The Physiome Projects are a diverse set of scientifically independent projects addressing integrative systems physiology and biology, conducted by individual investigators and teams from different countries. The emphasis in these projects is on medically related physiology and pharmacology. They gather modeling work, information processing methods and tools, databases, and other resources and make them available to a large research community.
Their name comes from the physiome, which is defined as the quantitative description of the functional behavior of a living organism. Because the physiome spans the scales of anatomy from gene to whole organism, from bacteria to man, and covers a time scale from embryogenesis to senescence and death (i.e., minutes for bacteria and a century for a few of Earth’s creatures), the goal of obtaining the physiome even for a single species is extremely ambitious and unrealizable in the near term. Nevertheless, it provides a definition for integrative efforts and a setting for strategizing and collaborating in science.
In this article, we present an overview of the status, accomplishments, and challenges of the Physiome Projects and outline the role played by multiscale modeling.
THE PHYSIOME PROJECTS
STATUS
The Physiome Projects (and the more focused efforts on modeling of the heart, kidney, lung, etc.) represent current efforts in integrative biology. Most such efforts are just beginning and have not achieved great depth, for many theoretical and technological issues have yet to be addressed. Most of these projects have gathered support from national health research agencies and/or international support.
Projects like E-cell, Virtual Cell, and the Physiome Projects [1] herald the importance of biological and physiological modeling. Exemplary of the new attitudes toward collaboration in the health sciences is the Interagency and Modeling Analysis Group (IMAG), which consists of 30 projects supported by the National Science Foundation (NSF) and some National Institutes of Health (NIH) institutes [2]. These groups span efforts ranging from molecular modeling and subcellular kinetics to large-scale finite element modeling of three-dimensional flows in airways and blood vessels and tissue mechanics; therefore, they could also be termed Physiome Projects.
A national effort that is showing outstanding success in relatively large-scale modeling is the “Leading Project for Biosimulation” sponsored by the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT). Each of MEXT’s several projects is a multi-investigator group effort: the Kyoto model centers on cardiac electrophysiology and its relationship to cardiac metabolism; the Keio project centers on oxygen handling and the metabolome of the erythrocyte; the Osaka project focuses on the effects of drugs on cardiac excitation and on structure-function relationships in the lung; and the Kobe project focuses on antidiabetic drugs. The plan is to integrate these projects so that the cardiac system can be understood to considerable depth.
Another large project is the Europhysiome Project. With initial funding from the European Economic Community, a large set of projects are being organized through multiuniversity collaborations, generated by sets of investigators with mutual interests. These funding opportunities are expanded through support to be provided by the various European health research agencies. In general, there appears to be more emphasis on biodevices and bio-instrumentation than on broader-scale biotechnology applications.
ACCOMPLISHMENTS
In the physiome research and related work there have been outstanding successes that are biophysical and biochemical in nature. These address subcellular scales where proteins, substrates and product solutes, and ions interact to provide quantitative descriptions of cell-level functions. Some provide linkages between circulating hormones and cellular responses (e.g., a signaling pathway [3]). Others provide kinetic descriptions linking cellular events to gene signaling and regulation of transcription and translation [4], [5]. At the higher physiological levels, the models are less biophysical and more descriptive. They are less precisely defined in terms of cellular responses, although it is in the cells where adaptive responses really occur, even when they are driven by higher-level signals.
CHALLENGES
DATA REPOSITORIES
A serious challenge in integrated biology and physiome-related efforts is the lack of suitable databases. Large models are generally composed of modular subsystems, each of which requires specific data. Although nationally supported databases for physiological models do not yet exist, the priority is to organize subsystem databases [similar to GenBank and Protein Data Bank (PDB) for genomics and proteomics]. Some steps in the efforts to organize suitable databases have been made through the establishment of the Systems Biology Markup Language (SBML), CellML, and JSim.
SBML [6] is an XML-based computer-readable format for representing models of biochemical reaction networks. It is used mainly for metabolic networks, cell-signaling pathways, and regulatory networks, and can be applied to describe continuous system models of many types (so long as they are restricted to using ordinary differential equations). SBML has been evolving since mid-2000 through the efforts of an international group of software developers and users, and it is supported by over 100 software systems, some of which translate the models into computable code.
CellML [7], [8] is another markup language based on XML being developed at the Bioengineering Institute at the University of Auckland. It is designed to store and exchange mathematical models and allow investigators to share models, even if they are using different model-building software. CellML also enables the reuse of components from one model in another, thus accelerating model building. CellML includes information about how the parts of a model are related as well as the equations describing the processes.
JSim [9], developed at the University of Washington, serves as both a model database format and a mathematical modeling language (MML) that can be directly computed via a parser-compiler using Java. JSim provides storage within project files for data sets and parameter sets, for graphics, for optimizer setting, and for a variety of numerical methods. JSim’s MML is easy for a human reader to comprehend, as its format differs very little from writing the equations in ASCII text. Since both CellML and SBML can be translated directly into JSim’s MML, its capacity to store different types of equations may make it a preferred vehicle for model archiving.
MODELING AND INTERPRETATION
Mathematical modeling of biological systems has only been attempted in the past half-century for biological systems, and many models have been single-scale. For instance, in 1972 the authors of [10] constructed a model of the cardiovascular system and the regulation of the blood pressure that was composed of many modules, most of which described functional relationships empirically. Although the model was composed of a single layer, it was nevertheless effective in describing an enormous amount of physiology, all the way from pressure pulse waveforms to whole-body water balance. This model is still in use and has been made available for medical teaching [11]. However, in the Physiome Projects, given the need to model different components and subsystems at different time and spatial scales, multiscale extensions of such solutions are needed.
MULTISCALE MODELING
Multiscale methods have been exploited in signal and image processing and in a wide range of other fields including biology, nanotechnology, materials, and aerodynamics. For example, the U.S. Department of Energy funded a series of workshops on multiscale problems that led to funding for a multiagency Multiscale Mathematics Initiative.
For multiple spatial scales, wavelet transforms and pyramidal approaches aim to discriminate between the local and global characteristics. For multiple time scales, the aim is to discriminate long-term dependencies from the immediate responses to local events, identify responses to short impulses (submilliseconds), and identify long-range time horizon effects (i.e. seconds, minutes, or much more).
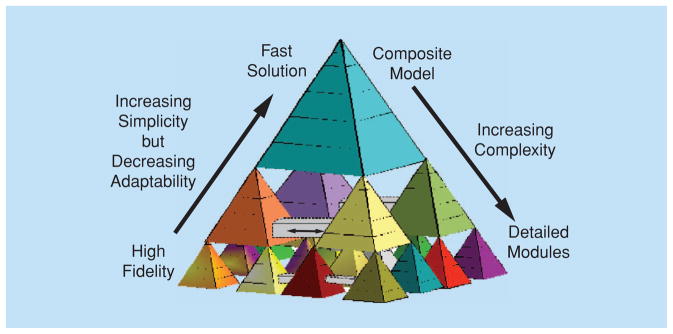
A block diagram of a multiscale model is illustrated in Figure 1. While such a model reflects the biology with increasing accuracy, the complexity of the model increases. At the same time, the experimental data required to validate such a model increases similarly.
Fig 1.
Block diagram of a multiscale model. Grey bars indicate connections between modules.
In Figure 1, the robustness, adaptability, and highest complexity are at the bottom level of the fully expanded model. Connectivity between modules, indicated by the grey bars connecting pairs of modules at low levels, must still be represented in the reduced-form modules at the middle levels, for there is information flow between modules. At the highest level, the model simplifications made for convenience and computational reasons have compromised the fidelity to the biology and the robustness of responses to changes in conditions. However, appropriate multiscale structuring of the modules should allow recognition of infidelity and the reparameterization of the high-level model to effect the behavioral modification that is actually governed at the lowest levels.
One of the obvious difficulties in multiscale modeling of biological systems using models such as that in Figure 1 is the signal interpretation step. This stems from the fact that a multiscale model has to be reduced to a computationally simpler form in order to be used in real time, and this necessarily results in a diminished capacity of the model to be able to adapt realistically to changing conditions. If such changes are directly observed, and if the model parameters are functions of the observed variables, then this desired adaptability can be maintained by having those functional relationships coded into the model.
EXAMPLE—TOWARD A MULTISCALE MODEL FOR CARDIOVASCULAR/RESPIRATORY PHYSIOME
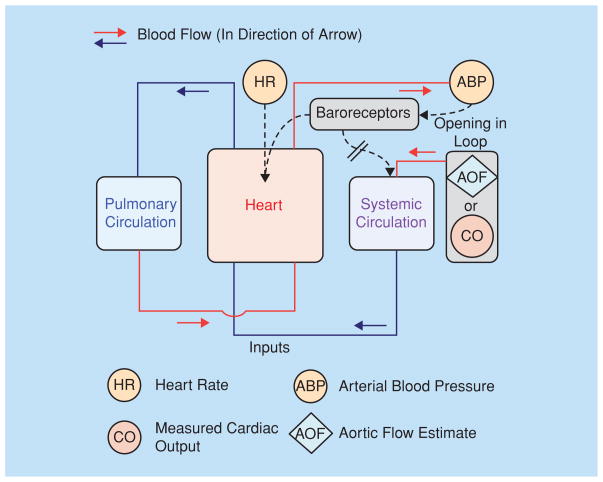
Consider the cardiovascular/respiratory model introduced in [12], as illustrated in Figure 2. The goal of using this model is to asses a patient’s status in a real-time setting. The model is composed of a circulatory system describing blood pressures throughout the systemic and pulmonary circulations. This includes details of their shape during each beat, the contractions of the cardiac chambers, and the operations of the cardiac valves, the blood pressure sensors in the carotid sinus and aortic arch, and the cerebral feedback control of heart rate and peripheral resistance, as well as the chemoreceptors of the carotid sinus and the feedback to control the rate and depth of respiration. This model gives appropriate responses to blood loss from the circulation, to changes in intrathoracic pressures by compressing the air in the lungs, and to changes in carbon dioxide.
Fig 2.
Cardiovascular/respiratory system model configured for open-loop analysis of blood pressure and heart rate signals and the continuous prediction of cardiac output and systemic blood volume.
Although the model contains components that belong to more than one level, the model itself is not a multiscale model. This model can serve as the basis for a truly multiscale-level model, if it is augmented by adding detailed modeling of cellular components that describe and define the responses at a deeper level. To understand such responses, let us recall that the model in [12] responds immediately to the feedback to change the heart rate. In actuality, when the heart rate is driven to a new rate (higher or lower), the strength of the contraction changes slowly, taking eight or so beats to reach a new level of contractile strength. In general, raising the heart rate causes the heart muscle to act somewhat as if it were being stimulated by sympathetic nerve stimulation; the heart contracts more quickly and more strongly, and at the end of systolic contraction relaxes more quickly, so that the ventricle can be refilled in time for the next beat. This response is mediated through a large set of cellular responses, greater calcium influx, greater release of intracellular calcium, a higher rate of adenosine triphosphate (ATP) use, and an increase in the fraction of the volume of the ventricle that is ejected with each beat. The control, and the robustness of the response mechanism, is at the level of ionic fluxes across the cell membranes and of the intracellular metabolism to produce ATP. All of this applies to the acute situation of a simple response to a change of heart rate. The onset of exercise requires substantially more adaptation, including changes in respiratory ventilatory rate and increases in cardiac output, the amount of blood pumped through the lungs and to the body per minute. Exercise induces changes down to the level of protein transcription, as is seen with exercise training and the development of larger, stronger muscles. Capturing all of these phenomena would result in a true multiscale model.
PHYSIOME MEETS SYSTEMS BIOLOGY
In the landscape of integrative biology efforts, it is important to make a distinction between Physiome Projects and related efforts on one hand and systems biology efforts on the other hand. Physiome efforts make use of top-down, quantitative biophysical and deterministic approaches to describing molecular, cellular, organ, and overall system behavior. The goal of these efforts is to elucidate the paths from genes to organisms. Systems biology efforts make use of bottom-up approaches to study intra- and intercellular dynamics using systems and signal-oriented approaches. The goal of these efforts is to elucidate the paths from genotype to phenotype, with emphasis on regulation, prediction and control, signals and information, mathematical modeling, and predicting behavior. The coalescence of physiome and systems biology interests is reflected in [13].
Acknowledgments
This work was supported by NSF Grant 04-607 and by National Institutes of Health (NIH) Grant T15 HL088516.
Biographies
James B. Bassingthwaighte (jbb@bio-eng.washington.edu) is a professor of bioengineering and radiology at the University of Washington. His research interests are in the areas of physiological transport and fractals in biology. He is a member of the U.S. National Academy of Engineering.
Howard Jay Chizeck (chizeck@ee.washington.edu) is a professor of electrical engineering and an adjunct professor of bioengineering at the University of Washington in Seattle. His research interests involve control engineering theory and the application of control engineering to biomedical and biologically inspired engineered systems. He is a Fellow of the IEEE.
Footnotes
More resources are available at the Physiome Organization, http://www.physiome.org and http://www.physiome.org.nz.
References
- 1.Bassingthwaighte JB. Strategies for the Physiome Project. Ann Biomed Eng. 2000;28(8):1043–1058. doi: 10.1114/1.1313771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Interagency opportunities in multi-scale modeling in biomedical, biological, and behavioral systems. National Science Foundation [Online] Available: http://www.nsf.gov/pubs/2004/nsf04607/nsf04607.htm.
- 3.Saucerman JJ, Brunton LL, Michailova AP, McCulloch AD. Modeling beta-adrenergic control of cardiac myocyte contractility in silico. J Biol Chem. 2003;278(48):47997–48003. doi: 10.1074/jbc.M308362200. [DOI] [PubMed] [Google Scholar]
- 4.Kuile BH, Westerhoff HV. Transcriptome meets metabolome: Hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 2001;500(3):169–171. doi: 10.1016/s0014-5793(01)02613-8. [DOI] [PubMed] [Google Scholar]
- 5.Herrgard MJ, Covert MW, Palsson BO. Reconstruction of microbial transcriptional regulatory networks. Curr Opin Biotechnol. 2004;15(1):70–77. doi: 10.1016/j.copbio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 6.SBML: Systems Biology Markup Language [Online] Available: http://sbml.org/index.psp.
- 7.CellML: Cell Modeling Markup Language [Online] Available: http://www.cellml.org/
- 8.Hunter PJ. Modeling human physiology: The IUPS/EMBS Physiome Project. Proc IEEE. 2006;94(4):678–691. [Google Scholar]
- 9.JSim: Java-based Simulation Platform for Data Analysis [Online] Available: http://www.physiome.org/jsim/
- 10.Guyton AC, Coleman TG, Granger HJ. Circulation: Overall regulation. Ann Rev Physiol. 1972;34:13–46. doi: 10.1146/annurev.ph.34.030172.000305. [DOI] [PubMed] [Google Scholar]
- 11.Abram SR, Hodnett BL, Summers RL, Coleman TG, Hester RL. Quantitative circulatory physiology: An integrative mathematical model of human physiology for medical education. Adv Physiol Educ. 2007;31:202–210. doi: 10.1152/advan.00114.2006. [DOI] [PubMed] [Google Scholar]
- 12.Neal ML, Bassingthwaighte JB. Subject-specific model estimation of cardiac output and blood volume during hemorrhage. Cardiovasc Eng. 2007;7:96–119. doi: 10.1007/s10558-007-9035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coatrieux JL, Bassingthwaighte JB. Proc IEEE (Special Issue on the Physiome and Beyond) 2006;94(4):671–677. [Google Scholar]