Abstract
To further refine our current nanoparticle-based HIV-1 p24 antigen assay, we investigated immune responses to p24 to identify diagnostically significant immune dominant epitopes (IDEs) in HIV-infected human sera, to address cross-reactivity of anti-p24 antibodies to different subtypes, and to identify new biomarkers that distinguish acute from chronic HIV infection for more accurate incidence estimation. We identified two major linear epitope regions, located in the CypA binding loop and adjacent helices and at the end of the C-terminal domain. Most sera (86%) from acutely HIV-1-infected individuals reacted with multiple peptides, while 60% and 30% of AIDS patient samples reacted with multiple and single peptides, respectively. In contrast, 46% and 43% of chronically HIV-1-infected individuals reacted with one and none of the peptides, respectively, and only 11% reacted with multiple p24 peptides, indicating a progression of immune responses from polyclone-like during acute infection to monoclone-like or a nonresponse to linear epitopes during chronic infection. Anti-p24 antibodies (subtype B) show broad cross-reactivity to different HIV-1 subtypes, and the synergistic action of different combinations of anti-HIV antibodies improves capture and detection of divergent HIV-1 subtypes. Our results indicate that the modified peptide immunoassay is sensitive and specific for the rapid identification of HIV-1 p24 IDEs and for investigation of immune responses to p24 during natural HIV-1 infection. The data provide the foundation for development and refinement of new assays for improved p24 antigen testing as future tools for rapid and accurate diagnosis as part of early intervention strategies and estimations of incidence.
Capsid protein (CA), or p24 antigen, of human immunodeficiency virus type 1 (HIV-1) is the most abundant viral protein, since each virus contains about 1,500 to 3,000 p24 molecules (30, 37). During early and late stages of HIV infection, it is always present at relatively high levels in the blood, making it a potential viral marker for diagnosis, blood donor screening, monitoring disease progression, and evaluating antiretroviral therapy (1, 5, 6, 25). However, conventional enzyme-linked immunosorbent assays (ELISAs) for HIV-1 p24 detection have relatively low sensitivity and have been replaced by nucleic acid testing (NAT) in the United States (29). Over the past decade, the performance of p24 assays has been improved significantly by implementing immune complex disruption methods (23, 26), using more effective lysis buffers (27), and incorporating tyramide-mediated signal amplification (TSA) (4). We showed that by using gold nanoparticles (NPs), the detection limit for p24 antigen could be reduced to 0.1 pg/ml (35) and the window period (the time between HIV exposure and detection of antibody seroconversion) could be shortened by at least 3 days (35). Antigen assays could also be useful for HIV diagnostics in pediatrics and for testing the blood supply in resource-limited settings where NAT is not available or practical. By using nanotechnology and nanoparticles, the sensitivity of the immunoassay could be improved while making it less expensive and simpler than current ELISA methods (32, 33, 35). However, assay accuracy depends on the quality of anti-p24 antibodies and their immune response to p24 antigen. To refine and develop a more sensitive HIV-1 p24 antigen assay, further study of immune responses to p24 antigen to identify the immune dominant epitopes (IDEs) in HIV-infected human sera is necessary, since B-cell epitopes of p24 that have been identified are based mainly on the characterization of immune responses to murine monoclonal anti-p24 antibodies (Los Alamos HIV Molecular Immunology Database [http://www.hiv.lanl.gov/content/immunology/maps/ab/p24.html]). Such studies are limited and show controversial results (9, 13, 15, 18).
The second issue to be addressed with p24 antigen testing is the cross-reactivity of anti-p24 antibodies with different viral subtypes due to the broad genetic diversity of HIV-1 (21, 24). Cross-reactivity has been evaluated with several commercially available HIV-1 assays (14, 19), but detailed information on the anti-p24 antibodies used was not provided. Finally, there is a need to identify new biomarkers for acute HIV infection to more accurately estimate incidence rates in order to monitor the utility of prevention measures. Several unique epitopes of HIV-1 p24 antigen have been found to be immunodominant and may be recognized early in the course of natural infection or associated with disease progression (12, 13). These results indicate that assays utilizing specific epitopes of p24 and anti-p24 antibodies may help in the diagnosis of recent or acute HIV infection.
Here we describe the characterization of major IDEs of HIV-1 p24, studies to evaluate the immune response profile during acute and chronic HIV-1 infection, and the cross-reactivities of monoclonal anti-p24 antibodies among different subtypes, as determined using a rapid, sensitive, NP-based immunoassay. The implications for p24 detection and assay development are also discussed.
MATERIALS AND METHODS
Antibodies, antigens, and europium nanoparticles.
The sources of monoclonal anti-HIV p24 antibodies are listed in Table 1. Polyclonal anti-HIV p24 antibodies were either made by the authors or purchased. Purified HIV-immune IgG (HIVIG) 3957 was obtained from the NIAID AIDS Research and Reference Reagent Program and prepared from pooled plasmas of asymptomatic, antibody-positive donors with high titers of anti-p24 antibody. Nine overlapping peptides with consensus sequences of HIV-1 p24 from major subtypes and recombinant forms of HIV-1 group M (Fig. 1) were synthesized and purified by high-performance liquid chromatography (HPLC) at CHI Scientific Inc. (Maynard, MA). Recombinant p24 protein was purchased from Virogen (Watertown, MA). Goat anti-human immunoglobulin (Ig) and biotinylated goat anti-rabbit and rabbit anti-human antibodies were purchased from Pierce (Rockford, IL). Biotinylated anti-mouse Ig light chains λ and κ and biotinylated anti-human Ig light chains λ and κ were purchased from BioLegend (San Diego, CA). Europium (Eu3+) NPs (107 nm in diameter) were obtained from Seradyn Inc. (Indianapolis, IN). The preparation and characterization of Eu3+ NPs coated with streptavidin (SA) have been described previously (11).
TABLE 1.
Characteristics of HIV-1-infected individuals and AIDS patients
| Parameter | Value for groupa |
||||
|---|---|---|---|---|---|
| Acutely infected individuals | AIDS patients | Chronically infected individuals |
|||
| United States | Cameroon | Total | |||
| Median (range) viral load (copies/ml) | 5,800 (390-750,000) | 325,000 (91,300-679,000) | 1,213 (50-683,333) | ND | NA |
| Median (range) CD4+ count (cells/ml) | ND | 43 (6-279) | 498 (235-1,016) | ND | NA |
| Median (range) time (days) to seroconversion | 15 (3-26) | NA | NA | NA | NA |
| No. of patients with current HAART therapy/total no. of patients | None | 2/10 | 13/20 | NA | NA |
| No. of patients with response/total no. of patients (%) | |||||
| Polyclone-like response | 6/7 (86) | 6/10 (60) | 2/20 (10) | 2/17 (12) | 4/37 (11) |
| Monoclone-like response | 0 | 3/10 (30) | 9/20 (45) | 8/17 (47) | 17/37 (46) |
| No response to peptides | 1/7 (14) | 1/10 (10) | 9/20 (45) | 7/17 (41) | 16/37 (43) |
ND, not done; NA, not applicable.
FIG. 1.
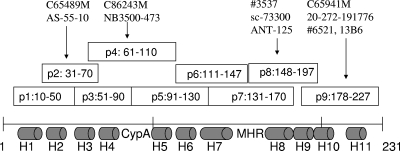
HIV-1 capsid protein, synthesized peptides, and anti-p24-reacting peptides. The HIV-1 capsid protein has 231 aa and consists of 11 helices (H1 to H11; gray cylinders) and two unique motifs, namely, the CypA binding loop and the major homology region (MHR). Nine overlapping peptides (p1 to p9; open squares), which extend from aa 10 to aa 227, were synthesized and used to identify the peptides that reacted with (indicated by black arrows) monoclonal anti-p24 antibodies (top).
Samples and viruses.
A total of 54 HIV-1-positive serum specimens were tested and included the following: 7 samples from consenting, acutely infected individuals, obtained from the American Red Cross; 10 samples from AIDS patients and 20 samples from HIV-1-positive individuals without AIDS who participated in research protocols approved by the National Cancer Institute Institutional Review Board and who gave informed consent; and 17 de-identified, asymptomatic HIV-1-positive samples from donors in Cameroon (Table 1). An FDA CBER HIV panel consisting of cell culture supernatants of 21 viruses, representing HIV-1 group M subtypes A, C, D, E, F, and G, CRF01_AE, CRF02_AG, HIV-1 groups O and N, and HIV-2, was also tested (16).
ENIA.
The Eu3+ NP-based immunoassay (ENIA) methodology and sensitive detection of HIV-1 p24 by ENIA have been described previously (32-34). ENIA is 30- to 100-fold more sensitive than conventional ELISA (32, 33). To identify epitopes of and characterize immune responses to p24, well plates coated with 2.5 to 5.0 μg/ml of p24 peptide or recombinant p24 protein were incubated with anti-HIV p24 antibodies or serum samples, followed by incubation with biotinylated rabbit anti-human or anti-mouse antibody or biotinylated goat anti-rabbit antibody and 107 SA-coated Eu3+ NPs at 37°C for 30 min. Finally, the wells were washed five times with 1× phosphate-buffered saline (PBS)-0.05% Tween 20. Fluorescence was measured by use of a Victor fluorometer (PerkinElmer, Waltham, MA) in a time-resolved mode. The cutoff (CO) value was the average signal intensity (S) of negative controls plus 3 standard deviations (SD). Samples with S/CO values of ≥1.00 were considered positive.
For the competitive inhibition experiment, wells coated with 2.5 μg/ml of p24 recombinant protein were incubated with 100 ng/ml of monoclonal anti-HIV p24 antibody or 1:100-diluted HIV-1-positive sera and 25 μg/ml of each of the nine peptides. The percentage of inhibition was calculated as follows: [(fluorescenceuninhibited − fluorescenceinhibited)/fluorescenceuninhibited] × 100.
To test the cross-reactivity of anti-p24 antibodies with different HIV subtypes, the above ENIA was modified by coating wells with 2 μg/ml of monoclonal anti-p24 antibodies or 2.5 μg/ml of HIVIG, followed by incubation with culture supernatants of different HIV-1 subtypes for 1 h at 37°C with shaking at 500 rpm and a 30-min incubation with rabbit anti-p24 antibody (4250), biotinylated goat anti-rabbit antibody, and SA-coated Eu3+ NPs at 37°C (with five washes between steps).
For detection of light chain isotypes, well plates coated with 1 μg/ml of goat anti-human Ig or 2 μg/ml of recombinant p24 were incubated with 1:100-diluted HIV-positive sera at 37°C for 1 h, followed by incubation with biotinylated anti-human λ and κ antibodies (200 ng/ml and 1 μg/ml, respectively) and SA-coated Eu+ NPs. The ratio was calculated by dividing fluorescence counts of λ by those of κ.
RESULTS
Antigenicity and IDEs of HIV-1 p24.
Polyclonal anti-HIV antibodies from different sources reacted well with recombinant p24 antigen and most of the nine peptides (data not shown), indicating the strong antigenicity of p24 and a broad range of epitopes within the antigen (15). As expected, each monoclonal anti-HIV antibody reacted with recombinant p24 and one or two overlapping peptides, although some antibodies did not react with any of the peptides (Fig. 1 and Table 2). The most active peptides were p2, p3, p4, p8, and p9, which cover the region of helix 2/3, helix 4/5, and the CypA binding loop in the N-terminal domain and the end of the C-terminal domain of CA (7, 15). Several monoclonal anti-HIV antibodies (ANT-152 and SC73300, C86243M and NB500-473, and C65690M and 20-511-241432) obtained from the same clones but produced by different companies were active against the same peptides (Table 2) and displayed the same cross-reactivities against various HIV-1 subtypes (see below). Interestingly, two additional antibodies (C86243M and NB500-473), produced by a synthetic peptide from the C-terminal domain, were found to react with the two N-terminal domain peptides p3 and p4 of p24, and it is not known whether the antibody source information from the vendors was incorrect. In any event, these results indicate that the peptide-based immunoassay is sensitive and specific for identifying immune responses and determining the major epitopes of p24. Four monoclonal anti-HIV antibodies (C65690M, 20-511-241432, 012-A, and 13G4) were captured by the recombinant p24 protein but not by any of the peptides, despite the overlapping sequences being designed to avoid not detecting epitopes that span two adjacent peptides. The reactivity of this group with the p24 protein could not be inhibited by coincubation with monoclonal anti-p24 antibodies and p24 peptides, suggesting that these antibodies may be produced against “discontinuous” or conformational epitopes of p24 (10, 15, 28).
TABLE 2.
Characterization of monoclonal anti-HIV-1 p24 antibodies
| Antibody | Vendor | Immunogen | Clone | Isotype | Reacting peptide (aa)a |
|---|---|---|---|---|---|
| 6521 | NIH AIDS Research and Reference Reagent Program | P24 (HXB-3) | 24-2 | IgG2bκ | p9 (178-227) |
| 13B6 | Institute of Human Virology, University of Maryland School of Medicine | Unknown | 13B6 | IgG1κ | p9 (178-227) |
| C65941M | Meridian Life Science, Inc. | Unknown | 491 | IgG1λ | p9 (178-227) |
| 20-272-19776 | GenWay Biotech, Inc. | Recombinant p24 | 473 | IgG1 | p9 (178-227) |
| 3537 | NIH AIDS Research and Reference Reagent Program | Unknown | 183-H12-5C | IgG1κ | p8 (148-197) |
| ANT-152 | ProSpec-Tany TechnoGene Ltd. | Recombinant p24 | YDHIV gp24 | IgG1 | p8 (148-197) |
| SC-73300 | Santa Cruz Biotechnology, Inc. | Recombinant p24 | YDHIV gp24 | IgG1 | p8 (148-197) |
| C86243M | Meridian Life Science, Inc. | C-terminal peptide | ND1 | IgG1 | p3/p4 (51-110) |
| NB500-473 | Novus Biologicals | C-terminal peptide | ND1 | IgG1 | p3/p4 (51-110) |
| C65489M | Meridian Life Science, Inc. | p24 protein | BDI489 | IgG1 | p2 (31-70) |
| AS55-10 | Microbix System, Inc. | Recombinant p24 | MX-0316 | IgG1 | p1/p2 (10-70) |
| 1103 | ImmunoDiagnostics, Inc. | HIV-1 IIIB p24 | Unknown | IgG | None |
| 012-A | Virogen | Recombinant p24 | 1A1 | IgG1 | None |
| 20-511-241432 | GenWay Biotech, Inc. | Unknown | BDI690 | IgG1 | None |
| C65690M | Meridian Life Science, Inc. | Unknown | BDI690 | IgG1κ | None |
| 13G4 | Institute of Human Virology, University of Maryland School of Medicine | Unknown | 13G4 | IgG1κ | None |
None, does not react to any of the peptides tested, but does react to HIV-1 p24 protein.
Cross-reactivity of anti-HIV antibodies with different HIV subtypes.
Cross-reactivities were determined by incubating HIV cell culture supernatants of different subtypes and groups in well plates coated with monoclonal anti-p24 antibodies or HIVIG. In general, monoclonal anti-p24 antibodies targeting different epitopes captured all of the HIV-1 subtypes tested, including subtypes A to G of group M and groups O and N, but not HIV-2 (Table 3). Several anti-p24 antibodies were unable to detect some HIV-1 subtypes. For example, antibody 1103 showed reactivity against subtypes B and A of HIV-1 but either did not react with or had a very low affinity for other subtypes. Antibodies C65690M, 20-511-241432, 3537, and ANT-152 showed no reactivity against several HIV subtypes. In particular, these four antibodies did not detect HIV-1 subtype D non-syncytium-inducing virus (D, NSI), and both 3537 and C65690M/20-511-241432 failed to detect HIV-1 subtype E (E, NSI). However, the combination of C65690M and 3537 or ANT-152 at a ratio of 3:1 displayed reactivity against HIV-1 subtypes and produced significantly higher signals. Interestingly, purified HIVIG interacted with all HIV-1 subtypes tested, although the pooled HIV Ig reacted with only a single peptide (p9). These results confirmed the strong cross-reactivity of subtype B-specific anti-p24 antibodies against different HIV-1 subtypes and the synergistic action of different combinations of anti-HIV antibodies targeting various epitopes, improving the capture and detection of divergent HIV-1 subtypes.
TABLE 3.
Cross-reactivities of anti-HIV-1 antibodies with different HIV subtypes
| HIV group, subtype, or recombinant form (isolate from CBER HIV panel) |
S/CO ratio with antibodya |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3957 | 3537 | C65690M | C65941M | ANT-152 | 1103 | 3537 + C65690M | ANT-152 + C65690M | PEb | ZMc | |
| A, NSI (UG/0229/92) | 35 | 26 | 56 | 16 | ND | 10 | ND | ND | 28 | 50 |
| B, NSI | 49 | 58 | 73 | 65 | 75 | 55 | 54 | 65 | 45 | 69 |
| B (MN) | 48 | 63 | 76 | 67 | 83 | 56 | 57 | 64 | 51 | 69 |
| C, SI (192431) | 10 | 3.5 | 17 | 5 | 5 | <1 | 42 | 58 | 7 | 15 |
| C, NSI (SE/364/90) | 20 | 13 | 36 | 3.3 | 17 | 1.3 | 52 | 68 | 13 | 22 |
| D, SI (UG/021/92) | 10 | 11 | <1 | <1 | 13 | <1 | 72 | 115 | 15 | 27 |
| D, NSI (UG/035/92) | 1.3 | <1 | <1 | <1 | <1 | 3.7 | 3.5 | 6.3 | 1.5 | 1.5 |
| E, SI (TH/1465/95) | 21 | 4.8 | 22 | <1 | 9 | 1.8 | 64 | 112 | 19 | 25 |
| E, NSI (TH/022/92) | 1.6 | <1 | <1 | <1 | 1.6 | <1 | 2.4 | 2.6 | <1 | <1 |
| F, SI (BZ/126/89) | 31 | 23 | 34 | 16 | 31 | <1 | 62 | 103 | 25 | 55 |
| F, NSI (BZ/161/90) | 2.2 | <1 | 4 | 1.2 | <1 | <1 | 9.3 | 19 | 1.8 | 3 |
| G (HH8793) | 24 | 17 | 56 | 17 | 32 | <1 | 7 | 58 | 27 | 54 |
| G (G3, Nigeria) | 2.9 | <1 | 7 | <1 | <1 | <1 | 13 | 25 | 6 | 4.8 |
| CRF01 (14th day) | 16 | 5 | 32 | <1 | 11 | 3 | 6 | 40 | 25 | 14 |
| CRF01 (7th day) | 23 | 6.5 | 40 | <1 | 6 | 3.4 | 6 | ND | 28 | 14 |
| CRF02 (2325) | 32 | 43 | 20 | 23 | 62 | 12 | 15 | 60 | 38 | 72 |
| CRF02 (NYU360, 14th day) | 21 | 14 | 65 | <1 | 27 | 1.8 | 41 | 51 | 37 | 65 |
| CRF02 (NYU360, 7th day) | 3.2 | <1 | 8 | <1 | <1 | <1 | 34 | 40 | 7 | 12 |
| O (GER_5267) | 13 | 6 | 41 | 3.3 | 11 | 1.2 | 5 | 39 | 18 | 32 |
| N (Sinnousi) | 19 | 8.7 | 36 | 3.4 | 14 | 1.5 | 5 | 39 | 21 | 33 |
| HIV-2 (B9) | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
Microtiter plates were coated with anti-HIV antibodies and incubated with 5 μl (1:20 dilution) of HIV from the CBER HIV panel. Cross-reactions were assessed by ENIA and are presented as S/CO ratios. S/CO ratios of <1 indicate no cross-reaction. ND, not done.
PerkinElmer HIV p24 EIA kit.
HIV p24 kit of ZeptoMatrix.
Immune responses to p24 during natural HIV-1 infection.
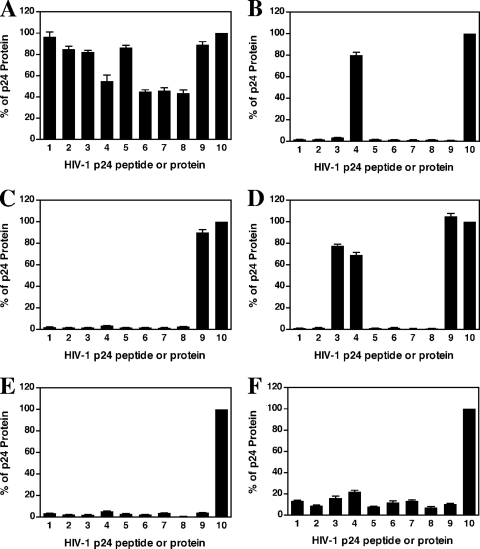
All 34 HIV-positive serum samples showed strong reactivity against the recombinant p24 protein, but they had significantly different affinities toward the p24 peptides (Fig. 2). Three immune response patterns were observed: polyclone-like responses (Fig. 2A), monoclone-like responses (Fig. 2B, C, and D), and responses against conformational epitopes (Fig. 2E and F). Like polyclonal anti-HIV antibodies, samples with a polyclone-like response showed strong reactivities toward multiple HIV-1 peptides, and this was seen in 30% (16/54 sera) of the sera tested, mainly from acutely infected individuals (86%) and some AIDS patients (60%) (Table 1; see Table S1 in the supplemental material). A monoclone-like response, observed in 37% (20/54 sera) of the samples tested, was clearly reactive toward one or two peptides and was predominant in chronically infected individuals (46%) and some AIDS patients (30%). Reactivity toward two different peptides was also observed in 7.4% (4/54 sera) of the infected sera (Fig. 2D). Finally, 33% (18/54 sera) of the samples were reactive only against recombinant p24 and either did not react (22.2% [12/54 sera]) (Fig. 2E) or reacted very weakly (11.1% [6/54 sera]) with the peptides (Fig. 2F). Among samples from acutely infected individuals, 86% (6/7 sera) reacted with multiple peptides and 14% (1/7 sera) did not react with any peptides (Table 1). However, 60% (6/10 sera) and 30% (3/10 sera) of the AIDS patient samples showed responses against multiple and single peptides, respectively, with 10% of the AIDS patient sera failing to interact with any of the p24 peptides. In contrast, only 11% (4/37 sera) of chronically HIV-infected individuals responded against multiple p24 peptides, and the rest had reactivity to either one (46% [17/37 sera]) or none (43% [16/37 sera]) of the peptides (Table 1; see Table S1 in the supplemental material). The differences in immune responses toward p24 peptides among the three groups were statistically significant (P < 0.001; Fisher's exact test [SAS; SAS Institute, Inc., Cary, NC]). These results indicate a clear switch and progression of immune responses toward the peptides, from a polyclone-like pattern during acute infection to a monoclone-like response or response against conformational epitopes during chronic infection. Similar to what we found for monoclonal anti-HIV p24 antibodies, among the 11 HIV-1-positive sera with a monoclone-like response, 85% (17/20 sera) reacted with peptide p9, 15% (3/20 sera) reacted with peptides p3 and p4, and 4 reacted with both p9 and p3/4. This suggests that peptides p3/4 and p9 harbor the predominant linear epitopes of p24 during natural HIV-1 infection. Another interesting finding was that purified HIVIG from pooled asymptomatic HIV-positive donors reacted with only one peptide, p9. This was consistent with the results from testing of asymptomatic, Cameroonian sera, which showed that most sera were also reactive only against p9. Thus, the C-terminal domain, in particular the helix 10/11 region, may contain dominant linear immunogenic B-cell epitopes that are active during chronic HIV-1 infection. Helix 4/5, including the CypA binding area, may be another dominant epitope region. It should be noted that for the nine sera that were reactive against recombinant p24 protein but not against any of the peptides, the interactions with p24 protein could not be inhibited by any of the nine peptides in the competitive inhibition experiment, suggesting the possibility of conformational epitopes.
FIG. 2.
Immune responses to HIV-1 p24 peptides and recombinant protein. Immune responses were quantified as described in Materials and Methods and were normalized by calculating the S/CO ratios relative to the level of HIV-1 recombinant p24 protein, which was arbitrarily set at 100%. The data were plotted in bar graphs, and error bars indicate standard deviations for three independent experiments. Numbers 1 to 9 and 10 on the x axis represent peptides p1 to p9 and p24 protein, respectively. (A) Polyclone-like response; (B, C, and D) monoclone-like response; (E and F) responses against conformational epitopes.
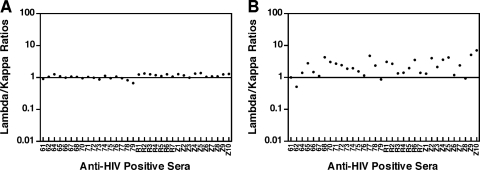
To further determine whether the monoclone-like immune response to p24 was caused by clonally expanded B-cell clones, the contribution of light chain isotypes (λ and κ) was measured in the 34 HIV-1-positive serum samples. Since total human IgG usually consists of equal amounts of λ and κ light chains, their ratio is around 1.0 (22). As expected, a 1:1 ratio (1.09 ± 016) was observed in the sample wells coated with anti-human IgG (Fig. 3 A). However, when recombinant p24 antigen was used to capture anti-p24 antibody, a skewed ratio (2.48 ± 1.46) resulted (Fig. 3B). The difference in ratios between total human IgG and anti-p24 was significant (P < 0.0001; paired t test [GraphPad Prism; GraphPadSoftware, Inc., San Diego, CA]). Only 5 (14.7%) of the 34 samples displayed a 1:1 ratio (1.08 ± 0.11) of λ/κ chains for anti-HIV p24 antibodies. Furthermore, the skewed ratios did not show significant differences between acutely infected donors, AIDS patients, and asymptomatic, HIV-positive Cameroonian individuals (P > 0.05; multiple comparison test [GraphPad Prism]). This confirmed the production of clonally restricted antibodies following HIV-1 infection (22), but it may not fully explain the monoclone-like immune response observed during chronic infection. The monoclone-like immune response may reflect immune responses to predominant immune epitopes and inhibition of nondominant epitopes of p24 during chronic infection.
FIG. 3.
Lambda/kappa ratios in HIV-1-positive sera. (A) Lambda/kappa ratios for whole human Ig. Microplates were coated with goat anti-human Ig and incubated with dilutions of HIV-1-positive sera. Lambda/kappa ratios for antibodies were calculated as described in Materials and Methods. Identification numbers of the sera are shown on the x axis. (B) Lambda/kappa ratios for anti-p24 antibodies. Microplates were coated with recombinant HIV-1 p24 and incubated with dilutions of HIV-1-positive sera. The assay was done as described for panel A.
DISCUSSION
We adapted a peptide-capture immunoassay to determine IDEs of HIV-1 p24 antigen and used monoclonal anti-p24 antibodies to investigate cross-reactivity with various HIV subtypes. By testing HIV-positive sera and monoclonal anti-p24 antibodies, we identified two major linear immunodominant regions in CA: (i) helix 4/5, including the CypA binding loop, within the N-terminal domain, and (ii) the end of the C-terminal domain. Similar results were reported by Liu et al. (18), who found that 62% (8/13 sera) of HIV-1-positive sera recognized DC-22 (amino acids [aa] 197 to 218), a peptide very similar to p9. In their study, none of 67 serum samples reacted with GA-12 (aa 94 to 104), which overlaps with our p4 and p5 peptides. However, GA-12 reacted with 84% of HIV-positive sera from Australia. Graham et al. found two short p24 peptides, 18 and 21 (which cover aa 171 to 190 and aa 201 to 215, respectively, and overlap with our p9 peptide), that reacted with 6 of 12 HIV-positive asymptomatic samples from Scotland (9). Janvier et al. (13) reported a dominant epitope sequence, AAEWDRVHP (aa 77 to 85), that is very close to the CypA binding loop and overlaps with our peptide p4. However, the most reactive peptide from their study was peptide 12, which covers aa 111 to 125 and overlaps with our peptide p5/6. The identification of dominant immune epitopes may help in the selection of antibodies for improved HIV assay development.
Also, different immune response profiles were observed for acute and chronic HIV-1 infections. Specifically, there was a significant shift in immune responses, from a polyclone-like pattern during acute infection to a monoclone-like pattern or nonresponse against linear epitopes during chronic infection. This may be due to a vigorous humoral immune response during acute HIV-1 infection which controls initial viral replication and depletes circulating p24 antigen. It is not clear whether a higher viral load, particularly during the acute phase of HIV-1 infection, could induce a polyclonal response to p24 antigen. During chronic infection, viral mutations result in the emergence of escape variants that avoid neutralizing antibodies and host immune responses. Although anti-p24 is not a neutralizing antibody, the monoclone-like response to p24 could be the outcome of immune selection. Our study of the immune responses to HIV-1 envelope protein (the possible neutralizing antibody) during acute and chronic infection with HIV-1 is ongoing. Therefore, immune dominant epitopes found in chronically infected individuals may be useful biomarkers of HIV variants that escape neutralizing antibodies and could serve as an aid in distinguishing recent from longstanding infections, through further discrimination of specific epitope motifs. Furthermore, in our study, almost 50% of the asymptomatic Cameroonian samples, in which the dominant virus is HIV-1 CRF02_AG (20), did not recognize the linear epitopes of p24 or reacted with very few specific epitopes. Similar results were found for HIV-1-positive U.S. individuals without AIDS. Liu et al. found that 80% of serum samples from HIV-1-positive individuals most likely infected with the predominant strains of CRF07_BC/CRF08_BC or CRF01_AE in Yunnan, China (38), did not respond to five p24 peptides tested (18). These results suggest that infection with different subtypes of HIV-1 may generate similar immune responses to HIV-1 p24 antigen, although further studies need to be performed to confirm these observations.
Finally, the cross-reactivity of anti-HIV p24 antibodies among various HIV-1 subtypes was evaluated. Although HIV is a highly divergent virus, with numerous groups, subtypes, and recombinant forms, subtype-specific anti-HIV p24 antibodies show broad cross-reactivity. Ly et al. (19) showed that two antigen-antibody combo assays were able to detect all 31 HIV-1 isolates (including subtypes A to D, F, and G, CRF01, CRF02, and HIV-1 group O) tested in their study. Similar results were reported by Kwon et al. (14), who evaluated fourth-generation (combination antigen-antibody) assays against the same panel of isolates. Furthermore, several anti-p24 antibodies that showed strong reactivity with the subtype B virus in our study have been reported to efficiently capture p24 in different assays (2, 3, 17), providing a foundation for the application of current HIV subtype B-specific antibodies in HIV diagnosis. However, variations in p24 monoclonal antibody reactivity among different subtypes have also been reported (31). Tersmette et al. found that 85% (55/65 isolates) of HIV isolates from Dutch and Belgian individuals (but only 4 of 9 Cameroonian isolates) were recognized by five monoclonal anti-p24 antibodies, although 55% (5/9 isolates) of isolates from Central Africa were not recognized by at least one of the antibodies (36). An HIV-1 antigen-antibody combo assay failed to detect 3/7 subtype B, 1/5 subtype C, 2/3 subtype F, 1 subtype G, 1/8 CRF01, and one HIV-1 group O sample. Impaired sensitivity for subtype C was also found in a p24 assay (19). These results may reflect the inability of monoclonal anti-p24 antibodies to efficiently capture p24 proteins from certain subtypes in a heterogeneous population of HIV-1 strains. Consequently, it may be necessary to use combinations of monoclonal antibodies against different epitopes to detect as many variants as possible and to achieve a high sensitivity. Even with fourth-generation immunoassays, there are HIV-1 mutants that escape detection (8). Our data indicate that purified HIV Ig may be useful for detecting p24 antigen, since HIV Ig from pooled plasma contains anti-p24 antibodies against different epitopes and clades and may be broader and more accurate. However, quality control and batch-to-batch variations in production of HIV Ig could lead to inconsistent results.
In conclusion, the peptide-based immunoassay described here was shown to be sensitive and specific for the rapid determination of HIV-1 p24 IDEs and for investigation of immune responses to p24 during natural HIV-1 infection. The identification of two major epitope regions may be of value for development of new assays. Knowledge of immune responses toward p24 is important to further improve our NP-based HIV detection platforms. Further studies to identify epitopes that distinguish between acute and chronic HIV-1 infections may generate new biomarkers and methods for incidence testing and early detection. The cross-reactivity of anti-p24 antibodies with numerous HIV-1 subtypes provides the foundation for development and refinement of new assays for detection of p24 antigen.
Supplementary Material
Acknowledgments
This study was funded by an interagency agreement with the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID), NIH, and was supported in part by the Intramural Research Program of the National Cancer Institute (NCI), NIH.
We acknowledge Robert Gorelick for providing p24 protein and Kathleen Wyvill for contributing samples from AIDS patients. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIVIG, obtained from NABI and NHLBI; HIV-1 p24 monoclonal antibody (3537 [clone 183-H12-5C]), obtained from Bruce Chesebro and Kathy Wehrly; HIV-1 p24 Gag monoclonal antibody (6521 [clone 24-4]), obtained from Michael H. Malim; and HIV-1SF2 p24 antiserum (4250). We thank Carol Weiss, Krishna Devadas, and Hira Nakhasi for their reviews of the manuscript.
The findings and conclusions in this article have not been disseminated formally by the FDA and should not be construed to represent any Agency determination or policy.
Footnotes
Published ahead of print on 9 June 2010.
Supplemental material for this article may be found at http://cvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Allain, J. P., Y. Laurian, D. A. Paul, F. Verroust, M. Leuther, C. Gazengel, D. Senn, M. J. Larrieu, and C. Bosser. 1987. Long-term evaluation of HIV antigen and antibodies to p24 and gp41 in patients with hemophilia. Potential clinical importance. N. Engl. J. Med. 317:1114-1121. [DOI] [PubMed] [Google Scholar]
- 2.Barletta, J., A. Bartolome, and N. T. Constantine. 2009. Immunomagnetic quantitative immuno-PCR for detection of less than one HIV-1 virion. J. Virol. Methods 157:122-132. [DOI] [PubMed] [Google Scholar]
- 3.Biancotto, A., B. Brichacek, S. S. Chen, W. Fitzgerald, A. Lisco, C. Vanpouille, L. Margolis, and J. C. Grivel. 2009. A highly sensitive and dynamic immunofluorescent cytometric bead assay for the detection of HIV-1 p24. J. Virol. Methods 157:98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrow, M. N., T. D. Harris, K. J. Shaughnessy, and G. J. Litt. 1989. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J. Immunol. Methods 125:279-285. [DOI] [PubMed] [Google Scholar]
- 5.de Wolf, F., J. M. Lange, J. T. Houweling, R. A. Coutinho, P. T. Schellekens, J. van der Noordaa, and J. Goudsmit. 1988. Numbers of CD4+ cells and the levels of core antigens of and antibodies to the human immunodeficiency virus as predictors of AIDS among seropositive homosexual men. J. Infect. Dis. 158:615-622. [DOI] [PubMed] [Google Scholar]
- 6.Fiebig, E. W., D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871-1879. [DOI] [PubMed] [Google Scholar]
- 7.Gallina, A., F. Rossi, M. Mariani, F. Bonelli, G. Achilli, E. Cattaneo, and G. Milanesi. 1990. Major antigenic domain recognized by monoclonal antibodies maps within the carboxy-terminal moiety of a recombinant human immunodeficiency virus-1 p24 protein. J. Med. Virol. 32:164-170. [DOI] [PubMed] [Google Scholar]
- 8.Gaudy, C., A. Moreau, S. Brunet, J. M. Descamps, P. Deleplanque, D. Brand, and F. Barin. 2004. Subtype B human immunodeficiency virus (HIV) type 1 mutant that escapes detection in a fourth-generation immunoassay for HIV infection. J. Clin. Microbiol. 42:2847-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, S., E. A. Follett, L. Wallace, U. Desselberger, and H. S. Marsden. 1992. Immunodominant epitopes of HIV-1 p17 and p24. AIDS Res. Hum. Retroviruses 8:1781-1788. [DOI] [PubMed] [Google Scholar]
- 10.Haaheim, L. R., J. P. Maskell, P. Mascagni, and A. R. Coates. 1991. Fine molecular specificity of linear and assembled antibody binding sites in HIV-1 p24. Scand. J. Immunol. 34:341-350. [DOI] [PubMed] [Google Scholar]
- 11.Harma, H., T. Soukka, and T. Lovgren. 2001. Europium nanoparticles and time-resolved fluorescence for ultrasensitive detection of prostate-specific antigen. Clin. Chem. 47:561-568. [PubMed] [Google Scholar]
- 12.Janvier, B., P. Archinard, B. Mandrand, A. Goudeau, and F. Barin. 1990. Linear B-cell epitopes of the major core protein of human immunodeficiency virus types 1 and 2. J. Virol. 64:4258-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janvier, B., A. Baillou, P. Archinard, M. Mounier, B. Mandrand, A. Goudeau, and F. Barin. 1991. Immune response to a major epitope of p24 during infection with human immunodeficiency virus type 1 and implications for diagnosis and prognosis. J. Clin. Microbiol. 29:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, J. A., S. Y. Yoon, C. K. Lee, C. S. Lim, K. N. Lee, H. J. Sung, C. A. Brennan, and S. G. Devare. 2006. Performance evaluation of three automated human immunodeficiency virus antigen-antibody combination immunoassays. J. Virol. Methods 133:20-26. [DOI] [PubMed] [Google Scholar]
- 15.Langedijk, J. P., J. J. Schalken, M. Tersmette, J. G. Huisman, and R. H. Meloen. 1990. Location of epitopes on the major core protein p24 of human immunodeficiency virus. J. Gen. Virol. 71:2609-2614. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S., O. Wood, R. E. Taffs, J. Hu, A. Machuca, A. Vallejo, and I. Hewlett. 2006. Development and evaluation of HIV-1 subtype RNA panels for the standardization of HIV-1 NAT assays. J. Virol. Methods 137:287-291. [DOI] [PubMed] [Google Scholar]
- 17.Li, J., W. Xie, N. Fang, and E. S. Yeung. 2009. Single-molecule immunosorbent assay as a tool for human immunodeficiency virus-1 antigen detection. Anal. Bioanal. Chem. 394:489-497. [DOI] [PubMed] [Google Scholar]
- 18.Liu, G., L. Yang, J. Wang, G. Zhang, X. Chen, and Y. Zheng. 2005. Immune responses to six synthetic peptides of capsid protein with sera from HIV-1 infected individuals. Cell. Mol. Immunol. 2:289-293. [PubMed] [Google Scholar]
- 19.Ly, T. D., S. Laperche, C. Brennan, A. Vallari, A. Ebel, J. Hunt, L. Martin, D. Daghfal, G. Schochetman, and S. Devare. 2004. Evaluation of the sensitivity and specificity of six HIV combined p24 antigen and antibody assays. J. Virol. Methods 122:185-194. [DOI] [PubMed] [Google Scholar]
- 20.Machuca, A., S. Tang, J. Hu, S. Lee, O. Wood, C. Vockley, S. G. Vutukuri, R. Deshmukh, B. Awazi, and I. Hewlett. 2007. Increased genetic diversity and intersubtype recombinants of HIV-1 in blood donors from urban Cameroon. J. Acquir. Immune Defic. Syndr. 45:361-363. [DOI] [PubMed] [Google Scholar]
- 21.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 22.Muller, S., H. Wang, G. J. Silverman, G. Bramlet, N. Haigwood, and H. Kohler. 1993. B-cell abnormalities in AIDS: stable and clonally-restricted antibody response in HIV-1 infection. Scand. J. Immunol. 38:327-334. [DOI] [PubMed] [Google Scholar]
- 23.Nishanian, P., K. R. Huskins, S. Stehn, R. Detels, and J. L. Fahey. 1990. A simple method for improved assay demonstrates that HIV p24 antigen is present as immune complexes in most sera from HIV-infected individuals. J. Infect. Dis. 162:21-28. [DOI] [PubMed] [Google Scholar]
- 24.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14(Suppl. 3):S129-S140. [PubMed] [Google Scholar]
- 25.Petersen, L. R., G. A. Satten, R. Dodd, M. Busch, S. Kleinman, A. Grindon, and B. Lenes. 1994. Duration of time from onset of human immunodeficiency virus type 1 infectiousness to development of detectable antibody. Transfusion 34:283-289. [DOI] [PubMed] [Google Scholar]
- 26.Schupbach, J., M. Flepp, D. Pontelli, Z. Tomasik, R. Luthy, and J. Boni. 1996. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. AIDS 10:1085-1090. [PubMed] [Google Scholar]
- 27.Schupbach, J., Z. Tomasik, M. Knuchel, M. Opravil, H. F. Gunthard, D. Nadal, and J. Boni. 2006. Optimized virus disruption improves detection of HIV-1 p24 in particles and uncovers a p24 reactivity in patients with undetectable HIV-1 RNA under long-term HAART. J. Med. Virol. 78:1003-1010. [DOI] [PubMed] [Google Scholar]
- 28.Spence, R. P., W. M. Jarvill, R. B. Ferns, R. S. Tedder, and D. Parker. 1989. The cloning and expression in Escherichia coli of sequences coding for p24, the core protein of human immunodeficiency virus, and the use of the recombinant protein in characterizing a panel of monoclonal antibodies against the viral p24 protein. J. Gen. Virol. 70:2843-2851. [DOI] [PubMed] [Google Scholar]
- 29.Stramer, S. L., S. Caglioti, and D. M. Strong. 2000. NAT of the United States and Canadian blood supply. Transfusion 40:1165-1168. [DOI] [PubMed] [Google Scholar]
- 30.Summers, M. F., L. E. Henderson, M. R. Chance, J. W. Bess, Jr., T. L. South, P. R. Blake, I. Sagi, G. Perez-Alvarado, R. C. Sowder III, D. R. Hare, et al. 1992. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundqvist, V. A., J. Albert, E. Ohlsson, J. Hinkula, E. M. Fenyo, and B. Wahren. 1989. Human immunodeficiency virus type 1 p24 production and antigenic variation in tissue culture of isolates with various growth characteristics. J. Med. Virol. 29:170-175. [DOI] [PubMed] [Google Scholar]
- 32.Tang, S., and I. Hewlett. 2009. Europium nanoparticle-based immunoassays for sensitive detection of pathogens. Chem. Today 27:50-52. [Google Scholar]
- 33.Tang, S., and I. Hewlett. 2010. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J. Infect. Dis. 201(Suppl. 1):S59-S64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, S., M. Moayeri, Z. Chen, H. Harma, J. Zhao, H. Hu, R. H. Purcell, S. H. Leppla, and I. K. Hewlett. 2009. Detection of anthrax toxin by an ultrasensitive immunoassay using europium nanoparticles. Clin. Vaccine Immunol. 16:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang, S., J. Zhao, J. J. Storhoff, P. J. Norris, R. F. Little, R. Yarchoan, S. L. Stramer, T. Patno, M. Domanus, A. Dhar, C. A. Mirkin, and I. K. Hewlett. 2007. Nanoparticle-based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J. Acquir. Immune. Defic. Syndr. 46:231-237. [DOI] [PubMed] [Google Scholar]
- 36.Tersmette, M., I. N. Winkel, M. Groenink, R. A. Gruters, R. P. Spence, E. Saman, G. Van Der Groen, F. Miedema, and J. G. Huisman. 1989. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV p24gag. Virology 171:149-155. [DOI] [PubMed] [Google Scholar]
- 37.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Y., L. Lu, L. Ba, L. Liu, L. Yang, M. Jia, H. Wang, Q. Fang, Y. Shi, W. Yan, G. Chang, L. Zhang, D. D. Ho, and Z. Chen. 2006. Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med. 3:e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.