Abstract
The structure of bacterial communities in first-year spring and summer sea ice differs from that in source seawaters, suggesting selection during ice formation in autumn or taxon-specific mortality in the ice during winter. We tested these hypotheses by weekly sampling (January–March 2004) of first-year winter sea ice (Franklin Bay, Western Arctic) that experienced temperatures from −9°C to −26°C, generating community fingerprints and clone libraries for Bacteria and Archaea. Despite severe conditions and significant decreases in microbial abundance, no significant changes in richness or community structure were detected in the ice. Communities of Bacteria and Archaea in the ice, as in under-ice seawater, were dominated by SAR11 clade Alphaproteobacteria and Marine Group I Crenarchaeota, neither of which is known from later season sea ice. The bacterial ice library contained clones of Gammaproteobacteria from oligotrophic seawater clades (e.g. OM60, OM182) but no clones from gammaproteobacterial genera commonly detected in later season sea ice by similar methods (e.g. Colwellia, Psychrobacter). The only common sea ice bacterial genus detected in winter ice was Polaribacter. Overall, selection during ice formation and mortality during winter appear to play minor roles in the process of microbial succession that leads to distinctive spring and summer sea ice communities.
Introduction
Arctic sea ice covers an area of about 15 × 106 km2 at its maximal extent during winter (Antarctic 18 × 106 km2; Fetterer and Knowles, 2002), providing an extensive (albeit shrinking; Serreze et al., 2007) habitat for microorganisms. During the annual lifetime of polar sea ice, it experiences wide ranges in environmental conditions, yet changes in its microbial (bacterial and archaeal) communities through the seasons are not well known (Mock and Thomas, 2005; Deming, 2009). Winter sea ice in the Arctic is an extreme environment characterized by limited light, very cold temperatures in its upper horizons (to −35°C) and correspondingly high salinity (37–237‰) in its brine inclusions, where the organisms have been observed to reside (Junge et al., 2001). The assumption that these microbial inhabitants are largely inactive during the winter has been tested infrequently. Working with first-year sea ice north of Barrow (Alaska) during the coldest month of the year (March), Junge and colleagues (2004) observed that a small percentage of cells (0.5–4.0% by CTC staining) were metabolically active to −20°C, the coldest ice horizon examined. Wells and Deming (2006b), working with landfast sea ice in Franklin Bay (Western Arctic, also in March), detected an increase in bacterial numbers (doubling time > 4 days) in one of the three ice-brine samples they incubated at −12°C (salinity of 160‰).
Relative to other marine environments, bacteria in sea ice have proven highly amenable to cultivation: up to 50% of total counts from Antarctic ice (Helmke and Weyland, 1995) and 62% from Arctic ice (Junge et al., 2002). The bacterial groups that have been cultured from polar sea ice consist primarily of Gammaproteobacteria of the orders Oceanospirillales and Alteromonadales, the marine Roseobacter clade of Alphaproteobacteria (Bowman et al., 1997; Brown and Bowman, 2001; Brinkmeyer et al., 2003; Buchan et al., 2005), and the Bacteroidetes (also known as Cytophaga–Flavobacter–Bacteroides). Betaproteobacteria and high G+C Gram-positives (Actinobacteria) have also been obtained in culture from Antarctic sea ice (Junge et al., 1998) and Baltic Sea ice (Kaartokallio et al., 2005). Results from culture-independent methods, including cloning and sequencing of 16S rRNA genes and fluorescent in situ hybridization (FISH), overlap remarkably well with culture-based results, confirming the prevalence of these bacterial groups in spring and summer sea ice (Brown and Bowman, 2001; Brinkmeyer et al., 2003). In contrast, Archaea were not known from sea ice until recently, even though they are prevalent members of Arctic pelagic communities (Galand et al., 2006; 2008b; Wells et al., 2006) and comprise a sizable fraction of the biomass in Antarcticfrazil ice in late winter (Delong et al., 1994). To date, Archaea have only been detected in Arctic winter sea ice using domain-level probes, where they comprised ≤ 3.4% of the total assemblage (Junge et al., 2004). They appear absent in sea ice of later seasons by both culture-based and culture-independent methods (Brown and Bowman, 2001; Brinkmeyer et al., 2003).
Possible shifts in community composition and richness in Arctic winter ice over time and increasingly severe conditions are not known. From cultivation studies of Antarctic sea ice, psychrophilic bacteria are thought to outlive psychrotolerant species during winter (Helmke and Weyland, 1995; Delille and Rosiers, 1996; Helmke and Weyland, 2004; Fiala et al., 2006), leaving them positioned to dominate during the biologically productive spring and summer months. Such common heterotrophic microorganisms may also begin their habitation of sea ice in relative abundance if attached to larger phytoplankton known to entrain selectively into ice by frazil ice scavenging (Grossmann, 1994). The dynamics of microbial communities in polar sea ice, whether during or after ice entrapment, have not been examined using phylogenetic approaches, although these techniques have been used to study winter succession of bacterial and archaeal communities in polar waters (Murray et al., 1998; Murray and Grzymski, 2007), including surface waters at our study site (Alonso-Sáez et al., 2008). The only ice examined by a PCR-based ‘fingerprinting’ technique for high-throughput analysis of microbial community structure (Osborn et al., 2000; Hewson et al., 2007; Fuhrman et al., 2008) is the warm (> −5°C) and thin (< 30 cm) ice of the brackish (5–6‰) Baltic Sea (Kaartokallio et al., 2008). There the dynamics of Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes over the short (2 month) lifetime of this ‘mild’ ice were linked to exchange processes at the ice–water interface and progression of the ice-algal bloom. These features do not pertain to upper horizons of much colder, thicker and light-limited Arctic winter sea ice, the subject of this study.
During the Canadian Arctic Shelf Exchange Study (CASES), when the CCGS Amundsen was immobilized in landfast sea ice of Franklin Bay (Western Arctic) through the winter, we first investigated the spatial heterogeneity and temporal dynamics of particulate matter, including microorganisms and particulate extracellular polymeric substances (pEPS), within the ice (Collins et al., 2008). The same ice field was cored repeatedly each week from January through March, a period when ice thickness increased from 90 to 200 cm and upper ice temperatures were well below −5°C, leaving the ice impermeable (Golden et al., 1998) and the microbial communities entrapped. We focused on the upper 70 cm of the ice, analysing three 10 cm horizons centred at 25, 45 and 65 cm below the ice surface (reserving subsamples for later phylogenetic analyses). The results indicated winter losses of 38% and 49% of the total number of bacteria in the two coldest ice horizons (Fig. S1), where in situ temperatures had ranged from −15°C to −26°C (25 cm) and −12°C to −22°C (45 cm; the range at 65 cm was −9°C to −18°C; Collins et al., 2008). At these temperatures brine salinities are also extreme, from 130‰ to 230‰. We attributed some of the bacterial losses to virally mediated mortality, given measurements of viral production in ice brines from the same ice field (Wells and Deming, 2006b). We attributed the persistence of the majority of bacteria throughout the ice to the generic cryoprotective effects of pEPS (Krembs and Deming, 2008), which had increased significantly in all three horizons during winter (Collins et al., 2008).
Here, we investigate effects of the increasingly severe conditions in the aforementioned sea ice on the structure and richness of the natural microbial communities found within it, beginning several weeks after ice formation. Because bacterial communities that thrive within spring and summer sea ice differ substantially from pelagic communities prior to freeze-up, we hypothesized that extreme conditions in winter sea ice would exert selective pressure on the microbial community, favouring survival and subsequent dominance of the easily cultured psychrophilic Bacteria already known from spring and summer sea ice. To test this hypothesis we performed community fingerprinting of Bacteria (by automated ribosomal intergenic spacer analysis, ARISA) and Archaea (by terminal restriction fragment length polymorphism, T-RFLP) on the entire sample set collected during the CASES overwintering expedition (Collins et al., 2008), also generating complementary clone libraries of bacterial and archaeal 16S rRNA genes from selected samples of winter sea ice and under-ice seawater. Although we were unable to test directly for selection during the autumn freezing process, our results also bear upon this concept.
Results
Community dynamics
Bacterial ARISA
Contrary to expectation, no changes in the community structure (Fig. 1) or richness (Fig. S1) of the sea ice bacterial community were detected through the winter. A large majority (77.5%) of the total ARISA signal intensity (i.e. global cumulative peak height) derived from 14 operational taxonomic units (OTUs) present in all 11 successfully analysed sea ice samples (Table 1). Clone libraries of the 16S-intergenic transcribed spacer (ITS)-23S region identified some of these common OTUs, including SAR11 clade Alphaproteobacteria, which made up 50% of the total signal intensity (Table 1), in agreement with the dominance of this clade in those clone libraries; OTUs best matching Polaribacter, a genus of Bacteroidetes, made up a further 12%. Several more OTUs persisting through the winter had best matches to various Alphaproteobacteria, Gammaproteobacteria and Flavobacteria (Table 1). Correlation analysis revealed no significant change (P < 0.05) in community structure similarity (Sørensen's index; 84 ± 7%, Fig. 1) or richness (range 19–29, Fig. S1) over time in any horizon. Clustering analysis also indicated no differences among communities by depth horizon, but the limited number and non-uniform distribution of successfully amplified samples precluded further statistical assessments as a function of ice depth. The under-ice seawater sample (88-SW), though lacking about 30% of the OTUs found in every ice sample, was nevertheless dominated by many of the same OTUs detected in the sea ice library (Table 1), yielding Sørensen's similarity index of 64.3% relative to ice sample 24-II.
Fig. 1.
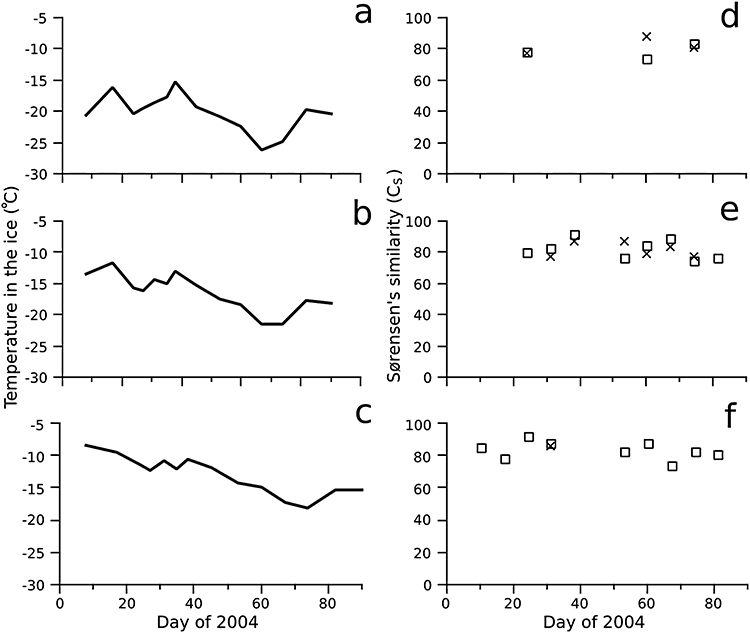
Temperature in the ice (A–C) from Collins and colleagues (2008), and pairwise similarities (D–F) for bacterial (×) and archaeal (□) communities in Arctic winter sea ice horizons I–III, representing depths of 25, 45 and 65 cm below the ice surface (top, middle and bottom panels respectively). Sørensen's similarity index (%) was calculated for each ARISA (Bacteria) or T-RFLP (Archaea) sample relative to the first sample in the time series, 24-II (Bacteria) or 17-II (Archaea), which each had self-similarities of 100% (unplotted). Pearson correlation coefficients of pairwise similarity over time were not significant (at P < 0.05) for either community in any horizon.
Table 1.
Bacterial ARISA OTUs in Franklin Bay (FB) sea ice horizons I–III (representing depths of 25, 45 and 65 cm below the ice surface) and under-ice seawater (SW).
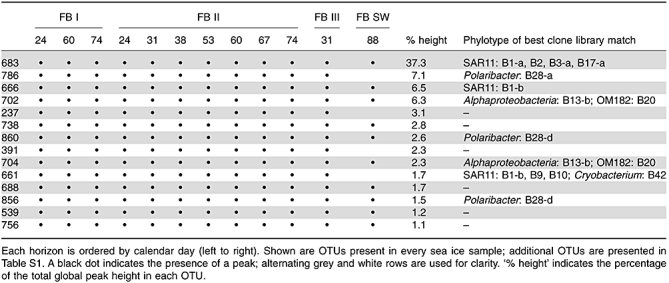 |
Several OTUs persisting in ice through the winter had no representatives in the bacterial clone library; these OTUs summed to 15% of the total signal, indicating that potentially important groups were missed by this approach. Nevertheless, overlap between clone library sequences and ARISA OTUs (Table 1, Table S1) was substantial. Of the 50 identifiable bacterial subtypes (members of the same phylotype, as identified by 16S rRNA gene sequence, with variable ARISA lengths) detected in the clone libraries, 32 subtypes had predicted ARISA lengths ± 1 bp of an ARISA OTU (Table 1, Table S1); 10 more subtypes had putative matches ± 2.5 bp.
Archaeal T-RFLP
Archaeal 16S rRNA genes were readily amplified from these winter sea ice samples, yielding a larger data set than for Bacteria, yet still no changes in community structure (Fig. 1) or richness (Fig. S1) of the archaeal communities were detected through the winter. A large majority (88%) of the total T-RFLP signal intensity derived from 17 OTUs present in all 21 successfully analysed sea ice samples (Table 2). Those OTUs included several matching the Marine Group I.1a Crenarchaeota, which comprised 45% of the total signal intensity (Table 2), in agreement with the dominance of this clade in the clone libraries (Table 3). OTUs best matching the Marine Group II.b Euryarchaeota made up a further 25% of the total signal intensity. Correlation analysis revealed no significant change (P < 0.05) over time in community structure similarity (81 ± 6%, Fig. 1) or richness (range 18–33, Fig. S1) in any horizon.
Table 2.
Archaeal T-RFLP OTUs in Franklin Bay (FB) sea ice horizons I–III (representing depths of 25, 45 and 65 cm below the ice surface), and under-ice seawater (SW).
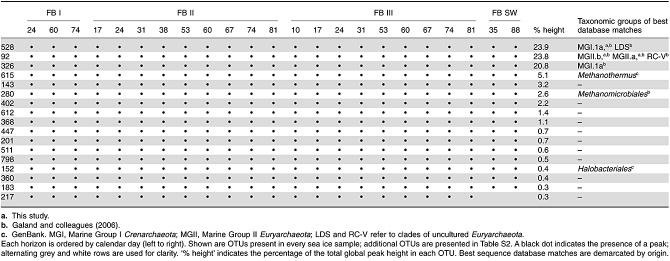 |
Table 3.
A summary of the bacterial and archaeal clone libraries from sea ice horizon I (25 cm depth) and under-ice seawater, including the abundance of major taxonomic groups.
| Sea ice | Seawater | |
|---|---|---|
| Day of year collected | 74 + 81 | 35 |
| Temperature at collection | −22°C | −1.7°C |
| Bacterial clone libraries | FB04bi | FB04bw |
| All Bacteria | 109 | 46 |
| Proteobacteria | ||
| Alphaproteobacteria | ||
| SAR11 clade | 76 | 22 |
| Other | 5 | 3 |
| Gammaproteobacteria | 12 | 2 |
| Betaproteobacteria | 2 | 1 |
| Deltaproteobacteria | 1 | 0 |
| Unclassified Proteobacteria | 1 | 0 |
| Bacteroidetes | ||
| Flavobacteria | 6 | 13 |
| Sphingobacteria | 2 | 3 |
| Other | ||
| Actinobacteria | 3 | 1 |
| Verrucomicrobia | 1 | 0 |
| ‘Marine Group A’ | 0 | 1 |
| Archaeal clone libraries | FB04ai | FB04aw |
| All Archaea | 52 | 45 |
| Crenarchaeota | ||
| Marine Group I | 46 | 41 |
| Euryarchaeota | ||
| Marine Group II | 6 | 4 |
No trend in spatial distribution was observed in the dominant members of the community, but several minor unidentified OTUs exhibited such patterns: best matches to Marine Group I.3a Crenarchaeota (515 bp) and Thermoplasma (228 bp) were found primarily in upper ice horizons; OTUs with best matches to Marine Group I.1c Crenarchaeota (499 bp), Methanobacteria (477 bp), RC-V Euryarchaeota (531 bp) and unknown groups (529 bp, 322 bp) were found primarily in lower ice horizons (Table 2, Table S2). The under-ice seawater samples, 35-SW and 88-SW, contained most of the same OTUs dominant in the sea ice library (Table 2, Table S2) and did not cluster separately from the ice samples, although some minor OTUs were present only in the seawater samples: 317 bp, 428 bp and 602 bp. Relative to ice sample 17-II, seawater samples 35-SW and 88-SW had Sørensen's similarity indices of 70.8% and 67.8% respectively.
All of the phylotypes detected in the archaeal clone libraries were also detected as T-RFLP OTUs, although many OTUs persisting through the winter had no representatives in our clone libraries, possibly as a result of the different primers used for cloning and fingerprinting. Several of these OTUs were putatively identified using a database of sequences from a recent study (Galand et al., 2006) conducted near the outflow of the Mackenzie River (due west of our study site; Fig. S2), including Methanomicrobiales, uncultured methanogen-associated groups LDS and RC-V, and uncultured members of the Marine Group I.3a and Marine Group I.3c Crenarchaeota. Other OTUs were putatively identified as halophiles (Halobacteriales, 152 bp) or thermophiles (e.g. Methanothermus, 615 bp and Thermoplasmata, 228 bp), based on a database of predicted terminal restriction fragment lengths of archaeal sequences from GenBank.
Community composition
Bacterial community
The dominant phylotypes in both sea ice and seawater libraries (70% and 48% of sequences respectively; Table 3) were associated with the common seawater clade of SAR11 Alphaproteobacteria (Fig. S3), consistent with their dominance in the ARISA analysis (Table 1). Two major SAR11 subtypes were detected; neither showed a differential distribution between the sea ice and seawater libraries (Table S3).
Overlap between the sea ice and seawater libraries was limited: 6 of 44 phylotypes were shared between ice (28 total phylotypes) and water (22 total phylotypes), but due to the common dominance of SAR11, no statistical difference was detected between the libraries using WebLIBSHUFF (p = 0.143; p = 0.183). Differences were evident in the relative occurrence of Gammaproteobacteria, which appeared primarily in the sea ice library, and of Bacteroidetes, found primarily in the seawater library (Table S3). The most abundant gammaproteobacterial phylotypes clustered with cultured oligotrophic bacterioplankton clades OM182 and OM60 (Fig. S4, Table S3). The Bacteroidetes sequences were dominated by polar marine Cryomorphaceae phylotypes, present only in the seawater library. Despite being represented by only five sequences in the libraries, the single Polaribacter phylotype consisted of four subtypes. Together, two of these subtypes accounted for 11.2% of the total signal intensity in the ARISA analysis (Table 1). Each library included a different OM43-clade Methylophilales phylotype, as well as several distinct marine Roseobacter phylotypes (Fig. S3, Table S3). Other unshared phylotypes belonged to the high G+C Gram-positive Actinobacteria (sea ice and seawater), Verrucomicrobia and Deltaproteobacteria (sea ice), and the uncultured ‘Marine Group A’ division (seawater). The seawater library had a greater Chao1 index of richness (174 for seawater, 39 for ice), likely underestimated due to lesser coverage (61% for seawater, 88% for ice), but the libraries had similar Shannon diversity indices (2.25 for seawater, 2.43 for ice).
Eukaryotic 18S rRNA gene sequences were detected only in the seawater library. These 13 sequences were closely related to uncultured marine stramenopile group MAST-1, clade NS1A, with worldwide distribution including Arctic and Antarctic surface waters (Lovejoy et al., 2006; Massana et al., 2006). The predicted ARISA fragment lengths for these sequences were less than 100 bp so their detection by ARISA was not likely.
Archaeal community
Archaeal sequences were present in both sea ice and seawater clone libraries; seven phylotypes had high similarities to existing Arctic archaeal clone sequences. The great majority (91%) of sequences from the archaeal libraries belonged to Crenarchaeota of the Marine Group I clade, most of which fell into a single phylotype of the Marine Group I.1a (Fig. 2, Table S4). The remaining crenarchaeal sequences were scattered among four more phylotypes within the Marine Group I.1a and Marine Group I.1c clades (Fig. 2). Two phylotypes belonged to uncultured Euryarchaeota of the Marine Group II.b clade clusters 5 and 7. The libraries had high coverage (> 95% each), similar richness (5 for ice, 6 for seawater) and similar indices of Shannon diversity (0.61 for ice, 0.76 for seawater).
Fig. 2.
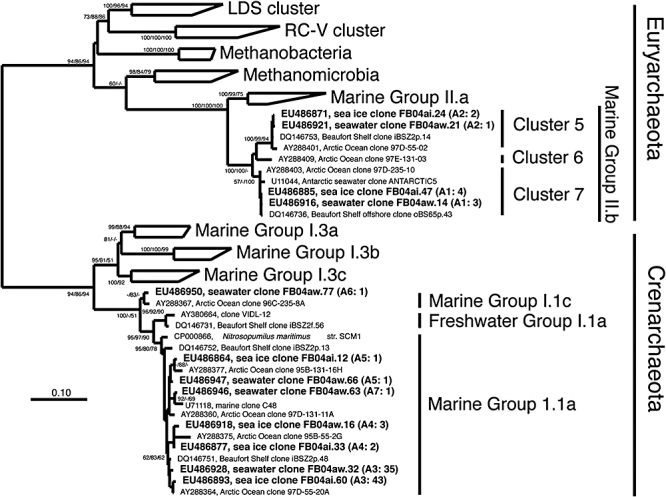
Phylogenetic tree of archaeal 16S rRNA gene sequences. Tree topology was defined by the consensus of 1000 maximum parsimony bootstrap replications utilizing 511 parsimony-informative nucleotides. Branch lengths were defined by Tamura-Nei distances calculated from 807 hypervariable-masked nucleotides. Node values indicate percentage of 10 000 distance, 1000 maximum parsimony and 100 maximum likelihood bootstrap replications respectively; only bootstrap values greater than 50% are shown. One sequence from each phylotype (defined by > 99% similarity) from each library in this study is shown in bold, followed in parentheses by the phylotype and number of sequences from each library within that phylotype.
Discussion
The expectation that selective losses of dominant bacterial and archaeal community members would occur under the extreme conditions of winter sea ice was not realized in this study. Although both bacterial growth and mortality had been inferred from changes in total bacterial counts (Wells and Deming, 2006b; Collins et al., 2008), microbial community succession was not detectable in the upper ice horizons we sampled. We found no significant change in bacterial or archaeal richness or community similarity over the 3-month period of our investigation (Fig. 1, Fig. S1). Persistence of the dominant members of the sea ice microbial community through winter was also evidenced by the high proportion of total signal associated with members present in every sea ice sample analysed by ARISA (Table 1) and T-RFLP (Table 2). Whereas culture-based studies of heterotrophic microbes had suggested forces at work in sea ice to select for psychrophilic over psychrotolerant strains (Kaneko et al., 1977; Delille and Rosiers, 1996; Delille et al., 1997; Fiala et al., 2006), not even the extreme conditions of the winter sea ice we studied exerted any more than very limited (statistically undetectable) selective pressure at the taxonomic level on the bacterial and archaeal communities entrapped in this ice. Techniques used with polar seawater to quantify differences in relative abundance of specific groups of microorganisms, like FISH (Alonso-Sáez et al., 2008), quantitative PCR of 16S rRNA and functional genes like amoA (Galand et al., 2009b), or massively parallel hypervariable tag sequencing (Huse et al., 2008; Galand et al., 2009a), have not yet been applied to sea ice. Their use on winter sea ice might reveal subtle patterns of succession in bacterial and archaeal communities that escaped the sensitivity limits of our methods.
Many Bacteria and Archaea may lie dormant in winter sea ice (Kaneko et al., 1977; Helmke and Weyland, 1995), surviving without reproducing and thus limiting the possible mechanisms of selection. Although Bacteria and Archaea were presumed to ‘survive’ if their DNA was present on our membrane filters, we have no data to demonstrate their continued viability in situ. Protistan bacterivores, which can selectively graze bacteria based on size and biochemical cues, are essentially absent from very cold Arctic winter ice (Krembs et al., 2002) and so are unlikely to contribute a selective effect, even if they may play a role in shaping the microbial community in warmer ice. The persistence of the ice microbial communities that we measured through winter is consistent with cells being relatively inactive in very cold sea ice and surviving in the absence of bacterivores, whereas more active under-ice seawater communities are subject to predation and do change over the course of winter (Murray and Grzymski, 2007; Alonso-Sáez et al., 2008). Complete preservation in the ice, however, is inconsistent with reports of actively respiring cells in Arctic winter sea ice (Junge et al., 2004) and, from the same ice we sampled, examples of bacterial mortality (Collins et al., 2008) and viral and bacterial production in experimental ice brines (Wells and Deming, 2006b). To reconcile the general absence of species-specific mortality reported here with the presumed presence of a mixture of active and inactive populations (whether taxonomically similar or not), both populations must share similar mortality rates. This assumption can be fulfilled if the primary mechanisms of mortality in the ice are taxonomically non-selective, despite the possibility that they may each affect active and inactive cells differently.
Two mechanisms to consider for non-selective mortality are virally mediated lysis and cell damage from co-occurring extremes in temperature and salinity. Modelled contact rates between viruses and bacteria in sea ice brines are extremely high (up to 600 times that in seawater; Wells and Deming, 2006b) and, under environmental stress, host specificity may give way to a broad range of infectable hosts (Wells and Deming, 2006a), linking both mechanisms under consideration. Viral production (whether by species-specific or generalist phages) may play a role in mortality of any taxonomic group that remains active in the ice, whereas dormant populations might be affected not by viral reproduction from within but ‘lysis from without’ due to a large number of attached viruses (Delbrück, 1940).
For a generalized protective mechanism for cells within sea ice, hydrated coatings of extracellular polymeric substances (EPS) that simultaneously buffer against external ice-crystal damage, osmotic shock and viral attack has been proposed (Krembs et al., 2002; Krembs and Deming, 2008). EPS are produced by many sea ice bacteria in culture (Mancuso Nichols et al., 2005; Marx et al., 2009), colonized by bacteria in sea ice (Meiners et al., 2004), and increased in abundance during winter in the ice we studied (Collins et al., 2008). Production of EPS by a subset of microorganisms entrained into sea ice might serve all entrapped within it, including common seawater species not adapted to life in ice, thereby limiting species-specific mortality.
The structure of these persistent winter sea ice communities is unique in resembling that of communities in autumn and winter seawater, measured in this study and reported by Alonso-Sáez and colleagues (2008) for our study site, rather than previously observed communities in spring and summer sea ice. For the first time, sea ice bacterial communities were observed (by both clone library and fingerprinting results) to be dominated by the common pelagic SAR11 clade Alphaproteobacteria, with only much smaller complements of the Alphaproteobacteria and Flavobacteriales well known from sea ice. While studies of later season sea ice have failed to detect Archaea, all of our winter ice (and seawater) samples contained them, with the dominant archaeon belonging to Marine Group I.1a Crenarchaeota. Clearly, these pelagic Bacteria and Archaea entrained into sea ice upon its formation and persisted through the winter. Our community structure results also indicate that microorganisms from other habitats, including terrestrial soil, riverine waters and marine sediment, entrain into sea ice and persist through the winter. Likely sources of the non-marine microorganisms include freshwater from the nearby Horton River, eroded soils from the Smoking Hills, and terrestrial organic matter from the Mackenzie River, the largest source of suspended particulates to the Beaufort Shelf (Macdonald et al., 1998).
All of the dominant SAR11 phylotypes detected in our winter ice samples were highly similar to sequences from the coastal Beaufort Sea. Regarding the dominant SAR11 phylotypes detected in our winter ice samples, all were highly similar to sequences from the coastal Beaufort Sea (Galand et al., 2008a), central Arctic Ocean (Bano and Hollibaugh, 2002; Malmstrom et al., 2007), and Antarctic surface waters (Murray and Grzymski, 2007), even though absent from summer sea ice (Brown and Bowman, 2001; Brinkmeyer et al., 2003) and highly productive autumn sea ice near Antarctica (Brinkmeyer et al., 2003). The only cultured representative of the SAR11 clade is ‘Candidatus Pelagibacter ubique’, an obligately oligotrophic alphaproteobacterium distributed widely throughout the waters of the global ocean (Morris et al., 2002). Although we detected two clades of SAR11 ITS sequences, no evidence was found to indicate ecotype differentiation (as suggested by García-Martínez and Rodríguez-Valera, 2000) between sea ice and seawater environments. Because sequences within the SAR11 clade made up a larger fraction of the late winter sea ice library (70%) than the under-ice seawater library (48%), SAR11 may nevertheless overwinter more successfully in ice than in seawater. This inference is consistent with quantitative FISH studies by Alonso-Sáez and colleagues (2008), who detected a seasonal decrease in the relative abundance of SAR11 in Franklin Bay surface waters from 36% of total DAPI counts at the time of freeze-up (December) to 18% by late winter (March).
The dominant archaeal phylotypes in our sea ice and seawater libraries, Marine Group I Crenarchaeota (Fig. 2, Table S4) have been identified from central Arctic seawater (Bano et al., 2004), Beaufort shelf nepheloid layers and riverine particles (Galand et al., 2006; 2008b;), and Antarctic seawater growing frazil ice (Delong et al., 1994), yet were absent from summer sea ice (Brown and Bowman, 2001; Brinkmeyer et al., 2003) and highly productive autumn sea ice near Antarctica (Brinkmeyer et al., 2003). These phylotypes clustered with the ‘Marine’ Group I Crenarchaeota rather than the ‘Freshwater’ Group I which are prominent in the Mackenzie River (Galand et al., 2008a,b;). The seasonally high relative abundance of the Marine Group I Crenarchaeota that we detected both by T-RFLP (45% of total signal intensity) and clone library sequencing (91% of sequences in both libraries) in Franklin Bay is consistent with FISH counts in surface waters at the same site, showing a winter high for this group (to 16% of DAPI counts) that decreased to undetectable levels by late summer (Alonso-Sáez et al., 2008). This trend is also consistent with results from Antarctic waters showing that the relative abundance of Marine Group I Crenarchaeota was highest during winter and inversely correlated with algal biomass (chlorophyll a) on seasonal time scales (Murray et al., 1998; Church et al., 2003). The rarer Marine Group II Euryarcheota we detected were also closely related to phylotypes found prevalently in archaeal communities from the coastal Beaufort Sea (Galand et al., 2006; 2008b;) and central Arctic Ocean (Bano et al., 2004). Although Archaea were identified by Junge and colleagues (2004) in winter sea ice, their FISH probes were generic for the domain Archaea so no further taxonomic assignment was possible.
Bacterial sequences from several frequently cultured groups of copiotrophic sea ice bacteria were absent from our winter clone libraries. Neither the bacterial sea ice nor seawater library harboured sequences from cold-adapted genera of the Oceanospirillales, Alteromonadales or Flavobacteriales commonly cultured and cloned from sea ice, including Colwellia, Glaciecola, Halomonas, Marinobacter, Pseudoalteromonas, Psychrobacter, Psychromonas and Shewanella (Fig. S4, Table S3). Instead, the majority of gammaproteobacterial phylotypes were related to recently cultured oligotrophic seawater clades OM60 and OM182 (Cho and Giovannoni, 2004). The OM60 clade is prominent in Arctic seawater (Bano and Hollibaugh, 2002; Kellogg and Deming, 2009) while the OM182 clade is seasonally abundant in Antarctic surface waters (Murray et al., 1998; Grzymski et al., 2006; Murray and Grzymski, 2007). A representative from the OM60 clade was reported in Arctic pack ice (Brinkmeyer et al., 2003), but no members of the OM182 clade have previously been identified from sea ice. Cultured isolates of sea ice Alphaproteobacteria generally cluster with marine Roseobacter, but only one of our phylotypes was most similar to a cultured representative (Sulfitobacter sp.). Others in the uncultured Roseobacter RCA cluster were detected infrequently in our libraries, consistent with their low abundance in seawater at our overwintering station (< 5%; Alonso-Sáez et al., 2008). Bacteroidetes are well known from sea ice and often make up a sizable fraction of the community in polar seawater (Bano and Hollibaugh, 2002; Wells and Deming, 2003; Malmstrom et al., 2007), including at our overwintering site (Alonso-Sáez et al., 2008), yet made up only a small fraction of the bacterial clones in our winter sea ice library. The implication is that these common sea ice bacteria only become dominant in sea ice after the winter season has passed.
Polaribacter, a genus of Bacteroidetes appearing in the ARISA analysis, provided a notable exception to the lack of representation by cultivated isolates in the winter sea ice we sampled (Table 1 and Table S1). Polaribacter spp., known for their production of gas vacuoles, have been cultured and identified as abundant Bacteroidetes from both Arctic and Antarctic sea ice (Gosink et al., 1998; Brown and Bowman, 2001; Junge et al., 2002; Brinkmeyer et al., 2003; Auman et al., 2006). The recent genome sequence of Antarctic seawater isolate Polaribacter irgensii strain 23-P (GenBank accession: AAOG00000000) may help elucidate their adaptations to sea ice, just as the genome sequence analysis of Polaribacter MED152 has revealed their adaptations to sunlit surface waters (e.g. rhodopsins; González et al., 2008). In this study, preferential entrainment of Polaribacter spp. into sea ice was inconclusive, because they were also detected in under-ice seawater by both cloning and ARISA, but the high intra-specific ITS variability we observed may prove useful in future tests of ecotype differentiation by these successful sea ice colonizing bacteria.
Overall, the dominant presence of common seawater microorganisms and the absence of many known sea ice microorganisms in the Arctic winter ice we studied indicate that species-specific mortality during the winter was rare. The degree of similarity between winter ice and seawater communities also implies that selection during the freezing process must have been relatively minor. The distinctive nature of well-known microbial communities in sea ice of the warmer biologically productive seasons must not be predetermined by selective survival of community members exposed to freezing and severe winter conditions, but rather as a result of competitive outgrowth by copiotrophs that overwinter below detection limit or arrive as immigrants once the warming ice becomes permeable.
Experimental procedures
Ice core and seawater sampling
Sampling location and procedures have been reported in detail by Collins and colleagues (2008), along with measurements of air and ice temperature, bulk ice and brine salinity, brine volume fraction, and content of total particulate matter, bacterial abundance and pEPS in the ice. Briefly, each week from 10 January (calendar day 10) to 28 March 2004 (day 88), three ice cores were drilled from a designated field of landfast first-year sea ice in Franklin Bay, Northwest Territories, Canada (at 70.0°N, 126.3°W; 16 km from the mouth of the Horton River) without reaching seawater to capture 10 cm depth horizons centred at 25, 45 and 65 cm from the ice surface and designated horizons I, II and III respectively. Freezing dates for horizons I and II were predicted to be 9–18 November and 26 November–5 December, respectively, while horizon III was observed to freeze from 14 to 20 December 2003.
The ice sections were cut aseptically in the field, placed into sterile Whirl-Pak bags and transported in an insulated cooler to a shipboard cold room set at 0°C, where they were processed within 24 h. To protect against osmotic shock and possible cell lysis, each section (after mechanical crushing) was melted into 0.22 µm filtered artificial brine solution (prepared as in Collins et al., 2008) at 0°C, using an ice : brine volume ratio of 1:2. After subsampling for other variables (Collins et al., 2008), the remainder of each melted sample was gently filtered onto a 47 mm diameter 0.22-µm-nitrocellulose filter (Millipore) and stored at −80°C for later DNA extraction.
Under-ice seawater samples were collected on calendar days 35 and 88 by lowering a hand-held 2 l Niskin bottle through a hole in the ice to the base of the ice sheet. Designated 35-SW and 88-SW, the seawater samples, each with a salinity of 30, were returned to the ship, filtered immediately, and the filters stored at −80°C, as for fully melted sea ice samples.
DNA extraction
Within 2 years of collection, each filter was removed from storage at −80°C and cut into small fragments with sterilized scissors. For horizons II and III, all three filters (one from each of the three ice cores) from each sampling day were combined in a single tube. In horizon I, due to low DNA yields, six filters – three from each of two sampling days in adjacent weeks – were combined in a single tube, i.e. from the following pairs of calendar sampling days: 10 and 17, 24 and 31, 60 and 67, and 74 and 81. Each tube then received, per filter, 800 µl of STE buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and 40 µl of 20% SDS. After incubation at 65°C for 20 min, each tube was vortexed, then centrifuged at 1400 g for 15 min. The supernate was transferred to a Centricon YM-100 centrifugal filtration device (Millipore) to concentrate and de-salt the genomic DNA, according to the manufacturer's recommendations. The recovered volume was increased to 600 µl with TE buffer before three rounds of phenol/chloroform extraction, followed by ethanol precipitation and re-suspension in 50 µl of TE buffer. Total DNA concentration was measured in a SpectraMaxM2 plate reader (Molecular Devices) using PicoGreen fluorescence (Invitrogen) according to the manufacturer's recommendations. Recovery of genomic DNA was < 1–33% based on cell counts (Collins et al., 2008), assuming 2.5 fg of DNA per bacterium (Button and Robertson, 2001). No attempt was made to separate the DNA of viable cells from that of dead cells.
Community fingerprinting
Bacterial DNA fragments were PCR-amplified for ARISA using fluorescently labelled (6-HEX) forward primer Uni1392F, and unlabelled reverse primer R23S-125R (sequences located in Table S5). Amplified bacterial DNA was pooled from two PCR amplifications, each containing 3–117 ng of total DNA. Partial archaeal 16S rRNA genes were PCR-amplified for T-RFLP using fluorescently labelled (6-FAM) forward primer Arch109F, and unlabelled reverse primer Arch915R. Amplified archaeal DNA from four PCR amplifications was pooled, then digested with restriction enzyme HpyCH4III at 37°C for 6 h, which was determined empirically to enable complete digestion without overdigestion. PCR amplifications using archaeal primers were generally more robust than with bacterial primers, even though archaeal abundance was likely only a few per cent of the bacterial abundance (Junge et al., 2004), a phenomenon which has also been observed in a highly saline Arctic spring system (T. Niederberger, pers. comm.). All DNA fragments were analysed on a MegaBACE1000 capillary gel electrophoresis instrument (Molecular Dynamics).
Electropherograms were analysed using DAx analysis software (v8.0, Van Mierlo Software Consultancy). A low-pass Fourier transform was applied to ARISA electropherograms to reduce noise and increase peak calling efficiency. Heights of saturated peaks in several T-RFLP electropherograms were estimated by fitting a Gaussian function to the non-saturated points, using the open-source statistical package R (R Development Core Team, 2008). For both ARISA and T-RFLP, a peak was called if its height was > 5× the baseline root-mean-square noise level (< 1.0% of the cumulative peak height for each profile). Profiles with cumulative peak heights less than 1 × 104 RFUs (ARISA) or 8 × 104 RFUs (T-RFLP) were removed from the analysis. The peaks in the remaining samples were binned using in-house software (http://rocaplab.ocean.washington.edu/cgi/dakster/index.html) and distance cut-offs of 1 bp for fragment lengths of 70–700 bp, 2 bp for 700–1200 bp and 4 bp for > 1200 bp. Each bin, representing a 16S-ITS-23S ribosomal DNA fragment (ARISA) or terminal restriction fragment (T-RFLP), was designated an OTU. The cumulative peaks heights of each OTU were used as gross measures of relative abundance to compare fingerprinting with clone libraries. A presence/absence matrix containing OTUs with at least one peak height greater than 1.0% (ARISA) or 0.25% (T-RFLP) of the sample's cumulative peak height was used for calculation of richness and pairwise similarity, and non-parametric multivariate analyses, performed with PRIMER v6 (Clarke and Gorley, 2006). Pairwise distances were calculated as Sørensen's similarity coefficient (C_s = 2C/(A + B), where A and B are the number of OTUs in each of two samples, and C is the number of shared OTUs between the two samples; Hughes et al., 2001); samples were then clustered by Group Average and the significance of each cluster was calculated by SIMPROF at the 95% confidence level. Pearson correlation coefficients for Sørensen's similarity with date were calculated in R, as were partial Pearson correlation coefficients for richness with date controlling for the cumulative peak height of each profile. A correlation function in the open-source plotting program Qtiplot was used to calculate the point of maximum covariance between ARISA RFUs and clone library phylotype frequencies to correct for different running rates of fluorescent labels 6-HEX and ROX (used for the ARISA ladder), resulting in an adjustment of ARISA OTU lengths by −7 bp, near the −5.5 bp difference determined by Hahn and colleagues (2001).
Clone library construction and sequencing
Four clone libraries were constructed for phylogenetic analysis, using the ribosomal RNA operon, from two samples: under-ice seawater from day 35, used to create libraries FB04bw and FB04aw; and winter sea ice from horizon I (combined days 74 and 81), used to create libraries FB04bi and FB04ai, where FB04 = Franklin Bay 2004, b = Bacteria, a = Archaea, w = water and i = ice. Bacterial 16S-ITS-23S ribosomal DNA was amplified using primer pair Uni515F/R23S-125R (Table S5). Archaeal 16S ribosomal DNA was amplified using primer pair Arch21F/Arch958R. Four separate amplifications were performed for each sample and each was reconditioned using 1/10 of the PCR product as template for an additional four cycles with fresh reaction mix (Thompson et al., 2002). The pooled reconditioned PCR product was cloned using the TOPO-TA Cloning Kit for Sequencing (Invitrogen) according to the manufacturer's recommendations. Bacterial clone libraries were subjected to dye-termination sequencing at the High-Throughput Genomics Unit (HTGU), Department of Genome Sciences, University of Washington. Archaeal clone libraries were sequenced on a MegaBACE1000 capillary gel electrophoresis instrument. Bi-directional double-stranded sequences were obtained for > 900 bp of the bacterial and archaeal 16S rRNA genes and the complete ITS region for Bacteria. Sequencher software (v4.6, Gene Codes Corp.) was used to call bases and construct contigs which were checked and edited manually as necessary. Most of the bacterial sequences included the 16S–23S ITS region, which we excluded from phylogenetic analyses but from which we calculated predicted ARISA fragment lengths and defined subtypes of 16S rRNA gene phylotypes. Seventeen sequences related to Stenotrophomonas maltophila were removed from the bacterial sea ice library as probable contaminants. ARISA fragment lengths predicted from these sequences (777 and 779 bp) overlapped with those predicted from other clone library sequences, but these fragments were observed only rarely in the ARISA data set. Bacterial sequences were deposited in GenBank with Accession No. EU836892–EU837057 and FJ753995–FB754002; archaeal sequences: EU486859–EU486955; and eukaryotic sequences: FJ753982–FJ753994.
Phylogenetic analysis
Small-subunit ribosomal rRNA gene sequences were aligned using the NAST aligner (Desantis et al., 2006) at Greengenes (http://greengenes.lbl.gov). All sequences were checked for chimeras with Bellerophon (Huber et al., 2004) and Mallard (Ashelford et al., 2006); none were detected. Sequence alignments were imported into ARB (Ludwig et al., 2004) and edited manually as necessary. Bootstrapped phylogenetic trees were constructed in paup* v4.10beta (Swofford, 2003). ModelTest was used to determine the optimal nucleotide substitution model for maximum likelihood tree construction (Posada and Crandall, 1998). In ARB, partial sequences were added to the tree by parsimony and assigned phylotypes based on their location within the tree. Distance matrices calculated in paup* using the Tamura and Nei (1993) model of nucleotide substitution were used with WebLIBSHUFF (Schloss et al., 2004) to compare clone libraries statistically, and with dotur (Schloss and Handelsman, 2005) to define phylotypes and calculate estimates of species richness and diversity using the Chao1 and Shannon indices respectively. Phylotypes were defined by > 98% similarity for bacterial sequences and > 99% for archaeal sequences. Subtypes were designated if multiple fingerprinting fragments were predicted within any single phylotype. The archaeal sequences were subsequently used to aid the choice of restriction enzymes for community profiling by T-RFLP (Collins and Rocap, 2007).
Acknowledgments
We thank the officers and crew of the CCGS Amundsen for logistical support, S. Carpenter, L.E. Wells, O. Owens and A. Langlois for field assistance, P. Galand and three anonymous reviewers for helpful comments on the manuscript, and H. Eicken for insightful discussion. This study was funded by NSF awards (OPP 0327244 to J. Deming and IGERT through DGE-980713), with additional support from the Washington Sea Grant Program and a NASA-ABI award (NCC2-1273); G. Rocap was supported by NSF OCE award 0220826. This article is a contribution from the international Canadian Arctic Shelf Exchange Study.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Bacterial abundance (A–C) from Collins and colleagues (2008), and the community richness (D–F) of Bacteria (×, 98% similarity cut-off) and Archaea (□, 99% similarity cut-off) in Arctic winter sea ice horizons I–III, representing depths of 25, 45 and 65 cm below the ice surface (top, middle and bottom panels, respectively). Dashed lines in (A) and (B) indicate robust linear best fit lines in horizons with statistically significant decreases (P < 0.05) in cell abundance over time. Pearson correlation coefficients of richness over time were not significant for either community in any horizon.
Fig. S2. Map of the vicinity of Franklin Bay, Northwest Territories, Canada, indicating Franklin Bay (arrows), the primary sampling site at the CCGS Amundsen overwintering station (o) and nearby Smoking Hills (x).
Fig. S3. Phylogenetic tree of alphaproteobacterial and betaproteobacterial 16S rRNA gene sequences. Tree topology was defined by the consensus of 1000 maximum parsimony bootstrap replications utilizing 291 parsimony-informative nucleotides. Branch lengths were defined by Tamura-Nei distances calculated from 558 hypervariable-masked nucleotides. Node values indicate percentage of 10 000 distance, 1000 maximum parsimony and 100 maximum likelihood bootstrap replications respectively; only bootstrap values greater than 50% are shown. One sequence from each phylotype (defined by > 98% similarity) from each library in this study is shown in bold, followed in parentheses by the phylotype and number of sequences from each library within that phylotype.
Fig. S4. Phylogenetic tree of gammaproteobacterial 16S rRNA gene sequences. Tree topology was defined by the consensus of 1000 maximum parsimony bootstrap replications utilizing 240 parsimony-informative nucleotides. Branch lengths were defined by Tamura-Nei distances calculated from 570 hypervariable-masked nucleotides. Other features as in Fig. S3.
Table S1. Bacterial ARISA OTUs in Franklin Bay (FB) sea ice horizons I–III (representing depths of 25, 45 and 65 cm below the ice surface) and under-ice seawater (SW). Each horizon is ordered by calendar day (left to right). Shown are OTUs that matched the predicted fragment length from a clone library sequence and OTUs present in at least 70% of the sea ice samples, excepting OTUs present in Table 1. A black dot indicates the presence of a peak; alternating grey and white rows are used for clarity. ‘% height’ indicates the percentage of the total global peak height in each OTU.
Table S2. Archaeal T-RFLP OTUs in Franklin Bay (FB) sea ice horizons I–III (representing depths of 25, 45 and 65 cm below the ice surface), and under-ice seawater (SW). Each horizon is ordered by calendar day (left to right). Shown are OTUs present in at least 50% of the sea ice samples, and those with at least one peak height greater than 0.25% of the sample’s cumulative peak height, excepting OTUs present in Table 2. A black dot indicates the presence of a peak; alternating grey and white rows are used for clarity. ‘% height’ indicates the percentage of the total global peak height in each OTU. Best sequence database matches are demarcated by origin: †this study; ‡ Galand and colleagues (2006); and §GenBank. MGI, Marine Group I Crenarchaeota; MGII, Marine Group II Euryarchaeota; LDS and RC-V refer to clades of uncultured Euryarchaeota.
Table S3. Bacterial phylotypes (defined by > 98% 16S rRNA gene similarity) in clone libraries from late winter sea ice (FB04bi) and under-ice seawater (FB04bw). Subtypes were designated by predicted ARISA lengths, calculated for primer pair Uni1392F and R23S-125R. ‘Unknown’ indicates partial sequence for which ARISA length could not be calculated.
Table S4. Archaeal phylotypes (defined by > 99% 16S rRNA gene similarity) in clone libraries from late winter sea ice (FB04ai) and under-ice seawater (FB04aw). Subtypes were designated by predicted TRF lengths, calculated for primer/enzyme combination Arch109F/HpyCH4III. ‘Unknown’ indicates partial sequence for which TRF length could not be calculated.
Table S5. Primers used in this study. Universal name as described by Alm and colleagues (1996).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Sáez L, Sánchez O, Gasol JM, Balagué V, Pedrós-Alió C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman AJ, Breezee JL, Gosink JJ, Kampfer P, Staley JT. Psychromonas ingrahamii sp. nov., a novel gas vacuolate, psychrophilic bacterium isolated from Arctic polar sea ice. Int J Syst Evol Microbiol. 2006;56:1001–1007. doi: 10.1099/ijs.0.64068-0. [DOI] [PubMed] [Google Scholar]
- Bano N, Hollibaugh JT. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol. 2002;68:505–518. doi: 10.1128/AEM.68.2.505-518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol. 2004;70:781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeekin TA. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmeyer R, Knittel K, Jugens J, Weyland H, Amann R, Helmke E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Environ Microbiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Bowman J. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO) FEMS Microbiol Ecol. 2001;35:267–275. doi: 10.1111/j.1574-6941.2001.tb00812.x. [DOI] [PubMed] [Google Scholar]
- Buchan A, González JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button DK, Robertson BR. Determination of DNA content of aquatic bacteria by flow cytometry. Appl Environ Microbiol. 2001;67:1636–1645. doi: 10.1128/AEM.67.4.1636-1645.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JC, Giovannoni SJ. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of plankton Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr. 2003;48:1893–1902. [Google Scholar]
- Clarke K, Gorley R. PRIMER v6: User Manual and Tutorial. Plymouth, UK: PRIMER-E; 2006. [Google Scholar]
- Collins RE, Rocap G. REPK: an analytical web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res. 2007;35:W58–W62. doi: 10.1093/nar/gkm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Carpenter SD, Deming JW. Spatial heterogeneity and temporal dynamics of particles, bacteria, and pEPS in Arctic winter sea ice. J Mar Sys. 2008;74:902–917. [Google Scholar]
- Delbrück M. The growth of bacteriophage and lysis of the host. J Gen Physiol. 1940;23:643–660. doi: 10.1085/jgp.23.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delille D, Rosiers C. Seasonal changes of Antarctic marine bacterioplankton and sea ice bacterial assemblages. Polar Biol. 1996;16:27–34. [Google Scholar]
- Delille D, Basseres A, Dessommes A. Seasonal variation of bacteria in sea ice contaminated by diesel fuel and dispersed crude oil. Microb Ecol. 1997;33:97–105. doi: 10.1007/s002489900012. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Wu KY, Prézelin BB, Jovine RV. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- Deming JW. Sea ice bacteria and viruses. In: Thomas DN, Dieckmann GS, editors. Sea Ice: An Introduction to Its Physics, Chemistry, Biology, and Geology. Oxford, UK: Blackwell Science; 2009. pp. 247–282. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetterer F, Knowles K. Sea ice index, updated 2008. 2002. Technical report, Boulder, CO, USA: National Snow and Ice Data Center.
- Fiala M, Kuosa H, Kopczynska EE, Oriol L, Delille D. Spatial and seasonal heterogeneity of sea ice microbial communities in the first-year ice of Terre Adelie area (Antarctica) Aquat Microb Ecol. 2006;43:95–106. [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Lovejoy C, Vincent WF. Remarkably diverse and contrasting archaeal communities in a large Arctic river and the coastal Arctic Ocean. Aquat Microb Ecol. 2006;44:115–126. [Google Scholar]
- Galand PE, Lovejoy C, Pouliot J, Garneau ME, Vincent WF. Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a stamukhi lake and its source waters. Limnol Oceanogr. 2008a;53:813–823. [Google Scholar]
- Galand PE, Lovejoy C, Pouliot J, Vincent WF. Heterogeneous archaeal communities in the particle-rich environment of an Arctic shelf ecosystem. J Mar Sys. 2008b;74:774–782. [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009a;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Galand PE, Lovejoy C, Hamilton AK, Ingram RG, Pedneault E, Carmack EC. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ Microbiol. 2009b;11:971–980. doi: 10.1111/j.1462-2920.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- García-Martínez J, Rodríguez-Valera F. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol Ecology. 2000;9:935–948. doi: 10.1046/j.1365-294x.2000.00953.x. [DOI] [PubMed] [Google Scholar]
- Golden KM, Ackley SF, Lytle VI. The percolation phase transition in sea ice. Science. 1998;282:2238–2241. doi: 10.1126/science.282.5397.2238. [DOI] [PubMed] [Google Scholar]
- González JM, Fernández-Gómez B, Fernández-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink JJ, Woese CR, Staley JT. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga–Flavobacterium–Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int J Syst Bacteriol. 1998;48:223–235. doi: 10.1099/00207713-48-1-223. [DOI] [PubMed] [Google Scholar]
- Grossmann S. Bacterial activity in sea ice and open water of the Weddell Sea, Antarctica: a microautoradiographic study. Microb Ecol. 1994;28:1–18. doi: 10.1007/BF00170244. [DOI] [PubMed] [Google Scholar]
- Grzymski JJ, Carter BJ, DeLong EF, Feldman RA, Ghadiri A, Murray AE. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl Environ Microbiol. 2006;72:1532–1541. doi: 10.1128/AEM.72.2.1532-1541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Wilhelm J, Pingoud A. Influence of fluorophor dye labels on the migration behavior of polymerase chain reaction-amplified short tandem repeats during denaturing capillary electrophoresis. Electrophoresis. 2001;22:2691–2700. doi: 10.1002/1522-2683(200108)22:13<2691::AID-ELPS2691>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Helmke E, Weyland H. Bacteria in sea ice and underlying water of the Eastern Weddell Sea in midwinter. Mar Ecol Prog Ser. 1995;117:269–287. [Google Scholar]
- Helmke E, Weyland H. Psychrophilic versus psychrotolerant bacteria – occurrence and significance in polar and temperate marine habitats. Cell Mol Biol. 2004;50:553–561. [PubMed] [Google Scholar]
- Hewson I, Meyers MEJ, Fuhrman JA. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, Southern California Borderlands. Environ Microbiol. 2007;9:923–933. doi: 10.1111/j.1462-2920.2006.01214.x. [DOI] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge K, Gosink JJ, Hoppe HG, Staley JT. Arthrobacter, Brachybacterium and Planococcus isolates identified from Antarctic sea ice brine. Description of Planococcus mcmeekinii, sp. nov. Syst Appl Microb. 1998;21:306–314. doi: 10.1016/S0723-2020(98)80038-6. [DOI] [PubMed] [Google Scholar]
- Junge K, Krembs C, Deming J, Stierle A, Eicken H. A microscopic approach to investigate bacteria under in situ conditions in sea-ice samples. Ann Glaciol. 2001;33:304–310. [Google Scholar]
- Junge K, Imhoff F, Staley T, Deming JW. Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperature. Microb Ecol. 2002;43:315–328. doi: 10.1007/s00248-001-1026-4. [DOI] [PubMed] [Google Scholar]
- Junge K, Eicken H, Deming JW. Bacterial activity at −2 to −20 degrees C in Arctic wintertime sea ice. Appl Environ Microbiol. 2004;70:550–557. doi: 10.1128/AEM.70.1.550-557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartokallio H, Laamanen M, Sivonen K. Responses of Baltic Sea ice and open-water natural bacterial communities to salinity change. Appl Environ Microbiol. 2005;71:4364–4371. doi: 10.1128/AEM.71.8.4364-4371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartokallio H, Tuomainen J, Kuosa H, Kuparinen J, Martikainen P, Servomaa K. Succession of sea-ice bacterial communities in the Baltic Sea fast ice. Polar Biol. 2008;31:783–793. [Google Scholar]
- Kaneko T, Atlas R, Krichevsky M. Diversity of bacterial populations in the Beaufort Sea. Nature. 1977;270:596–599. [Google Scholar]
- Kellogg C, Deming J. Comparison of free-living, particle- and aggregate-associated archaeal and bacterial diversity in the Laptev Sea. Aquat Microb Ecol. 2009;57:1–18. [Google Scholar]
- Krembs C, Deming JW. The role of exopolymers in microbial adaptation to sea ice. In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles: From Biodiversity to Biotechnology. New York, NY, USA: Springer-Verlag; 2008. pp. 247–264. [Google Scholar]
- Krembs C, Eicken H, Junge K, Deming JW. High concentrations of exopolymeric substances in Arctic winter sea ice: implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep Sea Res Part I Oceanogr Res Pap. 2002;49:2163–2181. [Google Scholar]
- Lovejoy C, Massana R, Pedrós-Alió C. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Appl Environ Microbiol. 2006;72:3085–3095. doi: 10.1128/AEM.72.5.3085-3095.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar AB, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RW, Solomon SM, Cranston RE, Welch HE, Yunker MB, Gobeil C. A sediment and organic carbon budget for the Canadian Beaufort Shelf. Mar Geol. 1998;144:255–273. [Google Scholar]
- Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL. Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aquat Microb Ecol. 2007;47:45–55. [Google Scholar]
- Mancuso Nichols C, Guezennec J, Bowman J. Bacterial exopolysaccharides from extreme environments with special consideration of the Southern Ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar Biotechnol. 2005;7:253–271. doi: 10.1007/s10126-004-5118-2. [DOI] [PubMed] [Google Scholar]
- Marx J, Carpenter S, Deming J. Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can J Microbiol. 2009;55:63–72. doi: 10.1139/W08-130. [DOI] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol. 2006;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Meiners K, Brinkmeyer R, Granskog MA, Lindfors A. Abundance, size distribution and bacterial colonization of exopolymer particles in Antarctic sea ice (Bellingshausen Sea) Aquat Microb Ecol. 2004;35:283–296. [Google Scholar]
- Mock T, Thomas DN. Recent advances in sea-ice microbiology. Environ Microbiol. 2005;7:605–619. doi: 10.1111/j.1462-2920.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Murray AE, Grzymski JJ. Diversity and genomics of Antarctic marine micro-organisms. Philos Trans R Soc Lond B Biol Sci. 2007;362:2259–2271. doi: 10.1098/rstb.2006.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, DeLong EF. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AM, Moore ER, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0. [Google Scholar]
- Schloss PD, Handelsman J. Introducing dotur, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serreze MC, Holland MM, Stroeve J. Perspectives on the Arctic's shrinking sea-ice cover. Science. 2007;315:1533–1536. doi: 10.1126/science.1139426. [DOI] [PubMed] [Google Scholar]
- Swofford DL. paup* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.10beta. Sunderland, MA, USA: Sinauer Associates; 2003. [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells LE, Deming JW. Abundance of Bacteria, the Cytophaga–Flavobacterium cluster and Archaea in cold oligotrophic waters and nepheloid layers of the Northwest Passage, Canadian Archipelago. Aquat Microb Ecol. 2003;31:19–31. [Google Scholar]
- Wells LE, Deming JW. Characterization of a cold-active bacteriophage on two psychrophilic marine hosts. Aquat Microb Ecol. 2006a;45:15–29. [Google Scholar]
- Wells LE, Deming JW. Modelled and measured dynamics of viruses in Arctic winter sea-ice brines. Environ Microbiol. 2006b;8:1115–1121. doi: 10.1111/j.1462-2920.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Wells LE, Cordray M, Bowerman S, Miller LA, Vincent WF, Deming JW. Archaea in particle-rich waters of the Beaufort Shelf and Franklin Bay, Canadian Arctic: clues to an allochthonous origin? Limnol Oceanogr. 2006;51:47–59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


