Abstract
Infections due to extraintestinal pathogenic Escherichia coli (ExPEC) are common in humans and animals and include urinary tract infections (from uropathogenic E. coli [UPEC]), septicemia, and wound infections. These infections result in significant morbidity and mortality and in high health care costs. In view of the increasing number of ExPEC infections and the ever-growing antibiotic resistance capability of ExPEC isolates, preventive measures such as an effective vaccine against ExPEC are desirable. An ExPEC vaccine may be cost-effective for select patient groups. Previous vaccine candidates consisted of single target proteins or whole ExPEC cells. Here we describe a subunit vaccine against ExPEC which is based on immunodominant epitopes of the virulence-associated ExPEC proteins FyuA, IroN, ChuA, IreA, Iha, and Usp. Using a novel approach of computer-aided design, two completely artificial genes were created, both encoding eight peptide domains derived from these ExPEC proteins. The recombinant expression of these two genes resulted in a protein vaccine directed against ExPEC but not against commensal E. coli of the gut flora. In mice, the vaccine was highly immunogenic, eliciting both strong humoral and cellular immune responses. Nasal application resulted in high secretory immunoglobulin A (sIgA) production, which was detectable on the mucosal surface of the urogenital tract. Finally, it conveyed protection, as shown by a significant reduction of bacterial load in a mouse model of ExPEC peritonitis. This study provides evidence that a novel vaccine design encompassing distinct epitopes of virulence-associated ExPEC proteins may represent a means for providing a protective and pathogen-specific vaccine.
Escherichia coli is among the most common bacterial species encountered in clinical microbiology laboratories. Although E. coli strains represent a significant part of the normal gut flora, distinct E. coli pathotypes may cause either diarrhea and gastroenteritis (intestinal pathogenic E. coli [IPEC]) or infections outside the gastrointestinal tract (extraintestinal pathogenic E. coli [ExPEC]) (41).
ExPEC strains can reside in the gut as part of the normal intestinal flora and can be isolated from 10 to 20% of healthy individuals (12). However, their entry into and colonization of extraintestinal sites result in a wide variety of infections, which occur in patients from the ambulatory, long-term-care, and hospital settings (23, 39). Diverse organs and anatomical sites are affected. Typical extraintestinal infections due to ExPEC include urinary tract infections (UTIs), surgical site infections, soft tissue infections, newborn meningitis, diverse intra-abdominal infections, and pneumonia. Among these, ascending urinary tract infection (pyelonephritis) most commonly leads to severe sepsis, which ranks as the 10th overall cause of death in the United States (13, 23, 31, 42). Since ExPEC strains are the major cause of most types of extraintestinal infection due to Gram-negative bacteria, prevention of ExPEC infections is a desirable goal from both medical and economic viewpoints (39).
In the past, ExPEC strains were usually highly susceptible to common antibiotics such as ampicillin and trimethoprim-sulfamethoxazole (SXT). However, in recent years, the prevalence of E. coli resistance to various classes of antibiotics has risen progressively, becoming a major concern in both hospitals and the community. For example, resistance to SXT, the traditional drug of choice for uncomplicated UTIs, has increased each year worldwide (17, 18). Moreover, many clinical ExPEC isolates have acquired genes encoding extended-spectrum β-lactamases (ESBLs), which confer resistance to extended-spectrum cephalosporins and aztreonam (50). ESBL-positive ExPEC strains frequently contain additional resistance determinants, e.g., for aminoglycosides and tetracyclines. Thus, emerging antimicrobial resistance likely will make the future management of extraintestinal E. coli infections more difficult and costly than ever. Furthermore, the incidence of serious extraintestinal infection due to E. coli increases with age (2, 30), and as the proportion of elderly patients increases, it is likely that so will the number of extraintestinal E. coli infections. Thus, a preventive strategy, such as vaccinations, is very desirable to counteract these infections.
An ideal vaccine target should be (i) exposed on the bacterial surface and (ii) widely distributed among clinical ExPEC isolates but not among commensal E. coli strains of the gut flora. Furthermore, it should (iii) possess epitopes that are conserved across diverse ExPEC strains and (iv) elicit a protective immune response. Other desirable characteristics of vaccine targets include (v) increased expression at the site of infection and (vi) a role in the pathogenesis of disease.
In the present study, we developed a novel multiepitope subunit vaccine against ExPEC infection which fulfils these criteria. We hypothesized that subunits of the E. coli outer membrane siderophore receptors FyuA, IroN, and IutA, the heme receptor ChuA, and the uropathogenic E. coli (UPEC)-specific protein UspA could be used as vaccine targets to prevent the majority of infections due to extraintestinal E. coli. The goals of this study were (i) to use computer-predicted immunogenic epitopes of the outer protein loops of these proteins in a rational vaccine approach to form two artificial chimeric polypeptides, (ii) to apply either of these two novel multiepitope subunit vaccines by the nasal route to elicit both strong humoral and cellular immune responses, and (iii) to provide a high degree of protection in a mouse model of ExPEC peritonitis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. The clinical ExPEC isolates included 40 strains from patients with pyelonephritis, 40 cystitis isolates, and 40 isolates from patients suffering septicemia. These clinical isolates were collected between 2002 and 2005 in the university hospitals of Munich, Germany, and Helsinki, Finland. Additionally, 40 E. coli isolates from stool samples of healthy volunteers were collected. All strains were cultured on LB agar plates or in LB broth with appropriate antibiotic concentrations (e.g., ampicillin at 100 μg/ml and kanamycin at 50 μg/ml). UPEC strain CFT073 was isolated from the urine and blood of a patient with acute pyelonephritis (32). Strain CFT073 for mouse challenge infections was grown in LB medium at 37°C with aeration for 24 h, harvested by centrifugation (4°C, 3,800 × g, 10 min), and resuspended in a 25% (vol/vol) glycerol solution. The aliquots were stored at −80°C, and CFU were determined by plating serial dilutions in triplicate. Before use, aliquots were thawed on ice, washed twice in 2 ml ice-cold phosphate-buffered saline (PBS) (by centrifugation at 2,655 × g for 3 min at 4°C), and resuspended in prewarmed PBS immediately before administration to reach 3 × 106 CFU per 200 μl. The CFU of the administered doses were verified by plating serial dilutions of the infection doses on LB agar in duplicate.
TABLE 1.
Bacterial strains, plasmids, oligonucleotides, and peptides used in this study
| Strain, plasmid, cosmid, oligonucleotide, or peptide (mass [Da]) | Relevant characteristic(s), description, or sequence (5′-3′)a | Reference(s) or source |
|---|---|---|
| Strains | ||
| Top10 | Laboratory strain for cloning | Invitrogen, Carlsbad, CA |
| M15/pREP4 | Expression strain with pREP4 | Qiagen, Hilden, Germany |
| SCS110 | Strain for cloning and expression of lac operon-based system, has chromosomally carried lac repressor | Stratagene, La Jolla, CA |
| BL21/pLysS | Expression strain with pLysS | Promega, Madison, WI |
| CFT073 | Used for challenge infection in the mouse model | 32 |
| Plasmids and cosmids | ||
| pQE30 Vol1 | Expression vector with Vol1 cloned in via BamHI and KpnI sites | This study |
| pQE30 Vol2 | Expression vector with Vol2 cloned in via BamHI and KpnI sites | This study |
| pQE30 DHFRm45 | Expression vector for mouse dihydrofolate reductase coupled with a C-terminal m45 tag and an N-terminal 6×His tag | This study |
| pQE30 iha | Expression vector with Iha gene cloned via BamHI and SalI sites, using PCR primers Iha.BamHI.for and Iha.SalI.rev | This study |
| pQE30 usp | Expression vector with usp gene cloned via SstI and PstI sites, added by PCR with the primers usp.SstI.for and usp.PstI.rev | This study |
| pASK.IBA 33+ FyuA | Expression vector with FyuA gene cloned via SstI and PstI sites, added by PCR using primers FyuA.SstI.for and FyuA.PstI.rev | This study |
| pASK-IBA37+ IroN | Expression vector with IroN gene cloned via SstI and PstI sites, using primers IroN.SstI.for and IroN.PstI.rev | This study |
| pBAD HisA ChuA | Expression vector with ChuA gene cloned via SstI and PstI sites, using primers ChuA.SstI.for and ChuA.PstI.rev | This study |
| Oligonucleotides | ||
| pQE40.PstI.for | AAGTACTGCAGGACTCCTGTTGATAGATCCA | This study |
| pQE40.m45.PstI.rev | TCAGATCTGCAGTTAcaggatgcgggtttcggtttcgaatggaggcagacggtcacgggaacgatccatTTTCTTCTCGTAGACTTCAAACTTATACT | This study |
| IroN.SstI.for | ATCAGAGCTCAGAATTAACAAAATCCTCTGGTC | This study |
| IroN.PstI.rev | AAACTGCAGGAATGATGCGGTAACTCCGGCATA | This study |
| FyuA.SstI.for | ATCAGAGCTCAAAATGACACGGCTTTATCCTCT | This study |
| FyuA.PstI.rev | AAACTGCAGGAAGAAATCAATTCGCGTATTGAT | This study |
| Usp.SstI.for | ATCAGAGCTCCTACTGTTCCCGAGTAGTGTGTTG | This study |
| Usp.PstI.rev | AAACTGCAGTCTCCTGTAGTGAATTTCATCATG | This study |
| ChuA.SstI.for | ATCAGAGCTCTTGTTGGCTTTGGCTGTTTCTGCC | This study |
| ChuA.PstI.rev | AAACTGCAGCCATTGATAACTCACGAAAATTTT | This study |
| Iha.BamHI.for | ATCAGGATCCCGAATAACCACTCTGGCTTCCGTA | This study |
| Iha.SaII.rev | AAAGTCGACGAACTGATAGTTCAGCGACATCCA | This study |
| Synthetic peptides | ||
| IreA 1 (3,254.70) | GIAKAFRAPSIREVSPGFGTLTQGGASIMYGN | This study |
| IreA 2 (4,169.56) | RRKSDDESLNGKSLKGEPLERTPRHAANAKLEWDYT | This study |
| IreA 3 (2,981.33) | LRDDSATGKKTTETQSVSIKQKAVFIE | This study |
| IutA 1 (3,757.03) | NRVDDFIDYTQQQKIAAGKAISADAIPGGSVDYDN | This study |
| IutA 2 (5,697.39) | FSQGVALPDPGKYYGRGIYGAAVNGHLPLTKSVNVSDSKLEGVKVDSYELGWR | This study |
Restriction sites are marked by underlining. Lowercase letters indicate the m45 tag sequence.
SDS-PAGE and Western blot analysis.
Discontinuous one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described by Ausubel et al. (3), using a Protran II Mini-Vertical unit (Bio-Rad, Munich, Germany). After electrophoresis, gels were either stained with Coomassie blue or transferred to a nitrocellulose membrane (Trans Blot cell; Bio-Rad, Munich, Germany). The blot was blocked with skim milk and incubated with mouse monoclonal antibodies against the m45 tag followed by goat anti-mouse immunoglobulin G (IgG)-peroxidase conjugate or goat anti-mouse IgA-peroxidase conjugate (both from Sigma-Aldrich). Blots were developed using enhanced chemiluminescence (ECL) detection reagents (Amersham Pharmacia Biotech). Dilutions of secondary antibodies used for blot delivery are stated in the legends to Fig. 2 and 3. For dot blot analysis, 10 μl of probe solution was pipetted and bound to nitrocellulose membranes directly. Blocking and detection were performed as described above.
FIG. 2.
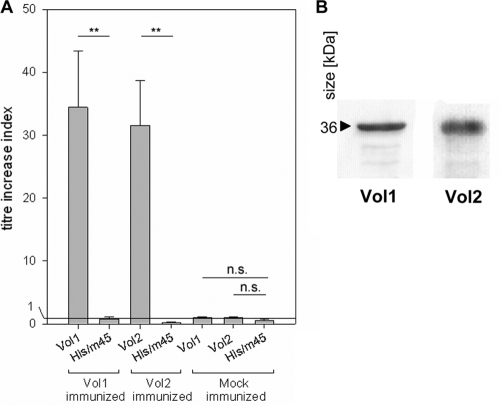
Titer increases of vaginal wash IgA antibodies specific for the vaccine proteins Vol1 and Vol2 after intranasal vaccination. (A) ELISA plates were coated with the Vol1 and Vol2 vaccine proteins. As a control, His-DHFR-m45 protein was used to exclude an antibody response against the N- or C-terminal tag sequences. (B) Western blot of the Vol1 and Vol2 vaccine proteins with the vaginal washes of mice immunized with Vol1 and Vol2, respectively. Vaginal washes were diluted 1:250, and the detection antibody, anti-mouse IgA-horseradish peroxidase, was diluted 1:1,000. **, P < 0.01; n.s., not significant.
FIG. 3.
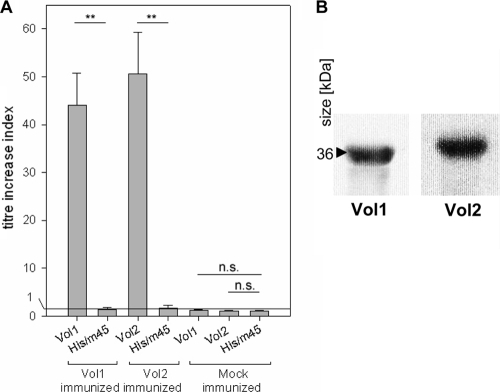
(A) Titer increases of serum IgG/IgM antibodies specific for the Vol1 and Vol2 vaccine proteins after intranasal vaccination. As a control, His-DHFR-m45 protein was used to exclude an antibody response against the N- or C-terminal tag sequences. (B) Western blot of the Vol1 and Vol2 vaccine proteins with sera of Vol1- and Vol2-immunized animals, respectively. Sera were diluted 1:2,500, and the detection antibody, anti-mouse IgG/IgM-horseradish peroxidase, was diluted 1:10,000. **, P < 0.01; n.s., not significant.
Prediction of structures of vaccine target proteins.
To identify the extracellular domains of the virulence factors, we used software which is available free of charge via the Internet. The results of the following four programs were taken into consideration: (i) TM BETA (http://psfs.cbrc.jp/tmbeta-net/), (ii) PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB/), (iii) HMM-B2 TMR (http://gpcr.biocomp.unibo.it/biodec/), and (iv) PROFtmb (http://cubic.bioc.columbia.edu/services/ProfTMB/index.html). Detailed results of the predictions are given in Fig. S1 to S4 in the supplemental material. Manual optimization of results was performed by inspection of the amino acid sequences and the results of each software algorithm as well as by comparison to homologous outer membrane siderophore receptor proteins with known structures, such as FepA and FhuA (6, 8).
Epitope prediction.
To ensure effective T-cell stimulation, we also predicted major histocompatibility complex class I (MHC-I) and MHC-II epitopes contained in the virulence factor proteins which would be recognized by our model organism, the BALB/c mouse. This inbred mouse strain carries the Kd, Ld, and Dd alleles for MHC-I and the Ld and Dd alleles for MCH-II (21). The simulation was performed using the hidden Markov model-based program Rankpep (35). This server-based application predicts peptides able to bind to MHC-I and MHC-II molecules from protein sequences or sequence alignments by using position-specific scoring matrices (PSSMs) (http://immunax.dfci.harvard.edu/Tools/rankpep.html). For MHC-II epitopes, nonamers were selected, whereas for MHC-I epitopes, groups of 8 to 11 amino acid (aa) residues were selected.
Proteasome cleavage site prediction.
To ensure effective presentation of the epitopes of the vaccine proteins in case of uptake, processing, and presentation of these extracellular antigens with MHC class I molecules to CD8 T cells (cross presentation), proteasome cleavage sites of the virulence factors and again of the multiepitope proteins were simulated with the computer programs Rankpep (35) and Net Chop (http://www.cbs.dtu.dk/services/NetChop/). Simulated MHC-I epitopes with a putative cleavage site on the C-terminal end were preferred (49).
Construction, expression, and purification of multiepitope proteins.
The multiepitope vaccine proteins were planned as protein sequences and then reversely translated into coding DNA sequences by using the optimized codon usage of Enterobacteriaceae included in Lasergene software (DNAStar Inc., Madison, WI). Optimization to exclude restriction enzyme sites and GC- or AT-rich repeats and to optimize mRNA substructure was performed manually. For mRNA structure prediction, we used bioinformatic tools such as GeneBee (Belozersky Institute, Moscow State University) and RNAFold (16). Further enhancement of the sequence to remove internal ribosomal entry sites or chi sites was performed at Geneart (Regensburg, Germany) with the proprietary software tool GeneOptimizer.
The genes coding for the multiepitope vaccine proteins were cloned into BamHI and SalI sites of the expression vector pQE-30 (Qiagen, Hilden, Germany). Plasmids were established in the E. coli laboratory strain SCS110 (Stratagene Europe, Amsterdam, Netherlands). After the sequence of the insert was confirmed by sequencing, the plasmid was transformed into E. coli strain M15(pREP4) for protein expression (Qiagen, Hilden, Germany). Induction was performed as described in the manufacturer's instructions. In brief, an overnight culture was inoculated into fresh medium and grown at 37°C until mid-log phase (optical density at 600 nm [OD600] of 0.5). Protein expression was then induced with IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were harvested by centrifugation and lysed in lysis buffer. The multiepitope proteins (Vol1 and Vol2), which were tagged with a 6×His tag at the N-terminal side, were purified using Ni-nitrilotriacetic acid (Ni-NTA) columns. Several wash steps were performed, and the protein was then gel filtered to ensure high purity. The concentration of vaccine protein in the final elution protein sample buffer (5 M urea, 100 mM NaCl, 20 mM Na2HPO4, pH 7.4), determined by the biuret method, was 6.2 mg/ml for Vol1 and 5.5 mg/ml for Vol2.
The control protein His-DHFR-m45 was expressed and purified exactly like the vaccine proteins. Its concentration in elution buffer was determined to be 4.8 mg/ml. All purification steps were performed under denaturing conditions in the presence of urea, as this was found to produce the highest protein yield as well as the highest level of purity.
Measurement of immune response in the mouse. (i) ELISA.
The antibody response was measured by an enzyme-linked immunosorbent assay (ELISA). In brief, 96-well microtiter plates (MaxiSorb; Nunc, Wiesbaden, Germany) were coated with 200 μl of 30-μg/ml recombinant vaccine protein Vol1 or Vol2 in coating buffer (60 mM carbonate buffer, pH 9.6). Afterwards, unspecific binding sites were saturated with 1% bovine serum albumin (BSA) in PBS for 1 h at 30°C. Sera and vaginal wash fluids of the mice were serially diluted in PBS with 3% BSA and incubated for 2 h in a wet chamber at 37°C. After intense washing with Tris-buffered saline-Tween 20 (TBS-T) buffer, the alkaline phosphatase (AP)-conjugated detection antibody, class-specific goat anti-mouse IgG/IgM (Sigma) for sera or goat anti-mouse IgA (Dianova) for vaginal washes, was added. The detection reaction was performed by incubating the sample with Sigma 104 substrate for 1 h at room temperature. Absorbance was measured with an ELISA reader (Sunrise remote; Tecan-Austria, Groeding, Austria) at a wavelength of 405 nm. Titers of mouse preimmune and hyperimmune sera (drawn at day −1 and day 30, respectively) were always measured on the same ELISA plate, and titer increases were determined by calculating the ratio of OD405 values before and after immunization, at a dilution of 1:500 for serum IgG or 1:100 for vaginal wash IgA. To exclude specific antibody responses against the N-terminal 6×His tag or the C-terminal m45 tag, titer increases against a control protein, dihydrofolate reductase coupled to His and m45 tags (6×His-DHFR-m45), were also determined.
(ii) Antibodies.
Mouse sera were obtained by bleeds from the tail vein. Secretory IgA (sIgA) antibodies were measured in vaginal washes obtained from anesthetized mice by washing the vaginal mucosa with 100 μl of PBS via a micropipette tip.
(iii) ELISPOT assay.
On day 31 after the first immunization, splenocytes were analyzed directly in vitro for T-cell responses by use of a gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay as described previously (15, 47). The assays were performed in 96-well Nunc silent screen plates (Nunc GmbH, Wiesbaden, Germany) with a nitrocellulose back coated with a rat anti-mouse IFN-γ monoclonal antibody (Biosource). Isolated splenocytes (1.5 × 106 per well) were prestimulated for 4 h in round-bottomed 96-well microtiter plates with 33 μg/ml vaccine protein Vol1 or Vol2 or with BSA. Additional controls included buffer solution (negative control) and 45 μg/ml concanavalin A (ConA) (positive control). Subsequently, activated cells (1 × 106 per well) were transferred to ELISPOT plates and incubated without vibration for 12 h at 37°C. ELISPOT plates were developed with a biotin-labeled rat anti-mouse IFN-γ monoclonal antibody (Pharmingen, San Diego, CA), horseradish peroxidase-streptavidin conjugate (Dianova, Hamburg, Germany), and aminoethylcarbazole dye solution. The frequency of antigen-specific cells was calculated as the number of spots per 1 × 106 splenocytes.
Active immunization and challenge.
Animal experiments were approved by the German authorities and were performed according to legal requirements. Seven-week-old pathogen-free female BALB/c mice were purchased from Harlan-Winkelmann (Borchen, Germany). For the experiments, the animals were housed in positive-pressure cabinets (Tecniplast, Hohenpeissenberg, Germany). Food and water were offered ad libitum. Each experiment was repeated at least twice. The mice were immunized intranasally with either recombinant vaccine protein (Vol1 or Vol2) or protein buffer. For intranasal vaccination, the mice were anesthetized with diethyl ether. Subsequently, using 20-μl Eppendorf GELoader tips, each nostril was inoculated with 10 μl of an aqueous solution containing either (i) vaccine protein Vol1 or Vol2 (15 μg/10 μl) plus cholera toxin (CT; Quadratech, Surrey, United Kingdom) (0.5 μg/10 μl) or (ii) CT (0.5 μg/10 μl) diluted in phosphate-buffered saline. The procedure was started on day 0 and repeated on days 3, 7, 10, and 24, in a booster scheme.
Challenge infection was performed on day 31 by intraperitoneal injection of E. coli strain CFT073 at a dose of 3 × 106 CFU suspended in 200 μl of prewarmed PBS. Forty-eight hours after the infection, livers and spleens of the mice were aseptically removed, homogenized in PBS supplemented with 0.2% Tergitol, and plated out in triplicate in serial dilutions to determine the bacterial load.
Statistical analyses.
Individual data sets were analyzed using SigmaStat software, version 3.5 (Systat, Erkrath, Germany). Tests used were the Mann-Whitney rank sum test or Student's t test, as appropriate. P values of <0.05 were considered statistically significant.
RESULTS
Identification and selection of vaccine targets.
The premise of the multiepitope subunit vaccine described in this study was to include virulence-associated proteins that are highly abundant among ExPEC strains but only rarely found among nonpathogenic E. coli strains of the commensal gut flora. In order to choose the most applicable target proteins as vaccine candidates, we determined the prevalence of putative virulence-associated genes among 120 clinical ExPEC strains isolated from different hospitals in Germany. Using an ExPEC-specific DNA microarray carrying probes for 100 ExPEC genes, we screened 120 ExPEC isolates and 40 E. coli isolates from stool samples of healthy volunteers for the presence of virulence-associated genes according to previously published protocols (4, 10). Besides the high prevalence among ExPEC strains (and scarcity among commensal strains), a further criterion for choosing the vaccine target proteins was their location at the bacterial cell surface. Based on these criteria, we selected 7 genes. The fyuA gene was present in 81% of ExPEC strains, and 72% of ExPEC strains carried the chuA gene. The other selected genes were usp (68%), iroN (58%), iutA (55%), ireA (40%), and ihaA (32%). Except for the uropathogenic-specific protein (Usp), all other targets represent outer membrane proteins involved in siderophore-mediated bacterial iron uptake or acquisition of host heme-derived iron. All of these siderophore receptors are surface-exposed outer membrane proteins and are strongly expressed during infection (5, 19).
Prediction of epitopes and in silico construction of two multiepitope vaccine proteins.
The aim of this study was to design two completely artificial multiepitope subunit vaccine proteins consisting of highly immunogenic surface-exposed fragments and several putative MHC-I and MHC-II epitopes. For this purpose, we first predicted the protein structures of all siderophore receptor proteins in silico by simulation on a computational basis, using different publicly available programs as described in Materials and Methods. The siderophore receptors are known to consist of transmembrane beta-barrel sheets crossing the outer membrane and several alpha-helical loops at the outer surface as well as in the periplasmic space. The classification of transmembrane or outer surface-exposed protein domains results primarily from the hydrophobicity and steric structure of the respective polypeptide. All predictions were further correlated with the known crystal structure of the siderophore receptors FhuA and FepA (6, 8) to improve the results and to differentiate between outer surface domains exposed to either the extracellular or periplasmic space. Figure 1 shows an example of the prediction of surface loops and transmembrane domains of the siderophore receptor FyuA, as derived from PROFtmp software (data from further algorithms are given in Fig. S1 in the supplemental material).
FIG. 1.
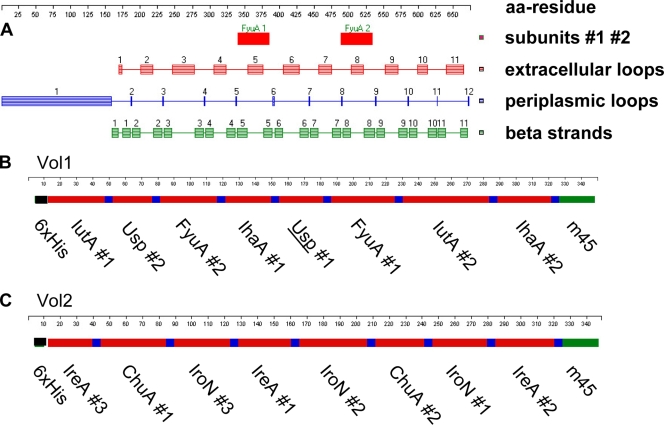
(A) Example of structure prediction of the FyuA protein, using PROFtmb software. The protein is drawn in linear form from left to right. The start codon is numbered 1. Schemes of the vaccine proteins Vol1 (B) and Vol2 (C) are shown, including peptide subunits (red), spacers (blue), the m45 tag (green), and His tag sequences (black).
In addition to the structure analysis, we performed predictions of putative MHC-I and MHC-II epitopes within the outer surface domains exposed to the extracellular space. These computer-aided predictions used the Rankpep algorithm (35) and were focused on the well-defined alleles of the BALB/c mouse, which was used in the following animal experiments (21). Finally, we simulated and predicted probable proteasome cleavage sites for each of the outer surface domains. In order to ensure an optimal presentation of the predicted epitopes of the vaccine target protein by the MHC-I pathway, those epitopes with C-terminal proteasome cleavage sites were preferred (49).
All the information was manually merged to identify subfragments of the virulence-associated outer membrane proteins which fulfilled all of the previously mentioned criteria. The short amino acid sequences selected were predominantly hydrophilic, putatively surface exposed, and rich in predicted MHC-II as well as MHC-I epitopes. The structure and origin of the respective subfragments are given in Table 2.
TABLE 2.
Composition of multiepitope vaccine proteins Vol1 and Vol2
| Multiepitope vaccine protein | Target protein | Subfragment no. | Position (aa) | No. of aa | No. of putative epitopes |
|
|---|---|---|---|---|---|---|
| MHC-I | MHC-II | |||||
| Vol1 | IutA | 1 | 438-472 | 35 | 3 | 2 |
| IutA | 2 | 493-545 | 53 | 5 | 3 | |
| IhaA | 1 | 206-233 | 28 | 4 | 1 | |
| IhaA | 2 | 593-625 | 33 | 5 | 1 | |
| FyuA | 1 | 345-383 | 39 | 6 | 2 | |
| FyuA | 2 | 496-530 | 35 | 3 | 1 | |
| Usp | 1 | 298-324 | 27 | 4 | 3 | |
| Usp | 2 | 86-109 | 24 | 1 | 2 | |
| Vol2 | IreA | 1 | 436-467 | 32 | 2 | 2 |
| IreA | 2 | 559-594 | 36 | 3 | 1 | |
| IreA | 3 | 364-390 | 27 | 2 | 1 | |
| ChuA | 1 | 340-379 | 40 | 3 | 2 | |
| ChuA | 2 | 497-526 | 30 | 3 | 2 | |
| IroN | 1 | 274-306 | 33 | 4 | 1 | |
| IroN | 2 | 480-520 | 41 | 3 | 1 | |
| IroN | 3 | 554-587 | 34 | 2 | 2 | |
Preparation of two multiepitope subunit vaccine proteins.
Each of the two multiepitope vaccine proteins was designed to consist of eight single subfragments derived from the virulence-associated proteins, separated by spacers (Fig. 1). Thus, two new multiepitope vaccine proteins, designated Vol1 and Vol2, with calculated molecular masses of 36,192 Da and 36,189 Da, respectively, were created. The proline-glycine linker elements used between the subfragments consisted of the amino acid sequence GPGPG. Due to their shape, these peptide linkers are unlikely to be presented on MHC receptors (44). Moreover, due to the steric rigidity of the prolines, the high proline content of the GPGPG motif efficiently breaks up artificial secondary protein structures, such as helices, between adjacent peptides. Fewer than 8% of predicted epitopes of the final vaccine proteins contained parts of the spacer (data not shown). For purification, the well-characterized 6×His tag was fused to the N terminus of each vaccine protein (Fig. 1). The larger molecule size than that of single peptides should result in improved presentation of the inherited epitopes carried on both of the target proteins. The multiepitope structure of the two vaccine proteins enabled the inclusion of different peptides of distinct target proteins (siderophore receptors) and should therefore cover a broader range of ExPEC strains than a single peptide-based vaccine. Lastly, the hydrophilic properties of the two novel multiepitope vaccine proteins facilitated handling and recombinant expression in bacteria, in contrast to the respective hydrophobic full-length siderophore receptor proteins. Endotoxin contamination in the proteins used for vaccination was determined and found to equal a lipopolysaccharide (LPS) load of 0.039 ng per mouse per administration for Vol1 and 0.0098 ng per mouse per administration for Vol2.
Vaccination with multiepitope subunit vaccine by nasal administration.
To elicit a strong mucosal immune response, we used intranasal inoculation of the vaccine target proteins together with the adjuvant cholera toxin. For this purpose, purified recombinant multiepitope proteins Vol1 (36,192 Da) and Vol2 (36,189 Da) were mixed with PBS and CT to concentrations of 1.5 mg/ml of vaccine protein and 0.05 mg/ml CT. Control groups were immunized with a mixture of CT (0.05 mg/ml), PBS, and protein sample buffer containing urea without any protein. After primary immunization on day 0, mice were given booster doses on days 3, 7, 10, and 24.
The specific antibody reactions against the vaccine proteins Vol1 and Vol2 were determined by ELISA, followed by calculating the titer increase ratio for serum IgG as well as for vaginal wash IgA antibodies. A strong increase of specific antibody titer against the vaccine protein could be documented for both IgA and IgG after vaccination with vaccine proteins Vol1 (median increase index, 36.4 for IgA and 45.9 for IgG) and Vol2 (median increase index, 29.1 for IgA and 52.7 for IgG) (Fig. 2 and 3) (all P values were <0.008). The specificity of the ELISA results was verified by Western blot analyses using the respective vaccine proteins. In contrast, no titer increases could be detected against the control protein His-DHFR-m45, which shares the His and m45 tags with Vol1 and Vol2. Control sera of mock-immunized mice, which received protein buffer and CT, showed no titer increase against either the vaccine proteins or the control protein His-DHFR-m45 (Fig. 2 and 3).
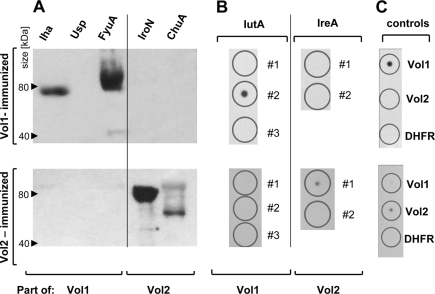
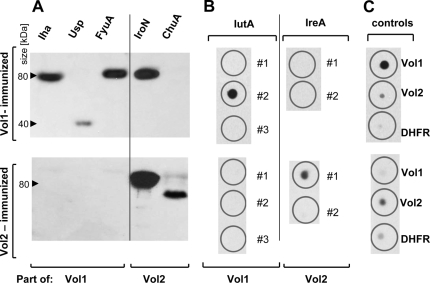
Next, we sought to identify those full-length target proteins bound by the antibodies evoked by vaccination with the recombinant vaccine proteins Vol1 and Vol2. For this purpose, Western blot analyses were performed with the full-length proteins Iha, Usp, FyuA, IroN, and ChuA. However, IutA and IreA could not be purified from E. coli. We therefore applied dot blot analysis to investigate the specific antibody responses against the IutA 1, 2, and 3 and IreA 1 and 2 peptides, which are included in the Vol1 and Vol2 vaccine proteins, respectively.
After vaccination with Vol1, IgA antibodies against the full-length proteins Iha and FyuA and against the IutA 2 peptide could be detected (Fig. 4). No cross-reactivity to Vol2 or His-DHFR-m45 was observed. However, serum IgA from Vol1- immunized mice reacted with neither the full-length Usp protein nor the IutA 1 and 3 peptides. As expected, no reactivity could be detected for Vol1-stimulated serum IgA against Vol2-specific targets, including the IroN and ChuA proteins and the IreA 1 and 2 peptides (Fig. 4).
FIG. 4.
IgA immunoblots of sera obtained from Vol1- and Vol2-immunized mice. (A) IgA binding to full-length target proteins for Vol1-immunized serum (upper part) and Vol2-immunized serum (lower part). (B) IgA binding to peptides derived from IutA and IreA. The peptides included in the vaccine proteins are numbered 1, 2, and 3. (C) Controls comprised full-length Vol1 and Vol2 proteins as well as the murine DHFR protein containing an N-terminal His tag and a C-terminal m45 tag to exclude an antibody response against the tag sequences.
The results for serum IgG of Vol1-immunized mice were similar. However, weak binding of serum IgG to Usp and cross-reactivity to the Vol2-specific protein IroN were discovered. Correspondingly, after Vol1 immunization, some cross-reactivity against the vaccine protein Vol2 was observed in the dot blot (Fig. 5).
FIG. 5.
IgG immunoblots of sera obtained from Vol1- and Vol2-immunized mice. (A) IgG binding to full-length target proteins for Vol1-immunized serum (upper part) and Vol2-immunized serum (lower part). (B) IgG binding to peptides derived from IutA and IreA. The peptides included in the vaccine proteins are numbered 1, 2, and 3. (C) Controls comprised full-length Vol1 and Vol2 proteins as well as the murine DHFR protein containing an N-terminal His tag and a C-terminal m45 tag to exclude an antibody response against the tag sequences.
After Vol2 immunization, serum IgA (Fig. 4) and IgG (Fig. 5) antibodies were detected that reacted with the (Vol2-associated) full-length IroN and ChuA proteins and the IreA 1 peptide (Fig. 4). No reaction against IreA 2 was seen. In contrast, no IgA or IgG cross-reactivity was observed with Vol1-specific full-length proteins or peptides or the Vol1 vaccine protein itself.
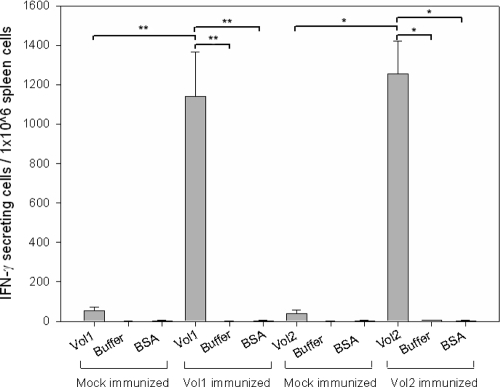
To investigate cellular immune responses, spleens of vaccinated animals were harvested on day 31 and reactive splenocytes were counted using ELISPOT in vitro restimulation assays as described in Materials and Methods. The average numbers of specific IFN-γ-secreting splenocytes in mice immunized with Vol1 and Vol2 were 1,148 (Vol1) and 1,277 (Vol2) per 1 million splenocytes. These represent significant increases compared to the mock-immunized groups, which received CT and protein buffer (P = 0.008 for Vol1 and P = 0.016 for Vol2) (Fig. 6). In all examined groups, protein buffer alone and BSA suspended in protein buffer resulted in activation values as low as those from nonstimulated splenocytes. Interestingly, mock-immunized mice exhibited slightly increased numbers of specific reactive splenocytes against Vol1 and Vol2 (Fig. 6).
FIG. 6.
IFN-γ-producing splenocytes in ELISPOT assay after intranasal immunization. In vitro restimulation was performed with the whole vaccine protein Vol1 or Vol2, as indicated, with protein buffer, or with protein buffer with BSA. Mock-immunized mice received CT with protein buffer intranasally. *, P < 0.05; **, P < 0.01.
Challenge in a mouse model of peritonitis.
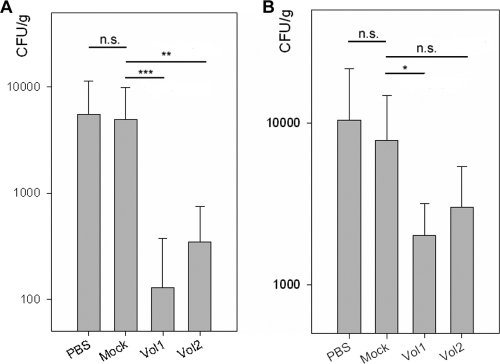
In order to determine the effect of multiepitope vaccination against ExPEC infections, challenge experiments were performed in a mouse model of peritonitis. After 30 days of immunization (with doses administered in a booster scheme on days 0, 3, 7, 10, and 24), mice were infected by intraperitoneal administration of 3 × 106 CFU of E. coli CFT073. Mice were sacrificed 48 h after infection, and the bacterial loads in spleens and livers were determined. Mock-immunized mice that received CT and protein buffer showed no significant reduction of bacterial loads compared to mock-immunized mice that received PBS (P = 0.872). In contrast, the groups of mice immunized with either Vol1 or Vol2 showed reduced bacterial loads in the spleen (for Vol1, P = 0.017; and for Vol2, P = 0.107) and, to an even greater degree (i.e., a 2-log reduction), in the liver (for Vol1, P = 0.001; and for Vol2, P = 0.002) (Fig. 7). Mice immunized with Vol1 exhibited a nonsignificant trend toward better protection than that for mice immunized with Vol2.
FIG. 7.
Bacterial load per organ weight for livers (A) and spleens (B) of mice 48 h after challenge infection. The PBS group was immunized with PBS alone, the mock-immunized group received CT with protein buffer, and the Vol1 and Vol2 groups received vaccine protein in protein buffer, with CT as adjuvant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
DISCUSSION
In this study, we developed a multiepitope, multiple-subunit vaccine against ExPEC by applying a computer-based approach. We used genomic data from E. coli to identify vaccine candidates that are (i) associated with ExPEC virulence, (ii) exposed on the outer surface of the bacterium, (iii) expressed in vivo, and (iv) highly conserved among different ExPEC isolates. After antigen selection and in silico composition of the amino acid sequence, we obtained two completely artificial vaccine proteins (Vol1 and Vol2), each comprising a concatenated series of predicted immunogenic peptides. These were derived from the ExPEC-associated outer membrane iron uptake receptors FyuA, IutA, ChuA, IhaA, IreA, and IroN and the uropathogen-specific protein Usp (27, 28). Immunization of mice with either of the two synthetic vaccine proteins elicited strong humoral and cellular immune responses and protected mice against challenge with an ExPEC strain in an intraperitoneal mouse model of sepsis.
In the past, there has been limited success in developing an efficacious vaccine against ExPEC infections. Past vaccines consisted of wild-type or genetically engineered, inactivated ExPEC (38, 43) or of ExPEC strains in combination with other inactivated uropathogenic bacteria (Urovaxom and Solco-Urovac/StroVac). Other ExPEC vaccines were based on single purified virulence factors, such as the type 1 fimbrial adhesin FimH (29) or hemolysin (34). These vaccine studies succeeded in animal models; however, most approaches failed to provide protection in clinical trials. More recently, attempts have been made to empirically identify single virulence-associated ExPEC proteins, e.g., siderophore receptors, as vaccine targets (1, 11). Despite these advances, a UTI vaccine that confers long-term protection against uncomplicated UTIs is currently lacking.
The present study focused on iron uptake proteins such as siderophore receptors, which have been reported to be highly virulence associated in ExPEC strains (22). Furthermore, up to six iron uptake systems coexist in ExPEC strains (7, 48). Despite the redundancy of these systems, the genes encoding the receptor proteins are among the most highly induced genes, as shown in transcription assays (45). The respective proteins are strongly expressed both in vitro, during growth in human urine, and in vivo, during infection (1, 36). Accordingly, these proteins elicit a humoral response during experimental UTI, as demonstrated by their reaction with antisera from mice infected with UPEC strain CFT073 (19). In spite of the diversity in iron uptake receptors present in distinct ExPEC strains, a further hallmark of these proteins is the presence of certain highly conserved amino acid sequence motifs.
Therefore, it is unsurprising that this group of proteins has been evaluated previously for use in vaccines (40, 42). The selection of the specific vaccine targets used in this study was based on our prevalence study of ExPEC-associated virulence genes by use of microarrays and on previous studies that identified siderophore receptors as promising vaccine targets (42). Additionally, another recent study demonstrated the relevance of iron uptake systems for vaccine development by investigating whole-protein vaccines based on different recombinant siderophore receptors (1).
However, all previously described vaccines were composed of whole proteins or ExPEC strains expressing different virulence-associated proteins (42). A recent attempt to use single peptides derived from the ExPEC siderophore receptors as a vaccine failed to elicit a humoral immune response in mice and to protect mice in a UTI infection model (1). In contrast, in the present study we focused on outer surface domains of ExPEC iron uptake receptors, which are exposed to the immune system and therefore should be highly accessible to humoral and cellular components of the host's immune system. For this purpose, we genetically combined different domains from distinct siderophore receptors and the Usp protein to build up a concatenated artificial vaccine protein.
The rationale for creating a subunit vaccine in the form of a concatenated polypeptide was to take into account the heterogeneity of ExPEC strains and to maximize the immunogenicity of the vaccine. Since little is known about the antigenicity of different outer membrane siderophore receptors, we further aimed to develop an improved ExPEC vaccine by including multiple different subunits for each target protein. In addition, the use of distinct subunits derived from virulence-associated proteins enables a flexible composition of vaccine proteins.
A further argument for the use of subunits was the anticipated better tolerability of multisubunit polypeptides than full-length outer membrane proteins or whole E. coli cells. In a pilot survey, we observed excessive deaths among mice vaccinated using full-length IroN and ChuA proteins compared to those among mice receiving the subunit vaccine (data not shown). This is in agreement with the recent study of Alteri et al., who observed a >30% lethality of mice after vaccination with the whole ChuA protein (1). Finally, the use of only the highly hydrophilic outer surface domains of the outer membrane siderophore receptors avoids integration of hydrophobic transmembrane domains. The resulting hydrophilic multiepitope subunit protein can therefore be expressed and purified more readily than the whole receptor proteins.
For each of the virulence-associated vaccine proteins selected, we searched for (i) surfaced-exposed protein domains and (ii) predictable immunogenic subunits by means of well-established computer-based algorithms. In parallel, the full-length target proteins were screened for putative MHC-I and MHC-II binding domains recognized by the BALB/c mouse, using the Rankpep tool (35). Besides MHC-II epitopes, we also included MHC-I epitopes, since recent data indicate the relevance of both humoral and T-cellular immune responses in the course of ExPEC infections. Thumbikat et al. demonstrated that T-cell-mediated protection against urinary E. coli infections is about as important as that mediated by antibodies (46). Finally, we focused on those subunits of the target proteins which appeared to be both extracellular and rich in MHC-I and -II epitopes.
In a modification of the protocol of Sette et al., we concatenated the selected epitope-rich, surface-exposed protein subunits in silico (44). According to the recommendation of these investigators, who described methods and processes to design multiepitope vaccines for infectious diseases and cancer, we separated each subunit by spacers. The spacers are unlikely to be presented on MHC molecules and therefore reduce the number of artificial epitopes comprised of two different subunits. However, we did not include merely short epitopes but also used epitope-rich peptide subunits of up to 53 aa. To our knowledge, this technique has not been used previously for protein vaccination against bacteria. Only recently have DNA-based vaccination strategies using similarly constructed artificial targets been described (14, 33). Using these DNA vaccines, strong cytotoxic T-lymphocyte (CTL)-mediated responses could be induced, but little attention was paid to antibody responses. In contrast, in the present study we analyzed the humoral response after vaccination with a multiepitope subunit vaccine and demonstrated the induction of antibodies directed against subunit peptides and full-length vaccine proteins alike.
Since most ExPEC infections rise from mucosal surfaces, an effective vaccine should confer immune protection not only systemically but also at the mucous membranes. Mucosal vaccination has been shown to elicit both strong humoral and cellular responses on mucous membranes as well as systemically. In this study, we chose intranasal immunization, since it induces immune responses at all relevant entry sites of ExPEC infections (20, 26). Here we used the adjuvant cholera toxin, which is highly efficient on mucosal surfaces in mice. However, it has not yet been studied extensively in humans, and the toxic potential remains unclear (9). In contrast to previous observations, we found in our study that coadministration of CT and protein antigens without previous cross-linking provided potent stimulation of immune responses (1).
We analyzed target protein-specific serum IgG and vaginal fluid IgA antibodies, as well as titer changes of antibodies against the vaccine proteins, as correlates of the systemic and mucosal humoral immune responses, respectively. IgA antibodies from vaginal fluids have previously been shown to be an important defense mechanism against UPEC, a prominent subgroup of ExPEC (24). We were able to detect strong antibody binding to most full-length virulence-associated proteins with both vaginal wash IgA and serum IgG. In contrast, a study using 30-aa polypeptides from siderophore receptors failed to evoke an antibody response of mucosal/urine IgA after nasal peptide vaccination (1). We thus concluded that the combination of different peptide domains in multiepitope proteins has a higher potential to evoke IgA antibody responses after nasal immunization. This might be due to the size of the multiepitope protein compared to single peptides or could be the result of the combination of different epitopes, which may serve as internal protein adjuvants.
The IgA and IgG antibody responses to IroN were the strongest in the Vol2-immunized mice. However, we also found IroN-specific antibodies after vaccination with Vol1, even though no vaccination was performed with IroN subunits in the Vol1 group. Apparently, there is a certain cross-reactivity after vaccination with Vol1. To exclude cross-reactivity to the N- or C-terminal tag sequences of Vol1 and Vol2, Western blot analysis and ELISA were performed with the control protein His-DHFR-m45. No binding to His-DHFR-m45 protein could be detected. Since the target proteins IutA and IreA could not be expressed in E. coli, the antibody responses to these proteins were checked by dot blot analysis involving the subunit peptides used in the vaccine proteins Vol1 and Vol2, respectively. In the case of IutA, only one of two peptides could be detected, whereas in IreA one of three was bound (Fig. 5). This suggests that only some of the predicted antibody binding sites are indeed relevant in vivo. In summary, we can conclude that even for the well-characterized BALB/c mouse, computer-based algorithms are able to predict only about 50% of relevant B-cell epitopes. This underscores the necessity (i) to include more epitopes in one vaccine protein in order to achieve a strong immune response or (ii) to iteratively refine the polypeptides empirically through repeated in vivo testing of the in silico-designed vaccine targets.
Aside from the humoral immune response and the local influx of neutrophils, the cellular immune system appears to be important for clearing UTIs (25, 37, 46). Published studies on ExPEC vaccine development have paid almost no attention to the role of T-cell-mediated responses against these infections. Only the very recent study conducted by Alteri and coworkers (1) provides some insight into T-cell-dependent cytokine release after vaccination. However, no quantification of reactive T cells has been performed. In the present study, to evaluate T-cell activation, we determined the numbers of IFN-γ-secreting splenocytes specific for the Vol1 and Vol2 vaccine proteins. In both immunized groups, we documented significantly increased numbers of specific IFN-γ-secreting T cells compared to those in the mock-immunized groups (buffer and CT). T-cell restimulation with BSA in vitro showed little reaction, as did restimulation with protein loading buffer. In addition, no significant cross-reactivity between the two vaccine proteins was detected in the restimulation assays (data not shown).
The multiepitope vaccine developed here is directed against ExPEC in general. This pilot study shows the efficacy of the vaccine in a mouse model of peritoneal sepsis. In this model, the vaccinated groups showed significantly reduced bacterial loads in the liver 24 h after infection, with a trend toward reduced splenic bacterial loads. The mock-immunized groups which received protein buffer together with CT showed no significant reduction compared to the buffer-immunized controls. Therefore, the protection can be attributed to immunization with the Vol1 and Vol2 vaccine proteins. This study reveals the high potential of the multiepitope vaccine against ExPEC. Further studies must be performed to elucidate the efficacy of the vaccine in a UTI model.
The present work describes the first single-protein, combined-subunit vaccine against ExPEC. The vaccine induces strong antibody and cellular responses in a mouse model and provides protection against peritoneal sepsis. This combined-subunit vaccine presents a promising example of rational vaccinology. Further studies are needed to define broader applications of this technique to gain an effective preventive measure against infections due to ExPEC and other pathogenic organisms.
Supplementary Material
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (SCHU1494/2-1) and the Bundesministerium für Bildung und Forschung (Kompetenznetzwerk Pathogenomik, Network of Excellence [NoE] Europathogenomic, and ERANET) to S.S. and from a FöFoLe grant (Förderung für Forschung und Lehre) of the University of Munich, LMU, to S.S., D.N., and A.W.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 24 May 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alteri, C. J., E. C. Hagan, K. E. Sivick, S. N. Smith, and H. L. Mobley. 2009. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 5:e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, D. C., W. T. Linde-Zwirble, J. Lidicker, G. Clermont, J. Carcillo, and M. R. Pinsky. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29:1303-1310. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2005. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 4.Bielaszewska, M., U. Dobrindt, J. Gartner, I. Gallitz, J. Hacker, H. Karch, D. Muller, S. Schubert, S. M. Alexander, L. J. Sorsa, and J. Zdziarski. 2007. Aspects of genome plasticity in pathogenic Escherichia coli. Int. J. Med. Microbiol. 297:625-639. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V. 2005. Bacterial iron transport related to virulence. Contrib. Microbiol. 12:210-233. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., M. Braun, and H. Killmann. 2000. Iron transport in Escherichia coli. Crystal structure of FhuA, an outer membrane iron and antibiotic transporter. Adv. Exp. Med. Biol. 485:33-43. [DOI] [PubMed] [Google Scholar]
- 7.Brzuszkiewicz, E., H. Bruggemann, H. Liesegang, M. Emmerth, T. Olschlager, G. Nagy, K. Albermann, C. Wagner, C. Buchrieser, L. Emody, G. Gottschalk, J. Hacker, and U. Dobrindt. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 9.Campos, E. A., J. Namikoshi, S. Maeba, M. Yamamoto, M. Fukumoto, and H. Yamamoto. 2003. Nasally administered cholera toxin A-subunit acts as a mucosal adjuvant. J. Oral Sci. 45:25-31. [DOI] [PubMed] [Google Scholar]
- 10.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durant, L., A. Metais, C. Soulama-Mouze, J. M. Genevard, X. Nassif, and S. Escaich. 2007. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli. Infect. Immun. 75:1916-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 13.Fluit, A. C., J. Verhoef, and F. J. Schmitz. 2001. Frequency of isolation and antimicrobial resistance of gram-negative and gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997-1998. Eur. J. Clin. Microbiol. Infect. Dis. 20:617-625. [DOI] [PubMed] [Google Scholar]
- 14.Gao, H., Y. Yue, L. Hu, W. Xu, and S. Xiong. 2009. A novel DNA vaccine containing multiple TB-specific epitopes casted in a natural structure (ECANS) confers protective immunity against pulmonary mycobacterial challenge. Vaccine 27:5313-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geginat, G., T. Nichterlein, M. Kretschmar, S. Schenk, H. Hof, M. Lalic-Multhaler, W. Goebel, and A. Bubert. 1999. Enhancement of the Listeria monocytogenes p60-specific CD4 and CD8 T cell memory by nonpathogenic Listeria innocua. J. Immunol. 162:4781-4789. [PubMed] [Google Scholar]
- 16.Gruber, A. R., R. Lorenz, S. H. Bernhart, R. Neubock, and I. L. Hofacker. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, K., T. M. Hooton, and W. E. Stamm. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 135:41-50. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, K., T. M. Hooton, C. L. Wobbe, and W. E. Stamm. 1999. The prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in young women. Int. J. Antimicrob. Agents 11:305-308. [DOI] [PubMed] [Google Scholar]
- 19.Hagan, E. C., and H. L. Mobley. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 75:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren, J., and C. Czerkinsky. 2005. Mucosal immunity and vaccines. Nat. Med. 11:S45-S53. [DOI] [PubMed] [Google Scholar]
- 21.Hood, L., M. Steinmetz, and B. Malissen. 1983. Genes of the major histocompatibility complex of the mouse. Annu. Rev. Immunol. 1:529-568. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J. R., M. A. Kuskowski, A. Gajewski, S. Soto, J. P. Horcajada, M. T. Jimenez de Anta, and J. Vila. 2005. Extended virulence genotypes and phylogenetic background of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J. Infect. Dis. 191:46-50. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, J. R., and T. A. Russo. 2002. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J. Infect. Dis. 186:859-864. [DOI] [PubMed] [Google Scholar]
- 25.Jones-Carson, J., E. Balish, and D. T. Uehling. 1999. Susceptibility of immunodeficient gene-knockout mice to urinary tract infection. J. Urol. 161:338-341. [PubMed] [Google Scholar]
- 26.Kantele, A. M., N. V. Palkola, H. S. Arvilommi, and J. M. Kantele. 2008. Distinctive homing profile of pathogen-specific activated lymphocytes in human urinary tract infection. Clin. Immunol. 128:427-434. [DOI] [PubMed] [Google Scholar]
- 27.Kurazono, H., M. Nakano, S. Yamamoto, O. Ogawa, K. Yuri, K. Nakata, M. Kimura, S. Makino, and G. B. Nair. 2003. Distribution of the usp gene in uropathogenic Escherichia coli isolated from companion animals and correlation with serotypes and size-variations of the pathogenicity island. Microbiol. Immunol. 47:797-802. [DOI] [PubMed] [Google Scholar]
- 28.Kurazono, H., S. Yamamoto, M. Nakano, G. B. Nair, A. Terai, W. Chaicumpa, and H. Hayashi. 2000. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb. Pathog. 28:183-189. [DOI] [PubMed] [Google Scholar]
- 29.Langermann, S., R. Mollby, J. E. Burlein, S. R. Palaszynski, C. G. Auguste, A. DeFusco, R. Strouse, M. A. Schenerman, S. J. Hultgren, J. S. Pinkner, J. Winberg, L. Guldevall, M. Soderhall, K. Ishikawa, S. Normark, and S. Koenig. 2000. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181:774-778. [DOI] [PubMed] [Google Scholar]
- 30.McBean, M., and S. Rajamani. 2001. Increasing rates of hospitalization due to septicemia in the US elderly population, 1986-1997. J. Infect. Dis. 183:596-603. [DOI] [PubMed] [Google Scholar]
- 31.Minino, A. M., R. N. Anderson, L. A. Fingerhut, M. A. Boudreault, and M. Warner. 2006. Deaths: injuries, 2002. Natl. Vital Stat. Rep. 54:1-124. [PubMed] [Google Scholar]
- 32.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molder, T., M. Adojaan, K. Kaldma, M. Ustav, and R. Sikut. 2009. Elicitation of broad CTL response against HIV-1 by the DNA vaccine encoding artificial multicomponent fusion protein MultiHIV—study in domestic pigs. Vaccine 28:293-298. [DOI] [PubMed] [Google Scholar]
- 34.O'Hanley, P., G. Lalonde, and G. Ji. 1991. Alpha-hemolysin contributes to the pathogenicity of piliated digalactoside-binding Escherichia coli in the kidney: efficacy of an alpha-hemolysin vaccine in preventing renal injury in the BALB/c mouse model of pyelonephritis. Infect. Immun. 59:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reche, P. A., J. P. Glutting, and E. L. Reinherz. 2002. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 63:701-709. [DOI] [PubMed] [Google Scholar]
- 36.Reigstad, C. S., S. J. Hultgren, and J. I. Gordon. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282:21259-21267. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, J. A. 1999. Unconventional immunology and urinary tract infection. J. Urol. 161:3. [PubMed] [Google Scholar]
- 38.Russo, T. A., J. M. Beanan, R. Olson, S. A. Genagon, U. Macdonald, J. J. Cope, B. A. Davidson, B. Johnston, and J. R. Johnson. 2007. A killed, genetically engineered derivative of a wild-type extraintestinal pathogenic E. coli strain is a vaccine candidate. Vaccine 25:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449-456. [DOI] [PubMed] [Google Scholar]
- 40.Russo, T. A., and J. R. Johnson. 2006. Extraintestinal isolates of Escherichia coli: identification and prospects for vaccine development. Expert Rev. Vaccines 5:45-54. [DOI] [PubMed] [Google Scholar]
- 41.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 42.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, R. Olson, and G. E. Wilding. 2003. The siderophore receptor IroN of extraintestinal pathogenic Escherichia coli is a potential vaccine candidate. Infect. Immun. 71:7164-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidhammer, S., R. Ramoner, L. Holtl, G. Bartsch, M. Thurnher, and C. Zelle-Rieser. 2002. An Escherichia coli-based oral vaccine against urinary tract infections potently activates human dendritic cells. Urology 60:521-526. [DOI] [PubMed] [Google Scholar]
- 44.Sette, A., M. Newman, B. Livingston, D. McKinney, J. Sidney, G. Ishioka, S. Tangri, J. Alexander, J. Fikes, and R. Chesnut. 2002. Optimizing vaccine design for cellular processing, MHC binding and TCR recognition. Tissue Antigens 59:443-451. [DOI] [PubMed] [Google Scholar]
- 45.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thumbikat, P., C. Waltenbaugh, A. J. Schaeffer, and D. J. Klumpp. 2006. Antigen-specific responses accelerate bacterial clearance in the bladder. J. Immunol. 176:3080-3086. [DOI] [PubMed] [Google Scholar]
- 47.Vijh, S., and E. G. Pamer. 1997. Immunodominant and subdominant CTL responses to Listeria monocytogenes infection. J. Immunol. 158:3366-3371. [PubMed] [Google Scholar]
- 48.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.York, I. A., S. C. Chang, T. Saric, J. A. Keys, J. M. Favreau, A. L. Goldberg, and K. L. Rock. 2002. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 3:1177-1184. [DOI] [PubMed] [Google Scholar]
- 50.Zahar, J. R., O. Lortholary, C. Martin, G. Potel, P. Plesiat, and P. Nordmann. 2009. Addressing the challenge of extended-spectrum beta-lactamases. Curr. Opin. Invest. Drugs 10:172-180. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.