Summary
The receptors for the second messenger InsP3 comprise a family of closely related ion channels that release Ca2+ from intracellular stores, most prominently the endoplasmic reticulum and its extension into the nuclear envelope. The precise sub-cellular localization of InsP3Rs and the spatial relationships among them are important for the initiation, spatial and temporal properties and propagation of local and global Ca2+ signals, but the spatial organization of InsP3Rs in Ca2+ stores is poorly characterized. Using nuclei isolated from insect Sf9 cells and freeze-dry rotary shadowing, we have addressed this by directly visualizing the cytoplasmic domain of InsP3R located on the cytoplasmic side of the nuclear envelope. Identification of ~15 nm structures as the cytoplasmic domain of InsP3R was indirectly supported by a marked increase in their frequency after transient transfections with cDNAs for rat types 1 and 3 InsP3R, and directly confirmed by gold labeling either with heparin or a specific anti-InsP3R antibody. Over-expression of InsP3R did not result in the formation of arrays or clusters with channels touching each other. Gold-labeling suggests that the channel amino terminus resides near the center of the cytoplasmic tetrameric quaternary structure. The combination of nuclear isolation with freeze-drying and rotary shadow techniques allows direct visualization of InsP3Rs in native nuclear envelopes and can be used to determine their spatial distribution and density.
Keywords: InsP3R, isolated nucleus, nuclear envelope, freeze-drying, calcium, channel
1. Introduction
Inositol 1,4,5 trisphosphate receptors (InsP3Rs) are ligand-gated channels through which calcium (Ca2+) is released from intracellular stores in many eukaryotic cells [1]. The channels mediate a wide variety of biological processes, such as proliferation, programmed cell death and gene expression [2]. They are assembled from four large subunits, each of ~2700 residues with a single InsP3 binding site. In mammals three different genes encode very similar subunits (types 1–3) that assemble as large homo- and hetero-tetrameric structures [3, 4]. The three types of InsP3Rs share approximately two thirds of their amino acid residues; however they have different affinities for InsP3 [5] and are differentially regulated by Ca2+ and ATP [6, 7]. The structure and domain organization of the full-length type 1 InsP3R have been determined by single particle analysis of purified receptors, mainly derived from brain cerebellum microsomes. The channel exhibits a four-fold symmetry and, in the presumably closed state in the absence of Ca2+, its large hydrophilic domain presents a square profile when viewed from the cytoplasm [3, 4, 8].
The precise sub-cellular localization and distribution of InsP3Rs are believed to be critical for the spatial and temporal properties of localized Ca2+ release and for the initiation and propagation of Ca2+ signals [3, 9, 10]. Accordingly, the physiological consequences of Ca2+ released are strongly influenced by the localization of InsP3Rs on Ca2+ stores, including the Golgi complex, sarco/endoplasmic reticulum (ER) and nuclear envelope [8, 10]. Optical imaging has revealed the presence of discrete Ca2+ release sites within cells [11], but the relationship between such sites and the localization of InsP3Rs is not well understood. For example, whereas spatially-segregated clusters of 10–100 InsP3Rs are believed to be responsible for the generation of so-called Ca2+ puffs in different cell types [9, 12], immunolocalization studies or imaging of GFP-tagged InsP3Rs suggests a more uniform distribution of the channels throughout the entire ER in many cell types [10, 13–17]. Furthermore, the number of channels and their spatial proximity determines the properties of localized Ca2+ release events such as puffs, but these features have been inferred and not directly examined.
Direct localization of InsP3Rs has been described in neuronal Purkinje cells of the cerebellum, where InsP3Rs, mainly type 1, are highly concentrated [18, 19]. In these cells, the ER forms stacks of cisternae decorated by semi-crystalline arrangement of InsP3Rs [19–23]. For other cells and cellular locations, however, the arrangement and density of InsP3Rs in membrane sites have been difficult to visualize and remain poorly characterized. Freeze-drying and rotary shadowing have been successfully used to determine the structure, position and arrangement of RyR Ca2+ release channels in isolated triads of skeletal muscle [22–24]. Here, we have used this technique, together with gold particle conjugation either to heparin or to specific InsP3R antibodies, to reveal the spatial disposition and density of InsP3Rs within the outer membrane of the nuclear envelope under native and over-expression conditions. The nuclear envelope is an InsP3-dependent Ca2+ store [25], whose outer membrane is continuous with the ER, and contains functional InsP3R channels [26–31]. The cytoplasmic domain of the nuclear envelope can be visualized in its entirety by freeze drying and rotary shadowing of isolated nuclei, facilitating observation of the spatial relationships among InsP3R channels.
2. Materials and methods
2.1. Cell culture and transfections
Spodoptera frugiperda (Sf9) cells (Invitrogen) were incubated at 27°C and maintained in serum-free SF900II medium (GIFCO). A baculovirus expression system was used to transiently express either rat types 1 or type 3 InsP3R. The chicken B lymphocyte-derived DT40 cell line with all three InsP3R isoforms genetically deleted (DT40-KO) [32] was maintained in suspension culture at 37°C (95/5% air/CO2) in RPMI 1640 medium (GIBCO/BLR) supplemented with 10% (v/v) FBS, 1% chicken serum, 2 mM glutamine, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin.
2.2. Western blotting
Cellular extracts were lysed in 100 µl ice-cold Cytobuster protein extraction reagent (Novagen) supplemented with a broad-spectrum protease inhibitor cocktail (Roche). Protein concentrations of the supernatants were determined with BSA as standard. Protein extracts were suspended in Laemmli buffer, separated in 5% SDS-polyacrylamide gels and transferred to PDVF membranes (Millipore). Blocking was at room temperature for 1 hr in 5% fat-free milk, and the membranes were incubated overnight with antibodies against either types 1 (recognizes the carboxyl-terminus; provided by Dr. S. Joseph, Thomas Jefferson Univ.) or 3 (recognizes the amino terminus; BD Transduction Laboratories) InsP3R. Immunoreactive proteins were detected using ECL reagents (Pierce Biotechnology) according to the manufacturer's instructions.
2.3. Sf9 and DT40 cell nuclear isolation
Nuclei were extracted from cells by a combination of hypotonic shock and mechanical disruption in a Dounce homogenizer, as previously described [33]. Briefly, both Sf9 and DT40 cells were pelleted, washed three times in phosphate-buffered saline and then re-suspended in hypotonic buffer (10 mM Tris-HCl, pH 7.8, 10 mM β-mercaptoethanol, 0.2 mM PMSF and protease inhibitors). After 5 min on ice, the swollen cells were broken in a Dounce homogenizer. The nuclei were separated by slow centrifugation (400 g for 7 min at 4°C), re-suspended in wash solution (10 mM Tris-HCl, pH 7.2, 110 mM KCl, 2.2 mM MgCl2 and protease inhibitors) and re-centrifuged (800 g for 5 min at 4°C). Finally, the isolated nuclei were re-suspended in 10 mM Hepes-Tris, pH 7.6, 110 mM KCl, 1 mM MgCl2 plus protease inhibitors.
2.4. Isolation of individual Xenopus oocyte nuclei
Ovary extraction from Xenopus laevis and oocyte nuclei isolation were performed as was described [34]. Briefly, nuclei were manually teased out of freshly isolated oocytes with fine forceps. The nuclei were cleaned of cytoplasmic material by gently sucking them up and down in a pipette in basic oocyte nucleus solution (BONS, containing 140 mM KCl, 10 mm HEPES, 3 mM MgCl2, 1 mM BAPTA, 0.543 mM CaCl2, pH 7.3). The isolated nuclei were then transferred to a glass coverslip previously treated with 0.1% poly-L-lysine for freeze-drying and replication.
2.5. Immunostaining, and confocal microscopy
Isolated nuclei were fixed in methanol at −20°C for 12 min, blocked in 1% BSA and incubated with the primary antibodies against types 1 or 3 InsP3R overnight at 4°C, with gentle rotation. The nuclei were washed with PBS/1%BSA, incubated with Alexa-488 secondary antibody (Molecular Probes) for 1 hr at room temperature, also rotating, mounted in Vectashield mounting medium (Vector Laboratories) and examined in a Zeiss Axiovert 510 LSM Pascal confocal microscope, using a high numerical aperture water immersion 63× objective.
2.6. Freeze-drying and replication
A suspension of freshly isolated nuclei was placed on fragments of glass coverslips previously treated with 0.1% poly-L-lysine, and allowed to attach for 5 min. The coverslips were then rinsed with 100 mM ammonium acetate, treated with 2% (w/v) uranyl acetate for 30–60 sec, and rinsed extensively with 40% (v/v) methanol. The solution was dried to a very thin film using the sandwich technique, and frozen in liquid nitrogen [35]. After mounting on the cold stage of a Balzer’s freeze-fracture apparatus, the nuclei were freeze-dried at 10−6 mbar pressure at −90°C for at least 30 min, and then re-cooled to −110°C, rotary shadowed with platinum at a 25° angle and replicated with carbon. Finally, the glass coverslips were dissolved with hydrofluoric acid and the replicas cleaned with bleach (6%) for 10 min, washed with water and mounted on an EM grid. Replicas were viewed and photographed in an electron microscope (Philips EM 410; Philips Technology, Cheshire, CT).
2.7. Heparin-gold and immuno-gold labeling
Shadowed replicas of gold-labeled nuclei were obtained as described [36], with modifications, using nuclei isolated from Sf9 cells transfected with rat type 3 InsP3R .
2.7.1. Heparin gold
Nuclei were incubated with 100 µg/ml heparin-biotin (Sigma) for 3 hr at 4°C while rotating, washed several times and then incubated with Alexa Fluor 488-streptavidin conjugated to 5 nm gold particles (Molecular Probes) for 1 hr. After the labeling, the isolated nuclei were fixed in 0.05% glutaraldehyde for 20 min and freeze-dried and replicated as above, except bleach was diluted to 0.5% and applied for 1 min, followed by three washes in water for 1 hr each. To avoid nonspecific interactions, all samples were treated for 1 hr with 0.05% avidin and 0.005% biotin before incubation with heparin-biotin. Streptavidin-gold incubation in the absence of heparin-biotin, as well as heparin-biotin incubation with DT40-KO cell nuclei, were used as negative controls.
2.7.2. Immunogold labeling
Nuclei were fixed with 4% formaldehyde and 1% glutaraldehyde for 20 min, blocked in 1% BSA for 1 hr, incubated overnight with anti-InsP3R-3 antibody, washed with PBS/1%BSA and incubated overnight with a gold-conjugated secondary antibody (Molecular Probes). After labeling, the nuclei were freeze-dried and replicated as described above. Incubation with the secondary antibody in the absence of primary antibody, and immunolabeling of DT40-KO cells were used as negative controls.
2.8 Image processing and analysis
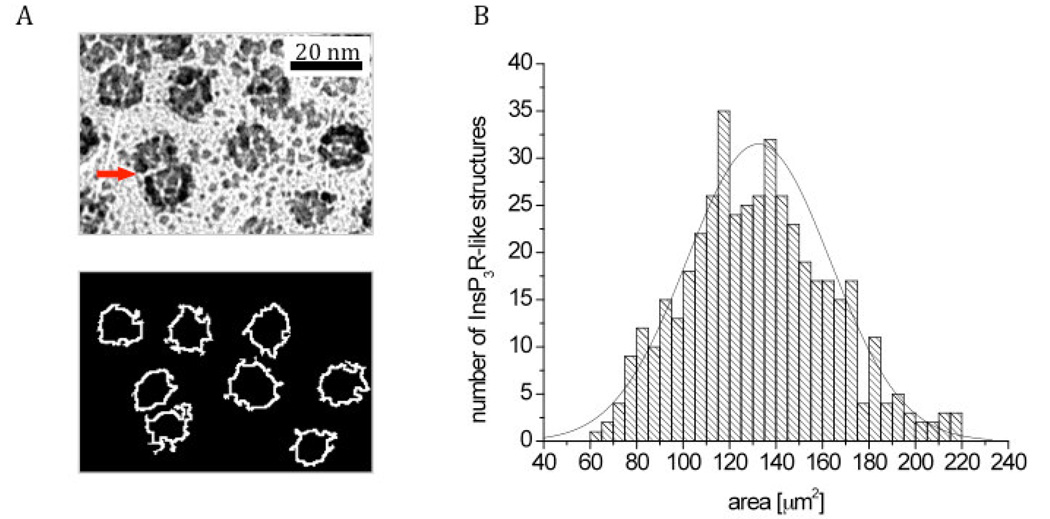
All image-processing routines describe below were developed in one of the author’s laboratories (www.scian.cl) on the basis of IDL 7.0 (Interactive Data Language, ITT, CO, USA), including interactive tools for the segmentation of InsP3R-like structures in the freeze drying images into regions of interest (ROIs), calculation of domain area, and visualization (see Figure 6A). A region of interest in this case is a delimited area that differs in gray scale from the background. Segmentation of the images was achieved by applying gradient filters and selecting threshold values of light intensity in the gradient histograms, resulting in a homogeneous definition of the borders of the InsP3R-like structures. Remaining holes inside the regions and small segmented signals outside the structures were corrected either by morphological filters or manually. The quality of the segmentation in terms of recognizing the particles that we define as InsP3R-like structures was controlled interactively by overlaying the original images with the segmented regions (see Figure 6A top and bottom). Although the gray scale threshold values varied between individual images, other segmentation criteria were kept constant for a total of N = 445 InsP3R-like structures from three different experiments (see Figure 6B). To determine if the surface area of the segmented regions corresponding to the InsP3R-like structure were from a single population, a Lilliefors test for normality was performed with MATLAB. The Lilliefors test estimates the population mean and variance of the data, computes the empirical cumulative distribution function (ECDF), and the cumulative distribution function (CDF) of the normal distribution using the estimated mean and variance. Finally, it calculates the maximum discrepancy between the ECDF and the CDF, and determines if the discrepancy is statistically significant leading to a rejection of the null hypothesis that the data is normally distributed.
Figure 6. Segmentation and area distribution of InsP3R-like structures as revealed by image analysis techniques.
(A) Electron micrographs of shadowed InsP3R-like structures before (top) and after (bottom) segmentation into Regions of Interest (ROIs) outlined by white borders. The areas of 445 ROIs were determined and analyzed for normal distribution by the Lilliefors test. Note that the two particles indicated by a red arrow have a smaller footprint because they partially overlap. Such images account for the smaller size footprints at the left of the distribution. (B) Frequency plot of the area distribution (columns) in combination with a normal distribution (line).
3. Results
3.1. Nuclear pore complexes in rotary shadowed images of isolated nuclei allow identification of nuclear envelope side
An initial survey of thin sections (not shown) indicated that isolated Sf9 cell nuclei had a surface that was devoid of cytoskeletal and/or cytoplasmic remains, thus enabling a clear view of the cytoplasmic surface of the nucleus in shadowed images.
The structures of the nuclear pore complex (NPC) on the nucleoplasm and cytoplasm sides of the nuclear envelope are quite different [37]. We used this well-established difference to identify nuclear surfaces in isolated nuclei. Rotary shadowed replicas of intact nuclei isolated from Sf9 cells showed NPCs characterized by a ring surrounding a central depression (Fig. 1A) indicating that the cytoplasmic side was shadowed. By comparison, fragments of manually isolated nuclei from Xenopus leavis oocytes, showed both cytoplasmic (Fig. 1B) and nucleoplasmic (Fig. 1C) sides of the envelope. The two NPC structures were easily differentiated: one side showed the ring structure, while the other showed a highly organized fibrous network forming the NPC basket [37]. Under the experimental conditions used in this work we only observed NPCs with the structural signature of the cytoplasmic side of the envelope, indicating that the nuclei were intact.
Figure 1. Identification of nuclear envelope surfaces in isolated nuclei by freeze-drying and rotary shadowing.
A. Cytoplasmic surface of a nucleus isolated from wild-type Sf9 cells. The nuclear pore complexes (NPC), arrows) are identifiable as annuli with a diameter of 100–110 nm, decorated by a ring of prominent particles. The structure is typical of the cytoplasmic side of the envelope [36] and intact nuclei always showed only this side The black square encloses a particle identified as a presumptive InsP3R based on evidence shown later. Scale bar = 100 nm. B and C. Details of freeze-dried rotary-shadowed NPCs in the cytoplasmic (B) and interior face (C) of nuclear envelope fragments from Xenopus oocytes Note that in B the NPC is dominated by a particulate ring, while in C the dominant structure is a fibrillar basket.
3.2. Small structures in the shadowed nuclear envelope might represent InsP3Rs
The membrane of the outer nuclear envelope in nuclei isolated from Sf9 cell is relatively featureless between individual NPCs, except for occasional small protrusions. Most of these were not identifiable, but a few were consistent with the appearance of the cytoplasmic domains of InsP3R, as shown in situ [20] and in isolated molecules [38–40]. Although somewhat variable in appearance, the small presumptive profiles of shadowed InsP3R fit into a square with an approximate size of 15×15 nm, some time had a central depression, and were clearly raised above the envelope surface (Fig. 1, black rectangle).
In order to establish the nature of these presumptive InsP3R profiles, we imaged the protein after over-expression in Sf9 cells and further labeling with specific ligands.
3.3. Small presumptive InsP3R- like structures are very frequent in nuclei of Sf9 cells after over-expression of rat types 1 and 3 channels
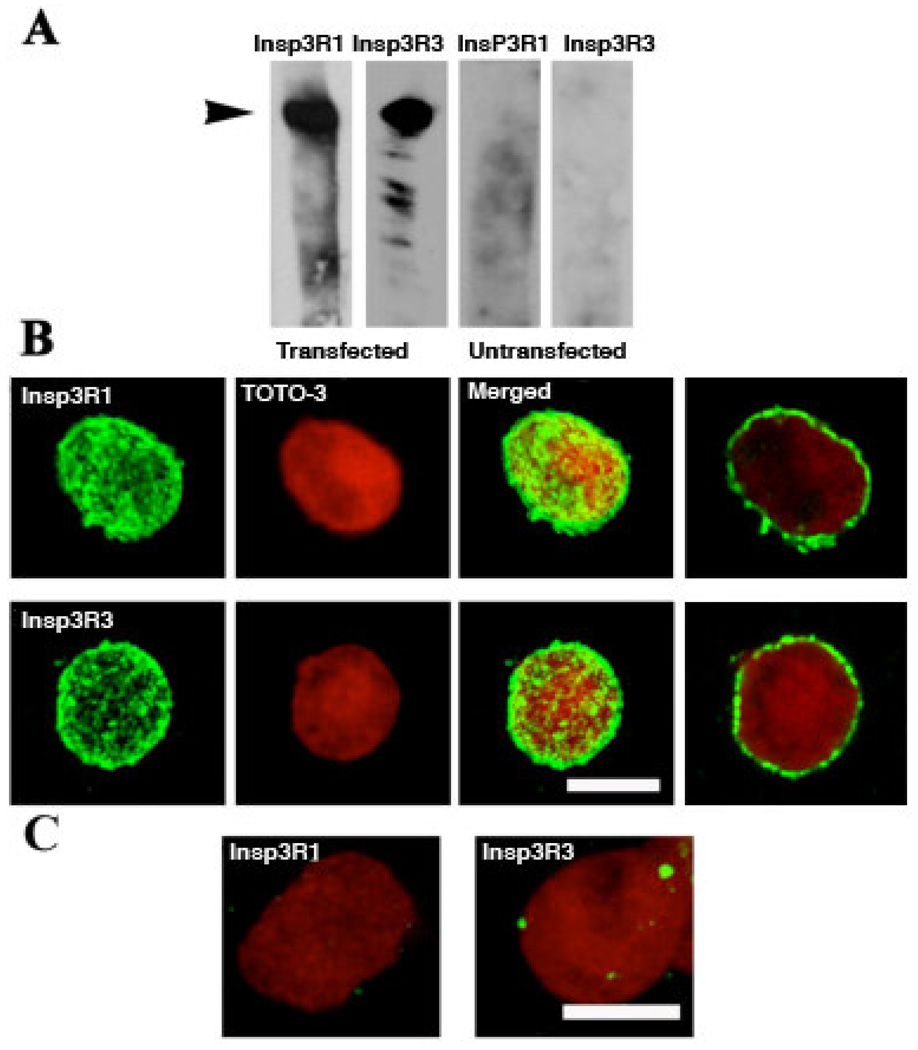
To provide a more definitive identification of the InsP3Rs, we over-expressed either the rat type 1 or type 3 InsP3Rs at very high levels in Sf9 cells using recombinant baculoviral infection, as evidenced by Western blot analysis of infected cell lysates (Fig. 2A). Because a pure population of nuclei could not be isolated for biochemical analysis, we verified that over-expressed recombinant InsP3R was present in the nuclear envelope by confocal microscopy of isolated nuclei immunolabeled with specific anti-rat types 1 and 3 InsP3R antibodies. In stacked serial optical sections of 0.5 µm thickness, both InsP3R isoforms were highly expressed in the nuclear envelope and had similar discontinuous distributions characterized by intense hot spots (Fig. 2B). Single optical sections across the middle of the nuclei (Fig 2B right) show that the spots are located at the nuclear periphery. The nuclei of native cells were totally negative with these antibodies, since the antibodies used to detect the rat channel isoforms do not recognize the endogenous Sf9 InsP3R (Fig. 2C). For that reason, the relative expression levels of the recombinant and endogenous channels in the transfected cells could not be directly quantified. However, baculovirus-mediated expression of recombinant InsP3R enhances the probability of recording InsP3R channels in electrophysiological experiments by over two orders of magnitude [43, 44] suggesting that the expression levels of the recombinant channels greatly exceed that of the endogenous InsP3R.
Figure 2. Heterologous expression of rat types 1 and 3 InsP3R in two separate sets of Sf9 cells.
A. Western blots of whole cell homogenates probed with specific anti-type 1 or -type 3 antibodies, revealing intense bands at ~240 kDa corresponding to the InsP3R specifically expressed in the infected cells. B. Images of isolated nuclei from either type 1 (IP3R1) or type 3 (IP3R3) InsP3R expressing cells, labeled with their respective antibodies. The first three figures in each row were produced by summing stacked confocal images of single optical sections ~ 0.5 µm thick. Both InsP3Rs (green) present a patchy distribution over the nucleus that was counterstained with TOTO-3 (red). Individual optical sections (at right) show that antibody labeling is at the periphery of the nucleus. C. Merged images, as in (B), but showing nuclei from untransfected cells. Scale bar, 10 µM.
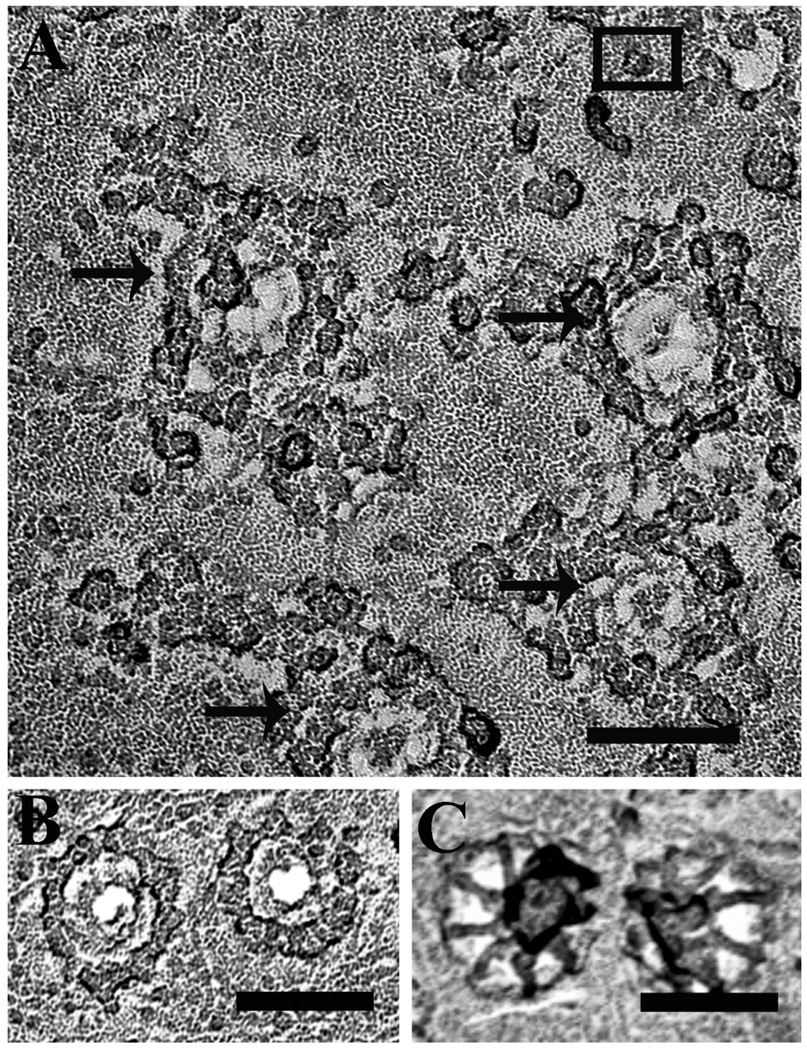
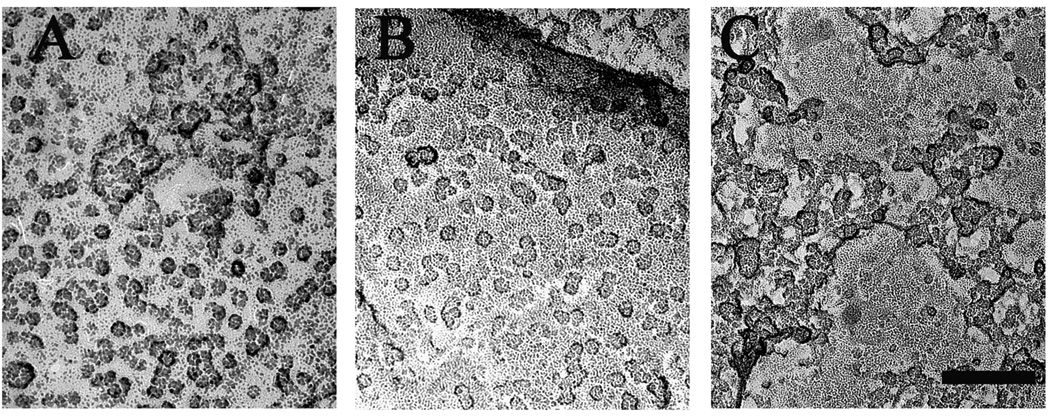
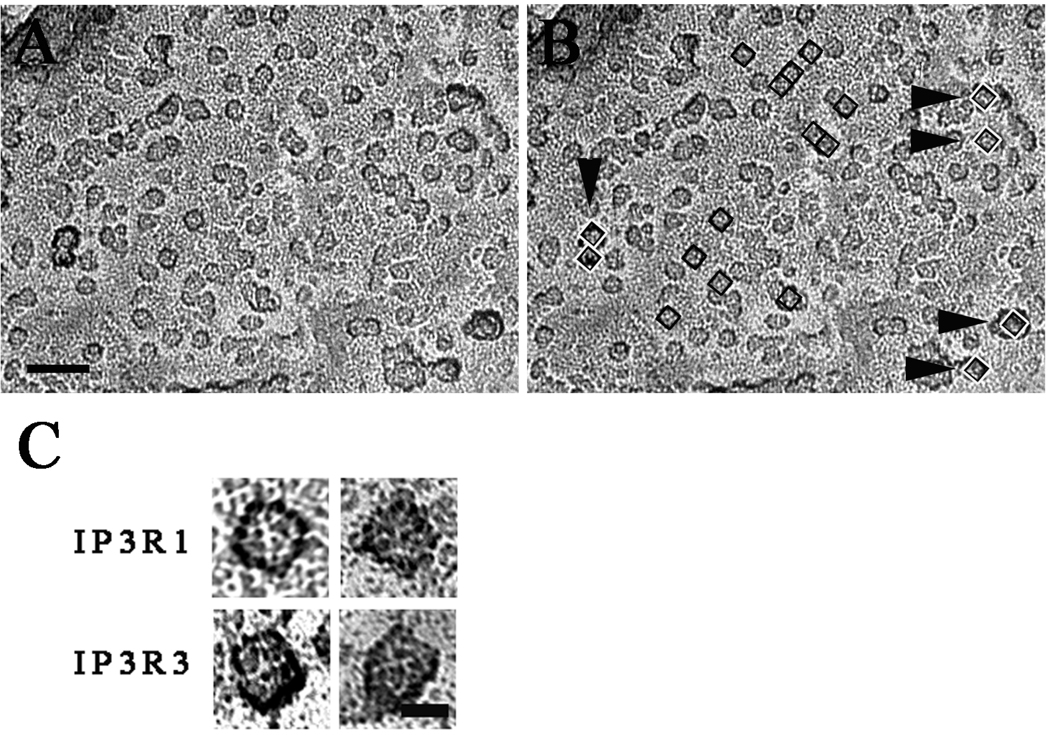
Images from freeze-dried and rotary-shadowed nuclei from the baculovirus-transduced cells revealed small structures in the nuclear envelope similar in appearance and approximate size to those observed in the non-transfected cells but present at a much a higher density in patches of nuclear envelope devoid of nuclear pores (Fig. 3A and B). Despite the high density of the protein structures, we did not detect a semi-crystalline arrangement such as shown for InsP3R in cerebellar Purkinje cells [20] and for RyRs in skeletal muscle [45].
Figure 3. Detection of InsP3R in rotary shadowed images of isolated nuclei.
Nuclei isolated from Sf9 cells expressing either rat type 1 IP3R (A), or type 3 IP3R (B), or Wolframin, a non-related ER membrane protein, (C) were freeze-dried and rotary shadowed. Nuclear membranes from type 1 and type 3 InsP3R-expressing cells showed large membrane patches occupied by a high density of small structures ~15 nm on the side similar to the rarer particles observed in the uninfected cell nuclear membranes. Nuclear pores were absent from these patches but visible elsewhere in the same nuclei. The small particles were not visible in cells transfected with cDNA for Wolframin. Scale bar, 100 nm.
Although the above data revealed a direct correlation between over-expression of InsP3R and the density in the nuclear envelope of InsP3R-like structures, nonspecific effects of the transfection, for example infection-induced over-expression of an unknown viral protein, had to be ruled out. To address this question, we infected Sf9 cells with either Wolframin, an unrelated ER membrane protein that assembles into high molecular weight complexes [46] or with the empty baculovirus vector. In both cases there was no increase in the appearance of the InsP3R-like structures (Fig. 3C for Wolframin; data not shown for the viral vector).
The high frequency of presumed InsP3R particles in the overexpressing nuclei strongly confirms their tentative structural identification.
3.4 Identifiable features of presumptive InsP3R particles in overexpressing cells
The majority of profiles in the large patches of InsP3R-overexpressing nuclear envelopes had several common structural features. Like the less frequent profiles on native nuclei, the profiles fit into a square of fairly uniform sixe (15×15 ± 2 nm, mean ± SD, from 300 particles) and some had a central depression (Fig. 4 A and B, black squares). The overall shape was approximately square, but the square had variably blunted corners. Most of the shape variations are to be expected by the superimposition of 3–4 nm size particles from the platinum shadow on the small molecule outline. The height of the particles was fairly uniform, as indicated by the intensity of the platinum shadow surrounding the particles in a given image. The intensity of the platinum shadow surrounding the particles is due to the fact that, since samples were rotary shadowed, each particle is surrounded by a ring of deposited platinum, and the taller the particle the larger the amount of platinum in the ring. In each cluster of particles, structures other than presumptive InsP3R particles were present at low frequency and were clearly distinguishable due to their larger and more variable size and height (Fig. 4B, white and black squares and arrowheads). High magnification views of the most clearly defined presumptive InsP3R particles revealed four equal components symmetrically disposed around a central depression, suggesting a tetrameric structure that measured ~15 nm on each side (Fig. 4C). These characteristics are consistent with previous descriptions of purified InsP3R [41, 47], reinforcing the idea that these structures truly correspond to the InsP3R.
Figure 4. Counting InsP3R-like structures.
A and B show electron micrographs of the same area of a nuclear envelope from a cell over-expressing type 3 InsP3R. In B, small squares with inside dimensions of 15×15 nm are superimposed on some of the presumed InsP3R profiles. The profiles thus selected, even though not exactly “square”, fit well within the square outlines and some have a central depression. Additionally they all have similar height as indicated by the equal intensity of the platinum shadow at their edges. Numerous other particles with similar characteristics are seen in the same image. Arrowheads point to profiles that are clearly different: they are mostly larger, with a more variable shape and a darker shadow outline, indicating taller structures. The 15×15 nm black and white squares superimposed on these structures cover them only partially. Scale bar= 50 nm. C) High magnification views of InsP3R-like structures from nuclei of cells over-expressing type 1 and 3 InsP3Rs. The four particles shown here have equal sizes and an approximate square outline, but this is emphasized by the shadow only in the two particles at left. The hint of four subunits and a small central depression are typical details of the cytoplasmic domain in InsP3Rs images obtained by single particle analysis [38,39,40]. Scale bar, 10 nm.
The observed variability of the shape of the presumed InsP3R on native nuclear envelopes is not an unexpected phenomenon, since exposed proteins tend to collapse to some degree once the supporting water is removed during the freeze-drying process. In addition the size of the platinum shadow grains is less than an order of magnitude smaller than the size of the molecule, thus affecting its appearance
It is important to note that both in the native nuclei and in those from overexpressing cells, the receptors are never associated with each other in a regular semi-crystalline disposition, even when they are in fairly close proximity. Occasional groups of two and short rows of 3–4 receptors are present in the overexpressing nuclei, but they represent less than 10% of the particles and do not show any precise arrangement.
3.5 Specific gold labeling identifies InsP3R in shadowed images
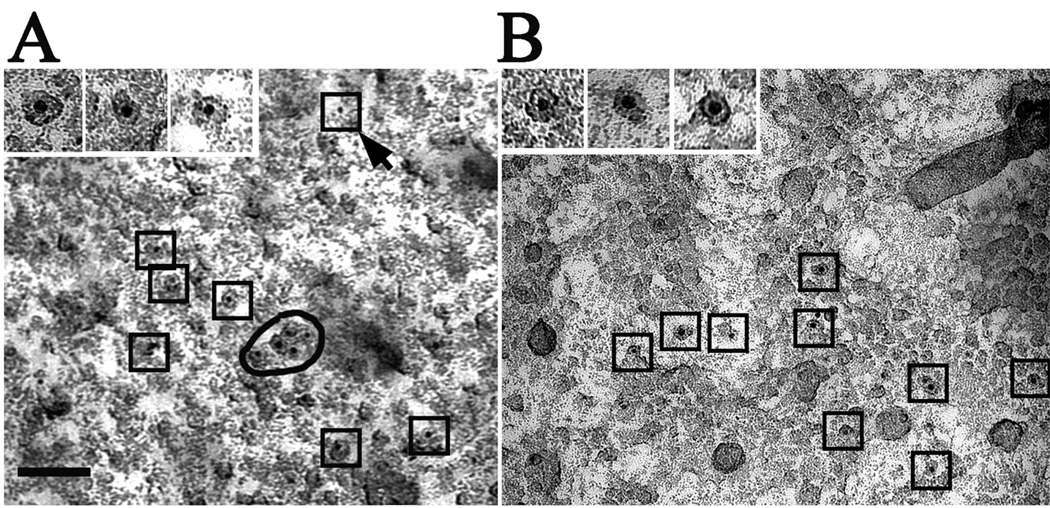
To further confirm the identity of the observed structures as InsP3Rs, we labeled the receptors with either heparin, a competitive InsP3 antagonist of the InsP3R, or a specific antibody, both followed by gold-conjugation and freeze-drying rotary shadowing. Both techniques require that the label be retained in the replica after limited cleaning in dilute bleach (0.5%) solution followed by repeated washings in water, and they exploit the fact that gold particles provide a strong contrast allowing their visualization after shadowing, even though the images are of lower quality due to remaining contamination from the poorly digested biological material [36, 48]. As recombinant type 1 and type 3 InsP3R did not appear to behave differently in terms of expression, or distribution, we performed the labelling experiments using nuclei isolated from cells over-expressing the recombinant type 3 isoform, for which the amino terminal-directed antibody has a high affinity.
With either label, the large majority of the gold particles were associated with small profiles of the appropriate size (Fig. 5A and B) and only a very small fraction adhered to apparently non-specific sites (e.g., see the outlined dot indicated by an arrow in Fig. 5A). Counts from 70 pictures in 6 different preparations (three labeled with heparin and three with antibody) showed 398 gold particles (92%) associated with InsP3R-like structures and 30 (8%) positioned over either unspecified structures or the background. This indicates a high specificity in the labeling. Omission of the heparin incubation step resulted in a complete absence of labeling by the streptavidin-gold conjugate (not shown). Importantly, neither incubation with heparin nor the antibody, followed by gold-conjugation, showed any labeling of nuclei isolated from the InsP3R-deficient DT40-KO cells (n = 3 preparations, data not shown).
Figure 5. Shadowed images of nuclear envelopes from Sf9 cells expressing rat type 3 InsP3R.
The nuclei were treated either with gold-labeled heparin (A) or with anti-type 3 InsP3R followed by gold-labeled secondary antibody (B) followed by freeze-drying and rotary shadowing (see Methods). In both cases, the 5 nm gold particles are clearly associated with the InsP3R-like structures, with the exception of 1 out of 11 gold particles in (A) (arrow). High magnification (insets) shows the gold particles mainly located in the central region of the structures. Note that the InsP3R-like structures labeled by the gold particles have similar sizes but variable shapes and overall resemble the presumptive InsP3R particles described above. The images are not as sharp as those in the previous figures because the technique used leaves some cellular debris associated with the shadowed replicas.
Both gold labels unequivocally identify InsP3R as particles that have the structural characteristics describe above,
3.6 Image analysis identifies the presumptive InsP3Rs as belonging to a single population
The question remains whether particles selected as InsP3R-like profiles by visual criteria as above represent a single population, despite their minor structural variability. This question can be explored by determining whether their “footprints” occupy an area of the image that is fairly well-defined. To do so, we performed image analysis in combination with Lilliefors test to determine whether the areas of the presumed InsP3Rs present a Gaussian distribution as would be expected for a single population of particles. Figure 6A (top) shows a sample shadowed image and Figure 6A (bottom) the outlines of the Regions of Interest (ROIs) that were segmented from the structures identified by us as presumptive InsP3R in the same image (see Materials and Methods). Figure 6B plots the area histogram for 445 segmented InsP3R–like structures with a mean size and standard deviation of 132 ± 31 nm2 from three images. Lilliefors test for each sample determined that at a 5% significance, footprints had a normal distribution.
The analysis indicated that despite some differences in shape, all structures tentatively identified as InsP3R are part of a single population in respect to their surface area.
It is noticeable that the ROI areas presented in the histogram of Fig. 6B are all smaller than the area subtended by a 15×15 nm square, This is for two main reasons, One is the fact that the particle outline does not entirely fill the square outline, the second is that the segmentation process follows the outline of the dense platinum grains surrounding the protein and these leave small short crevices in the outline that subtract from the overall area. In addition, the areas for 10–15% of footprints at the left of the histogram are quite small. These footprints represent profiles of particles that are not fully included in the image because they are very closely associated with other particles (see arrow in Fig 6 A).
3.6 Can shadowed images be used to estimate InsP3R density on the nuclear envelope?
The over-expression and gold labeling experiments clearly show that InsP3R can be identified on the shadowed surface of isolated nuclei. However, the appearance of the shadowed receptors has a small inherent variability, and in addition, other unidentified structures occupy the same nuclear membrane patches. We were interested in defining how accurately we could estimate receptor densities from shadowed images. To that effect we used images from over-expressing nuclei and in them identified presumptive InsP3R profiles using their fit into a 15×15 nm square in addition to an intensity of shadow indicating a fairly uniform height (Fig. 4 A and B). Most profiles in overexpressing nuclei clearly fit within the square outline, whereas a smaller number were questionable (question marks) and even fewer were simply undefined structures that did not fall into the InsP3R-like category (Fig. 4B, arrowheads). In the example shown in Figure 4B, for example, 40 profiles are quite clearly presumptive InsP3Rs, 17 profiles are equivocal and 10 are undefined structures. The disagreement in this assignation between two independent observers was less than 10%, providing an estimate of the possible error involved in such identifications. Counts of InsP3R-like structures in randomly collected images showing extended patches of particles in cells overexpressing InsP3R revealed an average density of 90.5 ± 7.4 µm−2 of nuclear envelope (exclusive of nuclear pores) for type 1 and 92.1 ± 6.2 µm−2 for type 3 (mean ± SD from 3 areas each in 6 different preparations). By contrast, the average density of similar structures was much lower in nuclei from native Sf9 cells: 3.2 ± 1.2 µm−2 (mean ± SD, from 3 areas each in 6 different preparations), that is ~30-fold lower than in the overexpressing cells. Despite the uncertainty level defined above, the structural counts are consistent with functional studies from Sf9 cell nuclear patch clamp recordings, where ~2 channels per ~ µm2 nuclear patch were detected on average in the same cells and a much higher level in overexpressing cells. [42, 43, 44].
4. Discussion
Using a direct shadowing technique and relying on the known structural signature of the InsP3R cytoplasmic domain, we have been able to directly visualize the channels in the outer membrane of the nuclear envelope of Sf9 cells. The InsP3R channels in this membrane are functional as shown by nuclear patch clamp electrophysiology [42]. The identity of the structures as InsP3R channels is strongly suggested by four observations. First, by the structural similarity of presumptive InsP3Rs in the images to native InsP3Rs in cerebellar Purkinje cells visualized by shadowing techniques [20] and to heterologously-expressed channels in Sf9 cells imaged by atomic force microscopy [49]; second, by their increased density upon InsP3R over-expression; thirdly, by population analysis confirming that the surface area of the InsP3R-like segmented structures is distributed normally; and finally, but most importantly by their specific labeling with both heparin and a specific anti-InsP3R antibody.
Gold-labeling before shadowing was first introduced by Pinto da Silva in 1984 [48] for freeze fracture samples to successfully study cytoskeletal networks [50, 51] and nuclear pores [36]. Labeling after fracture and shadowing has been effectively used for intrinsic membrane proteins [52, 53]. In both techniques it is essential that the digestion steps used to clean the tissue away from the shadowed replica allow retention of the label. We were successful using a brief exposure to a dilute bleach solution and obtained a high degree of specificity. The limited extent of labeling, which is clear in all above studies, is mostly related to the affinity of the specific primary and secondary probes for their ligands, rather than to a loss of the label during preparations.
The size of the exposed cytoplasmic domains of InsP3R has been reported to vary from a ~12×12 nm square in deep etched samples [20], to ~15×15 nm in our freeze dried preparations, to ~ 22×22 nm in single particle analysis [41], to ~30 nm in the atomic force images [49]. Obviously, the cytoplasmic domain of the InsP3R is highly deformable, which is perhaps not surprising given its highly hydrated structure. The larger size of the structures observed with AFM seems to be intrinsic to the technique, because AFM images of type 1 RyR show a size of 40–50 nm [54], almost double the size detected by rotary shadowing [55]. In the most recent reconstruction of the molecule [41], the space-filling outline of the cytoplasmic domain shows a compact structure with a square outline and barely distinguishable subunits, which would appear quite similar to our images if superimposed with randomly arranged 3–4 nm dots of platinum shadow.
Our studies confirm that in native nuclei InsP3R are located in the outer nuclear envelope, with the cytoplasmic domain projecting into the cytoplasm, supporting the concept of nuclei as InsP3-dependent Ca2+ stores [25]. However, the density in native nuclei of Sf9 cells is very low, and preliminary, unpublished data also show a very low density in nuclei from cultured DT40 and from native mouse cardiac and liver cells. The low density agrees with functional data from patch clamping studies [42]. Thus, this approach offers the first direct means of detecting receptor distribution and frequency in the nucleus, and may be useful in assessing InsP3R nuclear localization, including ligand-independent and -dependent clustering [14, 16, 17, 56–58]. Unfortunately, we could not visualize the inner nuclear envelope by this technique, and thus we cannot state whether InsP3Rs are located facing the nucleoplasm.
Two important observations were made in our studies. First, we note that recombinant InsP3R channels highly over-expressed in the nuclear envelope of Sf9 cells do not appear to organize into ordered semi-crystalline arrays, even when they are present at high levels and within small regions. By contrast, high density of InsP3Rs in the ER membrane natively expressed in cerebellar Purkinje cells [20], or over-expressed in COS cells [19], is associated with formation of flat cisternae that are decorated by semi-crystalline array of the channels. To understand this discrepancy, it is informative to compare observed InsP3R arrays with those formed by RyRs, Ca2+ release channels of ER/SR that share sequence similarity, large cytoplasmic domains and a tetrameric structure with InsP3Rs. RyRs form extensive arrays when they are either natively expressed in muscle cells [59], or expressed in a heterologous system [60], or assembled with lipids under in vitro conditions [61]. Within RyR arrays, the cytoplasmic domains of adjacent molecules are in intimate, identical contact at their four corners, so that it is logical to assume that direct intermolecular interactions are responsible for array formation, particularly when the purified protein is considered [62]. By contrast, InsP3Rs present in arrays in Purkinje cells are located within “touching” distance in one direction, but at a larger, even though perfectly regular, distance in another direction [20]. Thus, it seems likely that some component of cell, rather than a direct intermolecular contact between the channels, may be at least in part responsible for InsP3R array formation observed in some cells. This presumed organizing element must be absent from the nuclear envelope, so that InsP3Rs do not form arrays when over-expressed in the nuclear envelope of Sf9 cells (this work) and of Xenopus oocytes (unpublished results) even when the receptors are sufficiently crowded within the membrane to allow intermolecular interactions.
A second observation concerns the localization of the InsP3 binding sites within the channels’ tetrameric quaternary structure. It has been speculated that they reside either about half-way along the sides [40, 63] or near the center [38] of the large cytoplasmic region of the receptor. Heparin is a competitive inhibitor of InsP3R [64] that binds to the amino-terminal InsP3-binding region [65]. Colloidal gold-conjugated heparin has been previously used to detect InsP3R by electron microscopy of negatively stained preparations [47]. Peripheral binding of heparin-gold to immunopurified type 1 InsP3R suggested that the InsP3-binding domain is located far on the corners of the molecule [47]. However, in our studies with the channel in native ER membrane, heparin as well as the amino terminus antibody both marked sites very close to the center of the channel’s cytoplasmic domains, supporting a more central location. The channels in our study were not InsP3-liganded and were likely in a “closed” configuration. It is possible that InsP3 binding sites exist in different locations in different channel conformations, for example those associated with channel gating or binding to various ligands such as Ca2+. Only one gold particle was associated with each InsP3R, whereas each channel contains four subunits and thus four amino terminals. This is probably accounted for by steric hindrance by the large size of the gold particles that prohibited multiple-site binding.
5. Conclusions
In summary, we have described a system and procedures for visualizing the organization and spatial relationships of individual InsP3R channels in native nuclear envelope membranes. In the future, this technique may be valuable for quantification of channel distribution parameters that are critical parameters in models of InsP3R-mediated cellular Ca2+ signals.
Acknowledgments
We thank Dr. Suresh Joseph for the type 1 InsP3R antibody. Cesar Cardenas would like to thank the “Fundación Ciencias Para la Vida” (Chile). This work was support by a MDA grant to CFA and NIH GM065830 to JKF. SCIAN-Lab is a member of the German-Chilean Center of Excellence Initiative for Medical Informatics (DAAD) and the Advanced Imaging & Bioinformatics Initiative AI•BI (www.aibi.cl.ResearchinSCIAN-Lab (SH) is funded by FONDECYT 1090246, FONDEF (D07I1019), and ICM-P04-068-F (NEMO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Foskett JK, et al. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor CW, da Fonseca PC, Morris EP. IP(3) receptors: the search for structure. Trends Biochem Sci. 2004;29:210–219. doi: 10.1016/j.tibs.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Newton CL, Mignery GA, Sudhof TC. Co-expression in vertebrate tissues and cell lines of multiple inositol 1,4,5-trisphosphate (InsP3) receptors with distinct affinities for InsP3. J Biol Chem. 1994;269:28613–28619. [PubMed] [Google Scholar]
- 6.Mak DO, McBride SM, Foskett JK. Spontaneous channel activity of the inositol 1,4,5-trisphosphate (InsP3) receptor (InsP3R). Application of allosteric modeling to calcium and InsP3 regulation of InsP3R single-channel gating. J Gen Physiol. 2003;122:583–603. doi: 10.1085/jgp.200308809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mak DO, et al. Novel regulation of calcium inhibition of the inositol 1,4,5-trisphosphate receptor calcium-release channel. J Gen Physiol. 2003;122:569–581. doi: 10.1085/jgp.200308808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosanac I, et al. Structural insights into the regulatory mechanism of IP3 receptor. Biochim Biophys Acta. 2004;1742:89–102. doi: 10.1016/j.bbamcr.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Shuai J, et al. A kinetic model of single and clustered IP3 receptors in the absence of Ca2+ feedback. Biophys J. 2007;93:1151–1162. doi: 10.1529/biophysj.107.108795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermassen E, Parys JB, Mauger JP. Subcellular distribution of the inositol 1,4,5-trisphosphate receptors: functional relevance and molecular determinants. Biol Cell. 2004;96:3–17. doi: 10.1016/j.biolcel.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Smith IF, Parker I. Imaging the quantal substructure of single IP3R channel activity during Ca2+ puffs in intact mammalian cells. Proc Natl Acad Sci U S A. 2009;106:6404–6409. doi: 10.1073/pnas.0810799106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai J, Rose HJ, Parker I. The number and spatial distribution of IP3 receptors underlying calcium puffs in Xenopus oocytes. Biophys J. 2006;91:4033–4044. doi: 10.1529/biophysj.106.088880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreri-Jacobia M, Mak DO, Foskett JK. Translational mobility of the type 3 inositol 1,4,5-trisphosphate receptor Ca2+ release channel in endoplasmic reticulum membrane. J Biol Chem. 2005;280:3824–3831. doi: 10.1074/jbc.M409462200. [DOI] [PubMed] [Google Scholar]
- 14.Morita T, et al. Visualization of inositol 1,4,5-trisphosphate receptor type III with green fluorescent protein in living cells. Cell Calcium. 2002;31:59–64. doi: 10.1054/ceca.2001.0262. [DOI] [PubMed] [Google Scholar]
- 15.Petersen OH, Tepikin A, Park MK. The endoplasmic reticulum: one continuous or several separate Ca2+ stores? Trends Neurosci. 2001;24:271–276. doi: 10.1016/s0166-2236(00)01787-2. [DOI] [PubMed] [Google Scholar]
- 16.Tateishi Y, et al. Cluster formation of inositol 1,4,5-trisphosphate receptor requires its transition to open state. J Biol Chem. 2005;280:6816–6822. doi: 10.1074/jbc.M405469200. [DOI] [PubMed] [Google Scholar]
- 17.Tojyo Y, et al. The clustering of inositol 1,4,5-trisphosphate (IP(3)) receptors is triggered by IP3 binding and facilitated by depletion of the Ca2+ store. J Pharmacol Sci. 2008;107:138–150. doi: 10.1254/jphs.08021fp. [DOI] [PubMed] [Google Scholar]
- 18.Supattapone S, et al. Solubilization, purification, and characterization of an inositol trisphosphate receptor. J Biol Chem. 1988;263:1530–1534. [PubMed] [Google Scholar]
- 19.Takei K, et al. Inositol 1,4,5-trisphosphate receptor causes formation of ER cisternal stacks in transfected fibroblasts and in cerebellar Purkinje cells. Neuron. 1994;12:327–342. doi: 10.1016/0896-6273(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 20.Katayama E, et al. Native structure and arrangement of inositol-1,4,5-trisphosphate receptor molecules in bovine cerebellar Purkinje cells as studied by quick-freeze deep-etch electron microscopy. Embo J. 1996;15:4844–4851. [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto A, et al. Stacks of flattened smooth endoplasmic reticulum highly enriched in inositol 1,4,5-trisphosphate (InsP3) receptor in mouse cerebellar Purkinje cells. Cell Struct Funct. 1991;16:419–432. doi: 10.1247/csf.16.419. [DOI] [PubMed] [Google Scholar]
- 22.Block BA, et al. Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J Cell Biol. 1988;107:2587–2600. doi: 10.1083/jcb.107.6.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson DG, et al. Ordered arrays of Ca2+-ATPase on the cytoplasmic surface of isolated sarcoplasmic reticulum. Biophys J. 1985;48:597–605. doi: 10.1016/S0006-3495(85)83815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J Mol Biol. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 25.Malviya AN, Rogue P, Vincendon G. Stereospecific inositol 1,4,5-[32P]trisphosphate binding to isolated rat liver nuclei: evidence for inositol trisphosphate receptor-mediated calcium release from the nucleus. Proc Natl Acad Sci U S A. 1990;87:9270–9274. doi: 10.1073/pnas.87.23.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan RS, Hwang SY, Koulen P. Differential inositol 1,4,5-trisphosphate receptor signaling in a neuronal cell line. Int J Biochem Cell Biol. 2007;39:1852–1862. doi: 10.1016/j.biocel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Kusnier C, et al. Single-channel recording of inositol trisphosphate receptor in the isolated nucleus of a muscle cell line. Biol Res. 2006;39:541–553. doi: 10.4067/s0716-97602006000300015. [DOI] [PubMed] [Google Scholar]
- 28.Marchenko SM, et al. Spontaneously active and InsP3-activated ion channels in cell nuclei from rat cerebellar Purkinje and granule neurones. J Physiol. 2005;565:897–910. doi: 10.1113/jphysiol.2004.081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanishi S, et al. Immunohistochemical localization of inositol 1,4,5-trisphosphate receptors in non-neural tissues, with special reference to epithelia, the reproductive system, and muscular tissues. Cell Tissue Res. 1996;285:235–251. doi: 10.1007/s004410050641. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, et al. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116:675–682. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zima AV, et al. IP3-dependent nuclear Ca2+ signalling in the mammalian heart. J Physiol. 2007;584:601–611. doi: 10.1113/jphysiol.2007.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugawara H, et al. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. Embo J. 1997;16:3078–3088. doi: 10.1093/emboj/16.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardenas C, et al. Nuclear inositol 1,4,5-trisphosphate receptors regulate local Ca2+ transients and modulate cAMP response element binding protein phosphorylation. J Cell Sci. 2005;118:3131–3140. doi: 10.1242/jcs.02446. [DOI] [PubMed] [Google Scholar]
- 34.Mak DO, Foskett JK. Single-channel inositol 1,4,5-trisphosphate receptor currents revealed by patch clamp of isolated Xenopus oocyte nuclei. J Biol Chem. 1994;269:29375–29378. [PubMed] [Google Scholar]
- 35.Loesser KE, Franzini-Armstrong C. A simple method for freeze-drying of macromolecules and macromolecular complexes. J Struct Biol. 1990;103:48–56. doi: 10.1016/1047-8477(90)90085-q. [DOI] [PubMed] [Google Scholar]
- 36.Pante N, et al. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarnik M, Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J Struct Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]
- 38.da Fonseca PC, et al. Domain organization of the type 1 inositol 1,4,5-trisphosphate receptor as revealed by single-particle analysis. Proc Natl Acad Sci U S A. 2003;100:3936–3941. doi: 10.1073/pnas.0536251100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang QX, et al. Three-dimensional structure of the type 1 inositol 1,4,5-trisphosphate receptor at 24 A resolution. Embo J. 2002;21:3575–3581. doi: 10.1093/emboj/cdf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serysheva II, et al. Structure of the type 1 inositol 1,4,5-trisphosphate receptor revealed by electron cryomicroscopy. J Biol Chem. 2003;278:21319–21322. doi: 10.1074/jbc.C300148200. [DOI] [PubMed] [Google Scholar]
- 41.Ngo QT, Maxwell JT, Mignery GA, Chiu W, Ludtke SL, Serysheva II. Electron Cryomicroscopy of InsP3R1 Calcium Release Channel. 53rd Annual meeting, Biophysical Society; 2009. Abstract 496. [Google Scholar]
- 42.Ionescu L, et al. Graded recruitment and inactivation of single InsP3 receptor Ca2+-release channels: implications for quantal Ca2+ release. J Physiol. 2006;573:645–662. doi: 10.1113/jphysiol.2006.109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nosyreva E, et al. The high-affinity calcium-calmodulin-binding site does not play a role in the modulation of type 1 inositol 1,4,5-trisphosphate receptor function by calcium and calmodulin. Biochem J. 2002;365:659–667. doi: 10.1042/BJ20011789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu H, et al. Functional characterization of mammalian inositol 1,4,5-trisphosphate receptor isoforms. Biophys J. 2005;88:1046–1055. doi: 10.1529/biophysj.104.049593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franzini-Armstrong C, Protasi F, Ramesh V. Comparative ultrastructure of Ca2+ release units in skeletal and cardiac muscle. Ann N Y Acad Sci. 1998;853:20–30. doi: 10.1111/j.1749-6632.1998.tb08253.x. [DOI] [PubMed] [Google Scholar]
- 46.Hofmann S, et al. Wolfram syndrome: structural and functional analyses of mutant and wild-type wolframin, the WFS1 gene product. Hum Mol Genet. 2003;12:2003–2012. doi: 10.1093/hmg/ddg214. [DOI] [PubMed] [Google Scholar]
- 47.Hamada K, et al. Two-state conformational changes in inositol 1,4,5-trisphosphate receptor regulated by calcium. J Biol Chem. 2002;277:21115–21118. doi: 10.1074/jbc.C200244200. [DOI] [PubMed] [Google Scholar]
- 48.Pinto da Silva P, Kan FW. Label-fracture: a method for high resolution labeling of cell surfaces. J Cell Biol. 1984;99:1156–1161. doi: 10.1083/jcb.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suhara W, et al. Visualization of inositol 1,4,5-trisphosphate receptor by atomic force microscopy. Neurosci Lett. 2006;391:102–107. doi: 10.1016/j.neulet.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 50.Bailly M, et al. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isobe Y, Warner FD, Lemanski LF. Three-dimensional immunogold localization of alpha-actinin within the cytoskeletal networks of cultured cardiac muscle and nonmuscle cells. Proc Natl Acad Sci U S A. 1988;85:6758–6762. doi: 10.1073/pnas.85.18.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rash JE, et al. Freeze-fracture and immunogold analysis of aquaporin-4 (AQP4) square arrays, with models of AQP4 lattice assembly. Neuroscience. 2004;129:915–934. doi: 10.1016/j.neuroscience.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rash JE, Yasumura T. Direct immunogold labeling of connexins and aquaporin-4 in freeze-fracture replicas of liver, brain, and spinal cord: factors limiting quantitative analysis. Cell Tissue Res. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- 54.Kada G, et al. Recognition force microscopy/spectroscopy of ion channels: applications to the skeletal muscle Ca2+ release channel (RYR1) Ultramicroscopy. 2001;86:129–137. doi: 10.1016/s0304-3991(00)00070-x. [DOI] [PubMed] [Google Scholar]
- 55.Ferguson DG, Schwartz HW, Franzini-Armstrong C. Subunit structure of junctional feet in triads of skeletal muscle: a freeze-drying, rotary-shadowing study. J Cell Biol. 1984;99:1735–1742. doi: 10.1083/jcb.99.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chalmers M, Schell MJ, Thorn P. Agonist-evoked inositol trisphosphate receptor (IP3R) clustering is not dependent on changes in the structure of the endoplasmic reticulum. Biochem J. 2006;394:57–66. doi: 10.1042/BJ20051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taufiq Ur R, et al. Clustering of InsP3 receptors by InsP3 retunes their regulation by InsP3 and Ca2+ Nature. 2009;458:655–659. doi: 10.1038/nature07763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson BS, et al. Calcium-dependent clustering of inositol 1,4,5-trisphosphate receptors. Mol Biol Cell. 1998;9:1465–1478. doi: 10.1091/mbc.9.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franzini-Armstrong C. Myology. New York: McGrawHill; 2004. The membrane systems of muscle cells; pp. 232–256. [Google Scholar]
- 60.Takekura H, et al. Co-expression in CHO cells of two muscle proteins involved in excitation-contraction coupling. J Muscle Res Cell Motil. 1995;16:465–480. doi: 10.1007/BF00126431. [DOI] [PubMed] [Google Scholar]
- 61.Yin CC, Lai FA. Intrinsic lattice formation by the ryanodine receptor calcium-release channel. Nat Cell Biol. 2000;2:669–671. doi: 10.1038/35023625. [DOI] [PubMed] [Google Scholar]
- 62.Yin CC, D'Cruz LG, Lai FA. Ryanodine receptor arrays: not just a pretty pattern? Trends Cell Biol. 2008;18:149–156. doi: 10.1016/j.tcb.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Hamada K, Terauchi A, Mikoshiba K. Three-dimensional rearrangements within inositol 1,4,5-trisphosphate receptor by calcium. J Biol Chem. 2003;278:52881–52889. doi: 10.1074/jbc.M309743200. [DOI] [PubMed] [Google Scholar]
- 64.Ghosh TK, et al. Competitive, reversible, and potent antagonism of inositol 1,4,5-trisphosphate-activated calcium release by heparin. J Biol Chem. 1988;263:11075–11079. [PubMed] [Google Scholar]
- 65.Miyawaki A, et al. Structure-function relationships of the mouse inositol 1,4,5-trisphosphate receptor. Proc Natl Acad Sci U S A. 1991;88:4911–4915. doi: 10.1073/pnas.88.11.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]