Abstract
Objective
The etiology and pathogenesis of moyamoya disease remain unclear. Furthermore, the definitive diagnostic protein-biomarkers for moyamoya disease are still unknown. The present study analyzed serum proteomes from normal controls and moyamoya patients to identify novel serological biomarkers for diagnosing moyamoya disease.
Methods
We compared the two-dimensional electrophoresis patterns of sera from moyamoya disease patients and normal controls and identified the differentially-expressed spots by matrix-assisted laser desorption/ionization-time-of flight mass spectrometry and electrospray ionization quadruple time-of-flight mass spectrometry.
Results
We found and analyzed 22 differently-expressed proteomes. Two proteins were up-regulated. Twenty proteins were down-regulated. Complement C1 inhibitor protein and apolipoprotein C-III showed predominantly changed expressions (complement C1 inhibitor protein averaged a 7.23-fold expression in moyamoya patients as compared to controls, while apolipoprotein C-III averaged a 0.066-fold expression).
Conclusion
Although our study had a small sample size, our proteomic data provide serologic clue proteins for understanding moyamoya disease.
Keywords: Moyamoya disease, Proteome
INTRODUCTION
Moyamoya disease is defined by progressive stenosis and occlusion of the terminal portions of the bilateral internal carotid arteries24). Many studies have investigated the pathophysiology of moyamoya disease. However, the cause and pathogenesis of moyamoya disease are still unknown.
Previous studies have suggested a correlation between certain genetic factors and the pathogenesis of this disease4,5,25,26). Researchers performing CSF studies of moyamoya patients have commented on correlations between the presence of the disease and basic fibroblast growth factor (b-FGF), intercellular adhesion molecule Type-1 (ICAM-1), vascular cell adhesion molecule Type-1 (VCAM-1), and E-selectin level22,28).
Histopathologic studies have shown that the presence of mural thrombi is the most probable culprit in the generation of intimal lesions8), and other studies have suggested a correlation with inflammatory stimuli17). There have been no serologic studies of moyamoya disease, however. In our study, we analyzed the serum proteomes of both adult patients with moyamoya disease and normal controls, attempting to find the key proteins associated with moyamoya disease.
MATERIALS AND METHODS
Sampling of sera
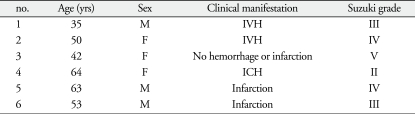
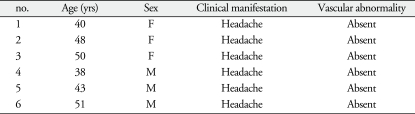
We collected venous blood from 6 moyamoya-disease patients and 6 normal controls. We recruited moyamoya patients with different clinical symptom and different Suzuki grade to avoid over- or down-regulated proteins are limited to a specific stage of moyamoya disease. Six normal controls had no vascular abnormalities, confirming the diagnoses via transfemoral cerebral angiography. To minimize individual variations, we matched the genders and ages of the moyamoya patients with the controls. Table 1, 2 summarize the profiles of the moyamoya patients and the controls.
Table 1.
Profiles of patients with moyamoya disease
F : female, IVH : intraventricular hemorrhage, ICH : intracerebral hemorrhage, M : male, yrs : years
Table 2.
Profiles of normal controls
F : female, M : male, yrs : years
Next, we separated out the sera and kept them frozen at -80℃ until use. We centrifuged the thawed samples at 4,500 rpm for 10 minutes to remove the fibrinogen and then filtered them (0.45 µm pore size, Millipore, Bedford, MA, USA).
The Ethics Committee of Chonbuk National University Medical School approved this study. The participants were fully informed, and all provided their written consent.
Two-dimensional electrophoresis
To desalt the samples, we used dialysis tubing (12k-Da molecular cutoff; Sigma, St. Louis, MO, USA). After desalting, each sample was mixed with a solution containing SDS (10% w/v) and DTT (2.3% w/v) and then heated to 95℃.
For each sample, 300 µg serum protein in rehydration solution [8 mol/L urea, 2% CHAPS, 0.5% immobilized pH gradient (IPG) buffer, 1% DTT, and a trace of bromophenol blue] was loaded onto an Immobiline Dry Strip (Pi 3-10, 24 cm) (Amersham Biosciences, Uppsala, Sweden) for 5 minutes.
We performed the first-dimension isoelectric focusing at 66,000 V/hr and 20℃, using an IPGPhor IEF system (Amersham Biosciences). Next, we equilibrated the gels 30 minutes each in equilibration buffer I [50 nmol/L Tris-Cl (Ph 8.8), 6 mol/L urea, 30% glycerol, 2% SDS, and 0.25% IAA].
We ran the second-dimension gel electrophoresis according to the Ettan DALT II system operating manual (Amersham Biosciences), which requires placing the IPG strips on the surface of the second-dimension gel (a 24 cm, 12.5% SDS-polyacrylamide slab gel) and then sealing the IPG strips with 0.5% agarose in SDS electrophoresis buffer (25 mmol/L Tris base, 192 mmol/L glycine, and 0.1% SDS). These gels were run overnight at 110 volts.
Silver staining
To silver stain the gels, we employed a modified silver staining protocol, using a Silver Stain PlusOne kit (Amersham Biosciences) and omitting the use of glutaraldehyde in the sensitization step and formaldehyde in the silver impregnation step. After electrophoresis, the gels were fixed with 40% methanol and 10% acetic acid for 30 minutes. The gels were sensitized by incubating them in sensitizing solution (0.2% sodium thiosulphate, 30% methanol, and sodium acetate, 68 g/L). We then rinsed the gels with 3 changes of distilled water at 5 minutes/rinse. After running, the gels were incubated in 0.25% silver nitrate for 20 minutes. Next, we discarded the silver nitrate, rinsed the gels twice with distilled water, 1 minute/rinse, and then developed them, with intensive shaking, in 2.5% sodium carbonate containing 0.15% formaldehyde. After we achieved the desired staining intensity, we terminated the gels' development using 1.46% ethylenediaminetetraacetic acid (EDTA).
Image analysis
We scanned the silver-stained two-dimensional electrophoresis (2-DE) gels, using LabScan software on an image scanner (Amersham Biosciences) and digitized and analyzed the information using an Image Master 2D (Amersham Biosciences), carrying out spot standardization on all the matched spots. Then we normalized each spot volume (intensity) as a percentage of the total spot volume, using only the spots that were present on all the gels.
Destaining
To destain the silver-stained proteins, we used chemical reducers as described Scheler et al.21), as follows. We prepared two stock solutions, 30 mM potassium ferricyanide and 100 mM sodium thiosulfate, dissolved in water. We mixed each working solution by mixing a 1 : 1 ratio prior to use. After we excised the gel sections containing the protein spots of interest, we added 30-50 mL working solution to cover these gels, vortexing them occasionally. We monitored stain intensity until the brownish color disappeared, then rinsed each gel with water a few times, to halt the reaction. Next, we covered each gel with 200 mM ammonium bicarbonate for 20 minutes and then discarded the solution. Subsequently, we cut each gel into small pieces, washed these with water, and dehydrated them repeatedly with fresh acetonitrile until the pieces became opaque white in color. Finally, the gel pieces were dried in a vacuum centrifuge for 30 minutes.
Trypsin digestion of proteins in gel
The enzymatic digestion was performed as described by Hellman and Shevchenko, as follows. The samples incubated overnight, at 37℃, in 5-10 ng/µL trypsin and 50 mM ammonium bicarbonate. Following this enzymatic digestion, we thrice extracted the resultant peptides with 10-20 µL of 5% trifluoroacetic acid in 50% acetonitrile and dried them 30 minutes in a vacuum centrifuge.
Identification of proteins
We analyzed the dried samples using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer [MALDI-TOF MS (Voyager-DE PRO)], for the peptide mass fingerprinting, and via electrospray ionization quadruple time-of-flight (ESI-Q-TOF) mass spectrometry analysis, for the peptide sequencing. The database searches employed MS-fit, which is accessible on the World Wide Web at http://kr.expasy.org and http://www.ncbi.nlm.nih.gov.
RESULTS
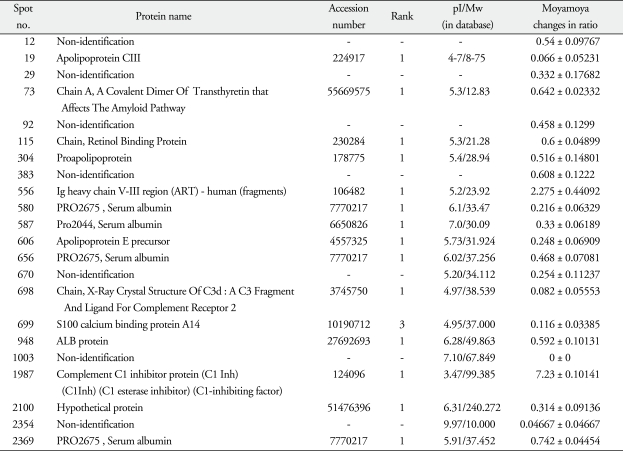
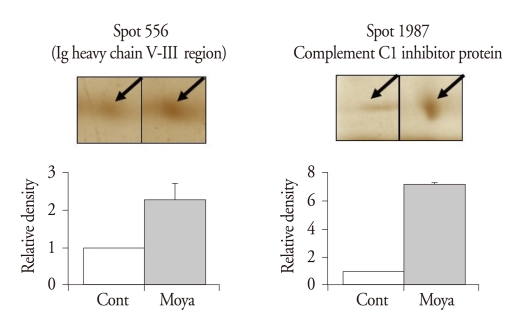
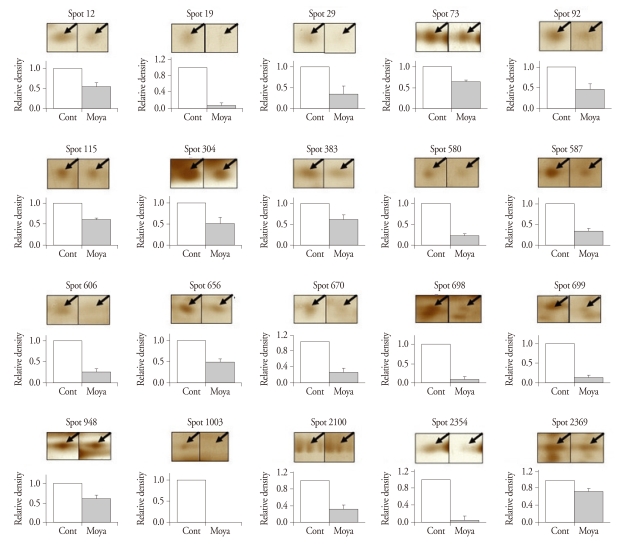
After routine silver staining procedures, we observed 2000 to 3000 protein spots of about 150 µg. We found 22 significantly different spots, 2 up-regulated and 20 down-regulated (Table 3). After these proteins digested overnight in trypsin, the MALDI-TOF MS and ESI Q-TOF MS analyses identified the up-regulated spots as an Ig heavy chain V-III region (spot 556, change of ratio in moyamoya : 2.275 ± 0.44092) and complement C1 inhibitor protein (spot 1987, 7.23 ± 0.10141) (Fig. 1) and the down-regulated spots as apolipoprotein C-III (spot 19, 0.066 ± 0.05231), a covalent dimer of transthyretin (spot 73, 0.642 ± 0.02332), a retinolbinding protein (spot 115, 0.6 ± 0.04899), a proapolipoprotein (spot 304, 0.516 ± 0.14801), PRO 2675 (spots 580, 0.216 ± 0.06329; spot 656, 0.468 ± 0.07081; spot 2369, 0.742 ± 0.04454), PRO 2044 (spot 587, 0.33 ± 0.06189), apolipoprotein E precursor (spot 606, 0.248 ± 0.06909), the X-ray crystal structure of C3d (spot 698, 0.082 ± 0.05553), S100 calcium binding protein A14 (spot 699, 0.116 ± 0.03385), ALB protein (spot 948, 0.592 ± 0.10131), a hypothetical protein (spot 2100, 0.314 ± 0.09136), and some unidentified proteins (spot 12, 0.54 ± 0.09767; spot 29, 0.332 ± 0.17682; spot 92, 0.458 ± 0.1299; spot 383, 0.608 ± 0.1222; spot 670, 0.254 ± 0.11237; spot 1003, 0 ± 0, and spot 2354, 0.04667 ± 0.04667) (Fig. 2).
Table 3.
The proteins significantly changed in sera of moyamoya patients
Mw : molecular weight
Fig. 1.
Up-regulated spots that were significantly and consistently changed in human sera of moyamoya patients compared to normal controls. In each pair of two-dimensional electrophoresis (2-DE) gels from a normal control and a moyamoya patient, the density of a spot in moyamoya gel was normalized to the density of the corresponding spot in the control gel.
Fig. 2.
Down-regulated spots that were significantly and consistently changed in human sera of moyamoya patients compared to normal controls. In each pair of two-dimensional electrophoresis gels from a normal control and a moyamoya patient, the density of a spot in moyamoya gel was normalized to the density of the corresponding spot in the control gel (characteristics of each spot is shown in Table 3).
DISCUSSION
Various studies have attempted to discover the pathophysiology of moyamoya disease. However, the cause and pathogenesis of moyamoya disease are still unknown.
Several previous reports have implicated genetic factors in moyamoya's pathogenesis4,5,25,26). Researchers have proposed chromosomes 3, 6, 8, 12, and 17 as possible locations of the responsible gene10,12,20,27). Ikeda and Yoshimoto11) reported that familial moyamoya disease patients have much lower mitochondrial deoxyhibonucleic acid (mtDNA) heterogeneity than that seen in the general Japanese population. Hong et al.7) documented HLA DRB1 and DQBI were elevated in familial moyamoya disease. Kang et al.13) suggested that the presence of a G/C heterozygous genotype at position-418 in the tissue inhibitor of metalloproteinases-2 (TIMP-2) promoter region could be a predisposing genetic factor for familial moyamoya disease.
Researchers performing a CSF study in moyamoya patients have commented on correlations with b-FGF, ICAM-1, VCAM-1, and E-selectin level22,28). Histopathologic studies suggested the existence of mural thrombi is the most probable candidate for the cause of intimal lesions8), and other studies have suggested a correlation with inflammatory stimuli17).
There have been no prior serologic studies of moyamoya disease. Our results show that complement C1 inhibitor protein was up-regulated (7.23 ± 0.10141 fold) in moyamoya patients. Complement C1 inhibitor protein reduces polymorphonuclear leukocyte accumulation and neuronal damage in focal ischemia and reperfusion1). The early inhibition of the classical complement activation pathway by complement C1 inhibitor protein may help in protecting the penumbra zone in focal cerebral ischemia6). In a model of cortical venous infarction, the infarct volume after complement inhibition (0.86 ± 0.23 mm3) was only 27.7% that of the group (3.09 ± 0.62 mm3). In ischemic mice receiving complement C1 inhibitor protein via intravenous injection, C1-INH significantly dampened the messenger ribonucleic acid (mRNA) expression of the adhesion molecules induced by the ischemic insult (P-selectin and ICAM-1). Also, it significantly decreased a pro-inflammatory cytokine [tumor necrosis factor (TNF)-alpha, interleukin (IL)-18], increased protective cytokine (IL-6, IL-10) gene expression, and markedly inhibited the activation and/or recruitment of microglia/macrophages23). In moyamoya patients, the over-expression of complement C1 inhibitor protein may be associated with protection from ischemia and progressive obstruction of distal internal carotid artery (ICA).
Our study also found the Ig heavy chain V-III region (ART) was up-regulated (2.275 ± 0.44092). No other moyamoya studies have commented on this protein. We need to study this protein to find its correlation with moyamoya disease.
Among down-regulated proteins in our result, apolipoprotein C-III was the lowest expressed protein (0.066 ± 0.05231). Apolipoprotein C-III induces both THP-1 cells and human peripheral monocytes to adhere to endothelial cells. In addition, ApoC-III is known to increase VCAM-1 and ICAM-1 protein expression in non-activated endothelial cells14). Study of Masuda et al.17) provided evidence that, in moyamoya, smooth muscle cells proliferate in intracranial major arteries' occlusive lesions. Colonization due to inflammatory stimuli may induce a proliferative response by smooth muscle and contribute to the formation of intracranial occlusive lesions in moyamoya disease17). We assume that the down-regulation of apolipoprotein C-III is a biological protection, reducing the formation of intracranial occlusive lesions.
We could not verify the other down-regulated proteins (a covalent dimer of transthyretin, a retinol-binding protein, a proapolipoprotein, PRO 2675, PRO2044, apolipoprotein E precursor, the X-ray crystal structure of C3d, S100 calcium binding protein A14, ALB protein, and a hypothetical protein) as having any relation to moyamoya disease.
Caillot et al.2) described how one potential use of transthyretin, in serological tests for the non-invasive diagnosis of liver fibrosis, opens new opportunities for better follow-up of hepatitis C virus-infected patients. Hybelová et al.9) pointed out the substantial clinical value to be found in MS patients' levels of CSF and serum transthyretin. Report of Chiang et al.3) demonstrated that the CSF transthyretin concentration of GBS patients was significantly higher than that of the multiple sclerosis, Alzheimer's disease, and viral meningitis patients and the controls.
Regarding the retinol-binding protein, study of Kwon et al.16) suggested that RBP4 is a serologic marker for disease severity in patients with chronic liver disease and could also be useful as an early marker of chronic liver disease and the relative success of antiviral therapy. Nobili et al.18) showed an inverse relationship between RBP4 levels and degree of liver damage. RBP4 therefore might be a novel, noninvasive marker of the severity of pediatric nonalcoholic fatty liver disease.
Kim et al.15) described how serum concentrations of proapolipoprotein A1 were higher in breast cancer patients. Park et al.19) reported that proteomic analysis of amniotic fluid can distinguish preeclampsia from chronic hypertension and identified the discriminatory proteins as proapolipoprotein A-I and SBBI42.
PRO 2675, PRO 2044, apolipoprotein E precursor, the X-ray crystal structure of C3d, S100 calcium binding protein A14, ALB protein, and the hypothetical protein have no reported serological significance.
Although our experimental group is small, each patient was at different stage of the disease and had different clinical symptom. Therefore, the over-expressed proteins and down-regulated proteins are not limited to a specific stage of moyamoya disease. This suggests that these proteins correlate to the general pathophysiology of moyamoya disease.
The proteins that were not readily identifiable in this study (spots 12, 29, 92, 383, 670, 1003, and 2354) may be novel proteins in the pathophysiology of moyamoya disease. We need to study further of these proteins.
CONCLUSION
We found 22 particular serum proteins in patients with moyamoya disease, as compared with normal controls. These proteins may provide clues regarding the pathophysiology of moyamoya disease.
References
- 1.Akita N, Nakase H, Kaido T, Kanemoto Y, Sakaki T. Protective effect of C1 esterase inhibitor on reperfusion injury in the rat middle cerebral artery occlusion model. Neurosurgery. 2003;52:395–400. doi: 10.1227/01.neu.0000043710.61233.b4. discussion 400-401. [DOI] [PubMed] [Google Scholar]
- 2.Caillot F, Hiron M, Goria O, Gueudin M, Francois A, Scotte M, et al. Novel serum markers of fibrosis progression for the follow-up of hepatitis C virus-infected patients. Am J Pathol. 2009;175:46–53. doi: 10.2353/ajpath.2009.080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang HL, Lyu RK, Tseng MY, Chang KH, Chang HS, Hsu WC, et al. Analyses of transthyretin concentration in the cerebrospinal fluid of patients with Guillain-Barré syndrome and other neurological disorders. Clin Chim Acta. 2009;405:143–147. doi: 10.1016/j.cca.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Fukui M, Kono S, Sueishi K, Ikezaki K. Moyamoya disease. Neuropathology. 2000;20(Suppl):S61–S64. doi: 10.1046/j.1440-1789.2000.00300.x. [DOI] [PubMed] [Google Scholar]
- 5.Fukuyama Y, Osawa M, Kanai N. Moyamoya disease (syndrome) and the Down syndrome. Brain Dev. 1992;14:254–256. doi: 10.1016/s0387-7604(12)80242-7. [DOI] [PubMed] [Google Scholar]
- 6.Heimann A, Takeshima T, Horstick G, Kempski O. C1-esterase inhibitor reduces infarct volume after cortical vein occlusion. Brain Res. 1999;838:210–213. doi: 10.1016/s0006-8993(99)01740-0. [DOI] [PubMed] [Google Scholar]
- 7.Hong SH, Wang KC, Kim SK, Cho BK, Park MH. Association of HLA-DR and -DQ genes with familial moyamoya disease in Koreans. J Korean Neurosurg Soc. 2009;46:558–563. doi: 10.3340/jkns.2009.46.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosoda Y, Ikeda E, Hirose S. Histopathological studies on spontaneous occlusion of the circle of Willis (cerebrovascular moyamoya disease) Clin Neurol Neurosurg. 1997;99(Suppl 2):S203–S208. doi: 10.1016/s0303-8467(97)00044-9. [DOI] [PubMed] [Google Scholar]
- 9.Hybelová M, Svatonová J, Sobek O, Adam P, Dolezil D, Adam D. Cerebrospinal fluid and serum prealbumin (transthyretin) in patients with multiple sclerosis (MS) : comparison of particular subgroups of MS patients. Folia Microbiol (Praha) 2009;54:173–176. doi: 10.1007/s12223-009-0027-4. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H, Sasaki T, Yoshimoto T, Fukui M, Arinami T. Mapping of a familial moyamoya disease gene to chromosome 3p24.2-p26. Am J Hum Genet. 1999;64:533–537. doi: 10.1086/302243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda H, Yoshimoto T. Specific genetic characteristics in patients with familial moyamoya disease. J Stroke Cerebrovasc Dis. 2005;14:244–250. doi: 10.1016/j.jstrokecerebrovasdis.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Linkage analysis of moyamoya disease on chromosome 6. J Child Neurol. 2000;15:179–182. doi: 10.1177/088307380001500307. [DOI] [PubMed] [Google Scholar]
- 13.Kang HS, Kim SK, Cho BK, Kim YY, Hwang YS, Wang KC. Single nucleotide polymorphisms of tissue inhibitor of metalloproteinase genes in familial moyamoya disease. Neurosurgery. 2006;58:1074–1080. doi: 10.1227/01.NEU.0000215854.66011.4F. discussion 1074-1080. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami A, Aikawa M, Alcaide P, Luscinskas FW, Libby P, Sacks FM. Apolipoprotein CIII induces expression of vascular cell adhesion molecule-1 in vascular endothelial cells and increases adhesion of monocytic cells. Circulation. 2006;114:681–687. doi: 10.1161/CIRCULATIONAHA.106.622514. [DOI] [PubMed] [Google Scholar]
- 15.Kim BK, Lee JW, Park PJ, Shin YS, Lee WY, Lee KA, et al. The multiplex bead array approach to identifying serum biomarkers associated with breast cancer. Breast Cancer Res. 2009;11:R22. doi: 10.1186/bcr2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon JH, Park ST, Kim GD, You CR, Kim JD, Woo HY, et al. [The value of serum retinol-binding protein 4 levels for determining disease severity in patients with chronic liver disease.] Korean J Hepatol. 2009;15:59–69. doi: 10.3350/kjhep.2009.15.1.59. [DOI] [PubMed] [Google Scholar]
- 17.Masuda J, Ogata J, Yutani C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke. 1993;24:1960–1967. doi: 10.1161/01.str.24.12.1960. [DOI] [PubMed] [Google Scholar]
- 18.Nobili V, Alkhouri N, Alisi A, Ottino S, Lopez R, Manco M, et al. Retinol-binding protein 4 : a promising circulating marker of liver damage in pediatric nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2009;7:575–579. doi: 10.1016/j.cgh.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Park JS, Oh KJ, Norwitz ER, Han JS, Choi HJ, Seong HS, et al. Identification of proteomic biomarkers of preeclampsia in amniotic fluid using SELDI-TOF mass spectrometry. Reprod Sci. 2008;15:457–468. doi: 10.1177/1933719108316909. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K, Yoshimoto T, Fukui M, et al. A novel susceptibility locus for moyamoya disease on chromosome 8q23. J Hum Genet. 2004;49:278–281. doi: 10.1007/s10038-004-0143-6. [DOI] [PubMed] [Google Scholar]
- 21.Scheler C, Lamer S, Pan Z, Li XP, Salnikow J, Jungblut P. Peptide mass fingerprint sequence coverage from differently stained proteins on two-dimensional electrophoresis patterns by matrix assisted laser desorption/ionization-mass spectrometry (MALDI-MS) Electrophoresis. 1998;19:918–927. doi: 10.1002/elps.1150190607. [DOI] [PubMed] [Google Scholar]
- 22.Soriano SG, Cowan DB, Proctor MR, Scott RM. Levels of soluble adhesion molecules are elevated in the cerebrospinal fluid of children with moyamoya syndrome. Neurosurgery. 2002;50:544–549. doi: 10.1097/00006123-200203000-00022. [DOI] [PubMed] [Google Scholar]
- 23.Storini C, Rossi E, Marrella V, Distaso M, Veerhuis R, Vergani C, et al. C1-inhibitor protects against brain ischemia-reperfusion injury via inhibition of cell recruitment and inflammation. Neurobiol Dis. 2005;19:10–17. doi: 10.1016/j.nbd.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki J, Takaku A. Cerebrovascular "moyamoya" disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol. 1969;20:288–299. doi: 10.1001/archneur.1969.00480090076012. [DOI] [PubMed] [Google Scholar]
- 25.Wakai K, Tamakoshi A, Ikezaki K, Fukui M, Kawamura T, Aoki R, et al. Epidemiological features of moyamoya disease in Japan : findings from a nationwide survey. Clin Neurol Neurosurg. 1997;99(Suppl 2):S1–S5. doi: 10.1016/s0303-8467(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi T, Houkin K, Tada M, Abe H. Familial occurrence of moyamoya disease. Clin Neurol Neurosurg. 1997;99(Suppl 2):S162–S167. doi: 10.1016/s0303-8467(97)00054-1. [DOI] [PubMed] [Google Scholar]
- 27.Yamauchi T, Tada M, Houkin K, Tanaka T, Nakamura Y, Kuroda S, et al. Linkage of familial moyamoya disease (spontaneous occlusion of the circle of Willis) to chromosome 17q25. Stroke. 2000;31:930–935. doi: 10.1161/01.str.31.4.930. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimoto T, Houkin K, Takahashi A, Abe H. Evaluation of cytokines in cerebrospinal fluid from patients with moyamoya disease. Clin Neurol Neurosurg. 1997;99(Suppl 2):S218–S220. doi: 10.1016/s0303-8467(97)00047-4. [DOI] [PubMed] [Google Scholar]