Abstract
To investigate the association of expression and promoter methyiation of tumor-suppressor genes with risk of ovarian cancer, we conducted a case-control study of 102 patients with serous epithelial ovarian cancer and 100 patients without ovarian cancers. We measured mRNA expression levels (by real-time reverse transcription polymerase chain reaction) and methyiation status (by methylation-specific polymerase chain reaction) of five candidate genes (BRCA1, BRCA2, hMLHl, MGMT, and DNMT3B) in tumors from the cases and normal ovaries from the controls. We found that mRNA expression levels of the five genes were decreased in tumors than in normal ovaries with 0.39-fold for BRCA1, 0.25-fold for BRCA2, 0.42-fold for hMLHl, 0.45-fold for MGMT, and 0.87-fold for DNMT3B, calculated by the 2-ΔΔCT method. Ovarian cancer risk (odds ratios, ORs) was associated with low expression of all genes (2.95 [95% confidence interval (CI), 1.51 - 5.78] for BRCA1, 3.65 (95% CI, 1.82 - 7.30) for BRCA2, 5.25 (95% CI, 2.52 - 10.96) for hMLHl, and 4.72 (95% CI, 2.32 - 9.62) for MGMT) but not DNMT3B. However, methyiation status was not associated with gene expression levels in the tumors, except for hMLHl whose mean (± SD) gene expression was significantly lower in methylated (13.0 ± 7.6) than in unmethylated (31.2 ± 44.8) tumors (P < 0.001). We concluded that low mRNA expression of these tumor-suppressor genes, likely due to molecular mechanisms in addition to the promoter methyiation in some instances, may be a biomarker for ovarian cancer risk in this study population. Larger studies are needed to validate our findings.
Keywords: Case-control study, DNA repair, epigenetics, molecular epidemiology, ovarian cancer
Introduction
Ovarian cancer is one of the most lethal malignancies in women worldwide [1, 2]. In the United States, ovarian cancer is the ninth most common malignancy and the fifth most common cause of death from female cancers. In 2009, the American Cancer Society estimated that 21,550 women will be diagnosed with ovarian cancer and that 14,600 women will lose their lives [3]. Because of the inability to detect ovarian cancer at its early stage that is highly treatable, more than two-third of patients are diagnosed with the advanced-stage disease, which leads to the survival rate essentially unchanged over the last decades. Although the molecular mechanisms leading to the development of ovarian cancer remain largely unknown, epigenetic alterations have been implicated. Therefore, further understanding epigenetic alterations underlying ovarian tumorigenesis may provide the basis for new tools for both identification of patients at risk and early diagnosis of ovarian cancer, which may ultimately reduce the incidence and mortality.
DNA methyiation at CpG sites in the promoter region of a gene can alter mRNA expression, which is one of the phenotypic characteristics of tumor development and progression [4-6]. The inactivation of tumor-suppressor genes due to aberrant methyiation of CpG islands has been implicated as one of the major pathways involved in the development of cancers, including ovarian cancer [6-8].
The importance of the role of aberrant methylation in the development of cancer has become increasingly apparent with the growing list of genes that has been shown to be susceptible to inactivation by promoter hypermethylation [9-15]. It has been observed that promoter methylation of specific genes in cancer occurs in both a tissue-specific and cell-specific manner, making the identification of methylation patterns a potentially useful tool for cancer diagnosis and management [9], particularly with the emerging high-throughput [16] and even genome-wide [13] technologies. It has been suggested that virtually all known cellular pathways contributing to carcino-genesis are more or less affected by epigenetic factors identified in cancer [13]. Because aberrant DNA methylation is frequently observed in early development of ovarian cancer, it has been predicted that such alterations can be detected in DNA circulating in the blood, potentially leading a non-invasive cancer detection test [17]. Specifically, frequent epigenetic inactivation of hMLHl, CDKN2A, and MG/W7were reported to be involved in ovarian carcinomas, using matched tumors and normal tissues from the same 18 patients [18], but another study showed a much less frequent methylation of hMLHl and MGMT in 13 ovarian cancer cell lines [19]. Such small studies often provide unstable estimates that are hard to replicate. In particular, the use of ovarian cancer cell lines without the control of normal ovaries from patients without ovarian cancer does not control for genetic effects on the carcinogenesis of normal ovaries.
In this study, we used a case-control design to investigate the association between ovarian cancer risk and mRNA expression levels and methylation of five candidate tumor-suppressor genes involved in DNA repair.
Materials and methods
Study subjects
Ovarian tumor tissues were obtained from patients with primary serous epithelial ovarian cancer newly diagnosed between January 2000 and March 2005 at The University of Texas M. D. Anderson Cancer Center. Tissues from normal ovaries, used as the control, were obtained from patients who underwent surgery during the same time period for conditions other than ovarian cancer. Informed consent was obtained from each patient, and the study was approved by M. D. Anderson's institutional review board. All samples were snap-frozen after surgical removal and then stored at −80°C in the Gynecologic Cancer Tumor Bank at M. D. Anderson Cancer Center until pathologic examination and testing. For this case-control study, we obtained 102 surgically-resected ovarian tumors and 100 apparently normal ovarian tissues and DNA and RNA were extracted from about 200 mg of fresh-frozen tissue specimens.
Real-time reverse transcription polymerase chain reaction for gene expression
In this study, we measured five tumor-suppressor genes involved in DNA repair: BRCA1, BRCA2, hMLHl, MGMT, and DNMT3B using GAPDH as the internal control. Total RNA was extracted with Tri-Reagent according to the manufacturer's protocol (Molecular Research Center, Cincinnati, OH). We assessed the quality of the extracted total RNA by 1% agarose gel electrophoresis for RNA degradation by visualizing the 18S and 28S RNA bands under ultraviolet light as shown previously with two clean bands [20]. The RNA concentration was determined with the Gene Quant Pro RNA/DNA Calculator (Amersham Pharmacia, Cambridge, England) before the detection of specific gene expression. The primers and probes for detecting mRNA levels of MGMT, hMLHl, and GAPDH were used as previously reported [20-22]. The cDNA sequences of BRCA1, BRCA2, and DNMT3B were referenced to design the primers and probes using express software from Applied Biosystems (Foster City, CA). All sequences of primers and probes are summarized in Table 1. Reverse transcription polymerase chain reaction (RT-PCR) was performed using TaqMan one-step RT-PCR Master Mix Reagents kit (Applied Biosystems) according to the manufacturer's protocol as previously described [20].
Table 1.
Oligonucleotide primer and probe sequences used in this study
| Gene | Primer/probe sequence* | Position/PCR† product size | |
|---|---|---|---|
| BRCA1 | Forward primer | TTTCTATTTGGATCCCTTCGAGG | 136 - 158 |
| Reverse primer | GTGAGCGCACTTCTGCCC | 185 - 202/67 bp | |
| Probe | FAM-CCCCCGTGGCTGTGGAACCC-TAMRA | 164 - 183 | |
| BRCA2 | Forward primer | TGCTGCAAGCAACCTCCA | 9587 - 9604 |
| Reverse primer | AGAAAAATCTCCAGCAAATAAAGTAAGAA | 9631 - 9659/73 bp | |
| Probe | FAM-TGGCGACCAGAATCCAAATCAGGC-TAMRA | 9606 - 9629 | |
| hMLH1 | Forward primer | GTTCTCCGGGAGATGTTGCATA | 1579 - 1600 |
| Reverse primer | TGGTGGTGTTGAGAAGGTATAACTTG | 1661 - 1681/ 102 bp | |
| Probe | FAM-CCTCAGTGGGCCTTGGCACAGC-TAMRA | 1627 - 1644 | |
| MGMT | Forward primer | CAATGAGAGGCAATCCTGTCC | 494 - 514 |
| Reverse primer | CACGGCTCCGCTGCTG | 546 - 561/ 68 bp | |
| Probe | VIC-CTCATCCCGTGCCACAGAGTGGTCT-TAMRA | 520 - 544 | |
| DNMT3B | Forward primer | TCTCCTATCGAAAAGCCATGTA | 1208 - 1229 |
| Reverse primer | GGGAAGGTCTTGCCAGC | 1258 - 1274/ 67 bp | |
| Probe | FAM-CATGCTCTGGAGAAAGCTAGGGTGC-TAMRA | 1231 - 1255 | |
| GAPDH | Forward primer | GAAGGTGAAGGTCGGAGTC | 131 - 149 |
| Reverse primer | GAAGATGGTGATGGGATTTC | 337 - 356/226 bp | |
| Probe | FAM-CAAGCTTCCCGTTCTCAGCC-TAMRA | 308 - 327 |
VIC, FAM, and TAMRA are different dyes that were used to label two ends of the probes.
The nucleotide (nt) position of the cDNAs with GenBank accession numbers XM_208274 for BRCA1, NM_000059.2 for BRCA2, U_07343 for hMLHl, M29971 for MGMT, NM_006892 for DNMT3B, and AK026525 for GAPDH.
Methylation-specific PCR
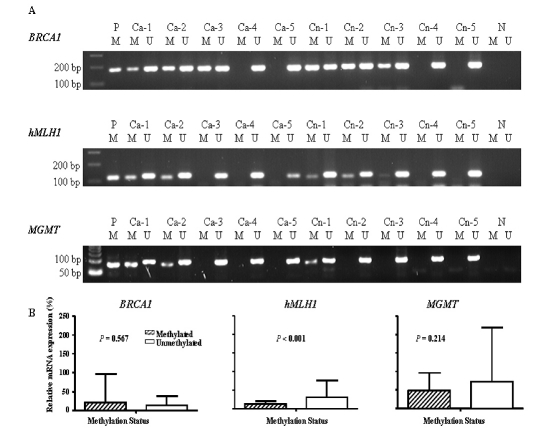
The methylation status of target genes were qualitatively analyzed as described previously [20]. Briefly, Genomic DNA samples were modified with sodium bisulfite. 1 μg of DNA was denatured by NaOH (50 μ1; final concentration, 0.2 M) for 10 min at 37°C, mixed with 30 μ1 of freshly prepared 10 mM hydroquinone (Sigma, St. Louis, MO) and 520 JLXI of 3 M, pH 5.0 sodium bisulfite (Sigma), and incubated under mineral oil at 55°C for 16 h. The DNA samples were desalted through Wizard columns (Promega, Madison, WI) and then desulphonated by NaOH treatment (final concentration, 0.3 M) for five minutes at room temperature followed by ethanol precipitation. DNA was resuspended in water and used shortly after reconstitution. For PCR amplification, the bisulfite-modified DNA (100 ng) was separately amplified using published primers specific for the methylated as well as the unmethylated sequences of genes including BRCA1 [23], MGMT [24], and hMLHl [25]. Since the BRCA2 gene is rarely methylated and there is no report on promoter methylation of the DNMT3B gene, we did not perform methylation-specific PCR assays for these two genes. CpGenome Universal Methylated DNA (Serologicals Corporation, Norcross, GA) was used as the positive control for amplification of methylated alleles, and water blanks without added DNA were included as the negative PCR controls in each assay. DNA amplification was carried out as previously described [20]. PCR products were analyzed on 2% agarose gels containing ethidium bromide (Figure 1A). Two researchers (JA and ZL) independently evaluated the results, and questionable assays were repeated to achieve complete agreement.
Figure 1.
A, Methylation-specific PCR analysis of the methylation status in BRCA1, hMLHl, and MGMT. Representative PCR products of the promoter region of these genes were amplified by the MSP method. P, positive control (CpGnome Universal Methylated DNA); Ca, ovarian cancer tissues; Cn, normal ovarian tissues; N, negative control (water blank); M, methylated; U, unmethylated. B, Relative mRNA expression levels of methylated and unmethylated BRCA1, hMLHl, and MGMT in the cases.
Statistical analysis
The 2-ΔΔCT method was used to calculate changes in candidate gene expression levels in tumor tissues normalized to the internal control GAPDH and relative to the normal tissues as reported [26-28]. The Student's t-test was used to compare differences in the relative expression levels to the internal control GAPDH for the subgroups, which were analyzed as a continuous variable between groups. Two-sided X2 test was used for the comparison of categorical variable distribution between two groups. For calculating odds ratios (ORs) and 95% confidence intervals (CIs), the median relative expression level of each gene in the controls was used as the cutoff point. Adjusted ORs were calculated by fitting logistic regression models with adjustment for age and ethnicity. All statistical analyses were performed with SAS software (version 9.1; SAS Institute Inc., Cary, NC).
Results
Demographic characteristics for the study population are summarized in Table 2. There was significant difference in age between the case and control groups. Controls (53%) were younger (<50 years) than patients (13%) (Table 2). The mean age of cases (62.3 ± 10.0 years [±SD]) was significantly higher than that of controls (50.7 ± 14.1 years) (P < 0.001), and ages ranged from 39 to 81 years for cases and from 23 to 85 years for controls (Table 3). About 78% of cases and 74% of controls were non-Hispanic whites. The other one-forth of subjects consisted of small numbers in minority groups including African-, Mexican-, and Asian-American and other ethnicities as shown in Table 2. All cases were diagnosed with high-grade serous ovarian tumors, which had 93%stage III or IV tumors (Table 2).
Table 2.
Distribution of demographic characteristics of patients with ovarian cancer (cases) and with normal ovarian tissues (controls)
| Cases (n = 102) | Controls (n = 100) | ||||
|---|---|---|---|---|---|
| Variable | No. | % | No. | % | P value* |
| Age (years) | <0.001 | ||||
| <50 | 13 | (12.8) | 53 | (53.0) | |
| ≥50 | 89 | (87.2) | 47 | (47.0) | |
| Ethnicity | 0.007 | ||||
| Non-Hispanic white | 80 | (78.4) | 74 | (74.0) | |
| African American | 5 | (4.9) | 7 | (7.0) | |
| Mexican American | 9 | (8.8) | 19 | (19.0) | |
| Others† | 8 | (7.9) | 0 | (0.0) | |
| Tumor grade | |||||
| High | 102 | (100.0) | |||
| Tumor stage | |||||
| I + II | 7 | (6.9) | |||
| III | 71 | (69.6) | |||
| IV | 24 | (23.5) | |||
Two-sided χ2 tests.
Others included five Asian cases and three cases with unknown ethnicity.
Table 3.
mRNA Expression of candidate genes in ovarian tumors and normal ovarian tissues
| Variable | Controls | Cases | P vlaue† | Stage I + II | Stage III | Stage IV | P value‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mean ± SD* | No. | Mean ± SD* | No. | Mean ± SD* | No. | Mean ± SD* | No. | Mean ± SD* | |||
| Age, range (years) | 100 | 50.7 ± 14.1, 23-85 | 102 | 62.3 ± 10.0, 39-81 | <0.001 | 7 | 64.0 ± 10.7, 48-78 | 71 | 63.5 ± 10.1, 39 - 81 | 24 | 58.4 ± 8.9 45-74 | 0.087 |
| mRNA Expression | ||||||||||||
| BRCA1 | 97 | 25.4 ± 31.5 | 98 | 17.0 ± 54.6 | 0.190 | 7 | 12.6 ± 28.6 | 69 | 17.9 ± 61.8 | 22 | 15.6 ± 34.3 | 0.963 |
| Methylated | 50 | 29.0 ± 37.1 | 45 | 20.7 ± 76.3 | 0.511 | 3 | 2.2 ± 0.6 | 27 | 29.0 ± 79.5 | 15 | 9.4 ± 34.3 | 0.674 |
| Unmethylated | 47 | 21.6 ± 24.1 | 53 | 13.8 ± 24.7 | 0.118 | 4 | 20.4 ± 38.1 | 42 | 10.7 ± 21.1 | 7 | 28.8 ± 52.7 | 0.173 |
| 0.246 | 0.567 | 0.409 | 0.341 | 0.380 | ||||||||
| BRCA2 | 99 | 12.9 ± 15.5 | 91 | 6.2 ± 9.6 | <0.001 | 7 | 11.7 ± 25.9 | 63 | 5.3 ± 5.5 | 21 | 7.1 ± 10.4 | 0.224 |
| hMLH1 | 97 | 47.4 ± 25.4 | 102 | 27.1 ± 40.3 | <0.001 | 7 | 12.8 ± 8.1 | 71 | 29.2 ± 43.3 | 24 | 25.0 ± 36.4 | 0.570 |
| Methylated | 33 | 43.7 ± 23.7 | 23 | 13.0 ± 7.6 | <0.001 | 2 | 12.1 ± 8.5 | 13 | 12.1 ± 8.2 | 8 | 14.6 ± 7.1 | 0.769 |
| Unmethylated | 64 | 49.3 ± 26.2 | 79 | 31.2 ± 44.8 | 0.003 | 5 | 13.1 ± 8.9 | 58 | 33.1 ± 47.0 | 16 | 30.3 ± 43.9 | 0.636 |
| 0.308 | <0.001 | 0.901 | 0.002 | 0.183 | ||||||||
| MGMT | 100 | 94.1 ± 91.2 | 101 | 66.9 ± 123.2 | 0.057 | 7 | 23.4 ± 12.8 | 70 | 77.0 ± 144.0 | 24 | 41.6 ± 46.5 | 0.314 |
| Methylated | 14 | 84.3 ± 95.3 | 33 | 48.4 ± 47.9 | 0.199 | 3 | 26.6 ± 16.4 | 24 | 57.2 ± 52.4 | 6 | 23.9 ± 22.8 | 0.227 |
| Unmethylated | 86 | 95.7 ± 91.0 | 68 | 72.9 ± 146.1 | 0.262 | 4 | 21.1 ± 11.5 | 46 | 87.4 ± 173.4 | 18 | 47.4 ± 51.3 | 0.479 |
| 0.667 | 0.214 | 0.622 | 0.282 | 0.294 | ||||||||
| DNMT3B | 97 | 6.2 ± 17.7 | 93 | 5.9 ± 6.6 | 0.895 | 7 | 4.6 ± 4.1 | 66 | 5.9 ± 7.0 | 20 | 6.5 ± 6.2 | 0.817 |
mRNA expression is the expression level relative to that of the GAPDH gene / 10.
The sample size in each gene is less than the total number because the assay failed or the expression values are out of the 90% confidence interval.
Two-sided Student's t-tests for the difference s in the means between cases and controls.
Analysis of variance tests for the differences among the stages within cases.
We conducted RT-PCR assays to assess the relative mRNA expression levels of BRCA1, BRCA2, hMLHl, MGMT, and DNMT3B in cases and controls (Table 3). We performed an analysis of variance for differences in the relative mRNA expression levels of these genes among subgroups of age and ethnicity in both cases and controls but did not find statistically significant differences. Therefore, we combined all ethnicity groups together in the following analysis. Overall, mean mRNA expression levels of BRCA2 and hMLHl were significantly lower in ovarian tumors than in normal ovaries (P < 0.001 for both genes), whereas the difference in expression levels of MGMT between cases and controls was approaching significant (P = 0.057). There were no statistically significant differences in the mean mRNA expression levels of BRCA1 and DNMT3B between cases and controls.
We then evaluated the association between the risk of ovarian cancer and mRNA expression levels of the five tumor-suppressor genes and found that the risk was associated with low levels of mRNA expression in all genes but DNMT3B. Specifically, using the control median as the cutoff value, low expression levels were associated with a 2.95-fold increased risk (95% CI, 1.51 - 5.78) for BRCA1, a 3.65-fold increase (95% CI, 1.82 -7.30) for BRCA2, a 5.25-fold increase (95% CI, 2.52 - 10.96) for hMLHl, and a 4.72-fold increase (95% CI, 2.32 - 9.62) for MGMT after adjustment for age and ethnicity. In contrast, no increased risk was associated with the mRNA expression level of DNMT3B (adjusted OR, 0.59; 95% CI, 0.31 - 1.12) (Table 4).
Table 4.
Crude and adjusted odds rations (ORs) and 95% confidence intervals (CIs) for the relative gene expression levels in ovarian tumors and normal ovarian tissues
| Expression level* | No. (%) of cases(N = 102) | No. (%) of controls (N = 100) | P value† | Crude OR (95% CI) | Adjusted OR‡ (95% CI) |
|---|---|---|---|---|---|
| BRCA1 | |||||
| High | 25 (25.5) | 49 (50.5) | 0.0003 | 1.00 | 1.00 |
| Low | 73(74.5) | 48 (49.5) | 2.98(1.63-5.45) | 2.95(1.51-5.78) | |
| BRCA2 | |||||
| High | 23 (25.3) | 50 (50.5) | 0.0004 | 1.00 | 1.00 |
| Low | 68(74.7) | 49 (49.5) | 3.02(1.63-5.58) | 3.65(1.82-7.30) | |
| hMLHl | |||||
| High | 17 (16.7) | 49 (50.5) | <0.001 | 1.00 | 1.00 |
| Low | 85 (83.3) | 48 (49.5) | 5.10(2.65-9.83) | 5.25 (2.52 -10.96) | |
| MGMT | |||||
| High | 20 (19.8) | 50 (50.0) | <0.001 | 1.00 | 1.00 |
| Low | 81 (80.2) | 50 (50.0) | 4.05(2.16-7.58) | 4.72(2.32-9.62) | |
| DNMT3B | |||||
| High | 59 (63.4) | 48 (49.5) | 0.053 | 1.00 | 1.00 |
| Low | 34 (36.6) | 49 (50.5) | 0.57(0.32-1.01) | 0.59(0.31-1.12) |
The median relative mRNA expression level in the controls was used as the cutoff point for each gene. The sar in each gene is less than the total number because the assay failed or the expression values are out of the 90% confidence interval.
Two-sided χ2-test
Adjusted for age (in years) and ethnicity (non-Hispanic whites versus others) in a logistic regression model.
Finally, we assessed whether low mRNA expression by these tumor-suppressor genes in ovarian tumors was due to altered promoter methylation status. The expression levels by methylation status in BRCA1, hMLHl, and MGMT in the cases are summarized in Figure 1B. We found that only methylated MGMT was significantly higher in the cases than in the controls (32.7% vs. 14.0%; P = 0.002) (data not shown). The stratification of mRNA expression levels by methylation status in ovarian tumors and normal ovarian tissues is presented in Table 3. Although in cases and controls, methylated MGMT and methylated hMLHl showed lower gene expression levels than their unmethylated counterparts did, the difference was statistically significant only for hMLHl in cases: the mean expression 13.0 ± 7.6 in 23 methylated tumors and 31.2 ± 44.8 in 79 unmethylated tumors (P < 0.001).
We also compared mRNA expression levels among tumor stages I and II, III, and IV but found no statistical differences or trends. However, methylated hMLHl in 13 stage III tumors had significantly lower expression_than unmethylated hMLHl did in 58 stage III tumors (P = 0.002) (Table 3).
Discussion
In this case-control study, we found that low levels of the relative mRNA expression of BRCA1, BRCA2, hMLHl, and MGMT, but not of DNMT3B, were associated with a significantly increased risk of ovarian cancer. However, except for hMLHl, the methylation status of the genes did not appear to explain the observed lower expression levels.
Inactivation of tumor-suppression genes BRCA1 and BRCA2 in ovarian tumors has been reported by other investigators. For example, promoter hypermethylation of the BRCA1 gene was found to be between 5% and 36% of tumors in primary ovarian carcinomas, a molecular event that has been proposed as a potential cause of the gene inactivation [8, 10, 23, 29-33]. We found in our study that the relative mRNA expression levels of BRCA1 were significantly lower in ovarian tumors than in normal ovaries of subjects without ovarian cancer; however, this difference was not attributable to the promoter methylation status in BRCA1. We detected a much higher methylation status of BRCA1 (46.6%) than previous reports did [8, 10, 23, 29-33], and we even found that BRCA1 methylation commonly existed in unaffected ovaries (51.5%) of the subjects with conditions other than ovarian cancer. It is known that BRCA1 and BRCA2 genes are involved not only with DNA repair but also with hormone regulation; therefore, the ovarian cell type and status may need to be strictly defined in such methylation studies, and the best controls may be the normal ovaries of subjects without hormonally related conditions or cancers other than ovarian cancer.
A correlation between hMLHl hypermethylation, loss of expression, and microsatellite instability has been demonstrated in colorectal, gastric, endometrial [25, 34-36], and ovarian cancers [31, 37, 38]. The frequencies of hMLHl promoter methylation have been reported to range from 9% to 39% [32, 33, 39, 40]. In our study, we observed lower hMLHl expression that was associated with increased risk of ovarian cancer, and among the 102 cases, hMLHl expression was significantly lower in 23 methylated tumors than that in 79 unmethylated tumors.
We also observed that MGMT mRNA expression was lower in ovarian tumors than in normal ovaries, with 31.1% of the promoters methylated in cases, a finding consistent with a recently published study that had a much smaller number of study subjects (only 18 ovarian carcinomas) [18] and reported a high frequency (48%) of methylation status of MGMT. MGMT hypermethylation was less frequently observed in one study using ovarian cancer cell lines (in only 23% of 13 cell lines) [19] and in another study of ovarian granulosa cell tumors (in 33% of 43 subjects) [41]; however, MGMT hypermethylation was not detected in a recent study of 120 patients with endometrial cancer [42]. Our results indicate that the incidence rate of MGMT promoter methylation was significantly higher in cases than in controls. However, abnormal promoter methylation of MGMT in serous ovarian tumors and normal ovaries did not predict mRNA expression levels in our study, suggesting that molecular mechanisms other than methylation may contribute to the altered MGMT mRNA expression observed in this study.
The strength in the present study is the use of fresh-frozen tissues in a relatively large number of study subjects and a population of cases with homogenous high-grade serous epithelial ovarian cancer. Also, we compared the relative mRNA expression levels and methylation status of candidate tumor suppressor genes of both serous ovarian tumors from cancer patients and unaffected ovaries from subjects without ovarian cancer. In the present study we did not observe an association between DNMT3B gene expression and ovarian cancer risk, which has not been reported to date. The results of DNMT3B gene expression may serve as an internal control in the present study, which suggest no systematic errors in the measurements of gene expression that may have occurred in the experiments of other genes. Therefore, the observed risk in the present study could not be biased by systematic errors in the assays, by sample collection method, or by possible experimental error. However, because ovarian cancer likely arises from the ovarian surface epithelium and can be contaminated with normal tissues, microdissection of ovarian tumor tissues may be required for future ovarian cancer association studies. Further detection of genetic mutations in ovarian tumor tissues may help explain the underlying mechanisms of reduced gene expression, with quantization of CpG methylation to provide a more accurate estimation of methylation status and correlation with gene expression levels.
Acknowledgments
We would like to thank Joseph Celestino for preparing the tissues; Zhibin Hu, Luo Wang and Kejing Xu for their laboratory assistance; and Tamara Locke for scientific editing. NIH Ovarian Specialized Programs of Research Excellence grants P50 CA-083639 (G. B. Mills), BLANTON-DAVIS Ovarian Cancer Research Development Award (L E. Wang), National Institutes of Health grant ES11740 (Q. Wei) and CA16672 (The University of Texas M. D. Anderson Cancer Center).
References
- 1.Hanna L, Adams M. Prevention of ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2006;20:339–362. doi: 10.1016/j.bpobgyn.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Sankaranarayanan R, Ferlay J. Worldwide burden of gynaecological cancer: the size of the problem. Best Pract Res Clin Obstet Gynaecol. 2006;20:207–225. doi: 10.1016/j.bpobgyn.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 5.Salozhin SV, Prokhorchuk EB, Georgiev GP. Methylation of DNA-one of the major epigenetic markers. Biochemistry (Mosc) 2005;70:525–532. doi: 10.1007/s10541-005-0146-8. [DOI] [PubMed] [Google Scholar]
- 6.Widschwendter M, Berger J, Muller HM, Zeimet AG, Marth C. Epigenetic downregulation of the retinoic acid receptor-beta2 gene in breast cancer. J Mammary Gland Biol Neoplasia. 2001;6:193–201. doi: 10.1023/a:1011360724350. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 9.Costello JF, Fruhwald MC, Smiraglia DJ, Rush U, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 11.Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/pl6/MTSl gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 13.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol. 2008;68:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 16.Butcher LM, Beck S. Future impact of integrated high-throughput methylome analyses on human health and disease. J Genet Genomics. 2008;35:391–401. doi: 10.1016/S1673-8527(08)60057-0. [DOI] [PubMed] [Google Scholar]
- 17.Barton CA, Hacker NF, Clark SJ, O'Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109:129–139. doi: 10.1016/j.ygyno.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Furlan D, Carnevali I, Marcomini B, Cerutti R, Dainese E, Capella C, Riva C. The high frequency of de novo promoter methylation in synchronous primary endometrial and ovarian carcinomas. Clin Cancer Res. 2006;12:3329–3336. doi: 10.1158/1078-0432.CCR-05-2679. [DOI] [PubMed] [Google Scholar]
- 19.Imura M, Yamashita S, Cai LY, Furuta J, Wakabayashi M, Yasugi T, Ushijima T. Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 Ovarian cancer cell lines. Cancer Lett. 2006;241:213–220. doi: 10.1016/j.canlet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Wang LE, Wang L, Lu KH, Mills GB, Bondy ML, Wei Q. Methylation and messenger RNA expression of pl5INK4b but not pl6INK4a are independent risk factors for ovarian cancer. Clin Cancer Res. 2005;11:4968–4976. doi: 10.1158/1078-0432.CCR-04-2293. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Wang LE, Strom SS, Spitz MR, Babaian RJ, DiGiovanni J, Wei Q. Overexpression of hMTH in peripheral lymphocytes and risk of prostate cancer: a case-control analysis. Mol Carcinog. 2003;36:123–129. doi: 10.1002/mc.10108. [DOI] [PubMed] [Google Scholar]
- 22.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, Karlan BY. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60:5329–5333. [PubMed] [Google Scholar]
- 24.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN, Shi YQ, Rhyu MG, Powell SM, James SP, Wilson KT, Herman JG, Meltzer SJ. Hypermethylation of the hMLHl gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090–1095. [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.ABI. 1997. Relative quantitation of gene expression. User bulletin No. 2. ABI prism 7700 Sequence Detection System. PE Applied Biosystems.
- 28.Hu N, Qian L, Hu Y, Shou J, Wang C, Giffen C, Wang Q, Wang Y, Goldstein AM, Emmert-Buck M, Taylor PR. Quantitative real-time RT-PCR validation of differential mRNA expression of SPARC, FADD, Fascin, C0L7A1, CK4, TGM3, ECM1, PPL and EVPL in esophageal squamous cell carcinoma. BMC Cancer. 2006;6:1–9. [Google Scholar]
- 29.Bianco T, Chenevix-Trench G, Walsh DC, Cooper JE, Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21:147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 30.Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18:1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 31.Strathdee G, Appleton K, Illand M, Millan DW, Sargent J, Paul J, Brown R. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiley A, Katsaros D, Chen H, Rigault de la Longrais IA, Beeghly A, Puopolo M, Singal R, Zhang Y, Amoako A, Zelterman D, Yu H. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107:299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 33.Dhillon VS, Aslam M, Husain SA. The contribution of genetic and epigenetic changes in granuiosa cell tumors of ovarian origin. Clin Cancer Res. 2004;10:5537–5545. doi: 10.1158/1078-0432.CCR-04-0228. [DOI] [PubMed] [Google Scholar]
- 34.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLHl promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, Palazzo JP, Fishel R, Goodfellow PJ. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 36.Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLHl promoter methylation and lack of hMLHl expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res. 1999;59:159–164. [PubMed] [Google Scholar]
- 37.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLHl promoter in loss of hMLHl expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 38.Geisler JP, Goodheart MJ, Sood AK, Holmes RJ, Hatterman-Zogg MA, Buller RE. Mismatch repair gene expression defects contribute to microsatellite instability in ovarian carcinoma. Cancer. 2003;98:2199–2206. doi: 10.1002/cncr.11770. [DOI] [PubMed] [Google Scholar]
- 39.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLHl methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Zhang S, Cui J, Zhang A, Shen L, Yu H. Expression and promoter methylation status of mismatch repair gene hMLHl and hMSH2 in epithelial ovarian cancer. Aust N Z J Obstet Gynaecol. 2008;48:505–509. doi: 10.1111/j.1479-828X.2008.00892.x. [DOI] [PubMed] [Google Scholar]
- 41.Dhillon VS, Shahid M, Husain SA. CpG methylation of the FHIT, FANCF, cyclin-D2, BRCA2 and RUNX3 genes in Granuiosa cell tumors (GCTs) of ovarian origin. Mol Cancer. 2004;3:33. doi: 10.1186/1476-4598-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimel BJ, Huettner P, Powell MA, Mutch DG, Goodfellow PJ. Absence of MGMT promoter methylation in endometrial cancer. Gynecol Oncol. 2009;112:224–228. doi: 10.1016/j.ygyno.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]