Abstract
The regulation of cellular processes by eukaryote-like serine/threonine kinases is widespread in bacteria. In the last 2 years, several studies have examined the role of serine/threonine kinases in Staphylococcus aureus on cell wall metabolism, autolysis, and virulence, mostly in S. aureus laboratory isolates in the 8325-4 lineage. In this study, we showed that the pknB gene (also called stk1) of methicillin-resistant S. aureus (MRSA) strain COL and the community-acquired MRSA (CA-MRSA) strain USA300 is involved in cell wall metabolism, with the pknB mutant exhibiting enhanced sensitivity to β-lactam antibiotics but not to other classes of antibiotics, including aminoglycosides, ciprofloxacin, bactrim, and other types of cell wall-active agents (e.g., vancomycin and bacitracin). Additionally, the pknB mutant of USA300 was found to be more resistant to Triton X-100-induced autolysis and also to lysis by lysostaphin. We also showed that pknB is a positive regulator of sigB activity, resulting in compromise in its response to heat and oxidative stresses. In association with reduced sigB activity, the expression levels of RNAII and RNAIII of agr and the downstream effector hla are upregulated while spa expression is downmodulated in the pknB mutant compared to the level in the parent. Consistent with an enhanced agr response in vitro, virulence studies of the pknB mutant of USA300 in a murine cutaneous model of infection showed that the mutant was more virulent than the parental strain. Collectively, our results have linked the pknB gene in CA-MRSA to antibiotic resistance, sigB activity, and virulence and have highlighted important differences in pknB phenotypes (virulence and sigB activity) between laboratory isolates and the prototypic CA-MRSA strain USA300.
The increasing morbidity and mortality of Staphylococcus aureus infections are raising concerns (15, 18). A major reason for this increase is the emergence of community-acquired methicillin-resistant S. aureus (CA-MRSA) strains that can affect previously healthy individuals outside the hospital setting. This contrasts with hospital-acquired MRSA (HA-MRSA) strains, which have been predominantly associated with immunocompromised hosts in hospitals.
CA-MRSA isolates can be distinguished from their HA-MRSA counterparts by their enhanced virulence and their spectrum of resistance to antibiotics. All MRSA strains harbor chromosomal cassette elements called staphylococcal cassette chromosome mec (SCCmec) elements, of which there are five types. CA-MRSA strains possess type IV and occasionally type V, while the larger types (I to III) are commonly found in HA-MRSA strains. Within all SCCmec elements is the invariant mecA, which encodes PBP2a and is the principal determinant of methicillin resistance in S. aureus.
Many CA-MRSA strains also carry additional virulence factors, such as Panton-Valentine leukocidin, and various staphylococcal enterotoxins. However, the role of these toxins in virulence is still debated. Recent reports have suggested that the differential expression levels of virulence factors in CA-MRSA strains may also contribute to the increased pathogenicity of these strains (19, 20).
In eukaryotic cells, many cellular processes are under the control of serine/threonine kinases. Recent studies have revealed the existence of eukaryotic serine/threonine kinase orthologs in bacteria (23). Bacterial homologues of these kinases have been shown to mediate a diverse range of functions, including cellular development, regulation, and pathogenesis (2, 23). The C termini of these kinases can bear three to five extracellular PASTA (for penicillin-binding protein [PBP] and serine/threonine kinase-associated) domains, which are penicillin-binding domains analogous to those found in penicillin-binding proteins while the N termini possess an intracellular kinase domain (23). Between these two domains is a single transmembrane segment. It is believed that the extracellular domain senses and transduces environmental signals by triggering the eukaryote-like N-terminal kinase domain to autophosphorylate and hence activate downstream signaling cascades. The S. aureus genome carries two putative kinase genes, one of which is pknB (locus SA1063 in S. aureus N315, also referred to as stk1). Using both laboratory and hospital S. aureus isolates, several groups have evaluated this gene (5, 9, 24). Despite some strain-to-strain variations in function of pknB, all three reports implicated that the kinase was involved in various aspects of cell wall metabolism, such as antibiotic resistance and autolysis; however, the precise mechanisms controlling these responses remain to be defined. Given the reported differences in antibiotic sensitivities between CA- and HA-MRSA strains, we sought to determine whether the pknB kinase gene has a major functional role in CA-MRSA.
In this report, we demonstrate that pknB of CA-MRSA strain USA300 is also involved in cell wall metabolism including antibiotic resistance. In particular, the pknB mutant was more sensitive to β-lactam antibiotics and became more resistant to the endopeptidase lysostaphin and also to Triton X-100-induced autolysis. In searching for the mechanism by which pknB affected antibiotic resistance, we discovered that the pknB gene was positively involved in controlling the activity of the alternative sigma factor SigB. Consequently, the pknB mutant was compromised in its response to heat and oxidative stresses. As the toxin-expressing phenotype related to agr (e.g., alpha toxin) has been shown to be enhanced in a sigB mutant (7), we also found that the expression levels of regulatory genes (e.g., sarA and agr) and selected target genes (e.g., hla) were upregulated in the pknB mutant compared with those in the parent USA300. The enhanced virulence of the mutant was also confirmed in a murine cutaneous model of infection. Together, our data suggest that the pknB kinase gene has an interesting phenotype of enhancing antibiotic resistance and SigB activity and downregulation of virulence in the most common CA-MRSA strain, USA300.
MATERIALS AND METHODS
Reagents, bacterial strains, and growth conditions.
All reagents were obtained from either Fisher or Sigma unless specified otherwise. The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus strains were cultured on either tryptic soy broth/agar (TSB/A) or Mueller-Hinton broth/agar. Escherichia coli strains were cultured on Luria-Bertani medium (LB). Bacterial growth was monitored at 650 nm, using 18-mm borosilicate glass tubes on a Spectronic 20D+ spectrophotometer (Spectronic Analytical Instruments, Garforth, England). Antibiotic selection and plasmid maintenance were provided at the following concentrations: for S. aureus, 2.5 μg/ml erythromycin, 10 μg/ml chloramphenicol, and 75 μg/ml kanamycin, and for E. coli, 100 μg/ml ampicillin and carbenicillin and at 34 μg/ml chloramphenicol. Xylose was provided at a concentration of 0.2% (wt/vol) for gene induction from the heterologous promoter of pEPSA5 as needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. aureus strains | ||
| RN4220 | Heavily mutagenized strain that accepts foreign DNA | |
| USA300 | Community-acquired MRSA strain | |
| COL | Hospital-acquired MRSA strain | |
| ALC6630 | USA300 ΔpknB | This work |
| ALC6634 | USA300 ΔpknB with pEPSA5 | This work |
| ALC6632 | USA300 ΔpknB complemented with pknBUSA300 in pEPSA5 | This work |
| ALC6625 | COL ΔpknB | This work |
| ALC6629 | COL ΔpknB with pEPSA5 | This work |
| ALC6627 | COL ΔpknB complemented with pknBCOL in pEPSA5 | This work |
| ALC6633 | USA300 ΔpknB complemented with pknBCOL in pEPSA5 | This work |
| ALC6628 | COL ΔpknB complemented with pknBUSA300 in pEPSA5 | This work |
| ALC6635 | USA300 ΔpknB with plasmid pALC2109 | This work |
| E. coli strains | ||
| XL1-Blue | General cloning strain | |
| BL21-DE3 | Strain used to express genes driven by the T7 promoter in the pET vector | |
| Plasmids | ||
| pMAD | E. coli-S. aureus shuttle vector containing a thermosensitive origin of replication, bgaB; Emr Apr | 1 |
| pMAD-Kn | pMAD with Knr | This work |
| pEPSA5 | E. coli-S. aureus shuttle vector containing a xylose inducible promoter; Apr Cmr | 13 |
| pALC2109 | pSK236 containing the tetR repressor and the xyl-tetO promoter driving sigB; Cmr | 4 |
| pET14b | E. coli expression vector to create N-terminal hexahistidine tags, containing an IPTG inducible T7 promoter; Apr Cmr | |
| pALC6640 | pET14b with pknBUSA300 | This work |
| pALC6641 | pET14b with pknBCOL | This work |
DNA manipulations.
A list of the oligonucleotides used for strain construction is available from the authors upon request. Routine cloning procedures were carried out according to standard laboratory protocols.
A markerless, in-frame deletion of pknB in strains USA300 and COL was constructed in accordance with a method described previously (23). Briefly, 1-kb regions flanking the gene of interest were amplified by PCR and joined by overlap extension. The resulting spliced product was inserted into either pMAD (COL) or pMAD-Kn (USA300) and transformed first into E. coli DH5α, then into RN4220 for proper methylation, and finally into either COL or USA300. Successive temperature shifts from 30°C to 42°C were used to promote the recombination of the pMAD constructs into the chromosome, resulting in the replacement of the native gene with the spliced allele. Correct clones were identified by colony PCR and confirmed by sequencing.
The pknB mutant strains were complemented by amplifying the native genes from either USA300 or COL genomic DNA and inserting them downstream of the xylose-inducible promoter of the S. aureus expression vector pEPSA5, followed by sequence confirmation.
His tag variants of pknB from both USA300 and COL were similarly amplified, inserted into pET14b, and transformed into E. coli DH5α, followed by confirmation with PCR and sequencing. For expression, the constructs were transformed into E. coli BL21-DE3.
Antibiotic susceptibility assays.
MICs of antibiotics were determined using Mueller-Hinton broth supplemented with 2% (wt/vol) NaCl, 25 μg/ml MgCl2, and 12.5 μg/ml CaCl2 in accordance with the microdilution procedure of the Clinical and Laboratory Standards Institute (9). Results were scored after 48 h and reported as modal values for at least 5 independent experiments. Susceptibility to imipenem was determined with the use of filter paper disks impregnated with 10 μg imipenem according to the disk diffusion procedure as outlined by the manufacturer (Remel). Zones of inhibition were measured after 24 h and reported as average values for 3 independent experiments.
Detection of pknB in whole-cell lysates.
E. coli BL21-DE3 cells harboring pET14b-pknBUSA300 or pET14b-pknBCOL were grown in 2× YT medium to an optical density at 650 nm (OD650) of 0.3, at which point heterologous protein expression was induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Equivalent samples of the cultures were taken at the 0-, 2-, and 4-h time points, resuspended in 1× SDS-PAGE sample buffer (containing 62.5 mM Tris-HCl [pH 6.8], 2% SDS, 0.01 mg/ml bromophenol blue, 10% glycerol, 10 mM dithiothreitol [DTT]), boiled for 10 min, resolved on 10% polyacrylamide SDS-PAGE gels, transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore), and analyzed according to standard Western blotting procedures. The primary antibody was mouse anti-His tag antibody (Novagen) used at a 1:5,000 dilution, and the secondary antibody was donkey anti-mouse IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA) used at a 1:10,000 dilution. The membrane was detected using the enhanced chemiluminescence (ECL) Western Blot system from GE Healthcare (Piscataway, NJ).
Autolysis, bacteriolysis, and lysostaphin sensitivity assays.
Static autolysis assays were conducted on exponential phase cells (OD650 = 0.7). In brief, cells were cultured in TSB plus 1 M NaCl, harvested, washed twice with ice-cold, sterile water, resuspended in 50 mM Tris-HCl (pH 7.5) plus 0.05% Triton X-100, and incubated at 30°C with shaking at 250 rpm, and the optical densities were serially monitored.
Bacteriolytic assays were performed using heat-killed cells as substrates and bacterial supernatants as the source of secreted autolysins. Heat-killed cells were prepared in phosphate-buffered saline (PBS) by heating the bacterial suspension at 65°C for 2 h and normalized to an OD650 of 5.0. Secreted autolysins in the culture supernatants were prepared from overnight cultures grown in TSB, normalized to cells at an OD650 of 10, and sterilized by passage through a 0.2-μm filter. Bacteriolytic activity was assessed by mixing 1 ml of heat-killed cells with 5 ml of supernatant (or TSB as a control) and incubating them at 37°C with shaking at 250 rpm, and the OD650 monitored. For some experiments, bacteria were cultured overnight in TSB plus 1 mM phenylmethanesulfonyl fluoride (PMSF) to inactivate serine proteases in the supernatant.
Lysostaphin sensitivity was assessed by adding lysostaphin (AMBI Products, LLC, Lawrence, NY) at a final concentration of 1.7 μg/ml (for USA300) or 2.5 μg/ml (for COL) to late-exponential-phase cells (OD650 = 1), followed by incubation at 37°C with shaking at 250 rpm. The optical densities of the cultures were then serially measured.
All data from the autolysis experiments described above are reported as percentages of the initial turbidity (at the zero time point) and are representative of 3 independent experiments.
Zymography.
Autolysin activity was also visualized on 8% SDS-polyacrylamide gels containing either heat-killed RN4220 or lyophilized Micrococcus lysodeikticus as described previously (27). Autolysins were extracted from exponential-phase cells (10 ml at an OD650 of 0.7) with 100 μl of 4% SDS, quantitated using a Pierce bicinchoninic acid (BCA) protein assay kit, applied to the gels, and resolved electrophoretically. After electrophoresis, the gels were washed extensively in water, incubated in 1% Triton X-100 in 25 mM Tris-HCl (pH 8) for 16 h at 37°C, and stained with 0.1% methylene blue. Zones of cell lysis were visualized as clear bands against a blue background. Data shown are representative of three independent experiments.
Transcriptional analyses.
RNA was extracted from approximately 1.2 × 1011 cells grown to the indicated growth phases using Trizol and a reciprocating shaker as described previously (26). For quantitative real-time PCR (qRT-PCR) analysis, the RNA was treated with a DNA-free kit (Ambion) to remove residual DNA, followed by reverse transcription using a Roche Transcriptor first-strand synthesis kit. qRT-PCR was performed with Maxima SYBR green qPCR master mix (Fermentas) in a Roche LightCycler 1.5 system. Absolute and relative quantification of the resulting amplicons was carried out using LightCycler software version 4.0. Data reported are normalized to the gyrB levels.
For Northern blot analysis, RNAs were separated on a 1.5% agarose-0.66 M formaldehyde gel in 20 mM morpholinepropanesulfonic acid, 10 mM sodium acetate, 2 mM EDTA (pH 7) and transferred to a Hybond XL membrane as described previously (23). PCR-generated probes were labeled with α-[32P]dCTP (GE Life Sciences) using a random primed DNA labeling kit (Roche). Following prehybridization, hybridization with the probe was conducted at 65°C overnight. The membranes were washed under high-stringency conditions and visualized by autoradiography. Bands were quantitated with ImageJ as needed. Images shown are representative of three blots.
Protease assay.
Supernatants from 30 ml of overnight cultures in TSB, normalized to an OD650 of 10, were concentrated ∼150 fold (Centricon; molecular weight cutoff, 3,000 [Millipore]). After the protein concentration was determined (BCA assay kit), protease assays of the supernatants were conducted with an Enz-Chek Bodipy fluorescence kit (Molecular Probes), with the expressed values standardized to 35 μg of total protein in the supernatants.
Stress resistance assays.
Temperature sensitivity was assessed by comparing the growth profiles of cells in TSB at 42°C with shaking at 250 rpm.
Sensitivity to hydrogen peroxide and ethanol was determined by adding these reagents to exponential-phase cells (OD650 = 0.7) to give final concentrations of 10 mM and 10% (vol/vol), respectively. Doubling times were calculated by determining the slope of the curve from 0.7 to 1.4 OD650 units.
Hemolysis assay.
Supernatants from overnight cultures grown with normalization to an OD650 of 10 were serially diluted 2-fold in a microtiter plate containing a suspension of 2% washed rabbit erythrocytes in PBS. The plate was incubated at 37°C for 1 h, and the hemolytic titers were determined as the reciprocal of the highest dilution resulting in 50% lysis, using 2% SDS as the positive control. Reported values are normalized to the protein content of the supernatants as determined by a Pierce BCA protein assay kit.
Murine model of subcutaneous infection.
Early-exponential-phase cells grown in TSB (OD650 = 0.35) were harvested, washed, and resuspended in normal saline to give a final concentration of 108 CFU/ml. The backs of 6-week-old male C57BL6 mice (Jackson Laboratories, Bar Harbor, ME) were shaved, sterilized with alcohol wipes, and injected with 100 μl of the cell suspension subcutaneously. This inoculum was previously determined with a titration assay, resulting in cutaneous infection in 90% of the mice. The general appearances, lesion sizes, and weights of the mice were monitored and reported as mean values. Mice were sacrificed after 3 days, and an 8-mm biopsy punch was made from the lesions. A portion of the biopsy sample was fixed in 10% formalin for histological analysis while the remainder was homogenized and used to determine the number of CFU/gm of tissue. Statistical significance was determined using the Kruskal-Wallace test, with a P value of <0.05 considered significant. For histological analysis, the fixed tissue samples were embedded, cut in 5-μm sections, and stained with hematoxylin and eosin (H&E) according to standard procedures. Representative sections from each group are shown.
RESULTS
Antibiotic susceptibility of a pknB mutant.
The pknB kinase gene is predicted to encode three extracellular penicillin-binding domains, called the PASTA domain. We thus speculated that this protein might be involved in methicillin resistance in MRSA isolates. To test this hypothesis, a markerless in-frame deletion of pknB was made in both COL (HA-MRSA) and USA300 (CA-MRSA) and the antibiotic susceptibilities of each mutant were compared to those of the parent and complemented mutants (Tables 2 and 3). In both strain backgrounds, a pknB mutation led to increased susceptibilities to many β-lactams, including the carbapenem imipenem, with the USA300 pknB mutant being more sensitive to a wider range of β-lactams than the pknB mutant of COL. Provision of pknB in trans with the plasmid pEPSA5 to both mutant strains resulted in either a full or a partial restoration of the MIC to wild-type levels.
TABLE 2.
Effect of pknB on the susceptibility of Staphylococcus aureus strain USA300 to β-lactam antibioticsa
| Antibiotic | MIC for USA300 |
|||
|---|---|---|---|---|
| Wild type | pknB | pknB/pEPSA5 | pknB/pknB+ | |
| Oxacillin | 32 | 1 | 0.5 | 8 |
| Cloxacillin | 1 | 0.125 | 0.25 | 2 |
| Penicillin | 16 | 0.25 | 0.25 | 16 |
| Carbenicillin | 64 | 1 | 2 | 16 |
| Nafcillin | 32 | 1 | 0.5 | 16 |
| Ceftriaxone | 64 | 8 | 8 | 16 |
| Cephalothin | 32 | 1 | 0.5 | 16 |
| Cefazolin | 32 | 0.5 | 0.5 | 32 |
| Imipenem | 22 | 37 | 40 | 27 |
All MICs, with the exception of those for imipenem, are presented in μg/ml. Imipenem sensitivity was measured as zones of clearing in mm.
Both sets of mutants displayed equivalent susceptibilities to representative drugs from other antibiotic classes, including aminoglycosides, ciprofloxacin, trimethoprim-sulfamethoxazole, and other cell wall-active agents, such as cycloserine, vancomycin, and bacitracin, compared with their respective parents, indicating that the antibiotic sensitivity of the pknB mutants was limited mostly to the β-lactam class of antibiotics.
Role of the PASTA domain in antibiotic sensing.
Examination of the pknB genes of USA300 and COL revealed that both sequences were identical, with the exception of a G-to-T transversion at position 1165 in COL. This point mutation translates into a stop codon, prematurely truncating the kinase protein in COL (Fig. 1). This region of COL was subsequently resequenced to confirm the transversion.
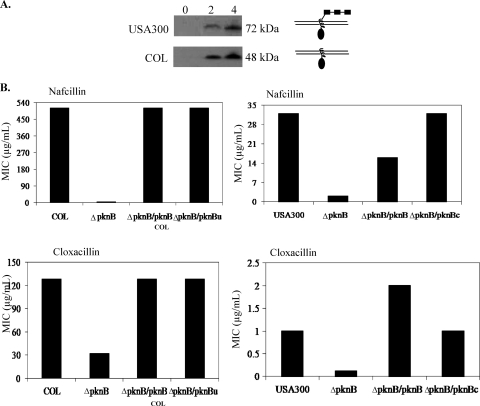
FIG. 1.
Structural differences between the PknB kinase proteins of USA300 and COL. (A) Western blot analysis of PknB from USA300 (top panel) and COL (bottom panel). The pknB gene from each strain was cloned behind an N-terminal His tag. Expression of the fusion proteins after IPTG induction at 0, 2, and 4 h was monitored by Western blot analysis using a primary antibody directed toward the hexahistidine moiety. The schematic on the right shows the probable architecture of each PknB kinase protein on the basis of the estimated sizes of each fusion protein. Each of the PASTA domains is depicted as rectangles, while the kinase domain is represented by an oval. (B) Role of the PASTA domain in antibiotic resistance. COL and USA300 pknB mutants were complemented and cross-complemented with either their own or the heterologous kinase gene. The designations pknBu and pknBc correspond to the pkn genes of USA300 and COL, respectively. The antibiotic susceptibilities of the resultant strains were then compared between the parent and mutant strains by performing MIC assays using nafcillin and cloxacillin.
For expression, the pknB genes from both COL and USA300 were cloned behind the N-terminal His tag of pET14b. After IPTG induction, the resulting fusion proteins were detected by Western blot analysis using an anti-His tag antibody (Fig. 1A). The USA300 pknB construct produced a 72-kDa His tag protein, in agreement with the predicted size of pknB, while the His tag PknB protein of COL was only 48 kDa, corresponding to the truncated size of PknB without the PASTA domains. Therefore, it appeared that translation of the COL PknB protein terminated prematurely, as predicted from the G-to-T transversion.
To determine whether the PASTA domains of pknB made any contribution to antibiotic resistance, we performed a cross-complementation experiment. The truncated kinase gene from COL was provided in trans to the USA300 pknB mutant and vice versa. Remarkably, both the truncated and full-length kinase genes were able to complement the pknB mutation in both strains to equivalent degrees with respect to sensitivity to β-lactams (Fig. 1B), suggesting that the kinase domain alone was sufficient to restore antibiotic resistance to the pknB mutants. In light of these data, we chose to pursue further work on pknB in USA300.
Cell wall phenotype of the pknB mutant.
Mutants with altered expression of penicillin-binding proteins (PBPs) are frequently sensitive to a range of β-lactam antibiotics (21, 22, 24). Therefore, we speculated that the sensitivity of the pknB mutants to antibiotic may have been due to altered levels of penicillin-binding proteins. However, Northern blot and real-time PCR analyses of gene transcription revealed that the transcript levels of five genes encoding penicillin-binding proteins (mecA, pbp1, pbp2, pbp3, and pbp4) were equivalent in both the pknB mutant and the USA300 parent strain, suggesting that the antibiotic sensitivity of the pknB kinase mutant was not due to alterations in pbp transcription. Transmission electron microscopy also revealed that the cellular morphologies (i.e., size, cell wall thickness, and septum formation) of all three strains (parent, mutant, and complemented mutant) were virtually indistinguishable, supporting the notion that PBP activity was unaltered in the pknB mutant compared with that in the parent (data not shown).
We next examined the peptidoglycan cross-bridging of the pknB mutant by determining its sensitivity to the glycyl-glycine endopeptidase lysostaphin. Remarkably, the pknB mutant exhibited greater resistance to lysostaphin. After 24 min of lysostaphin exposure, only 60% of the pknB cells exhibited lysis whereas cells of the parent USA300 and complemented strains were almost completely lysed in an identical time period (Fig. 2A). As the fem genes encode enzymes necessary for the addition of glycine to the pentaglycine bridge, we analyzed femA and femB transcription in the pknB mutant by real-time PCR and found no differences between the mutant and the parent and the complemented strain, implying that the synthesis of the glycyl-glycine peptidoglycan cross-bridges was not significantly altered in the pknB mutant. Additionally, evaluation of peptidoglycan cross-linking by high-performance liquid chromatography (HPLC) analysis of muropeptides also did not divulge any significant differences between the parent and the mutant (data not shown). We thus hypothesized that the peptidoglycan in the pknB mutant may have been cross-linked aberrantly and hence prevented lysostaphin from recognizing and/or accessing its substrates.
FIG. 2.
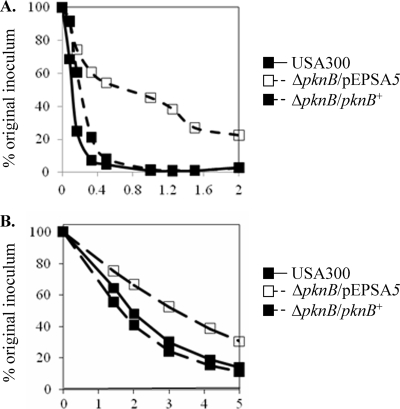
Cell wall phenotypes of the USA300 pknB mutant. Sensitivity of the pknB mutant to lysostaphin (A) and Triton X-100 (B). The pknB mutant was complemented with pEPSA5 containing pknB. The negative control for the mutant is the vector alone.
Autolysis phenotype of the pknB mutant.
A lack of gross morphological changes in the cell wall of the pknB mutant led us to consider autolysis as an indicator of altered cell wall metabolism. The pknB mutant exhibited a greater resistance to autolysis under the static condition, with only 70% of the initial inoculum lysed after 5 h at 30°C, compared to ∼90% lysis for the either the parent or the complemented mutant strain (Fig. 2B). This phenotype was also observed when the bacteria were cultured for 1 h in the presence of nafcillin prior to the autolysis assay (data not shown).
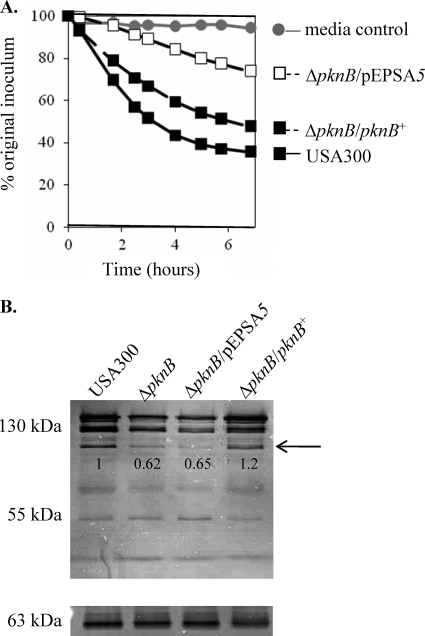
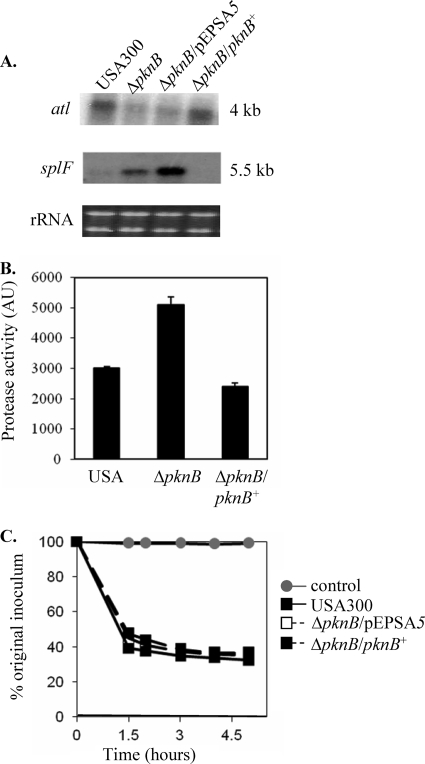
A comparison of the activities of the secreted autolysins derived from the parent, mutant, and complemented mutant disclosed a defect in the ability of the supernatant from the pknB mutant to lyse cells. After a 6-h incubation period, only 22% of heat-killed USA300 cells were lysed by the pknB supernatant, compared to 63% and 49% by the supernatants of the parent and the complemented mutant, respectively (Fig. 3A). To determine if there was a defect in the translocation of autolysins from the cells to the culture supernatants, zymographic analysis was performed using autolysins prepared from whole-cell extracts. The autolytic profile of the pknB mutant displayed a decrease in the activity of the bands corresponding to the proenzyme of the major autolysin Atl (130 kDa) as well as the processed Atl (110 kDa) (Fig. 3B). Northern blot analysis also verified that the transcription of the atl gene was approximately halved in the pknB mutant in comparison to the level in the parent and complemented mutant strains (Fig. 4A), which may account for its reduced autolytic activity. We also investigated the possibility that the autolysins of the pknB mutant might be degraded by serine proteases as has previously been reported (14). Notably, the supernatants derived from the pknB mutant demonstrated almost twice as much protease activity than the wild-type or complemented mutant strains (Fig. 4B), corroborating the evidence for an increase in the level of transcription of the serine protease operon splA-splF (Fig. 4A). Inactivation of the bacterial serine proteases through the addition of PMSF to the culture medium restored the autolytic activity of the pknB supernatant to wild-type levels (65% and 68% cells lysed, respectively, after 5 h) (Fig. 4C), thus implicating the role of proteases in reducing the autolytic activity of the pknB mutant.
FIG. 3.
Decreased autolysin activity of the pknB mutant. (A) Bacteriolytic activities of secreted autolysins in the supernatants of stationary phase cultures, using heat-killed USA300 cells as the substrate. (B) Zymographic analysis of autolytic activity of the pknB mutant of USA300 using either M. lysodeikticus (top panel) or S. aureus 4220 (bottom panel) as the substrate. Note the decrease in the activity of the proenzyme for major autolysin Atl (130 kDa) and the processed autolysin (110 kDa; arrow) while the activities of the glucosaminidase (55 kDa) and amidase (63 kDa) cleaved from Atl are relatively unchanged. Numbers underneath the gel panels indicate the relative band intensities as determined by ImageJ.
FIG. 4.
Decreased atl transcription and increased protease activity in the pknB mutant contribute to its autolytic phenotype. (A) Northern blot analysis of stationary-phase cells showing the transcriptions of atl and spl (carrying a serine protease operon) in the pknB mutant in comparison with the levels in the parent and the complemented strain. (B) Protease activity in the supernatants of stationary-phase cells as assessed by an Enz-Chek Bodipy fluorescence kit. AU, arbitrary units. (C) Bacteriolytic activity of supernatants taken from stationary-phase cultures of different constructs grown in the presence of 1 mM PMSF using heat-killed USA300 as the substrate.
Role for pknB in cell stress.
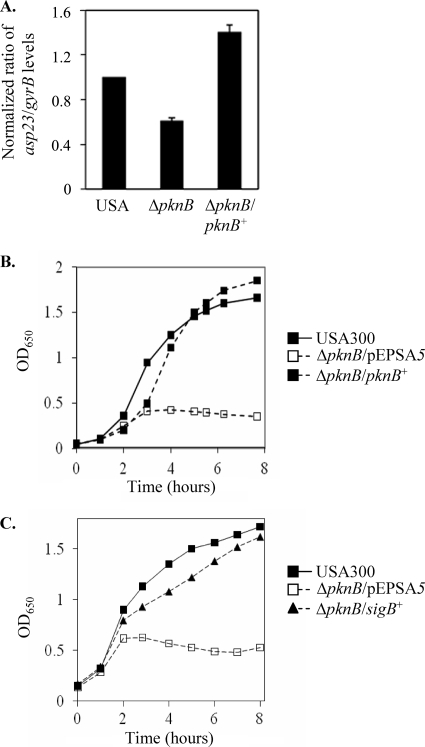
Since the pknB mutant did not exhibit any gross morphological defects in the cell wall that may have been the underlying cause of its hypersusceptibility toward β-lactams, we next sought alternative explanations for the observed phenotype. Recent literature has demonstrated that bactericidal antibiotics, such as β-lactams, exert their killing effects through the production of reactive oxygen species (29). Thus, we investigated the potential involvement of the kinase in the stress response by examining the activity of its primary mediator, the alternative sigma factor SigB, in the pknB mutant. Quantitative real-time PCR analysis of the SigB-dependent alkaline shock protein gene asp23 revealed that the pknB mutant had approximately half the amount of transcript that the parent and complemented mutant had (Fig. 5A). Correspondingly, the pknB mutant was compromised when it was grown in the presence of 10 mM hydrogen peroxide or 10% (vol/vol) ethanol, compared to both the parental strain USA300 and the complemented mutant (Table 4). The most dramatic growth defect, however, was observed when the strains were grown at a temperature of 42°C, with the pknB mutant severely compromised at the elevated temperature (Fig. 5B). Remarkably, provision of sigB in trans restored the growth of the mutant to the wild-type level, confirming that the growth defect was indeed due to reduced SigB activity in the pknB mutant (Fig. 5C).
FIG. 5.
Decreased SigB activity in the pknB mutant. (A) Real-time PCR analysis of asp23 transcription in USA300, the pknB mutant, and the complemented mutant. (B) Growth of the pknB mutant at 42°C in comparison with that of the parent and complemented strains. (C) Growth of the pknB mutant complemented with the sigB gene in trans at 42°C in comparison with that of the parent and the mutant.
TABLE 4.
Doubling times of the pknB mutant in 10 mM hydrogen peroxide and 10% ethanola
| Condition | Doubling time (h) for: |
||
|---|---|---|---|
| USA300 | pknB/pEPSA5 | pknB/pknB+ | |
| TSB | 1.5 | 1.9 | 1.7 |
| H2O2 | 2.4 | 3.1 | 2.4 |
| EtOH | 4.6 | 6.5 | 4.9 |
We defined the doubling time as the time in hours taken to increase the OD650 from 0.7 to 1.4. EtOH, ethanol.
The antibiotic susceptibility of the pknB mutant cross-complemented with sigB was then tested to ascertain whether the antibiotic sensitivity of the pknB mutant could be attributed to reduced activity of SigB. The susceptibilities of pknB and its cross-complemented mutant to β-lactams were identical, suggesting that reduced SigB activity was not involved in the β-lactam sensitivity of the mutant (data not shown).
Involvement of pknB in expression of virulence determinants.
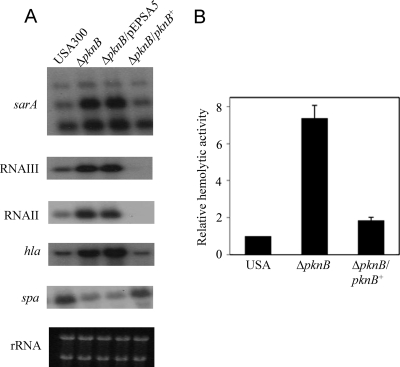
A number of reports have demonstrated an inverse relationship between SigB activity and virulence (4, 6, 7, 16, 25). Therefore, we sought to determine whether reduced SigB expression and augmented protease expression in the pknB mutant could be linked to a more general virulence phenotype. Northern blot analyses of several genes linked to the agr regulatory cascade revealed elevated expression of agr and related genotypes in the kinase mutant compared to the levels in the parent. Both RNAII and RNAIII, as well as sarA expression, were upregulated in the pknB mutant in comparison to the levels for the parent and the complemented mutant strain (Fig. 6A). Additionally, the transcription of genes regulated by agr, such as hla (α hemolysin gene) and spa (protein A gene) in the mutant was altered in a manner consistent with enhanced agr expression, displaying upregulation of hla and downregulation of spa in the mutant compared to the levels for the parent and the complemented mutant (Fig. 6A). Notably, the pknB mutant demonstrated a 7-fold increase in hemolytic activity compared with the parental strain (Fig. 6B); these data suggest that the pknB mutant likely possesses a hypervirulent phenotype.
FIG. 6.
Expression of virulence determinants of the pknB mutant. (A) Northern blot analysis of sarA, RNAIII, RNAII, hla, and spa gene transcription in USA300, the pknB mutant, and the complemented mutant. (B) Relative hemolytic titers of the pknB mutant with sheep red blood cells in comparison to those of the parent and complemented strains.
Enhanced virulence of the pknB mutant in a murine cutaneous model of infection.
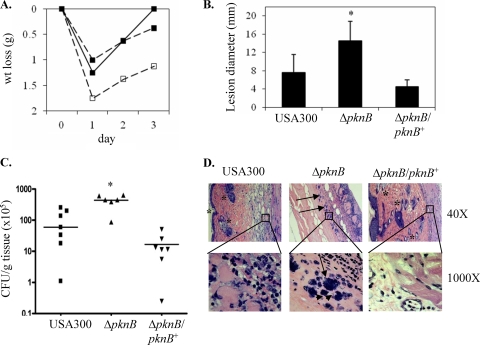
To evaluate whether the above-mentioned regulatory and phenotypic changes bore any clinical significance, we assessed the virulence of the pknB mutant in a murine model of cutaneous infection. Twenty-two C57B6 mice were injected subcutaneously with 100 μl of PBS containing 107 CFU (8 mice each with the wild-type and the complement strains and 6 mice with the mutant). The weights of the mice were also determined pre- and postinfection as an indicator of their general health (Fig. 7A). The greatest weight loss was observed on the first day postinfection, with the mice infected with the pknB mutant losing on average 1.75 g and the mice in the wild-type and complement groups losing on average ∼1.25 g and 1 g, respectively. One day after injection, all of the mice infected with the pknB mutant developed areas of redness surrounding the injection sites while inflammation was observed only for 4 of the mice in the wild-type group. By the second day after infection, all of the mice had developed erythematous cutaneous lesions. The cutaneous lesions of the mice infected with the pknB mutant were the most severe, reaching an average diameter of ∼14.5 mm on the third and final day of the experiment. The average sizes of the cutaneous lesions in the mice infected with the parent and complemented strains were 7.6 mm and 4.5 mm, respectively (Fig. 7B). A greater number of CFU per gram of tissue was recovered from the wound sites of the mice in the pknB mutant group (4.5 × 107) than from those infected with either the USA300 parent strain (9.9 ×106) or the complemented mutant (1.2 ×106) (Fig. 7C). Histological sections of the wound site stained with H&E stain from mice infected with the pknB mutant revealed a more severe form of infection than the mice from the other two groups, as indicated by the greater numbers of bacteria in the mice infected with the pknB mutant than in those from the other two groups. In addition, there was a complete loss of the outer dermal layer and a complete lack of anatomic definition and cellular structures in the epidermal layer of the mice infected with the pknB mutant, compared with those seen for the parent (Fig. 7D).
FIG. 7.
Ability of the pknB mutant to cause murine skin infections. Six-week-old C57BL6 mice were injected with USA300, the pknB mutant, or the complemented strain. Weight changes during the course of the experiment were noted (A). After 3 days, the mice were sacrificed and their skin lesions were examined for size (B), bacterial burden (C), and histological changes in punch biopsy samples (D). In panels B and C, an asterisk represents P values of <0.05 as determined by the Kruskal-Wallace test. (D) Note the absence of the dermis and other subcellular structures (*) and the presence of bacteria (→) in the sample taken from the mouse infected with the pknB mutant.
DISCUSSION
Protein phosphorylation of serine and threonine residues on effector proteins is a common regulatory event in eukaryotes. Analyses of an increasing number of bacterial genomes have divulged eukaryote-like Ser/Thr protein kinase and phosphatase genes in prokaryotes (3, 23). In S. aureus, the kinase gene pknB, lying immediately downstream of the phosphatase gene stp1, is transcribed together with the phosphatase gene as an operon (10). Similar to other serine/threonine kinases in Gram-positive bacteria, the PknB protein of S. aureus has three domains, including an amino-terminal kinase domain, a transmembrane segment, and three PASTA domains at the C terminus. However, in analyzing the pknB genes of strains USA300 and COL, we found that the PknB protein in strain USA300 is intact while its counterpart in COL is truncated due to a G-to-T transversion at position 1165, leading to a premature stop codon and hence lacking the PASTA domains (Fig. 1). Western blot analysis indeed confirmed that the His tag PknB protein in strain COL is ∼48 kDa, much smaller than the full-length His tag PknB protein of USA300 (at 72 kDa). As the PASTA domain has previously been linked to the binding of β-lactams (2, 30), we proceeded to determine the sensitivity of the pknB mutant of USA300 to these antibiotics. Similar to what has been found with N315 (5), the pknB mutants of strains USA300 and COL, representing CA- and HA-MRSA, respectively, were more sensitive to an assortment of β-lactam antibiotics, including a carbapenem (i.e., imipenem), than the respective parents. Interestingly, there were no differences in sensitivity to other cell wall-active agents, including vancomycin, cycloserine, and bacitracin, between the pknB mutant and the corresponding parent, thus implying restricted sensitivity to common β-lactams and carbapenem and not to other cell wall-active agents.
Prior studies suggested that the PASTA domain may sense the presence of β-lactam antibiotics, leading to activation of the kinase domain (30). However, despite not having any of the PASTA domains, the pknB gene of COL was able to restore the MICs of the pknB mutant of USA300 for nafcillin and cloxacillin to parental levels in cross-complementation experiments (Fig. 1B). Given that PknB proteins can autophosphorylate (5), this finding implied that the sensing of β-lactam by the PASTA domain and the ensuing activation of the kinase domain may be more intricate than previously thought.
Despite the sensitivity of the pknB mutant of USA300 to β-lactam, we did not observe any gross morphological cell wall changes in the mutant on transmission electron microscopy, nor did we discern any significant changes in cross-linking in the HPLC analysis of muropeptides in the mutant, compared with what was observed for the parent (data not shown). However, subtle changes in the peptidoglycan cross-bridge likely occur because the pknB mutant exhibited increased resistance to lysis by lysostaphin, a glycyl-glycine endopeptidase targeting the pentaglycine cross-bridge between peptidoglycan strands. Besides resistance to lysostaphin-mediated lysis, the pknB mutant of USA300 also displayed greater resistance to autolysis in 0.05% Triton X-100 under static conditions at 30°C when autolysins are still active while cell wall synthesis is slowing down. The effect of pknB on resistance to autolysis in strain USA300 may be 2-fold. First, the transcription of atl, the major autolysin gene, was decreased in the mutant compared to the level in the parent. This effect of pknB on atl was supported by recent microarray studies by Donat et al., demonstrating that atl and lytM expression is reduced in the pknB mutant of 8325 while transcription of lytR and lgrA, which are repressors of autolysis, is elevated (12). Second, the effect of autolysin may be decreased due to enhanced serine protease activity in the mutant (Fig. 4B). The latter observation was corroborated by the restoration of autolytic activity of the pknB mutant upon the addition of PMSF to the culture medium.
Perturbations in cell wall metabolism represent a unique form of cell stress. For this reason, we explored if pknB modulates the activity of SigB, a transcription factor intimately involved in the stress response of S. aureus. Analysis of the SigB-dependent alkaline shock protein gene asp23 revealed decreased sigB activity in the pknB mutant, but this activity was restored in the complemented mutant. Consistent with the defect in SigB activity were the slower growth at the elevated temperature (42°C) and the longer generation times of the mutant upon exposure to 10 mM hydrogen peroxide or 10% ethanol than of the parent. These findings are also in concordance with the observation by Truong-Bolduc et al. in which they proposed that additional factors related to sigB and pknB may contribute to the regulatory effect of MgrA on the expression of the norA promoter (28). At this junction, we do not know whether the effect of pknB on SigB is direct or indirect. Whether PknB phosphorylates an antisigma factor, such as RsbU, is not known. Nevertheless, the linkage between PknB and stress response may provide a pathway whereby PknB contributes toward pathogenesis.
In previous studies, it has been shown that sigB mutants exhibit an enhanced agr response (7, 16, 17, 25). To confirm this phenotype in the pknB mutant, we assayed for RNAII and RNAIII expression and the downstream effector genes. As expected from an enhanced agr phenotype, the expression levels of RNAII, RNAIII, and hla were elevated in the pknB mutant compared with the levels in the parent while spa expression was downmodulated. The increased expression of proteolytic activity in the pknB mutant was also consistent with an elevated agr response.
On the basis of increased agr expression, we hypothesized that the pknB mutant may be more virulent than the parental strain USA300. A murine model of cutaneous infection has confirmed our hypothesis. Notably, our findings are in stark contrast to the studies by Débarbouillé et al., who showed that the kinase mutant of strain SH1000 had reduced survival in the kidney in a murine bacteremic pyelonephritis model (10). The discrepancy between our studies and theirs could be due to several factors: (i) USA300 is a clinical CA-MRSA isolate whereas SH1000 is a laboratory strain, (ii) ours was a murine subcutaneous model while theirs was a bacteremic model looking at kidney abscesses, and (iii) data from Truong-Bolduc et al. indicated that additional factors related to sigB and pknB may contribute to the regulatory effect of virulence determinants (28).
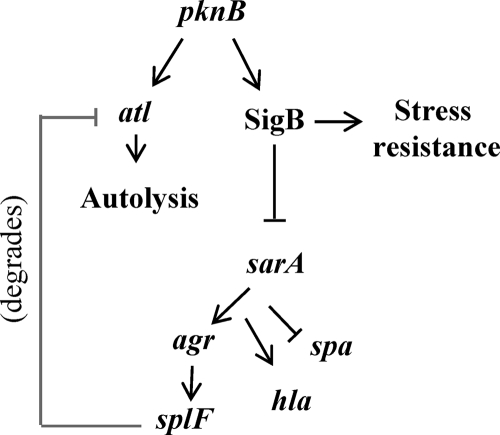
On the basis of the above observations on USA300, we have proposed a model (Fig. 8) whereby the PknB protein regulates SigB activity and also stress resistance. Activation of SigB would downregulate agr and its effector genes, including hla and splA-splF, leading to reduced virulence of S. aureus. Additionally, the reduced effect of protease on Atl in a pknB+ strain may promote normal autolysis in a growing S. aureus cell. Besides agr, it also appears that PknB positively regulates sarA transcription. It has been known that SarA can downregulate its own promoter (8). Moreover, recent studies by Didier et al. have described the phosphorylation of SarA by PknB (11). Whether phosphorylated SarA binds differently to the sarA promoter compared to its unphosphorylated counterpart is not known. The above-mentioned scenarios, coupled with the finding that SarA is also a positive regulator on agr, implied that the effect of PknB on target genes such as hla and spa is complex and may involve a combination of factors, including SarA and agr. The relationship among PknB, SarA, and agr with respect to virulence, autolysis, stress response, and antibiotic resistance in community-acquired strains of S. aureus is currently under investigation in our laboratory.
FIG. 8.
Proposed model of gene regulation by pknB. In USA300, pknB acts as an activator of both atl and sigB. Its actions upon sigB are predicted to make the cell more resistant to harsh environmental conditions and less virulent based on the role of sigB in cell stress and its actions upon sarA and agr. In addition to reduced virulence, the decreased expression of the downstream effector splF in the pknB+ strain is predicted to reduce the degradation of the Atl proenzyme.
TABLE 3.
Effect of pknB on the MICs of β-lactam antibiotics against COL
| Antibiotic | MIC (μg/ml) for COL |
|||
|---|---|---|---|---|
| Wild type | pknB | pknB/pEPSA5 | pknB/pknB+ | |
| Oxacillin | 256 | 64 | 64 | 256 |
| Cloxacillin | 512 | 2 | 2 | 512 |
| Carbenicillin | 128 | 32 | 32 | 128 |
| Nafcillin | 256 | 64 | 64 | 256 |
| Ceftriaxone | 1,024 | 512 | 512 | 1,024 |
| Cephalothin | 128 | 64 | 64 | 128 |
Acknowledgments
This work was supported in part by NIH grants AI37142 and AI47441 to A.L.C.
Editor: A. Camilli
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 3.Bakal, C. J., and J. E. Davies. 2000. No longer an exclusive club: eukaryotic signaling domains in bacteria. Trends Cell Biol. 10:32-38. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, B. Y., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in S. aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltramini, A. M., C. D. Mukhopadhyay, and V. Pancholi. 2009. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 77:1406-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Nishina, and A. C. Manna. 2008. SarA of Staphylococcus aureus binds to the sarA promoter to regulate gene expression. J. Bacteriol. 190:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Débarbouillé, M., S. Dramsi, O. Dussurget, M. A. Nahori, E. Vaganay, G. Jouvion, A. Cozzone, T. Msadek, and B. Duclos. 2009. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191:4070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didier, J. P., A. J. Cozzone, and B. Duclos. 2010. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol. Lett. 306:30-36. [DOI] [PubMed] [Google Scholar]
- 12.Donat, S., K. Streker, T. Schirmeister, S. Rakette, T. Stehle, M. Liebeke, M. Lalk, and K. Ohlsen. 2009. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191:4056-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowler, V. G., Jr., J. M. Miro, B. Hoen, C. H. Cabell, E. Abrutyn, E. Rubinstein, G. R. Corey, D. Spelman, S. F. Bradley, B. Barsic, P. A. Pappas, K. J. Anstrom, D. Wray, C. Q. Fortes, I. Anguera, E. Athan, P. Jones, J. T. van der Meer, T. S. Elliott, D. P. Levine, and A. S. Bayer. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012-3021. [DOI] [PubMed] [Google Scholar]
- 16.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahl, B. C., G. Belling, P. Becker, I. Chatterjee, K. Wardecki, K. Hilgert, A. L. Cheung, G. Peters, and M. Herrmann. 2005. Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 73:4119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 19.Li, M., B. A. Diep, A. E. Villaruz, K. R. Braughton, X. Jiang, F. R. DeLeo, H. F. Chambers, Y. Lu, and M. Otto. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loughman, J. A., S. A. Fritz, G. A. Storch, and D. A. Hunstad. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Memmi, G., S. R. Filipe, M. G. Pinho, Z. Fu, and A. L. Cheung. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreillon, P., Z. Markiewicz, S. Nachman, and A. Tomasz. 1990. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob. Agents Chemother. 34:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohlsen, K., and S. Donat. 2010. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int. J. Med. Microbiol. 300:137-141. [DOI] [PubMed] [Google Scholar]
- 24.Pinho, M. G., S. R. Filipe, H. de Lencastre, and A. Tomasz. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt, K. A., N. P. Donegan, W. A. Kwan, Jr., and A. Cheung. 2004. Influences of sigmaB and agr on expression of staphylococcal enterotoxin B (seb) in Staphylococcus aureus. Can. J. Microbiol. 50:351-360. [DOI] [PubMed] [Google Scholar]
- 26.Tamber, S., and A. L. Cheung. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 77:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trotonda, M. P., Y. Q. Xiong, G. Memmi, A. S. Bayer, and A. L. Cheung. 2009. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J. Infect. Dis. 199:209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong-Bolduc, Q. C., Y. Ding, and D. C. Hooper. 2008. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190:7375-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X., and X. Zhao. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27:438. [DOI] [PubMed] [Google Scholar]