsin, FCS, L-glutamine, and penicillin/streptomycin were obtained from PAA Laboratories. All other reagents were obtained from the following sources: TGF-α (PeproTech); LPS, FITC-dextran, and myeloperoxidase (Sigma-Aldrich); PMA (EMD); anti-CD3, anti-CD28, anti–CD4-FITC, anti–B220-FITC, and anti–L-selectin–PE (BD); anti–phospho-STAT3 and anti–cyclin-D1 (Cell Signaling Technology); and horseradish peroxidase–coupled anti–goat antibody (Thermo Fisher Scientific). The anti-ADAM10 and anti-ADAM17 antisera were a gift from S. Weber (Institute of Biochemistry, Christian-Albrechts-University, Kiel, Germany).
Generation of ADAM17ex/ex knockin mice.
We created hypomorphic and conditional knockout mice of ADAM17. The ADAM17 gene is composed of 18 exons. We designed the targeting vector in a way that exon 11, which encodes the catalytic zinc-binding domain, is flanked by two loxP sites and the FTR-flanked neo cassette. A cryptic acceptor splice site within the intron 11 of ADAM17 was predicted using the NetGene2 program. Hypomorphic expression of ADAM17 was achieved by inserting a loxP site together with a cryptic noncanonical donor splice site (GAG-GTAATT instead of the canonical sequence A/CAG-GTAAGT, where the hyphen denotes the exon-intron border) downstream of the cryptic acceptor splice site into intron 11 and thereby generating an additional, artificial, exon 11a (Fig. 1 A). Alternative splicing/use of the artificial exon 11a led to premature disruption of ADAM17 protein translation as a result of the in-frame stop codon TAG in exon 11a. If needed, deletion of exon 11 could be achieved by tissue-specific cre recombinase–mediated deletion of exon 11 of the ADAM17 gene. The ADAM17 targeting plasmid was assembled in pBR322 using standard cloning techniques. In brief, the 5′ADAM17 small homology was followed by the first loxP site, the FTR-neomycin-FTR cassette, partial intron 10, exon 11, and the first half of intron 12 (including the cryptic but canonical splice acceptor site), the second loxP site, an artificial noncanonical splice donor site, and the 3′ADAM17 large homology. Mouse ADAM17 is located on the minus strand of chromosome 12 from 21,329,378 to 21,379,454 bp (from VEGA annotation of National Center for Biotechnology Information Build 37). To simplify annotation of the ADAM17 sequences subcloned into our targeting plasmid, we defined the adenine base from the start codon ATG of the ADAM17 gene as position 1. The subcloned fragments were: 5′ADAM17 small homology from position 35,826 to 36,574, partial intron 10, exon 11, and the first half of intron 12 from position 36,575 to 37,976 and 3′ADAM17 large homology from position 37,977 to 43,640. The ADAM17 gene fragments were subcloned into pBR322 from a genomic lambda clone. The resulting plasmid was named pBR322-ADAM17-targeting. 129 SvEv ES cells were transfected with the ADAM17 targeting vector to obtain clones with homologous recombination. Targeted embryonic stem cells were injected into C57BL/6 blastocysts to produce chimeras, and germline-transmitted ADAM17exneo/+ mice were obtained by mating chimeras with C57BL/6 mice (InGenious Targeting Laboratory). Because ADAM17exneo/exneo mice were not viable, presumably because of the presence of the neomycin gene cassette, we deleted this cassette by crossing with ACTB:FLPe mice expressing FLP recombinase under the control of the β-actin promoter (Rodríguez et al., 2000). These mice were obtained from F. Stewart (BioInnovation Zentrum, Technical University Dresden, Dresden, Germany). Homozygous ADAM17ex/ex mice were viable but showed drastically reduced ADAM17 protein levels. To verify whether this strategy may be generally applicable, we introduced a DNA sequence containing the same noncanonical acceptor splice site and donor splice site into the single intron of the human CNTF gene. This approach led to the same result seen for the mouse ADAM17 gene. The introduced exon was only partly used in transfected HepG2 cells, leading to inefficient translation of the CNTF protein. We therefore concluded that the EXITS strategy should be applicable to other genes to generate hypomorphic phenotypes.
Southern blotting.
10 µg of genomic DNA was digested by HindIII, separated on 0.8% agarose gels, and transferred to a nylon membrane, which was hybridized at 65°C overnight with the α-32P-dATP–labeled probe as indicated in Fig. 1 A. The membrane was washed at 65°C and signals were detected by phosphoimaging.
RT-PCR.
RNA was isolated from different organs and 2 µg was reverse transcribed into cDNA using Reverse Aid M-MuLV reverse transcription (Fermentas). ADAM17 transcripts were subsequently amplified by PCR using the following primer pair: 3′exon 12 (5′-TAATACCCGGGTCACACTCC-3′) and 5′exon 10 (5′-CTTATTACAACCCAACTGTG-3′).
Induction of colitis and determination of clinical scores.
Colitis was induced by administration of 2% DSS (mol wt, 40,000; TdB Consultancy) in the drinking water for 5 d followed by 5 d of regular drinking water. A high-resolution mouse video endoscopic system was used (HOPKINS Optik 64019BA; KARL STOLZ AIDA VET). Mouse endoscopic index of colitis severity scores (MEICS) and disease activity index (DAI) were obtained as previously described (Fantini et al., 2006). In some experiments, mice were given daily doses (15 µg/mouse) of recombinant TGF-α, EGF, or PBS by intraperitoneal injection.
FITC-dextran and BrdU administration.
Intestinal permeability was assessed by administration of nonmetabolized FITC-dextran 4000 (Sigma-Aldrich). FITC-dextran was administered by oral gavage (60 mg/100 g of body weight) 4 h before killing. Whole blood was obtained by cardiac puncture and serum was obtained by centrifugation at 5,000 rpm for 15 min. Dilutions of FITC-dextran 4000 in PBS were used as a standard curve, and absorption of 100 µl of serum or standard was measured at 488 nm. To study proliferation of epithelial cells, 50 mg BrdU/kg of body weight (EMD) was injected intraperitoneally 2 h before sacrifice.
Colon organ culture and ELISA.
A segment of the distal colon was removed, cut open longitudinally, and washed in PBS containing penicillin and streptomycin. The colon was then further cut into segments of 1 cm2, placed, and incubated in 6-well flat-bottom culture plates containing 2 ml of fresh DME supplemented with penicillin and streptomycin at 37°C for 24h. Culture supernatants were then harvested and assayed for KC, MCP-1, IL-10, IL-11, IL-12, IL-17, IL-21, and TNF levels by ELISA assays (R&D Systems) according to the manufacturer’s protocols.
Myeloperoxidase (MPO) activity measurement.
MPO activity was measured in colon tissue taken from the distal colon. After freezing in liquid nitrogen, equal amounts of tissue samples (30 µl of buffer/mg of colon tissue) were homogenized in MPO buffer (0.05 KPO4 and 0.5% hexadecyl trimethyl ammonium bromide), incubated at 60°C for 2 h, and centrifuged for 5 min at 10,000 rpm. 10 µl of the supernatant was mixed with 50 µl peroxidase substrate (BM blue POD; Roche). After 10–20 min, the reaction was stopped by 50 µl H2SO4 and absorbance was read at 450 nm.
Western blotting.
To monitor protein expression, tissue samples were homogenized in lysis buffer (150 mM NaCl, 2 mM EDTA, 50 mM tris-HCl, pH 7.4, 1% Triton X-100, and 1% NP-40) containing complete protease inhibitor (Roche), and equivalent amounts of homogenate were used for SDS-PAGE. Proteins were transferred to PVDF membranes (GE Healthcare) incubated with primary and secondary antibodies and visualized by chemiluminescence.
Flow cytometric analysis.
107 mouse splenocytes were stimulated with 100 nM PMA for 2 h, 1 µg/ml LPS for 24 h, or 5 µg/ml anti-CD3 and 1 µg/ml anti-CD28 for 24 h and subsequently washed twice with FACS buffer (PBS, 1% BSA, and 0.01% NaN3). To block Fc receptors, the cell suspension was incubated with mouse Fc block CD16/32 mAb (BD) for 10 min and further stained with fluorescence-coupled antibodies against the B cell marker B220, T cell marker CD4, or L-selectin. Cells were washed with 500 µl FACS buffer and analyzed by FACS (FACS-Canto; BD). Data were acquired from 50,000 gated events per sample and further analyzed with the FACS Canto Diva Software.
Brain histology.
After perfusion with 6% glutaraldehyde in phosphate buffer, brains from WT and ADAM17ex/ex mice were serially sectioned at 100 µm on a vibratome (1200 S; Leica) and stained with 0.1% methylene blue.
Animal treatment.
All experiments were performed in accordance with the animal care guidelines of the University of Kiel (acceptance no.: V 742–72 241.121-3 (20–2/04) and (76–7/00)). Mice were maintained in a 12-h light–dark cycle under standard conditions and were provided with food and water ad libitum. Procedures involving animals and their care were conducted in conformity with national and international laws and policies. Blood was drawn from the mice by cardiac puncture under general anesthesia.
Histological examination and immunohistochemistry.
Tissues were fixed in 4% formalin or 4% PFA (colon tissue) and embedded in paraffin. Sections of 5 µM were stained with H&E using standard procedures. pSTAT3, ADAM17, and cyclin D1 staining were performed using an anti–mouse pSTAT3 mAb (1:3,000), an anti–rabbit cyclin D1 mAb (1:100), or an anti–rabbit ADAM17 (1:100) diluted in sample diluents (Dako), and the signal was amplified using the Tyramide Signal Amplification kit (PerkinElmer) and developed with AEC substrate (Dako). To visualize BrdU incorporation, sections were stained with a 1:500 dilution of mouse anti-BrdU mAb (Roche) overnight. Then the sections were incubated for 1 h with biotinylated anti–mouse secondary antibody (1:50; Dako) and developed with AEC substrate. Counterstaining was performed with Shadon Gill III hematoxylin (Thermo Fisher Scientific). The TGF-α staining was performed using a goat anti–mouse TGF-α mAb (1:50; R&D Systems). Staining was visualized after incubation with AEC substrate and counterstaining was performed with Gill III hematoxylin. For immunofluorescence staining, cryosections were fixed in ice-cold acetone for 10 min, followed by sequential incubation with blocking reagent (Dako) to eliminate unspecific background staining. Slides were then incubated for 1 h at room temperature with primary antibodies against CD3 (1:50; Abcam) and CD68 (1:100; Santa Cruz Biotechnology, Inc.). After three times washing with PBS, slides were incubated for 45 min at room temperature with Cy3- or FITC-labeled secondary antibodies (Jackson ImmunoResearch Laboratories). Before examination, nuclei were counterstained with DAPI.
Microarray analysis.
Total RNA was extracted and processed as previously described (Häsler et al., 2009) and hybridized to a mouse gene 1.0 ST array (Affymetrix) according to the manufacturer’s protocol. Data were normalized using RMA (AGCC; Affymetrix), and signals that were not stronger than the 95th percentile of all intron probes on the array were excluded from further analysis. Experimental and analytical part of the microarray analysis was performed according to the MIAME standards. The fold change was calculated based on the ratios of the two medians (WT vs. EX). Inclusion criteria for the genes in the Gene Ontology analysis and the cluster were a fold change >+2 or <−2, with a rank-sum difference ≥ 9. Only characterized genes (no ESTs and no predicted genes) were included. Gene Ontology analysis was performed as previously published (Tavazoie et al., 1999) by comparing regulated genes versus detected genes. Biological processes associated with regulated transcripts were retrieved from the Gene Ontology Consortium (www.geneontology.org). The hierarchical cluster was generated based on regulated genes using the correlation as the similarity measure and UPGMA (unweighted mean) as the clustering method (Spotfire Decision Site 9.1.1). The microarray data can be accessed at http://www.ncbi.nlm.nih.gov/geo/ under the accession no. GSE21691.
Statistical analysis.
All parametric data are presented as the means ± SD. Data from two groups were analyzed for significance using the Student’s t test. Differences were considered to be statistically significant if P ≤ 0.05.
Online supplemental material.
Fig. S1 shows Southern blots and Western blots of mice demonstrating homologous recombination of the ADAM17 ex allele and lack of ADAM17 protein expression in brain, liver, and spleen. Fig. S2 shows the open eye, skin, heart, and brain phenotype of ADAM17ex/ex mice. Fig. S3 demonstrates the endoscopic score and disease activity of mice after DSS treatment. Fig. S4 depicts a gene expression analysis in the intestine of WT and ADAM17ex/ex mice. Fig. S5 demonstrates increased intestinal inflammation in DSS-treated ADAM17ex/ex mice. Table S1 demonstrates segregation of offspring from ADAM17ex/ex mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092366/DC1.
Acknowledgments
We thank Manfred Blessing, Karl-Joseph Kallen, Jacques Peschon, and Jürgen Müllberg for their help during the initial phase of the project. The contribution of Dr. Krzysztof Paliga to Southern blotting and genotyping is gratefully acknowledged. Paulina Kulig helped with immunological analysis. Francis Stewart is thanked for transgenic ACTB-FLPe mice. We thank Silvio Weber for the gift of ADAM10 and ADAM17 antisera. We are grateful to Elsbeth Schulz, Inez Götting, Kaja Köhler, Angelika Zoons, and Birgit Rau for expert technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, Bonn, Germany (SFB415, projects B5, B8, B9; to J. Scheller, S. Rose-John, S. Schreiber, P. Saftig, and K. Reiß), SFB877, the European Union (EU) 6FP SP6MTKD-CT-2006-042586 (J. Cichy), the Bundesministerium für Forschung und Technologie (Berlin, Germany) Network Intestinal infection WP5 and the NGFNplus IGs Genomics of chronic inflammation/GMC (P. Rosenstiel and S. Schreiber), the Interuniversity Attraction Poles Program P6/58 of the Belgian Federal Science Policy Office and EU 6FP DeZnit (EU-FP VI; P. Saftig) and the Cluster of Excellence Inflammation at Interfaces (P. Rosenstiel, S. Schreiber, K. Reiß, J. Scheller, P. Saftig, and S. Rose-John).
All authors declare no conflicting financial interests.
Footnotes
Abbreviations used:
- ADAM
- a disintegrin and metalloprotease
- AR
- amphiregulin
- DSS
- dextran sulfate sodium
- EGF-R
- epidermal growth factor receptor
- EXITS
- exon-induced translational stop
References
- Atreya R., Mudter J., Finotto S., Müllberg J., Jostock T., Wirtz S., Schütz M., Bartsch B., Holtmann M., Becker C., et al. 2000. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat. Med. 6:583–588 10.1038/75068 [DOI] [PubMed] [Google Scholar]
- Berasain C., Perugorria M.J., Latasa M.U., Castillo J., Goñi S., Santamaría M., Prieto J., Avila M.A. 2009. The epidermal growth factor receptor: a link between inflammation and liver cancer. Exp. Biol. Med. 234:713–725 10.3181/0901-MR-12 [DOI] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Breckwoldt M.O., Chen J.W., Stangenberg L., Aikawa E., Rodriguez E., Qiu S., Moskowitz M.A., Weissleder R. 2008. Tracking the inflammatory response in stroke in vivo by sensing the enzyme myeloperoxidase. Proc. Natl. Acad. Sci. USA. 105:18584–18589 10.1073/pnas.0803945105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalaris A., Rabe B., Paliga K., Lange H., Laskay T., Fielding C.A., Jones S.A., Rose-John S., Scheller J. 2007. Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood. 110:1748–1755 10.1182/blood-2007-01-067918 [DOI] [PubMed] [Google Scholar]
- Egger B., Procaccino F., Lakshmanan J., Reinshagen M., Hoffmann P., Patel A., Reuben W., Gnanakkan S., Liu L., Barajas L., Eysselein V.E. 1997. Mice lacking transforming growth factor alpha have an increased susceptibility to dextran sulfate-induced colitis. Gastroenterology. 113:825–832 10.1016/S0016-5085(97)70177-X [DOI] [PubMed] [Google Scholar]
- Egger B., Carey H.V., Procaccino F., Chai N.N., Sandgren E.P., Lakshmanan J., Buslon V.S., French S.W., Büchler M.W., Eysselein V.E. 1998. Reduced susceptibility of mice overexpressing transforming growth factor alpha to dextran sodium sulphate induced colitis. Gut. 43:64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini M.C., Becker C., Tubbe I., Nikolaev A., Lehr H.A., Galle P., Neurath M.F. 2006. Transforming growth factor beta induced FoxP3+ regulatory T cells suppress Th1 mediated experimental colitis. Gut. 55:671–680 10.1136/gut.2005.072801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.-Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., et al. 2009. IL-6 and STAT3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler R., Begun A., Freitag-Wolf S., Kerick M., Mah N., Zvirbliene A., Spehlmann M.E., von Wurmb-Schwark N., Kupcinskas L., Rosenstiel P., Schreiber S. 2009. Genetic control of global gene expression levels in the intestinal mucosa: a human twin study. Physiol. Genomics. 38:73–79 10.1152/physiolgenomics.00010.2009 [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Kimura T., Miyamoto T., Takaishi H., Okada Y., Toyama Y., Blobel C.P. 2007. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 179:2686–2689 [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Kimura T., Miyamoto T., Miyamoto K., Akiyama H., Takaishi H., Morioka H., Nakamura T., Okada Y., Blobel C.P., Toyama Y. 2009. Conditional inactivation of TACE by a Sox9 promoter leads to osteoporosis and increased granulopoiesis via dysregulation of IL-17 and G-CSF. J. Immunol. 182:2093–2101 10.4049/jimmunol.0802491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Murata T., Suzuki T., Shindoh M., Nakajima K., Imai K., Yoshida K. 2006. Requirement of STAT3 activation for maximal collagenase-1 (MMP-1) induction by epidermal growth factor and malignant characteristics in T24 bladder cancer cells. Oncogene. 25:1195–1204 10.1038/sj.onc.1209149 [DOI] [PubMed] [Google Scholar]
- Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. 2005. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 123:291–304 10.1016/j.cell.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Kenny P.A. 2007. TACE: a new target in epidermal growth factor receptor dependent tumors. Differentiation. 75:800–808 10.1111/j.1432-0436.2007.00198.x [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Moss M.L., Sklair-Tavron L., Nudelman R. 2008. Drug insight: tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 4:300–309 10.1038/ncprheum0797 [DOI] [PubMed] [Google Scholar]
- Müllberg J., Althoff K., Jostock T., Rose-John S. 2000. The importance of shedding of membrane proteins for cytokine biology. Eur. Cytokine Netw. 11:27–38 [PubMed] [Google Scholar]
- Murphy G. 2008. The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 8:932–941 10.1038/nrc2459 [DOI] [PubMed] [Google Scholar]
- Peschon J.J., Slack J.L., Reddy P., Stocking K.L., Sunnarborg S.W., Lee D.C., Russell W.E., Castner B.J., Johnson R.S., Fitzner J.N., et al. 1998. An essential role for ectodomain shedding in mammalian development. Science. 282:1281–1284 10.1126/science.282.5392.1281 [DOI] [PubMed] [Google Scholar]
- Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., et al. 2009. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206:1465–1472 10.1084/jem.20082683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F., Leach M.W., Mauze S., Menon S., Caddle L.B., Coffman R.L. 1994. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1:553–562 10.1016/1074-7613(94)90045-0 [DOI] [PubMed] [Google Scholar]
- Rodland K.D., Bollinger N., Ippolito D., Opresko L.K., Coffey R.J., Zangar R., Wiley H.S. 2008. Multiple mechanisms are responsible for transactivation of the epidermal growth factor receptor in mammary epithelial cells. J. Biol. Chem. 283:31477–31487 10.1074/jbc.M800456200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Rose-John S., Scheller J., Elson G., Jones S.A. 2006. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J. Leukoc. Biol. 80:227–236 10.1189/jlb.1105674 [DOI] [PubMed] [Google Scholar]
- Sternlicht M.D., Sunnarborg S.W., Kouros-Mehr H., Yu Y., Lee D.C., Werb Z. 2005. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 132:3923–3933 10.1242/dev.01966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281–285 10.1038/10343 [DOI] [PubMed] [Google Scholar]
- van Dullemen H.M., van Deventer S.J., Hommes D.W., Bijl H.A., Jansen J., Tytgat G.N., Woody J. 1995. Treatment of Crohn’s disease with anti-tumor necrosis factor chimeric monoclonal antibody (cA2). Gastroenterology. 109:129–135 10.1016/0016-5085(95)90277-5 [DOI] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., Neurath M.F. 2007. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2:541–546 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]