Abstract
In this contribution we investigate the applicability of different methods from the field of independent component analysis (ICA) for the examination of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) data from breast cancer research. DCE-MRI has evolved in recent years as a powerful complement to X-ray based mammography for breast cancer diagnosis and monitoring. In DCE-MRI the time related development of the signal intensity after the administration of a contrast agent can provide valuable information about tissue states and characteristics. To this end, techniques related to ICA, offer promising options for data integration and feature extraction at voxel level. In order to evaluate the applicability of ICA, topographic ICA and tree-dependent component analysis (TCA), these methods are applied to twelve clinical cases from breast cancer research with a histopathologically confirmed diagnosis. For ICA these experiments are complemented by a reliability analysis of the estimated components. The outcome of all algorithms is quantitatively evaluated by means of receiver operating characteristics (ROC) statistics whereas the results for specific data sets are discussed exemplarily in terms of reification, score-plots and score images.
Keywords: DCE-MRI, (Topographic) independent component analysis, Tree-dependent component analysis, Breast cancer research
1. Introduction
Nowadays, one out of nine women will become affected by breast cancer during their lifetime, making it one of the most common malignancies [1]. As an adjunct to X-ray mammography, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has become more and more important under a variety of indications [2]. Even though classical MRI provides high contrast with respect to soft tissue, the differentiation of benign and malignant lesions based on MRI turned out to be rather difficult. In this context, the acquisition of a MRI sequence after the administration of a contrast agent has shown to provide powerful options for lesion detection and sub-classification [3]. In such an image sequence, which could also be considered as a multivariate image, each voxel can be related to a short time series which conveys information about, e.g. tissue vascularity.
In DCE-MRI of the breast, the analysis of the shape of the signal time curve according to Kuhl et al. [5], has become a widely accepted method for the differential diagnosis of enhancing lesions. Nevertheless, the analysis of different diagnostic criteria based on the time related development of the signal or on lesion morphology is still an important issue. For most of them a considerable overlap can be observed with respect to benign and malignant lesions [2,4].
For the visual inspection of DCE-MRI data, most commonly two different approaches are employed [22]. Using difference images, changes in the signal intensity over time can be identified, e.g. in order to detect lesions which typically exhibit a strong signal uptake in the early post-contrast phase. However, since a difference image can only account for signal intensity changes between two images of a sequence, multiple difference images have to be computed and analyzed in order to assess the complete signal characteristics. An alternative approach are pharmacokinetic models which are utilized for the computation of parameter maps from the signal time curves such as the three-time-point method [6]. Based on a three-compartment tissue model this approach allows for the generation of pseudo-colored images reflecting pathophysiological features such as extracellular volume fraction or microvascular permeability.
Next to the methods described above, artificial neural networks and machine learning techniques based on supervised [7,8] and unsupervised learning [9–13] received increased interest for the analysis of DCE-MRI data at voxel level.
Even though supervised learning is a highly promising approach for the classification of lesion tissue based on the time related development of the signal, the applicability of this concept is often limited because of the heterogeneity and size of the data sets available for adaptation processes.
In this context, unsupervised learning techniques such as independent component analysis (ICA) are an interesting alternative [14]. ICA has been introduced to the field of DCE-MRI in a preliminary study for the detection and characterization of breast lesions [13]. Similar to its application in fMRI [23], ICA was used as a blind source separation technique for estimating the source signals of the observed data (which could be subject to noise and may result from a combination of effects originating from vasculature). For the data employed in their study, Yoo et al. were able to reliably identify signal components which resembled typical signal patterns from a benign or malignant lesion.
Similar observations have been made in a recent study about the applicability of ICA in breast MRI [21]. Motivated by the hypothesis that different contrast enhancements patterns might be identified in the data by means of ICA, a spatial FastICA variant was applied to the data of six patients. For all cases, characteristic signals could be estimated while a high qualitative agreement between tumor outlines drawn by radiologists and ICA based representations was observed.
In the following, we will evaluate the utility of three different techniques for feature extraction and data integration in DCE-MRI. Thereby, next to ICA, Topographic ICA and tree-dependent component analysis (TCA) are considered for the analysis of the short time series at voxel level.
For a quantitative evaluation of the employed algorithms with respect to their utility for lesion detection, a receiver operating characteristics (ROC) analysis is carried out. In this context, not only different parameterizations of the employed algorithms are considered, but for ICA also the reliability of the estimated components is investigated using clustering and visualization techniques. In order to assess their utility for tissue differentiation, the results are exemplarily discussed in terms of reification, score plots and score images.
2. Material
In order to evaluate the utility of (Topographic) ICA and TCA for the examination of DCE-MRI data, twelve image sequences were subject to an analysis. All scans contain histological verified breast lesions, whereas the lesions of the first six sequences were diagnosed as benign, the remaining six as malignant (see Table 1).
Table 1.
Histopathologically verified diagnosis for all cases employed in this study. The lesions in the first six studies where diagnosed as benign, the remaining six as malignant.
| Case | Diagnosis | Case | Diagnosis |
|---|---|---|---|
| I | Fibroadenom | VII | Ductal carcinoma |
| II | Fibrous mastopathy | VIII | Scirrhous carcinoma |
| III | Scar | IX | Ductal carcinoma in situ |
| IV | Lymph node | X | Multilocullar recurrent ductal carcinoma (Status post mastectomy) |
| V | Granduloma with signs of inflammation | XI | Ductal papillomatosis, transition into papillary carcinoma |
| VI | Chronic mastitis | XII | Ductal carcinoma |
In the employed imaging setup, scans of both breasts were acquired simultaneously by means of a MRI system with a field strength of 1.5 T (Magnetom Vision, Siemens, Erlangen Germany) and dedicated surface coils. The imaging protocol can briefly described as follows (see also Ref. [11]). In a first step, for the acquisition of transversal images, a short T1 inversion recovery (STIR) sequence with TR = 5600 ms, TE = 60 ms, FA = 90° and TI = 150 ms was employed. Thereafter, a dynamic T1 weighted gradient echo sequence was carried out with TR = 12 ms, TE = 5 ms and FA = 25°. The matrix size was 256 × 256 with a resolution of 1.37 mm and 32–34 slices with a thickness of 4 mm. The first image in the dynamic study was acquired before the administration of a contrast agent and the second directly thereafter. The following images were obtained in steps of 110 s. Gadopentetate dimeglumine (Magnevist, Schering, Berlin, Germany) was used as a paramagnetic contrast agent with a dose of 0.1 mmol/kg body weight. In order to ensure the minimization of motion artifacts, all patients were placed in a prone position. Since none or only minor motion artifacts were observed in the data, no additional image registration was carried out.
The location and the extent of a lesion in the scans were determined by a radiologist based on the visual examination of difference images, revealing changes in the signal intensity between the pre-contrast and one of the post-contrast images. Additionally, data from corresponding X-ray mammograms was considered.
In order to ease the evaluation, the analysis was further restricted to breast tissue while the background and the area behind the thorax was removed automatically. For the data in this study, an appropriate segmentation could be achieved already by means of a thresholding technique [15], using the signal intensities of the individual scans. In order to fill small gaps, subsequently a morphological closing operator [16] with a ball structuring element was employed. This operation comprises the subsequent application of a dilation and erosion. The dilation enlarges segments and fills small gaps while the erosion removes boundaries and segments which are not covered by the structuring element.
3. Methods
In the following the main concepts of ICA, Topographic ICA and TCA are briefly introduced. For a throughout discussion see Refs. [17–19].
3.1. Independent component analysis
In the literature, the term ICA refers to a broad range of methods which make use of different objective functions and optimization strategies [17]. However, most of these approaches are closely related and can be considered as a special case of Projection Pursuit.
The idea of ICA is usually motivated by a blind source separation problem for a generative data model of the form X = AS. Here, S and X are random variables taking values in ℝp and ℝq respectively. In this context, the Si can be interpreted as source signals or noise while the Xi represent the observed signals. Hence, the Xi is given by a linear combination of the original signals, defined by a mixing matrix A.
The goal of ICA is the estimation of the source signals Si (respectively the corresponding scores) and the demixing matrix W such that
| (1) |
It can be shown that for mutual independent and non-Gaussian source signals Si, the demixing matrix can be estimated based on the observed signals only. Here, two or more scalar valued random variables Si are statistically independent if their joint probability density function p(S) can be expressed as the product of the marginal densities pi(Si):
| (2) |
For estimating W, in the following the FastICA algorithm was employed [17]. The method relies on an approximation of negentropy in order to quantify non-gaussianity and finally statistical independence. Hence, ICA might provide not only independent but also interesting data representations. Using a fixed- point algorithm, the rows of W are estimated such that the non-gaussianity of is maximized for all i. Thereby, the wi can be computed either one by one or in parallel as in this study.
3.1.1. Validating independent components via clustering and visualization
The FastICA approach is commonly attributed good properties with regard to consistency, asymptotic variance, and robustness [20]. However, being a statistical technique, the results of this method will depend on initialization conditions, the parameterizations of the algorithm and the sampling of the data set. Therefore, the outcome of a single application of the FastICA algorithm should be treated carefully and methods for the reliability analysis of the estimated components should be taken into account.
In this context, an approach for the estimation of the algorithmic and statistical reliability of independent components called Icasso was recently proposed [24]. It makes use of an repeated application of the ICA algorithm under varying conditions whereas reliable components are identified in a subsequent step by means of clustering and visualization techniques.
Formally, a single application of the FastICA algorithm will result in an unmixing matrix Ŵi, while the outcome for several runs can be considered as a matrix . Using this notation, the relation between a pair of independent components can be expressed in terms of a correlation coefficient ri j with
| (3) |
where C is the sample covariance matrix of the original data [24]. Using, e.g. |ri j| as a similarity measure, a broad range of clustering and visualization techniques can be employed in order to investigate the stability of the entire analysis. Using, e.g. agglomerative clustering, groups of similar components can be identified whereas the centroids of the component clusters might be considered as reliable estimates of the independent components. By means of visualization techniques such as multidimensional scaling (MDS), the pairwise similarities of the components can also be represented in terms of spatial relationships. Therefore, the outcome of such a process could also provide insight into the number of independent components, a quantity that normally has to be defined prior to the application of the FastICA algorithm.
3.2. Topographic independent component analysis
Topographic ICA can be considered a generalization of classical ICA [18] since it combines aspects of ICA with those from topographic mappings.
In Topographic ICA the assumption of statistical independence is relaxed and higher-order correlations are taken into account for estimating the components of the model. Using, e.g. correlation of energies, this leads to a topographic ordering of the components in terms of a lattice with a predefined topology. Here, the distance between to components is related to their degree of dependence.
Topographic ICA makes use of a modified generative data model. In order to describe the dependence between the source variables it introduces a set of common variance variables. The variables are given by a non-linear function φ(·), which depends on a linear combination of a neighborhood function h(i, k), introducing topography, and a variance generating random variable Uk. Given a set of random variables Zi with the same distribution as Si, the sources can then be described as
| (4) |
The demixing matrix can be estimated, e.g. using gradient descent techniques (for details see Ref. [18]).
3.3. Tree-dependent component analysis
In contrast to Topographic ICA, which is based on a topography that has to be defined in advance, TCA aims at estimating source signals which could be described by a tree-structured graphical model [19]. Dependent components can be identified by means of connected nodes while components from nodes without any connecting edges are mutually independent.
The basic idea TCA is derived from the approximation of n-dimensional discrete probability distributions using second-order component distributions. An approximation of a discrete random variable X variable of the form
| (5) |
is called a probability distribution of first-order tree dependence. Here, (m1, …, mn) is a random permutation of the integers 1, 2, …, n while P(Xi|X0) is defined as P(Xi). The distribution of first-order dependence which minimizes the mutual information with respect to the original distribution can be considered as optimal.
Using mutual information I(Xi, X j(i)) as a branch weight between the nodes, it can be shown that the optimal approximation is given by the maximum-weight dependence tree. This tree has the maximal sum of all possible first-order dependence trees, i.e.
| (6) |
For topographic ICA one finally obtains the following objective function
| (7) |
where I denotes the mutual information and S factorizes in a tree t. For details see Ref. [19].
4. Results
In order to evaluate the three approaches discussed above, they were applied to the individual data sets presented in Section 2. While ICA related techniques were originally introduced for the analysis of temporal independent signals, in multivariate image analysis usually the concept of spatial ICA is adopted. Here, the data is unfolded into a p × n matrix X where n denotes the number of voxels or pixels and p the number of variables respectively scans. Although often similar results are obtained with spatial and temporal ICA, the outcome of both approaches can differ when strong correlations in time or space exist [25]. However, since n is usually much larger than p, a spatial analysis is computationally more efficient and was pursued in the following.
Since a DCE-MRI study comprises only a small number of scans, except from a centering of the data no additional preprocessing was carried out as it is usually the case in, e.g. fMRI [11]. For each data set the maximum number of six components was computed according to the underling model, while different parameterizations were used in the adaptation process. For the FastICA algorithm four different contrast functions were employed as approximations for negentropy [17]. In order to apply Topographic ICA, additionally a neighborhood function and topography has to be chosen. For simplicity, in this study the analysis was restricted to an one-dimensional lattice with a neighborhood function that included only directly adjacent nodes. For TCA three different empirical contrast functions have been selected [19]. For all algorithms default parameters have been used as given in the corresponding literature.
The outcome of all algorithms was further quantitatively evaluated by means of ROC statistics. As the discussed algorithms make use of stochastic adaptation processes, each algorithm was applied ten times to each data set in order to investigate the reproducibility of the results. Furthermore, for FastICA, Icasso was employed for estimating stable components. Fig. 1 illustrates the outcome of such an analysis for case XII (see Table 1). Here, similarities between estimated components are given in terms of spatial relationships using MDS. The convex hulls indicate a grouping of the components computed by means of agglomerative clustering (group average) [24]. Even though this representation suggests the presence of several clusters, it also reveals some variability with respect to the estimated components.
Fig. 1.
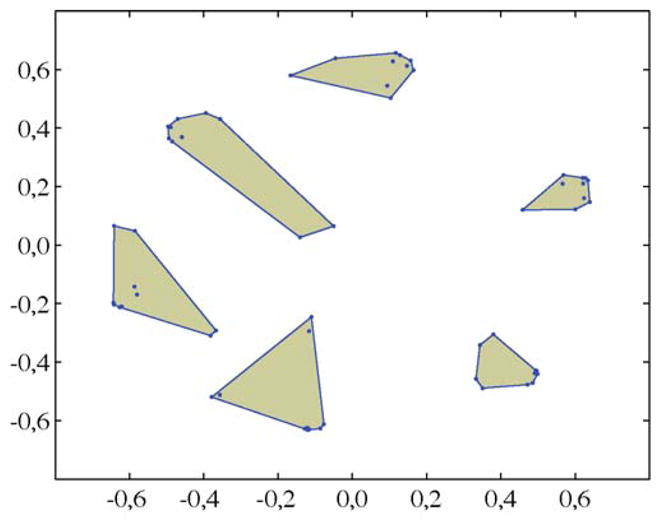
MDS based representations of similarities between components estimated in different FastICA applications. Convex hulls indicate six clusters computed by means of agglomerative clustering using 1 − |ri j| as a dissimilarity measure. The centroids of the clusters were considered as stable estimates.
4.1. ROC analysis
By means of the presented methods, different techniques can be employed to investigate the DCE-MRI sequences. In Ref. [13], the rows of a demixing matrix were interpreted as reference waveforms. For visualization purposes those voxels were depicted where the correlation between the signal time course and the waveforms exceeded a certain level. Contrary, in the following application, the estimated components Si, i = 1, …, 6 themselves were subject to an analysis. In order to measure the correspondence between the components and the label provided by the radiologist, ROC statistics was employed [27]. ROC analysis is an established method for accessing the performance of a system in signal detection theory. It is not limited to a specific threshold but depends on the ranking of the data. Therefore, in the context of this application, the scores for the voxels were interpreted as confidence values. In order to evaluate the utility of the different algorithms, for each component the area under the ROC curve (AUC) was computed, a quantity with a range of [0.5, …, 1.0].
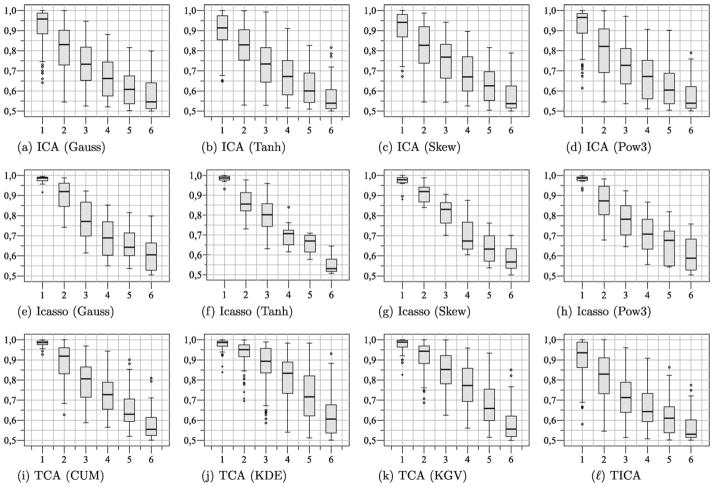
The AUC values achieved for all methods on the twelve data sets are summarized in Fig. 2. Since for ICA, different from Principal Component Analysis (PCA), there is no inherent order among the components, in this study a ranking of the components based on the corresponding AUC values was carried out.
Fig. 2.
Results achieved for FastICA, Icasso, TCA and Topographic independent component analysis (TICA). The box-plots depict the AUC values for ten repeated applications of these algorithms on all data sets. Since no natural ordering of the components exists for ICA, a ranking of the components based on AUC values was carried out. (a) ICA (Gauss), (b) ICA (Tanh), (c) ICA (Skew), (d) ICA (Pow3), (e) Icasso (Gauss), (f) Icasso (Tanh), (g) Icasso (Skew), (h) Icasso (Pow3), (i) TCA (CUM), (j) TCA (KDE), (k) TCA (KGV), and (l) TICA.
Following Fig. 2, with respect to AUC values, in most experiments at least one component could be identified which is indicative for a lesion. The application of TCA resulted consistently in a component with a high AUC value. Furthermore, for the FastICA algorithm in combination with Icasso, a considerable improvement with respect to the obtained AUC values could be observed which might be attributed to the fact that unstable components where removed from the analysis.
4.2. Data exploration
The AUC values discussed above provide a measure for the correspondence between the location and extent of a lesion as given by the radiologist and the outcome of the employed methods. For the differentiation of benign and malignant lesions, the interpretation of the coefficient components (aka reification) and the visual representation of the data in terms of so-called score plots and score images can provide additional information.
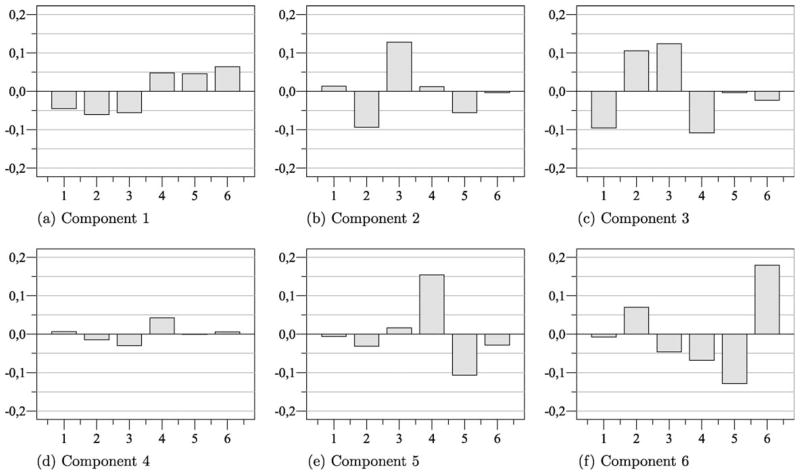
In Fig. 3, the coefficients of the components estimated from case XII based on FastICA (Tanh) using Icasso are depicted exemplarily. Even though the highest AUC values were obtained by means of TCA, the signal characteristics of the lesion become especially apparent in this experiment as several components with rather high AUC values were obtained. Here, the coefficients of the first component can be considered as some sort of contrast between the pre-resp. early post-contrast phase and the late post-contrast phase. The coefficients of the third component resemble an uptake-washout pattern. Hence, voxels which are related to an increasing signal intensity over time or a decreasing signal intensity in the late post-contrast phase, will most likely exhibit high scores with respect to S1 or S3.
Fig. 3.
Coefficients of six independent components computed for case XII using Icasso (Tanh). The first component is indicative for a signal enhancement over time while the third component resembles the typical uptake-washout pattern of a malignant lesion. (a) Component 1. (b) Component 2. (c) Component 3. (d) Component 4. (e) Component 5. (f) Component 6.
According to Kuhl et al. [5], a steady signal increase is often observed within benign lesions whereas a decreasing signal intensity during the intermediate and late post-contrast phase is highly indicative for a malignant lesion. Therefore, it could be expected that a representation of the data with respect to the components S1 and S3 will facilitate the analysis of the corresponding image sequence.
Fig. 4, depicts a corresponding scatter plot matrix. Here, the scores for samples from normal and lesion tissue are depicted with regard to the three components with the highest AUC value. The heterogeneous signal characteristic of the lesion can be clearly identified in a plot of S1 against S3.
Fig. 4.
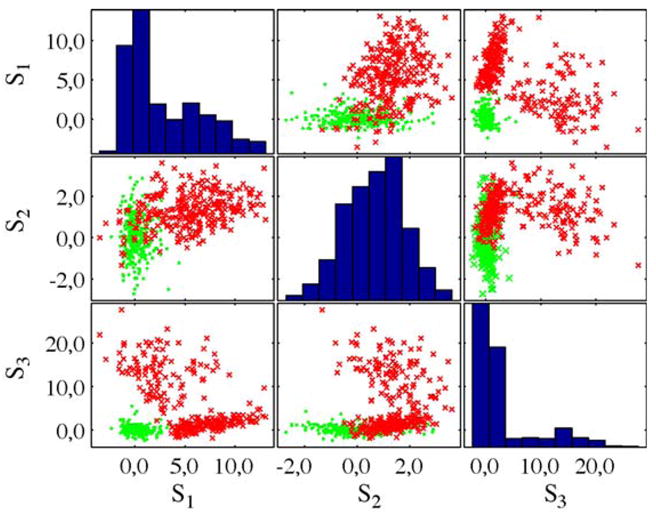
Scatter plot matrix for the three components with the highest AUC value for case XII (see Table 1), using Icasso (Tanh). Samples from normal tissue are given in green while samples from lesions are depicted in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
While the samples from the normal tissue constitute a compact cluster, two other regions become apparent which are related to the different compartments of the lesion. The distribution of scores for S1 and S3 further suggests that statistical measures might be employed for an automated identification of interesting components as proposed in context of remote sensing by means of kurtosis [26]. However, as indicated by S2 this might not be always the case [28].
The fused, respectively score images for the six components are given in Fig. 5. These images provide information about the spatial distribution of the signals. For visualization purposes not only the breast tissue but also the entire data of the DCE-MRI scan is represented. In the first score image the left part of the lesion becomes clearly visible as well as blood vessels with a similar signal characteristic. The third score image reveals the second compartment of the lesion that is related to a fast increasing signal intensity followed by a washout pattern that is highly indicative for a malignant lesion. While for the second component (resembling a delayed enhancement pattern) a high AUC value was achieved, a corresponding structure is hardly visible in the score image.
Fig. 5.
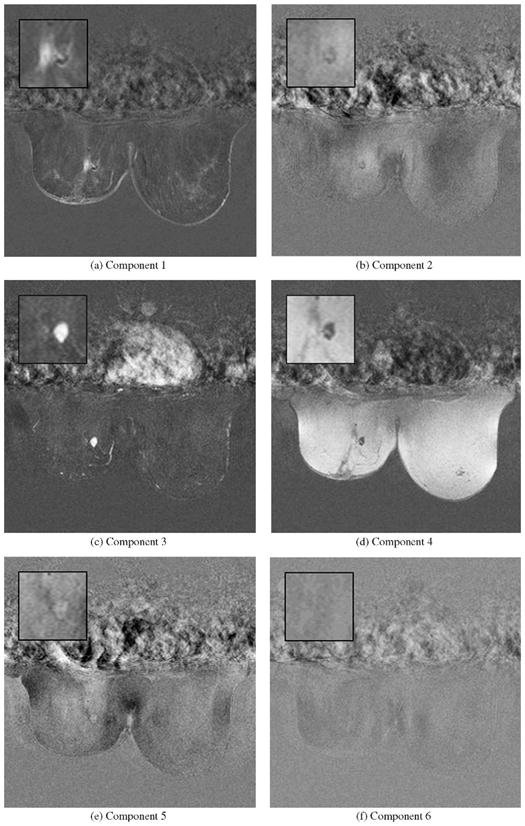
Slice based representation of six score images for case XII based on Icasso (Tanh). In the first image the location and extent of one lesion compartment could be identified which exhibits a continuously increasing signal whereas blood vessels with a similar characteristic become also visible. In the third image another compartment of the lesion stands out which shows a typical uptake-washout pattern which is highly indicative for malignant lesions. (a) Component 1. (b) Component 2. (c) Component 3. (d) Component 4. (e) Component 5. (f) Component 6.
5. Discussion
In this contribution we conducted a comparative study about the applicability of different ICA related algorithms for the analysis of DCE-MRI data in breast cancer research. These algorithms can be considered as powerful techniques for the decomposition and analysis of DCE-MRI data based on spatio-temporal characteristics.
Using ICA, Topographic ICA and TCA, in most experiments at least one component could be estimated which allowed for lesion detection in the considered data. Thereby, for TCA the highest average AUC value of 0.98 was achieved, making it a highly promising approach. This shows that a pre-specified topology as in case of the Topographic ICA does not lead to favorable results. While some variations in terms of AUC values were observed for all algorithms, for the FastICA approach a considerably improvement was achieved by means of a stability analysis. Unfortunately, these concepts could not be directly extended to the other algorithms as the underlying topology would have to be taken into account. Even though in the discussed example somewhat smaller AUC valueswere obtained for Icasso than for TCA, this could be attributed to the fact that by means of Icasso different compartments of the lesion were identified. Therefore, a final evaluation of the utility of the employed methods with regard to tissue differentiation remains difficult.
Future research should include larger data sets and address a detailed comparison with other techniques from the field of feature extraction and dimensionality reduction. Since in other applications ICA-like techniques have shown promising results even for the detection of objects at subpixel level [26], a more detailed investigation of the influence of the lesion size on the analysis would be also desirable. In this context, an Icasso-like approach might be employed in order to measure these effects.
In summary, this initial study demonstrated that ICA related techniques could reveal voxels associated with the tumor and provide insight into vascularity as well as imaging artifacts. These techniques have the potential to provide an improved computer-aided diagnosis by providing consistent and objective results.
Acknowledgments
This research was supported by NIH Grant 5K25CA106799-03.
References
- 1.Schnall MD. Application of magnetic resonance imaging to early detection of breast cancer. Breast Cancer Research. 2001;3(1):17–21. doi: 10.1186/bcr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szabó BK, Aspelin P, Kristoffersen Wiberg M, Boné B. Dynamic MR imaging of the breast: analysis of kinetic and morphologic diagnostic criteria. Acta Radiologica. 2003;44(4):379–386. doi: 10.1080/j.1600-0455.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 3.Heywang SH, Wolf A, Pruss E, Hilbertz T, Eiermann W, Permaneter W. MR imaging of the breast with Gd-DTPA: Use and limitations. Radiology. 1989;171(1):95–103. doi: 10.1148/radiology.171.1.2648479. [DOI] [PubMed] [Google Scholar]
- 4.Degenhard A, Tanner C, Hayes C, Hawkes DJ, Leach MO. Comparison between radiological and artificial neural network diagnosis in clinical screening. Physiological Measurements. 1999;23(4):727–739. doi: 10.1088/0967-3334/23/4/311. [DOI] [PubMed] [Google Scholar]
- 5.Kuhl CK, Mielcareck P, Klaschik S, Leutner C, Wardelmann E, Gieseke J, Schild H. Dynamic breast MR imaging: Are signal intensity time course data useful for differential diagnosis of enhancing lesions. Radiology. 1999;221:101–110. doi: 10.1148/radiology.211.1.r99ap38101. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein D, Strano S, Cohen P, Fields S, Gomori JM, Degani H. Breast fibroadenoma: Mapping of pathophysiologic features with three-time-point, contrast-enhanced MR imaging-pilot study. Radiology. 1999;210:233–240. doi: 10.1148/radiology.210.1.r99ja18233. [DOI] [PubMed] [Google Scholar]
- 7.Lucht R, Delorme S, Brix G. Neural network-based segmentation of dynamic MR mammographic images. Magnet Resonance Imaging. 2002;20(2):147–154. doi: 10.1016/s0730-725x(02)00464-2. [DOI] [PubMed] [Google Scholar]
- 8.Twellmann T, Saalbach A, Mueller C, Nattkemper TW, Wismüller A. Detection of suspicious lesions in dynamic contrast-enhanced MRI data. Proceedings of the 26th International Conference of the IEEE Engineering in Medicine and Biology Society; San Francisco. 2004. pp. 454–457. [DOI] [PubMed] [Google Scholar]
- 9.Varini C, Degenhard A, Nattkemper TW. Visual exploratory analysis of DCE-MRI data in breast cancer by dimensional data reduction: A comparative study. Biomedical Signal Processing and Control. 2006;1(1):56–63. [Google Scholar]
- 10.Saalbach A, Lange O, Nattkemper TW, Meyer-Baese A. Analysis of breast MRI data based on (topographic) independent and tree-dependent component analysis. Proceedings of SPIE, vol. 6576, Independent Component Analyses, Wavelets, Unsupervised Smart Sensors, and Neural Networks V; 2007. [Google Scholar]
- 11.Wismüller A, Lange O, Dersch DR, Leinsinger GL, Hahn K, Pütz B, Auer D. Cluster analysis of biomedical image time-series. International Journal of Computer Vision. 2002;46(2):103–128. [Google Scholar]
- 12.Twellmann T, Saalbach A, Gerstung O, Leach MO, Nattkemper TW. Image fusion for dynamic contrast-enhanced magnetic resonance imaging. BMC BioMedical Engineering OnLine. 2004;3(1):35. doi: 10.1186/1475-925X-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo SS, Gil Choi B, Han JY, Hee Kim H. Independent component analysis for the examination of dynamic contrast-enhanced breast magnetic resonance imaging data: preliminary study. Investigative Radiology. 2002;37(12):647–654. doi: 10.1097/00004424-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Meyer-Baese A, Lange O, Wismüller A, Leinsinger G. Computer-aided diagnosis in breast MRI based on ICA and unsupervised clustering techniques. Proceedings of SPIE, vol. 5818, Independent Component Analyses, Wavelets, Unsupervised Smart Sensors, and Neural Networks III; 2005. pp. 38–49. [Google Scholar]
- 15.Otsu N. A threshold selection method from gray level histograms. IEEE Transactions on Systems, Man and Cybernetics. 1979;9:62–66. [Google Scholar]
- 16.Jähne B. Digital Image Processing. Springer; Berlin: 2002. [Google Scholar]
- 17.Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Transactions on Neural Networks. 1999;10(3):626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 18.Hyvärinen A, Hoyer PO, Inki M. Topographic independent component analysis. Neural Computation. 2001;13(7):1527–1558. doi: 10.1162/089976601750264992. [DOI] [PubMed] [Google Scholar]
- 19.Bach F, Jordan M. Finding clusters in independent component analysis. Proceedings of the 4th International Symposium on Independent Component Analysis and Signal Separation; 2003. [Google Scholar]
- 20.Hyvärinen A. Survey on independent component analysis. Neural Computing Surveys. 1999;2:94–128. [Google Scholar]
- 21.San Koh T, Hua Thng C, Ho JTS, Hoon Tan P, Rumpel H, Khoo JBK. Independent component analysis of dynamic contrast-enhanced magnetic resonance images of breast carcinoma: A feasibility study. Journal of Magnetic Resonance Imaging. 2008;28(1):271–277. doi: 10.1002/jmri.21391. [DOI] [PubMed] [Google Scholar]
- 22.Nattkemper TW, Degenhard A, Twellmann T. In: Cancer Imaging—Lung and Breast Carcinomas. Hayat MA, editor. Academic Press; 2007. pp. 309–324. [Google Scholar]
- 23.McKeown MJ, Sejnowski TJ. Independent component analysis of fMRI data: examining the assumptions. Human Brain Mapping. 1998;6:368–372. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<368::AID-HBM7>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himberg J, Hyvärinen A, Esposito F. Validating the independent components of neuroimaging time-series via clustering and visualization. NeuroImage. 2004;22(3):1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Human Brain Mapping. 2001;13(1):43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang SS, Chang CI, Ginsberg IW. Unsupervised subpixel target detection for hyperspectral images using projection pursuit. IEEE Transactions on Geoscience and Remote Sensing. 2001;39(7):1380–1391. [Google Scholar]
- 27.Egan JP. Signal Detection Theory and ROC Analysis. Academic Press; New York: 1975. (Series in Cognition and Perception) [Google Scholar]
- 28.Saalbach A. Exploratory Analysis of Multivariate Image Data. Bielefeld University; 2006. [Google Scholar]