Abstract
Salmonella enterica serotype Typhi is the cause of typhoid fever and a human-restricted pathogen. Currently available typhoid vaccines provide 50 to 90% protection for 2 to 5 years, and available practical diagnostic assays to identify individuals with typhoid fever lack sensitivity and/or specificity. Identifying immunogenic S. Typhi antigens expressed during human infection could lead to improved diagnostic assays and vaccines. Here we describe a platform immunoaffinity proteomics-based technology (IPT) that involves the use of columns charged with IgG, IgM, or IgA antibody fractions recovered from humans bacteremic with S. Typhi to capture S. Typhi proteins that were subsequently identified by mass spectrometry. This screening tool identifies immunogenic proteins recognized by antibodies from infected hosts. Using this technology and the plasma of patients with S. Typhi bacteremia in Bangladesh, we identified 57 proteins of S. Typhi, including proteins known to be immunogenic (PagC, HlyE, OmpA, and GroEL) and a number of proteins present in the human-restricted serotypes S. Typhi and S. Paratyphi A but rarely found in broader-host-range Salmonella spp. (HlyE, CdtB, PltA, and STY1364). We categorized identified proteins into a number of major groupings, including those involved in energy metabolism, protein synthesis, iron homeostasis, and biosynthetic and metabolic functions and those predicted to localize to the outer membrane. We assessed systemic and mucosal anti-HlyE responses in S. Typhi-infected patients and detected anti-HlyE responses at the time of clinical presentation in patients but not in controls. These findings could assist in the development of improved diagnostic assays.
Salmonella enterica serotype Typhi is a human-restricted pathogen that is the primary cause of enteric fever. It is estimated that S. Typhi infects over 20 million individuals and kills approximately 200,000 people globally each year (4). Currently, commercially available typhoid vaccines provide approximately 50 to 75% protection for 2 to 5 years (21), although an anti-typhoid Vi conjugate vaccine demonstrated 90% protection in 2- to 5-year-old children in a large field trial (23). Available and practical diagnostic tests for typhoid fever lack sensitivity and/or specificity (28). Identifying immunogenic S. Typhi antigens expressed during human infection could lead to improved diagnostic assays and vaccines.
Infection with S. Typhi begins with the ingestion of contaminated water or food. The bacteria invade the gastrointestinal mucosa, translocate to the lymphoid follicles, where they survive and replicate within macrophages, and then disseminate via the bloodstream to the liver, spleen, and intestinal lymph nodes (14). The incubation period is typically 8 to 14 days (22), and symptoms include fever, abdominal pain, anorexia, weakness, potential complications of intestinal perforation, encephalopathy, and gastrointestinal bleeding (14, 34). Clinical studies demonstrate that S. Typhi infection stimulates both an intestinal mucosal and systemic humoral and cellular immune response (14, 34). S. Typhi is a facultative intracellular pathogen of macrophages, and both cellular and antibody-mediated immune responses are known to play roles in controlling and clearing S. Typhi infection (37). Despite this, there are limited data on antigen-specific cellular responses during wild-type S. Typhi infection in humans. Analyses of cellular immune responses during S. Typhi infection have largely used whole-cell preparations or flagellar antigens and have focused predominately on measuring immune responses in recipients of oral live attenuated typhoid vaccines, not in individuals with wild-type disease (24, 25, 40-42, 49).
Antibody responses during wild-type infection have been better studied but have focused largely on a relatively small number of antigens, including O antigen (lipopolysaccharide [LPS]), H antigen (flagellar component), polysaccharide capsular antigen (Vi antigen), heat shock proteins such as GroEL, and outer membrane proteins such as OmpC and -F (13, 34). In addition, gut-derived IgA antibody-secreting cells that recognize LPS, a membrane preparation, or whole-killed S. Typhi organisms can be detected in the peripheral blood following natural S. Typhi infection or oral typhoid vaccination (16, 43, 50, 54). These cells eventually return home to the gastrointestinal mucosa, where they secrete secretory IgA antibody (36, 43).
A number of immunoaffinity-based techniques that screen protein libraries of pathogens to identify immunogenic antigens have been developed (12, 17, 38), and we have previously reported using one such approach, in vivo-induced-antigen technology (IVIAT), to identify immunogenic S. Typhi antigens expressed during human infection (12). Another previously described technique, proteomics-based expression library screening (PELS), involves using antibody-charged columns to capture antigens produced by an Escherichia coli-based expression system containing an inducible library of a pathogen of interest, with subsequent elution and identification of bound proteins using mass spectrometric analysis (17). Here we describe using a modification of this approach that we have termed immunoaffinity proteomics-based technology (IPT). IPT involves directly screening the pathogen of interest using columns charged with IgG, IgM, or IgA antibody fractions recovered from the blood of infected humans. We applied IPT to S. Typhi to gain further insights into immunogenic antigens expressed in patients bacteremic with S. Typhi in Bangladesh.
MATERIALS AND METHODS
Bacterial strains and lysate preparation.
We obtained wild-type S. Typhi CT18 from the Salmonella Genetic Stock Centre, University of Calgary, Calgary, Alberta, Canada. To maximize the protein expression profile of S. Typhi prior to applying bacterial preparations to antibody-charged columns, we separately grew CT18 to mid-log phase and stationary phase at 37°C, with aeration in two media: Luria broth and low-magnesium (10 μM MgCl2)-containing minimal medium. The latter medium/condition is known to induce the PhoP regulon, involved in the intramacrophage survival of S. Typhi; we chose this condition to maximize the expression of antigens expressed in vivo during human infection (3). We resuspended cell pellets in Tris-buffered saline (TBS)-2% n-octyl-β-d-glucopyranoside (Sigma)-2 mM Mg-2× EDTA-free Roche Complete protease inhibitor (pH 8.0) and lysed cells using 0.1 mM silica beads (MP Biomedical, Solon, OH) and a Mini-Bead-Beater-8 (BioSpec Products, Bartlesville, OK).
Study subject selection, sample collection, and processing.
Individuals (3 to 59 years of age) presenting to the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), with fever of 3 to 7 days' duration (≥39°C) who were suspected of having enteric fever, who lacked an obvious focus of infection, and for whom we lacked an alternate diagnosis were eligible for enrollment. We collected venous blood (5 ml from children <5 years old and 10 ml from all others) at enrollment (day 0), on day 6, and on day 20. We cultured 3 to 5 ml of day 0 blood by using a BacT/Alert automated system, subculturing positive bottles on MacConkey agar, and identifying colonies using standard biochemical tests and reaction with Salmonella-specific antisera (51). Following collection of blood, all patients were initially treated with oral ciprofloxacin or cefixime or injectable ceftriaxone; these were continued for up to 14 days at the discretion of the attending physician. Using this approach, we identified individuals with S. Typhi bacteremia (n = 10). We also collected acute- and convalescent-phase control plasma from Bangladeshi patients with documented Vibrio cholerae infection (n = 5) and single blood samples from North American volunteers with no history of Salmonella infection or vaccination (n = 3).
To analyze mucosal IgA responses, we recovered peripheral blood mononuclear cells (PBMCs) from typhoid (days 0, 6, and 20) and cholera (days 2 and 7) patients. Activated mucosal lymphocytes migrate from intestinal tissue and circulate within peripheral blood before returning to mucosal tissues (15, 43). This migration peaks 1 to 2 weeks after intestinal infection and may be measured by using PBMCs in an antibody-secreting cell (ASC) assay or in supernatants recovered from harvested PBMCs (the “antibody-in-lymphocyte supernatant” [ALS] assay) (36, 43). To recover PBMCs, we diluted heparinized blood in phosphate-buffered saline (PBS; 10 mM [pH 7.2]) and isolated PBMCs by density gradient centrifugation on Ficoll-Isopaque (Pharmacia, Uppsala, Sweden). We resuspended isolated PBMCs to a concentration of 107 cells/ml in RPMI 1640 complete medium (Gibco, Gaithersburg, MD) with 10% heat-inactivated fetal bovine serum (HyClone, Ogden, UT), 100 U of penicillin/ml, 100 μg of streptomycin/ml, 100 mM pyruvate, and 200 mM l-glutamine (Gibco) (43). We incubated cells for 48 h at 37°C with 5% CO2, collected supernatants containing secreted antibodies, and added a protease inhibitor solution as previously described (43).
This study was approved by the human studies committees of the ICDDR,B and Massachusetts General Hospital.
Plasma preparation for antibody enrichment, coupling of antibodies to HiTrap NHS-activated HP columns, and capture of S. Typhi proteins.
Equal volumes of acute-phase (day 0) and convalescent-phase (day 20) plasma from four Bangladeshi patients with culture-confirmed S. Typhi bacteremia were pooled to create a total volume of 1 ml of acute- and 1 ml of convalescent-phase samples. We diluted each pooled plasma sample 1:1 with PBS, pH 7.4, and recovered IgG fractions using GE Healthcare HiTrap protein G high-performance (HP) columns, IgA fractions using GenWay Seppro human IgA-IgY microbeads, and IgM fractions using GE Healthcare HiTrap protein IgM HP columns, all according to the manufacturers' instructions. We bound recovered antibody fractions to HiTrap N-hydroxysuccinimide (NHS)-activated HP columns, applied bacterial lysates prepared as described above, and eluted bound proteins as previously described (17, 20). To assess nonspecific binding, we also eluted lysate proteins bound to columns that had been blocked but to which we had not loaded antibody.
Sample preparation for MS analysis.
Following elution from each HiTrap NHS column, protein samples were concentrated using a Vivaspin 5-kDa 2-ml spin filter according to the manufacturer's instructions. To each concentrated sample, we added 50 μl of 8 M urea-2% SDS-150 mM NH4HCO3-10 mM dithiothreitol (DTT)-1× lithium dodecyl sulfate (LDS), pH 8.5, and incubated the mixtures at 37°C for 60 min. We next cooled the alkylated samples with iodoacetamide, quenched them with excess DTT, and separated samples on a 10% Bis-Tris NuPAGE MOPS (morpholinepropanesulfonic acid) gel (Invitrogen). We fixed gels in destain (50% methanol and 7.5% acetic acid), rehydrated them, stained them with Simply Blue Safestain (Invitrogen), cut them horizontally into slices, and destained them until transparent. We rinsed gel samples with three alternating washes of 50 mM ammonium bicarbonate and acetonitrile, cooled them, resuspended each gel slice in trypsin (5.5 μg/ml in 50 mM ammonium bicarbonate-10% acetonitrile), and incubated them at 37°C for 24 h for digestion of proteins. We extracted peptides with one rinse of 50 mM ammonium bicarbonate-10% acetonitrile, followed by one rinse of 50% acetonitrile-0.1% formic acid, and prepared samples for mass spectrometry by lyophilization and rehydration in 20 μl 5% acetonitrile-0.2% formic acid.
LC-MS analysis.
We loaded eluted and digested samples into 96-well plates for mass spectrometry (MS) analysis on an LTQ-Orbitrap XL (Thermo Fisher Scientific). For each run, we injected 10 μl of each reconstituted sample using a Thermo Scientific MicroAutosampler. We performed reverse-phase chromatographic separation using 3-μm Hypersil Gold C18 medium packed into a fused-silica, 75-μm-inner-diameter, 20-cm-long column running at 250 nl/min from a Surveyor MS pump with a flow splitter. A gradient of 5 to 40% acetonitrile in 0.2% formic acid was produced and the over 150 min. The LTQ-Orbitrap was run in a top 8 configuration at a resolution of 60,000 for a full scan, with monoisotopic precursor selection enabled +1 and unassigned charge states rejected. The analysis on the LTQ-Orbitrap instrument was carried out with collision-induced dissociation (CID) fragmentation.
Peptide identification and statistical analysis.
We identified peptides using SEQUEST (Thermo Fisher Scientific) through the Bioworks browser, version 3.3.1 SR1. Tandem MS (MS/MS) data were searched using 10-ppm mass accuracy for precursor m/z and a 0.5-Da window for fragment ions. Fully enzymatic tryptic searches with up to three missed cleavage sites were allowed. Oxidized methionines were searched as a variable modification, and alkylated cysteines were searched as a fixed modification. Salmonella databases for CT18 were downloaded from the EMBL-EBI database and supplemented with common contaminants. We employed a reverse database strategy (5) using concatenating reversed protein sequences for each database entry in SEQUEST. We filtered peptides for each charge state to a false discovery rate (FDR) of 1% and then grouped peptides into proteins using Occam's razor logic. We used spectral counting to compare changes in protein abundance in fractions eluted from columns charged with antibody fractions to those in fractions eluted from blocked columns not containing antibody, and we required that proteins be associated with at least 3 spectral counts to be included in our analyses. After normalizing results of duplicate samples, we averaged total spectral counts and used a G-test (45) while controlling for a positive false-discovery rate (47) to test for significant differential protein detection between samples. Protein functional classification was based on J. Craig Venter Institute annotations (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi).
Detection of HlyE-specific antibodies in plasma and mucosal ALS fluid by ELISA.
We confirmed the screening results obtained by IPT by using standard enzyme-linked immunosorbent assays (ELISAs), with hemolysin E (HlyE) as a model antigen. HlyE was purified as previously described (55). We measured anti-HlyE IgG and IgA responses using a microtiter plate ELISA format and plasma from culture-confirmed typhoid and cholera patients, as well as North American volunteers. We also measured IgA responses in antibody-in-lymphocyte supernatant (ALS) fractions recovered from circulating mucosal lymphocytes from infected patients (43). To detect antigen-specific responses, we added 100 μl of plasma samples (diluted 1:200 for IgG and IgA) in PBS-Tween 20-0.1% bovine serum albumin (BSA) or 100 μl (1:2 dilution for IgA) of ALS specimens to plates coated with 100 ng of HlyE/well and detected responses using horseradish peroxidase-conjugated human IgA or IgG antibodies (Jackson Laboratories, Bar Harbor, ME) (1:1,000 in PBS-Tween 20-0.1% BSA for IgA and IgG isotypes). We developed and read plates as previously described (43). To compare across plates, we divided readings of unknown samples by readings of an in-house pooled standard, multiplied by 100, and expressed results as ELISA units. We used paired t tests to determine if there was a statistically significant difference between results for samples.
RESULTS
Proteins identified by IPT.
Using mass spectrometry, we identified 57 S. Typhi proteins grown under various conditions whose capture by affinity-purified IgG, IgM, and IgA antibody fractions from acute- and convalescent-phase plasma of patients with S. Typhi bacteremia was significantly increased compared to the capture by the column not containing antibody (IgG, column 29; IgA, column 2; and IgM, column 44) (Table 1 and see Table S1 in the supplemental material). These proteins could be grouped into a number of functional categories (Table 2). The most highly represented groups included proteins for energy metabolism and nutrient acquisition, protein and amino acid biosynthesis, and cellular processes, including pathogenesis and virulence, as well as proteins located in and involved in the synthesis of the cell envelope. Among the identified proteins were a number under the control of the PhoP regulon, including PagC, HlyE, and PhoN (6, 8, 9, 27). We also identified heat shock proteins GroEL and DnaK, outer membrane proteins, including OmpA, and other proteins involved in pathogenesis and virulence, including CdtB, PltA, and STY1364.
TABLE 1.
S. Typhi proteins bound by acute- and convalescent-phase serum antibody fractions from bacteremic patients and identified by mass spectrometric analysisa
| Protein group and Swiss-Prot ID | Protein name(s) | Gene locus | Protein description | IgG |
IgA |
IgM |
|||
|---|---|---|---|---|---|---|---|---|---|
| A | C | A | C | A | C | ||||
| Cellular processes, virulence, and pathogenesis | |||||||||
| Q8Z7B8 | STY1364 | Hypothetical periplasmic protein | X | X | |||||
| Q8Z727 | HlyE | STY1498 | Hemolysin E | X | X | X | X | ||
| Q8Z6B2 | PagC | STY1878 | Outer membrane invasion protein | X | X | ||||
| Q8Z6A7 | CdtB | STY1886 | Cytolethal distending toxin subunit B homolog | X | X | ||||
| Q8Z6A4 | PltA | STY1890 | Putative pertussis-like toxin subunit | X | X | ||||
| Cell envelope | |||||||||
| Q8Z939 | STY0351 | Possible outer membrane adhesion | X | X | |||||
| Q8XH17 | OmpX | STY0872 | Outer membrane protein X | X | X | ||||
| Q8Z7S0 | OmpA | STY1091 | Outer membrane protein A | X | X | ||||
| Q8Z5I6 | RfbH | STY2300 | Putative dehydratase RfbH | X | X | ||||
| P67913 | HldD | STY4085 | ADP-l-glycero-d-manno-heptose-6-epimerase | X | |||||
| Amino acid biosynthesis | |||||||||
| Q8Z9A8 | DapD | STY0236 | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate | X | X | ||||
| Q8Z7N9 | WrbA | STY1155 | Flavoprotein WrbA | X | X | X | |||
| Q8Z3B6 | MetE | STY3594 | 5-Methyltetrahydropteroyltriglutamate-homocysteine | X | |||||
| Q8Z381 | IlvC | STY3648 | Ketol acid reductoisomerase | X | X | ||||
| Q8Z123 | ArgI | STY4807 | Ornithine carbamoyltransferase | X | X | ||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | |||||||||
| P66039 | RibH | STY0456 | 6,7-Dimethyl-8-ribityllumazine synthase | X | |||||
| P0A2E2 | GlyA | STY2802 | Serine hydroxymethyltransferase | X | X | X | X | ||
| Central intermediary metabolism | |||||||||
| Q8Z3Q5 | STY3330 | Possible oxidoreductase | X | X | |||||
| Q934J6 | PhoN | STY4519 | Nonspecific acid phosphatase | X | |||||
| P65749 | Ppa | STY4773 | Inorganic pyrophosphatase | X | X | ||||
| Energy metabolism | |||||||||
| Q8Z9F0 | AceE | STY0175 | Pyruvate dehydrogenase E1 component | X | |||||
| Q8Z9E9 | AceF | STY0176 | Dihydrolipoamide acetyltransferase component | X | |||||
| Q8Z9E8 | LpdA | STY0177 | Dihydrolipoamide dehydrogenase | X | |||||
| P66870 | SucC | STY0781 | Succinyl-CoA ligase | X | |||||
| Q8XF12 | TrxB | STY0956 | Thioredoxin reductase | X | X | X | |||
| Q8XGV7 | PflB | STY0973 | Formate acetyltransferase 1 | X | X | ||||
| Q8Z7F1 | Adh | STY1302 | Alcohol dehydrogenase | X | |||||
| Q8Z747 | YdcW | STY1467 | Gamma-aminobutyraldehyde dehydrogenase | X | |||||
| P0A1P1 | GapA | STY1825 | Glyceraldehyde 3-phosphate dehydrogenase | X | X | ||||
| Q8Z5J3 | Gnd | STY2290 | 6-Phosphogluconate dehydrogenase, decarboxylating | X | X | ||||
| Q8Z4T0 | TalA | STY2710 | Transaldolase A | X | |||||
| P64077 | Eno | STY3081 | Enolase | X | X | ||||
| Q8XFG7 | Fba | STY3226 | Fructose 1,6-bisphosphate aldolase | X | X | X | |||
| P65703 | Pgk | STY3227 | Phosphoglycerate kinase | X | X | X | |||
| P65693 | PrkA | STY3809 | 6-Phosphofructokinase | X | |||||
| Q8Z2D5 | AldB | STY4116 | Aldehyde dehydrogenase B | X | |||||
| Q8Z1T0 | Qor | STY4441 | Quinone oxidoreductase | X | |||||
| Q8XFR3 | AspA | STY4685 | Aspartate ammonia-lyase | X | |||||
| Q8Z0U3 | DeoC | STY4918 | Deoxyribose-phosphate aldolase | X | |||||
| Fatty acid and phospholipid metabolism | |||||||||
| Q8Z7C7 | FabI | STY1352 | Enoyl-(acyl carrier protein) reductase (NADH) | X | |||||
| Protein fate | |||||||||
| Q8Z9R1 | DnaK | STY0012 | Chaperone protein DnaK | X | |||||
| Q8Z934 | PepD | STY0361 | Aminoacyl-histidine dipeptidase | X | X | ||||
| Q7AMH5 | ClpB | STY2849 | Chaperone protein ClpB | X | |||||
| P0A1D4 | GroEL | STY4690 | 60-kDa chaperonin | X | X | X | |||
| Protein synthesis | |||||||||
| P67564 | SerS | STY0961 | Seryl-tRNA synthetase | X | |||||
| Q8XGK9 | RpsA | STY0981 | 30S ribosomal protein S1 | X | |||||
| Q56112 | AsnS | STY1004 | Asparaginyl-tRNA synthetase | X | |||||
| Q8Z5W1 | AspS | STY2109 | Aspartyl-tRNA synthetase | X | |||||
| P0A1H4 | FusA | STY4352 | Elongation factor G | X | |||||
| Q8Z118 | ValS | STY4814 | Valyl-tRNA synthetase | X | |||||
| P0A1H6 | TufA, TufB | STY4353 STY3739 | Elongation factor Tu | X | X | X | X | ||
| Biosynthesis of purines, pyrimidines, nucleosides, and nucleotides | |||||||||
| Q8Z4Q3 | GuaA | STY2751 | GMP synthase | X | |||||
| Regulatory function | |||||||||
| Q8Z3E9 | SspA | STY3523 | Stringent starvation protein A | X | |||||
| Transcription | |||||||||
| Q8Z320 | RpoB | STY3732 | DNA-directed RNA polymerase subunit beta | X | |||||
| P0A7Z8 | RpoA | STY4383 | DNA-directed RNA polymerase subunit alpha | X | |||||
| Transport and binding | |||||||||
| Q8Z1Y0 | Bfr | STY4355 | Bacterioferritin | X | X | ||||
| Unclassified | |||||||||
| Q8Z5C3 | MetG | STY2384 | Methionyl-tRNA synthetase | X | |||||
ID, identifier; A, bound by immunoglobulin in acute-phase samples; C, bound by immunoglobulin in convalescent-phase samples; X, presence; CoA, coenzyme A.
TABLE 2.
Functional classification of S. Typhi proteins captured by acute- and/or convalescent-phase-antibody columns
| Functional classification of captured proteins | No. of proteins |
|---|---|
| Energy metabolism | 19 |
| Protein synthesis | 7 |
| Amino acid biosynthesis | 5 |
| Cellular processes, virulence, and pathogenesis | 5 |
| Cell envelope | 5 |
| Protein fate | 4 |
| Central intermediary metabolism | 3 |
| Transcription | 2 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 2 |
| Fatty acid and phospholipid metabolism | 1 |
| Biosynthesis of purines, pyrimidines, nucleosides, and nucleotides | 1 |
| Transport and binding proteins | 1 |
| Regulatory functions | 1 |
| Unclassified | 1 |
Comparison of proteins bound by acute- or convalescent-phase antibody fractions, by isotype.
When considering proteins bound on the IgM columns, we identified 40 proteins bound by acute- but not convalescent-phase plasma IgM, including GroEL and DnaK, and 4 proteins bound by both acute- and convalescent-phase IgM samples, including HlyE. When considering proteins bound on the IgG columns, we identified 29 proteins in total, including 27 proteins bound by acute-phase samples, 18 proteins bound by convalescent-phase samples, and 16 proteins bound by both acute- and convalescent-phase samples. Proteins bound by the IgG columns included HlyE, PagC, PhoN, CdtB, PltA, OmpX, OmpA, outer membrane adhesin STY0351, GroEL, and bacterioferritin. The only proteins bound by the IgA columns were the flavoprotein WrbA, a protein involved in tryptophan biosynthesis (57), and elongation factor protein TufA.
Confirmation of immune responses to HlyE by standard techniques.
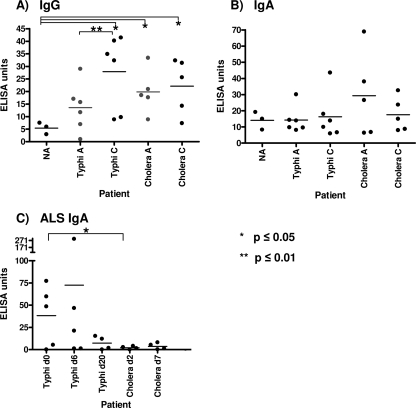
To further characterize the immunogenicity of a protein identified in our screening procedure, we measured anti-HlyE responses in plasma and ALS fluid of S. Typhi-infected patients and controls. We detected significant increases in anti-HlyE IgG, but not IgA, in the convalescent-phase plasma compared to levels in the acute-phase plasma of patients infected with S. Typhi (P ≤ 0.01) but not V. cholerae (Fig. 1 A and B). We also detected significantly higher anti-HlyE IgG titers in the convalescent-phase plasma of S. Typhi patients than in North American controls (P < 0.05) (Fig. 1A). We detected a significantly elevated anti-HlyE IgA level at the time of clinical presentation using the mucosal ALS assay (P < 0.05) in S. Typhi patients compared to the level in cholera patients (Fig. 1C).
FIG. 1.
Characterization of anti-HlyE immune responses. Anti-HlyE IgG (A) and IgA (B) responses in the plasma of North American volunteers (NA) and acute-phase (day 0) and convalescent-phase (day 20) plasma of patients infected with S. Typhi or V. cholerae (control). (C) Anti-HlyE IgA ALS responses in day 0, day 6, and day 20 samples of patients infected with S. Typhi and in day 2 and day 7 samples of patients infected with V. cholerae (control).
DISCUSSION
A number of high-throughput immunoaffinity screens have been developed to evaluate host-pathogen interactions during human infection, including IVIAT and PELS (11, 12, 17, 38, 39). Here we describe a further advance of such approaches, IPT, and used this to identify 57 S. Typhi proteins recognized by antibodies of various isotypes from individuals bacteremic with S. Typhi in Bangladesh. In comparison to other techniques, IPT has the advantage of directly using the pathogen of interest for screening of antibody responses and of being able to characterize these responses by isotype class. Antibody isotype analysis may provide insight into the stage of infection during which antigenic presentation occurs for pathogens like S. Typhi, which have both mucosal and systemic phases of infection.
Although an inducible expression library can be used to generate the proteins used in immunoaffinity screening, the generation of this library in an E. coli-based system is complicated by difficulties in definitively attributing the captured peptides to the pathogen's proteins or to a homolog in E. coli (33). For these reasons, we used S. Typhi itself as the source of our protein pool for the IPT modification of the PELS technique. To maximize the number of S. Typhi proteins present in the pool used in our analysis, we grew S. Typhi under a variety of growth conditions, including those known to induce the PhoP regulon, that mimic conditions encountered in the intracellular environment during human infection (3).
Another important issue for any immunoaffinity screening assay is the optimal negative control. The fact that many S. Typhi proteins share homology with corresponding E. coli proteins not only complicates attribution of immunoreactive peptides by mass spectrometric analysis but also complicates selection of optimal control plasma for use in immunoaffinity comparisons. We therefore compared the immunoreactivities of S. Typhi proteins identified using columns charged with antibody from infected humans to reactivities with blocked columns not charged with antibody. We also compared proteins captured using columns charged with antibody fractions recovered from the same patient at two different phases of infection: the acute and convalescent stages. Using these approaches, we identified a number of S. Typhi proteins known to be immunogenic, including HlyE, PagC, and GroEL, supporting the validity of our approach.
Hemolysin E (HlyE), also referred to as cytolysin A (ClyA) or SheA, is a pore-forming toxin that contributes to the cytotoxicity and invasion of epithelial cells and also affects bacterial growth within human macrophages (6, 7). HlyE is expressed under the control of the PhoP regulon (6) and shares >90% amino acid identity with ClyA in E. coli K-12 (30). ClyA has cytotoxic activity for both murine and human macrophages (19). Of the Salmonella serovars, the gene for HlyE was initially thought to be uniquely found within the S. Typhi and S. Paratyphi A genomes (30, 55); however, a recent study by Fuentes et al. suggests that it is found in several other S. enterica serovars known to cause systemic infection humans (7). PagC is also expressed under the control of the PhoP regulon (8, 27), and we previously identified PagC in our IVIAT screen of immunoreactive proteins following S. Typhi infection, including demonstrating a specific anti-PagC antibody response in the plasma of Bangladeshi patients infected with S. Typhi (12). In the current study, we also identified PhoN, a PhoP-regulated phosphatase, as well as heat shock proteins GroEL and DnaK, as immunoreactive. Overall, heat shock proteins are often immunogenic (32, 52), and immunization of mice with purified S. Typhi GroEL induces an IgG response and confers 70 to 90% protection against lethal intraperitoneal challenge with S. Typhi and S. Typhimurium in a murine model (31).
A number of studies have shown that S. Typhi outer membrane proteins are immunogenic during human infection (2, 29), and we identified OmpX and OmpA in our assay. OmpA has previously been shown to be immunogenic in mice following S. Typhimurium infection (18, 35, 44, 48). We also identified STY0351, an outer membrane adhesin with an OmpA-like transmembrane domain and with homology to PagC.
We also identified CdtB (STY1886), PltA (STY1891), and STY1364, all of which are present in S. Typhi and S. Paratyphi A but rarely found in other Salmonella serovars (26). CdtB (STY1886) is a homolog of the active subunit of a cytolethal distending toxin found in a number of bacterial pathogens, including E. coli, Shigella dysenteriae, Haemophilus ducreyi, Actinobacillus actinomycetemcomitans, and Helicobacter hepaticus (10). CdtB expression is upregulated intracellularly and induces cell cycle arrest of host cells by causing DNA damage leading to distention of cells and enlargement of nuclei (10). Interestingly, CdtB forms a complex with PltA (STY1890) and PltB (STY1891), pertussis-like toxins that mediate its delivery into host cells (46). STY1364, a putative periplasmic protein, shares homology with PltB (STY1891, 30% identity) and ArtB, a putative ADP-ribosyltransferase toxin of S. Typhimurium DT104 (73% identity). All 3 proteins (CdtB, PltA, and STY1364) are upregulated in minimal media, an in vitro condition that approximates that of the intracellular macrophage (1).
With regard to the immunoreactivities of antibodies of various isotypes, we found that the IgM fraction collected during the acute stage of illness recognized the largest number of S. Typhi proteins, including HlyE, GroEL, and DnaK. Such a result is consistent with early-stage infection and subsequent isotype maturation. The next largest group of S. Typhi proteins reacted with either acute- or convalescent-phase IgG antibody, including PagC, HlyE, CdtB, PltA, PhoN, and GroEL. The fact that both acute- and convalescent-phase antibody recognized many of these antigens may reflect the probable 2- to 3-week period between ingestion of infecting S. Typhi organisms, the onset of symptoms, and the collection of samples. Consistent with an evolving immune response, we identified several S. Typhi proteins that reacted only with convalescent-phase IgG, including the PhoP-regulated protein PhoN. We also identified an IgG response against bacterioferritin, a protein involved in iron homeostasis (53).
To validate the results of the anti-S. Typhi responses identified in our screening assay, we assessed anti-HlyE responses by standard ELISA using purified protein and plasma from Bangladeshi patients with documented S. Typhi bacteremia and compared them to patients with V. cholerae infection or plasma from North American controls. von Rhein et al. have previously shown that anti-HlyE serum antibodies can be detected in S. Typhi-infected patients by Western blotting (56). We used an ELISA-based format and plasma from Bangladeshi patients whose samples were not used in the screening IPT assay and confirmed anti-HlyE IgG, but not IgA, responses. We also detected significant increases in anti-HlyE IgG responses from the acute to the convalescent phase of illness in patients infected with S. Typhi but not in the negative-control cholera patients.
To test for a potential mucosal immune response to HlyE, we also evaluated anti-HlyE IgA responses using ALS samples. We recently found that performing an IgA-based ALS assay using an S. Typhi crude membrane preparation during acute-stage illness could be used to differentiate patients with typhoid from those with other illnesses (43). We were thus intrigued by our detection of an anti-HlyE IgA response in ALS fluid from patients with typhoid, which was absent in patients with cholera in Bangladesh, a setting where both infections are endemic. The identification of a specific anti-S. Typhi serological response at the time of clinical presentation of patients with typhoid fever could be the foundation for an improved diagnostic assay.
In summary, IPT is a rapid screening tool that can be used to identify proteins that elicit a serological immune response in infected hosts. Using this technology, we identified a number of immunogenic proteins of S. Typhi, including a subset previously known to be immunogenic (supporting the validity of the approach), a subset under the control of the PhoP regulon involved in intracellular survival, and a subset present in the genomes of human-restricted S. Typhi and S. Paratyphi A but largely absent from zoonotic and wide-host-range Salmonella bacteria. Many of the proteins are components of the outer membrane and involved in energy metabolism, protein synthesis, and iron homeostasis or are biosynthetic and metabolic enzymes; a significant subset are involved in virulence and intracellular survival. Our identification of S. Typhi proteins by IPT may facilitate targeted analysis of subsets of S. Typhi antigens in cellular immunologic studies. Additionally, our identification of HlyE by IPT, confirmation of differential anti-HlyE reactivities in convalescent- versus acute-phase plasma of patients with S. Typhi bacteremia, and detection of a mucosal anti-HlyE immune response present at the time of the clinical presentation of patients with typhoid fever but absent in control patients are of potential diagnostic significance.
Supplementary Material
Acknowledgments
This work was supported by the ICDDR,B, grants from the National Institutes of Health, including the National Institute of Allergy and Infectious Diseases (AI072599 [E.T.R.], AI077883 [E.T.R.], AI058935 [R.C.C. and S.B.C.], NS059429 [J.B.H.]), the PneumoADIP Project of the Johns Hopkins Bloomberg School of Public Health (W.A.B.), an ICDDRB-US Centers for Disease Control and Prevention Cooperative Agreement (W.A.B.), a Training Grant in Vaccine Development from the Fogarty International Center, including an ARRA supplement (TW05572 [A.S., M.S.B., and F.Q.] and TW05572-09S1 [R.C.C.]), Career Development Awards (K01) from the Fogarty International Center (TW007409 [J.B.H.], TW07144 [R.C.L.]) and (K08) from the NIAID (AI089721 [R.C.C.]), a Physician Scientist Early Career Award from the Howard Hughes Medical Institute (R.C.L.), and a grant from the Deutsche Forschungsgemeinschaft (LU 842/1-1 [A.L.]).
Footnotes
Published ahead of print on 23 June 2010.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Ansong, C., H. Yoon, A. D. Norbeck, J. K. Gustin, J. E. McDermott, H. M. Mottaz, J. Rue, J. N. Adkins, F. Heffron, and R. D. Smith. 2008. Proteomics analysis of the causative agent of typhoid fever. J. Proteome Res. 7:546-557. [DOI] [PubMed] [Google Scholar]
- 2.Calderon, I., S. R. Lobos, H. A. Rojas, C. Palomino, L. H. Rodriguez, and G. C. Mora. 1986. Antibodies to porin antigens of Salmonella typhi induced during typhoid infection in humans. Infect. Immun. 52:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charles, R. C., J. B. Harris, M. R. Chase, L. M. Lebrun, A. Sheikh, R. C. LaRocque, T. Logvinenko, S. M. Rollins, A. Tarique, E. L. Hohmann, I. Rosenberg, B. Krastins, D. A. Sarracino, F. Qadri, S. B. Calderwood, and E. T. Ryan. 2009. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica serovar Typhi versus Typhimurium. PLoS One 4:e6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. World Health Organ. 82:346-353. [PMC free article] [PubMed] [Google Scholar]
- 5.Elias, J. E., and S. P. Gygi. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4:207-214. [DOI] [PubMed] [Google Scholar]
- 6.Faucher, S. P., C. Forest, M. Beland, and F. Daigle. 2009. A novel PhoP-regulated locus encoding the cytolysin ClyA and the secreted invasin TaiA of Salmonella enterica serovar Typhi is involved in virulence. Microbiology 155:477-488. [DOI] [PubMed] [Google Scholar]
- 7.Fuentes, J. A., N. Villagra, M. Castillo-Ruiz, and G. C. Mora. 2008. The Salmonella Typhi hlyE gene plays a role in invasion of cultured epithelial cells and its functional transfer to S. Typhimurium promotes deep organ infection in mice Res. Microbiol. 159:279-287. [DOI] [PubMed] [Google Scholar]
- 8.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145:473-480. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haghjoo, E., and J. E. Galan. 2004. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. U. S. A. 101:4614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang, L., M. John, M. Asaduzzaman, E. A. Bridges, C. Vanderspurt, T. J. Kirn, R. K. Taylor, J. D. Hillman, A. Progulske-Fox, M. Handfield, E. T. Ryan, and S. B. Calderwood. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 100:8508-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, J. B., A. Baresch-Bernal, S. M. Rollins, A. Alam, R. C. LaRocque, M. Bikowski, A. F. Peppercorn, M. Handfield, J. D. Hillman, F. Qadri, S. B. Calderwood, E. Hohmann, R. F. Breiman, W. A. Brooks, and E. T. Ryan. 2006. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect. Immun. 74:5161-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornick, R. B., S. E. Greisman, T. E. Woodward, H. L. DuPont, A. T. Dawkins, and M. J. Snyder. 1970. Typhoid fever: pathogenesis and immunologic control. 2. N. Engl. J. Med. 283:739-746. [DOI] [PubMed] [Google Scholar]
- 14.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 15.Kantele, J. M., H. Arvilommi, S. Kontiainen, M. Salmi, S. Jalkanen, E. Savilahti, M. Westerholm, and A. Kantele. 1996. Mucosally activated circulating human B cells in diarrhea express homing receptors directing them back to the gut. Gastroenterology 110:1061-1067. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick, B. D., M. D. Bentley, A. M. Thern, C. J. Larsson, C. Ventrone, M. V. Sreenivasan, and L. Bourgeois. 2005. Comparison of the antibodies in lymphocyte supernatant and antibody-secreting cell assays for measuring intestinal mucosal immune response to a novel oral typhoid vaccine (M01ZH09). Clin. Diagn. Lab. Immunol. 12:1127-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kudva, I. T., B. Krastins, H. Sheng, R. W. Griffin, D. A. Sarracino, P. I. Tarr, C. J. Hovde, S. B. Calderwood, and M. John. 2006. Proteomics-based expression library screening (PELS): a novel method for rapidly defining microbial immunoproteomes. Mol. Cell. Proteomics 5:1514-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuusi, N., M. Nurminen, H. Saxen, M. Valtonen, and P. H. Makela. 1979. Immunization with major outer membrane proteins in experimental salmonellosis of mice. Infect. Immun. 25:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, X. H., I. Arencibia, A. Johansson, S. N. Wai, J. Oscarsson, S. Kalfas, K. G. Sundqvist, Y. Mizunoe, A. Sjostedt, and B. E. Uhlin. 2000. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect. Immun. 68:4363-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRocque, R. C., B. Krastins, J. B. Harris, L. M. Lebrun, K. C. Parker, M. Chase, E. T. Ryan, F. Qadri, D. Sarracino, and S. B. Calderwood. 2008. Proteomic analysis of Vibrio cholerae in human stool. Infect. Immun. 76:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine, M. M., C. Ferreccio, P. Abrego, O. S. Martin, E. Ortiz, and S. Cryz. 1999. Duration of efficacy of Ty21a, attenuated Salmonella typhi live oral vaccine. Vaccine 17(Suppl. 2):S22-S27. [DOI] [PubMed] [Google Scholar]
- 22.Levine, M. M., C. O. Tacket, and M. B. Sztein. 2001. Host-Salmonella interaction: human trials. Microbes Infect. 3:1271-1279. [DOI] [PubMed] [Google Scholar]
- 23.Lin, F. Y., V. A. Ho, H. B. Khiem, D. D. Trach, P. V. Bay, T. C. Thanh, Z. Kossaczka, D. A. Bryla, J. Shiloach, J. B. Robbins, R. Schneerson, and S. C. Szu. 2001. The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 344:1263-1269. [DOI] [PubMed] [Google Scholar]
- 24.Lundgren, A., J. Kaim, and M. Jertborn. 2009. Parallel analysis of mucosally derived B- and T-cell responses to an oral typhoid vaccine using simplified methods. Vaccine 27:4529-4536. [DOI] [PubMed] [Google Scholar]
- 25.Lundin, B. S., C. Johansson, and A. M. Svennerholm. 2002. Oral immunization with a Salmonella enterica serovar Typhi vaccine induces specific circulating mucosa-homing CD4+ and CD8+ T cells in humans. Infect. Immun. 70:5622-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 27.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, S. J., J. Pruckler, W. Bibb, T. M. Nguyen, M. T. Tran, T. M. Nguyen, S. Sivapalasingam, A. Gupta, T. P. Phan, T. C. Nguyen, V. C. Nguyen, D. C. Phung, and E. D. Mintz. 2004. Evaluation of rapid diagnostic tests for typhoid fever. J. Clin. Microbiol. 42:1885-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz, V., A. Isibasi, E. Garcia-Ortigoza, and J. Kumate. 1989. Immunoblot detection of class-specific humoral immune response to outer membrane proteins isolated from Salmonella typhi in humans with typhoid fever. J. Clin. Microbiol. 27:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oscarsson, J., M. Westermark, S. Lofdahl, B. Olsen, H. Palmgren, Y. Mizunoe, S. N. Wai, and B. E. Uhlin. 2002. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars Typhi and Paratyphi A. Infect. Immun. 70:5759-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paliwal, P. K., A. Bansal, S. S. Sagi, S. Mustoori, and I. Govindaswamy. 2008. Cloning, expression and characterization of heat shock protein 60 (groEL) of Salmonella enterica serovar Typhi and its role in protective immunity against lethal Salmonella infection in mice. Clin. Immunol. 126:89-96. [DOI] [PubMed] [Google Scholar]
- 32.Panchanathan, V., B. R. Naidu, S. Devi, A. Di Pasquale, T. Mason, and T. Pang. 1998. Immunogenic epitopes of Salmonella typhi GroEL heat shock protein reactive with both monoclonal antibody and patients sera. Immunol. Lett. 62:105-109. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 34.Parry, C. M., T. T. Hien, G. Dougan, N. J. White, and J. J. Farrar. 2002. Typhoid fever. N. Engl. J. Med. 347:1770-1782. [DOI] [PubMed] [Google Scholar]
- 35.Puohiniemi, R., M. Karvonen, J. Vuopio-Varkila, A. Muotiala, I. M. Helander, and M. Sarvas. 1990. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella typhimurium bacteria. Infect. Immun. 58:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qadri, F., E. T. Ryan, A. S. Faruque, F. Ahmed, A. I. Khan, M. M. Islam, S. M. Akramuzzaman, D. A. Sack, and S. B. Calderwood. 2003. Antigen-specific immunoglobulin A antibodies secreted from circulating B cells are an effective marker for recent local immune responses in patients with cholera: comparison to antibody-secreting cell responses and other immunological markers. Infect. Immun. 71:4808-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravindran, R., and S. J. McSorley. 2005. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology 114:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rollins, S. M., A. Peppercorn, L. Hang, J. D. Hillman, S. B. Calderwood, M. Handfield, and E. T. Ryan. 2005. In vivo induced antigen technology (IVIAT). Cell. Microbiol. 7:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Rollins, S. M., A. Peppercorn, J. S. Young, M. Drysdale, A. Baresch, M. V. Bikowski, D. A. Ashford, C. P. Quinn, M. Handfield, J. D. Hillman, C. R. Lyons, T. M. Koehler, S. B. Calderwood, and E. T. Ryan. 2008. Application of in vivo induced antigen technology (IVIAT) to Bacillus anthracis. PLoS One 3:e1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salerno-Goncalves, R., M. Fernandez-Vina, D. M. Lewinsohn, and M. B. Sztein. 2004. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 173:5852-5862. [DOI] [PubMed] [Google Scholar]
- 41.Salerno-Goncalves, R., M. F. Pasetti, and M. B. Sztein. 2002. Characterization of CD8(+) effector T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 169:2196-2203. [DOI] [PubMed] [Google Scholar]
- 42.Salerno-Goncalves, R., T. L. Wyant, M. F. Pasetti, M. Fernandez-Vina, C. O. Tacket, M. M. Levine, and M. B. Sztein. 2003. Concomitant induction of CD4+ and CD8+ T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain CVD 908-htrA. J. Immunol. 170:2734-2741. [DOI] [PubMed] [Google Scholar]
- 43.Sheikh, A., M. S. Bhuiyan, F. Khanam, F. Chowdhury, A. Saha, D. Ahmed, K. M. Jamil, R. C. LaRocque, J. B. Harris, M. M. Ahmad, R. Charles, W. A. Brooks, S. B. Calderwood, A. Cravioto, E. T. Ryan, and F. Qadri. 2009. Salmonella enterica serovar Typhi-specific immunoglobulin A antibody responses in plasma and antibody in lymphocyte supernatant specimens in Bangladeshi patients with suspected typhoid fever. Clin. Vaccine Immunol. 16:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, S. P., Y. U. Williams, S. Miller, and H. Nikaido. 2003. The C-terminal domain of Salmonella enterica serovar typhimurium OmpA is an immunodominant antigen in mice but appears to be only partially exposed on the bacterial cell surface. Infect. Immun. 71:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokal, R. R., and F. J. Rohlf (ed.). 1994. Biometry: the principles and practice of statistics in biological research, 3rd ed. Freeman, New York, NY.
- 46.Spano, S., J. E. Ugalde, and J. E. Galan. 2008. Delivery of a Salmonella typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30-38. [DOI] [PubMed] [Google Scholar]
- 47.Storey, J. D. 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Statist. 31:2013-2035. [Google Scholar]
- 48.Svenson, S. B., M. Nurminen, and A. A. Lindberg. 1979. Artificial Salmonella vaccines: O-antigenic oligosaccharide-protein conjugates induce protection against infection with Salmonella typhimurium. Infect. Immun. 25:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sztein, M. B. 2007. Cell-mediated immunity and antibody responses elicited by attenuated Salmonella enterica serovar Typhi strains used as live oral vaccines in humans. Clin. Infect. Dis. 45(Suppl. 1):S15-S19. [DOI] [PubMed] [Google Scholar]
- 50.Tacket, C. O., M. B. Sztein, G. A. Losonsky, S. S. Wasserman, J. P. Nataro, R. Edelman, D. Pickard, G. Dougan, S. N. Chatfield, and M. M. Levine. 1997. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect. Immun. 65:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talawadekar, N. N., P. J. Vadher, D. U. Antani, V. V. Kale, and S. A. Kamat. 1989. Chloramphenicol resistant Salmonella species isolated between 1978 and 1987. J. Postgrad. Med. 35:79-82. [PubMed] [Google Scholar]
- 52.Tang, S. W., S. Abubakar, S. Devi, S. Puthucheary, and T. Pang. 1997. Induction and characterization of heat shock proteins of Salmonella typhi and their reactivity with sera from patients with typhoid fever. Infect. Immun. 65:2983-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velayudhan, J., M. Castor, A. Richardson, K. L. Main-Hester, and F. C. Fang. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63:1495-1507. [DOI] [PubMed] [Google Scholar]
- 54.Viret, J. F., D. Favre, B. Wegmuller, C. Herzog, J. U. Que, S. J. Cryz, Jr., and A. B. Lang. 1999. Mucosal and systemic immune responses in humans after primary and booster immunizations with orally administered invasive and noninvasive live attenuated bacteria. Infect. Immun. 67:3680-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Rhein, C., S. Bauer, E. J. Lopez Sanjurjo, R. Benz, W. Goebel, and A. Ludwig. 2009. ClyA cytolysin from Salmonella: distribution within the genus, regulation of expression by SlyA, and pore-forming characteristics. Int. J. Med. Microbiol. 299:21-35. [DOI] [PubMed] [Google Scholar]
- 56.von Rhein, C., K. P. Hunfeld, and A. Ludwig. 2006. Serologic evidence for effective production of cytolysin A in Salmonella enterica serovars Typhi and Paratyphi A during human infection. Infect. Immun. 74:6505-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, W., L. Ni, and R. L. Somerville. 1993. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc. Natl. Acad. Sci. U. S. A. 90:5796-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.