Abstract
Autoinducer 2 (AI-2) is widely recognized as a signal molecule for intra- and interspecies communication in Gram-negative bacteria, but its signaling function in Gram-positive bacteria, especially in Staphylococcus aureus, remains obscure. Here we reveal the role of LuxS in the regulation of capsular polysaccharide synthesis in S. aureus NCTC8325 and show that AI-2 can regulate gene expression and is involved in some physiological activities in S. aureus as a signaling molecule. Inactivation of luxS in S. aureus NCTC8325 resulted in higher levels of transcription of capsular polysaccharide synthesis genes. The survival rate of the luxS mutant was higher than that of the wild type in both human blood and U937 macrophages. In comparison to the luxS mutant, a culture supplemented with chemically synthesized 4,5-dihydroxy-2,3-pentanedione (DPD), the AI-2 precursor molecule, restored all the parental phenotypes, suggesting that AI-2 has a signaling function in S. aureus. Furthermore, we demonstrated that the LuxS/AI-2 signaling system regulates capsular polysaccharide production via a two-component system, KdpDE, whose function has not yet been clarified in S. aureus. This regulation occurred via the phosphorylation of KdpE binding to the cap promoter.
Quorum sensing (QS) is a cell-cell communication mechanism in which bacteria secrete and sense small diffusible molecules called autoinducers (AIs) to coordinate social activities, such as bioluminescence, biofilm formation, swarming behavior, antibiotic production, and virulence factor secretion (7, 23, 38, 59). Many QS mechanisms have evolved among bacteria. In general, Gram-negative bacteria use acylated homoserine lactones (AHLs) as AIs, and Gram-positive bacteria use oligopeptide AIs, which act through two-component phosphorelay cascades. Studies have shown that one QS mechanism is shared by both Gram-positive and Gram-negative bacteria and involves the production of autoinducer 2 (AI-2) (4, 38, 59, 60), which is synthesized by the LuxS enzyme in a metabolic pathway known as the activated methyl cycle (50, 57, 61). AI-2 is not a single compound but a family of interconverting compounds derived from 4,5-dihydroxy-2,3-pentanedione (DPD), which cyclizes spontaneously to form two epimeric furanoses, (2R,4S)- and (2S,4S)-2,4-dihydroxy-2-methyldihydrofuran-3-one (R- and S-DHMF), respectively. Hydration of R- and S-DHMF produces (2R, 4S)- and (2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R- and S-THMF), respectively (40). In contrast to the other autoinducers that are usually involved in intraspecies communication, AI-2 is widely present in bacteria, leading to the suggestion that it is a universal language for interspecies communication (50, 64).
The LuxS/AI-2 system is a more recently described QS system which was first identified in Vibrio harveyi, where it functions as part of a complex multilayered QS system to regulate bioluminescence (9, 10). LuxS plays a metabolic role in the activated methyl cycle, and one molecule of DPD is formed as a by-product every one cycle. This gives DPD the ability of being an ideal signal to connect the metabolic activity to population density (12, 61-64). The LuxS/AI-2 QS system is known to be involved in the regulation of a range of behaviors in diverse bacteria, such as biofilm formation in Escherichia coli, virulence-associated traits in Vibrio cholerae, antibiotic susceptibility in Streptococcus anginosus, and motility in enterohemorrhagic E. coli (1, 9, 29, 39, 57). However, in many luxS-containing bacteria, the functions that are controlled by LuxS and whether LuxS is involved in the QS pathway still remain matters of debate.
Staphylococcus aureus is a major nosocomial pathogen with the ability to cause a variety of infectious diseases, from relatively benign skin infections to potentially fatal systemic disorders (3, 32). The pathogenicity is determined by surface-associated adhesins, superantigens, exoenzymes, and exotoxin, which are regulated by a wide range of regulatory systems (14, 18, 41). Among these regulatory elements, the Agr (the accessory gene regulator) system is the only characterized QS system in S. aureus and controls the expression of approximately 150 genes (20). Interestingly, S. aureus also possesses a functional luxS gene and has the ability to produce AI-2, and purified LuxS protein from S. aureus exhibited the catalytic activity of AI-2 production (19, 61). However, no potential AI-2 receptor has been found by searching for established AI-2 receptors (i.e., the LuxPQ receptor of V. harveyi and the Lsr ABC transporter of Salmonella enterica serovar Typhimurium) in S. aureus by genomic analysis (47). Due to the dual function of LuxS and the absence of genomic evidence of established AI-2 receptors, the AI-2 quorum-sensing function in S. aureus has been intangible, until now. In contrast, the function of the LuxS/AI-2 system in Staphylococcus epidermidis was investigated more clearly. The luxS gene is functional in S. epidermidis and impacts biofilm formation via transcriptional regulation of the polysaccharide intercellular adhesin-synthesized gene ica, and AI-2 has a signaling function controlling virulence-associated gene expression (30, 65). It is of great importance to explore whether AI-2 can function as a QS signal to regulate physiological functions in S. aureus.
Among the many virulence factors that are produced by S. aureus, capsular polysaccharide (CP) is an important cell wall component that can interact with the host immune system during the invasive process, allowing the organism to resist uptake and killing by phagocytes. More than 90% of S. aureus strains produce 1 of 11 CPs, and most strains colonizing and infecting humans produce either CP5 or CP8 (13, 27, 42, 56). CP5 and CP8 have been used as targets for vaccine development, and specific antibodies against CP5 and CP8 have been shown to be protective against S. aureus infections (21, 28). The capsular polysaccharide produced in S. aureus NCTC8325 is CP5, which is encoded by the cap operon which contains 16 closely linked genes, cap5A through cap5P, transcribed in one orientation (49). Existing experimental evidence shows that the transcription of the cap operon in S. aureus NCTC8325 can be modulated by a range of regulatory elements, such as yabJ-spoVG, arlRS, agr, sbcDC, ccpA, mgr, sae, and sarA (17, 33-36, 51, 53). In addition, it has been indicated that the expression of CP in S. aureus can also be regulated by various environmental cues (27).
In bacteria, there are many two-component systems involved in the regulation of gene expression. In general, these two-component systems consist of two proteins, a sensor histidine kinase and a response regulator, and function by sensing the environmental signals and initiating phosphorelay cascades (6, 45). KdpD together with KdpE constitutes a two-component signal transduction system, which was first characterized in E. coli. In this organism, proteins KdpD and KdpE regulate the production of Kdp-ATPase, which is an inducible high-affinity K+ transporter that is synthesized under conditions of severe K+ limitation or osmotic upshift (2, 26). A BLAST search with Kdp protein sequences shows that the Kdp-ATPase system is widely distributed among bacteria (5). Recently, several lines of evidence have shown that the KdpDE system is involved in virulence in some bacteria. For instance, in Mycobacterium tuberculosis, deletion of kdpDE resulted in increased virulence. Mice infected with the M. tuberculosis kdpDE mutant died more rapidly than those infected with wild-type bacteria (44). Although several reports have shown that in S. aureus the transcript level of kdpDE changes under certain environmental stresses (exposure to neutrophil microbicides or growth under biofilm conditions) (11, 43), information about the role of KdpDE in S. aureus and how it functions remains incomplete.
In the present study, we investigated the role of the S. aureus LuxS/AI-2 system by construction and analysis of an allelic replacement mutant of S. aureus NCTC8325. Our results show that luxS regulates the gene transcription of CP5 in S. aureus NCTC8325 by the AI-2 QS pathway. In addition, we demonstrate that KdpE regulates the transcription of cap and acts as an important connector linking AI-2 quorum sensing to CP5 production and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus and E. coli were grown in Luria-Bertani (LB) or tryptic soy broth (TSB; soybean-casein digest medium USP; Oxoid) medium with the appropriate antibiotics for plasmid selection or maintenance (ampicillin at 100 mg liter−1 and kanamycin at 50 mg liter−1 for E. coli; erythromycin at 2.5 mg liter−1 and chloramphenicol at 15 mg liter−1 for S. aureus).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| NCTC8325 | Wild type | NARSAa |
| RN4220 | 8325-4 r− | NARSA |
| SX1 | 8325 luxS::ermB | This study |
| SX2 | 8325 luxS::ermB pLIluxS | This study |
| SX8 | 8325 kdpDE::ermB | This study |
| SX9 | 8325 kdpDE::ermB pLIkdpDE | This study |
| SX10 | 8325 kdpE::ermB | This study |
| SX11 | 8325 kdpE::ermB pLIkdpE | This study |
| SX12 | 8325 kdpE::ermB pLIkdpEM | This study |
| E. coli | ||
| DH5α | Clone host strain, supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| BL21 | F−ompT hsdSB(rB− mB−) gal dcm(DE3) | Laboratory stock |
| V. harveyi BB170 | Sensor 1 negative, sensor 2 positive | B. L. Bassler |
| Plasmids | ||
| pEasy-TB | Clone vector, Kanr Apr | TransGen |
| pET28a(+) | Expression vector with hexahistidine tag, Kanr | Novagen |
| pGkdpE | pET28a(+) with kdpE, Kanr | This study |
| pEC1 | pBluescript derivative, source of ermB gene, Apr | Bruckner |
| pBT2 | Shuttle vector, temp sensitive, Apr Cmr | Bruckner |
| pBTluxS | pBT2 containing 500-bp upstream and 500-bp downstream fragments of luxS and ermB gene, for luxS mutagenesis, Apr Cmr Emr | This study |
| pBTkdpDE | pBT2 containing 600-bp upstream and 600-bp downstream fragments of kdpDE and ermB gene, for kdpDE mutagenesis, Apr Cmr Emr | This study |
| pBTkdpE | pBT2 containing 600-bp upstream and 700-bp downstream fragments of kdpE and ermB gene, for kdpE mutagenesis, Apr Cmr Emr | This study |
| pLI50 | Shuttle cloning vector, Apr Cmr | Addgene |
| pLIluxS | pLI50 with luxS and its promoter, Apr Cmr | This study |
| pLIkdpDE | pLI50 with kdpDE and its promoter, Apr Cmr | This study |
| pLIkdpE | pLI50 with kdpE and the promoter of kdp operon, Apr Cmr | This study |
| pLIkdpEM | pLIkdpE with a deletion mutation | This study |
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
DNA manipulation.
Genomic DNA of S. aureus NCTC8325 was prepared by a standard protocol for Gram-positive bacteria. Plasmid DNA was extracted using a plasmid purification kit (Promega), according to the manufacturer's instructions. Taq and Pfu DNA polymerases were purchased from Promega, and PrimeSTAR DNA polymerase was purchased from TaKaRa. Restriction enzymes were obtained from New England Biolabs. S. aureus was transformed by electroporation, as described previously (24).
Construction of S. aureus mutant strains.
To construct the luxS and kdpDE deletion mutants from S. aureus NCTC8325, two sets of primers (Table 2) were used to amplify the upstream and downstream fragments (about 600 bp) of each target gene. The primer pairs were as follows: up-luxS-f-BamHI/up-luxS-r-SalI and down-luxS-f-PstI/down-luxS-r-HindIII for luxS deletion and up-kdpDE-f-BamHI/up-kdpDE-r-SalI and down-kdpDE-f-HindIII/down-kdpDE-r-XbaI for kdpDE deletion. The sequences of the amplified fragments were verified and cloned into shuttle plasmid pBT2, such that the upstream and downstream fragments flanked the erythromycin resistance gene amplified from pEC1 with the relevant primers (Em-f-PstI/Em-r-SalI for luxS deletion and Em-f-HindIII/Em-r-SalI for kdpDE deletion) and had the same orientation as they do in the chromosome to create pBTluxS and pBTkdpDE. For kdpE deletion, the kdpE gene was amplified from S. aureus NCTC8325 with primers DEL-kdpE-f-XbaI and DEL-kdpE-r-BamHI, and the fragment was inserted into a gene for Ermr amplified from pEC1 with primers Em-f-NdeI and Em-r-HindIII and cloned into pBT2 to create pBTkdpE. The resulting plasmids were used for allele replacement, as described previously (15). All PCR primers used in this study were generated using the Primer (version 5.0) software and are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Oligonucleotide (5′-3′)a |
|---|---|
| up-luxS-f-BamHI | GCGggatccTTGTTCGACTGCTTTTCTGA |
| up-luxS-r-SalI | GCGgtcgacGAATGTTGAAAGTTTCAATT |
| down-luxS-f-PstI | GCGctgcagCGCTTATGATTTGTTGCATTATAA |
| down-luxS-r-HindIII | GCGaagcttTATAGCTGGCCCGTCAAGTC |
| up-kdpDE-f-BamHI | GCGggatccTCTACTCTTGACGATTGCAC |
| up-kdpDE-r-SalI | GCGgtcgacATCTAAATATT |
| down-kdpDE-f-HindIII | ATTCaagcttTAGATAACGG |
| down-kdpDE-r-XbaI | GCGtctagaCACCAAATGTGGTGAGTATA |
| DEL-kdpE-f-XbaI | GCGtctagaTCAATTTGATTGAAAATGCA |
| DEL-kdpE-r-BamHI | GCGggatccAACGCATTTCTACAGAGTTG |
| Em-f-NdeI | GCGcatatgGATACAAATTCCCCGTAGGC |
| Em-f-PstI | GCGctgcagGATACAAATTCCCCGTAGGC |
| Em-f-HindIII | GCGaagcttGATACAAATTCCCCGTAGGC |
| Em-r-HindIII | GCGaagcttGAAATAGATTTAAAAATTTCGC |
| Em-r-SalI | GCGgtcgacGAAATAGATTTAAAAATTTCGC |
| c-luxS-f-HindIII | GCGaagcttAAAGCATCGTATTCTGCTAA |
| c-luxS-r-EcoRI | GCGggatccTGAAAAATACAATCAATCTA |
| c-kdpDE-f-EcoRI | GCGgaattcCATTGTTAGAAACAAAATTATTTC |
| c-kdpDE-r-BamHI | GCGggatccCCACCAAATGTGGTGAGTAT |
| p-kdpDE-f-EcoRI | GCGgaattcATATTTTCAAATCTAGTAAATATTA |
| p-kdpDE-r-SmaI | GCGcccgggAACCTTCACCTCGATAGCAA |
| g-kdpE-f-SmaI | GCGcccgggATGCAATCTAAAATATTGATAATT |
| g-kdpE-r-BamHI | GCGggatccTTATTTCTCTTTCCACTGCA |
| M-kdpEAsp-f | TTAGGTTTACCAGATAAAGATGGAT |
| M-kdpEAsp-r | TAATAAAATGACATCTGGTTTATCA |
| p-cap-f | TCTATCTGATAATAATCATCTAACT |
| p-cap-r | TATTTACCTCCCTTAAAAAT |
| g-kdpE-f-NcoI | GCGccatggTGCAATCTAAAATATTGATAATTGAAG |
| g-kdpE-r-XhoI | GCGctcgagTTTAATATCATCTCATAAGT |
| rt-16S-f | CGTGGAGGGTCATTGGA |
| rt-16S-r | CGTTTACGGCGTGGACTA |
| rt-capA-f | CAGTTAAAGTCGCACCAA |
| rt-capA-r | GAACCCAATACAGGCAAT |
| rt-capC-f | CGTCATTAGCGGGTATTT |
| rt-capC-r | TCTTGTTGTGGCATTCGT |
| rt-capE-f | TTTCCAGTTGAGGCAGTG |
| rt-capE-r | TTCTTGATTTGGCTACGA |
| rt-capG-f | TTGAAGTTCCCTGGTGTC |
| rt-capG-r | CATCGGTTCGTTATTGTT |
| rt-capI-f | AAGGAAGCGAATGTTAT |
| rt-capI-r | AGCTGATGAAGCAATAA |
| rt-capK-f | GCTGCCCTATGTTTAGTC |
| rt-capK-r | ATGTTATTGATGCGTGTT |
| rt-capN-f | GACCTCGTTCAAAGATTA |
| rt-capN-r | CATAGTTGGTCGTAGGGT |
| rt-kdpA-f | ATTGTTCGGTTTATTGTCC |
| rt-kdpA-r | CATCATACTGCCCATTTCT |
| rt-kdpD-f | AGTCCAGGTGTTGGTAAA |
| rt-kdpD-r | TATTGTCGGCGATTCTTC |
| rt-kdpE-f | CTACAGCCGACAATGCCACA |
| rt-kdpE-r | TGCCCGAAGCTCATCAACA |
| rt-rna3-f | GGTTATTAAGTTGGGATGG |
| rt-rna3-r | GAGTGATTTCAATGGCACA |
| rt-agrA-f | AAAGTTGCAGCGATGGATTT |
| rt-agrA-r | ATGGGCAATGAGTCTGTGAG |
| rt-sarA-f | GACATACATCAGCGAAAA |
| rt-sarA-r | TACGTTGTTGTGCATTAA |
| rt-saeR-f | AAGTGGCGACCATTACAT |
| rt-saeR-r | CATTATTGCCTCAAATACGT |
| rt-mgrA-f | AGTACAATCTAACATACC |
| rt-mgrA-r | TTGCGATAAAGAAGAAGC |
| rt-ccpA-f | TCGTGGACTTGAAGATAT |
| rt-ccpA-r | CCATTTGTTCCTGATACT |
| rt-arlR-f | GCTGGGCTTGATTACGGT |
| rt-arlR-r | GCGCCATTTACCGTCACT |
| rt-sbcD-f | TTTTATACACTCCCTTATGC |
| rt-sbcD-r | GATGTCTTTCCACCTTGA |
| rt-spoVG-f | ACTCTGGCTTGTTCGTTG |
| rt-spoVG-r | CTTCTGATGTAGCGTTTT |
The sequences in lowercase letters refer to the restriction endonuclease recognition sites.
Complementation of mutants.
The deleted genes and their promoters from S. aureus NCTC8325 were amplified by PCR with primers c-luxS-f-HindIII and c-luxS-r-EcoRI for luxS and primers c-kdpDE-f-EcoRI and c-kdpDE-r-BamHI for kdpDE. The PCR products were cloned into pLI50 (Addgene) to create plasmids pLIluxS and pLIkdpDE. For pLIkdpE construction, before it was cloned into pLI50, the kdpE gene fragment was amplified with primers g-kdpE-f-SmaI and g-kdpE-r-BamHI and ligated to the kdpDE promoter fragment (amplified with primers p-kdpDE-f-EcoRI and p-kdpDE-r-SmaI). Site-directed mutagenesis by PCR was used to delete the Asp phosphorylation site of the KdpE protein. For example, to create plasmid pLIkdpEM, plasmid pLIkdpE was used as the template for PCR, and the primers for constructing a deletion mutation were M-kdpEAsp-f and M-kdpEAsp-r. The DNA fragment (6.5 kb, containing the full length of pLIkdpE except the 3-bp nucleotide bases) was amplified by PCR with PrimeSTAR DNA polymerase. The PCR products were digested with DpnI, phosphorylated, self-ligated, and transformed into E. coli DH5α. The positive clones with mutational plasmids yielded pLIkdpEM. The plasmids were transformed by electroporation into S. aureus RN4220 and subsequently transferred to their mutant strains. The AI-2 precursor molecule, DPD, was purchased from Omm Scientific Inc., TX.
Measurement of AI-2 activity.
The AI-2 activity of the culture supernatants was determined using the V. harveyi reporter strain BB170 as described previously (8, 55). Briefly, the supernatants were diluted 1:10 in fresh autoinducer bioassay medium containing V. harveyi BB170 (inoculated 1:5,000 from an overnight culture) to give a final volume of 200 μl. The culture was shaken (180 rpm) at 30°C, and the luminescence was determined at 30-min intervals for at least 8 h. AI-2 activity is reported as the fold induction of bioluminescence of the reporter strain over the bioluminescence of the background. The reported values represent the average bioluminescence stimulated by three independent preparations of each strain's supernatant.
Total RNA isolation, cDNA generation, and microarray processing.
Overnight cultures of S. aureus were diluted 1:100 in LB medium and grown to the late exponential phase (optical density at 600 nm [OD600] = 2.1). Cells were collected and resuspended in TE (Tris-EDTA) buffer (pH 8.0) containing 10 g liter−1 lysozyme and 40 mg liter−1 lysostaphin. After incubation at 37°C for 5 min, S. aureus cells were prepared for total RNA extraction using the Trizol method (Invitrogen), and residual DNA was removed with DNase (RNase free; TaKaRa). cDNAs were synthesized and labeled according to the manufacturer's suggestions for S. aureus antisense genome arrays (Affymetrix Inc., Santa Clara, CA). Further preparation, hybridization, and scanning were conducted by Biochip Company of Shanghai, China. Real-time reverse transcription-PCR (RT-PCR) was performed with a PrimeScript 1st Strand cDNA synthesis kit and SYBR Premix Ex Taq (TaKaRa) using a StepOne real-time PCR system (Applied Biosystems). The quantity of cDNA measured by real-time PCR was normalized to the abundance of 16S cDNA. Microarray data were analyzed with Microarray Suite software, version 5.1 (Affymetrix Inc.), and a four-comparison survival method.
Purification of KdpE.
The His6-tagged KdpE was cloned and purified using standard procedures. The full-length kdpE gene fragment was amplified by PCR with primers g-kdpE-f-NcoI and g-kdpE-r-XhoI from S. aureus NCTC8325 genomic DNA, cloned into expression vector pET28a(+) (Novagen), and transformed into E. coli BL21. The transformant was grown in LB medium at 37°C to an OD600 of 0.4, transferred to 16°C, and induced overnight with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside. Cells were harvested and lysed by sonication in lysis buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl). The His6-tagged KdpE protein was purified with nickel-nitrilotriacetic acid agarose solution (Invitrogen). The column was washed with 100 volumes of washing buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl) containing a linear gradient of imidazole (5 mM to 100 mM). Bound protein was eluted with elution buffer (250 mM imidazole, 20 mM Tris-HCl, pH 8.0, 1 M NaCl, 10% glycerol). The imidazole in the eluant was removed by using a Centrifuge Biomax-5 column (Millipore), and then the KdpE protein solution was stored at −80°C until use. The purity of the protein was analyzed by SDS-PAGE, and the protein concentration was determined by the Bradford assay with bovine serum albumin as the standard.
Gel-shift assay.
The DNA fragment containing the cap promoter was amplified from the S. aureus NCTC8325 chromosome with primers p-cap-f and p-cap-r by PrimeSTAR DNA polymerase (TaKaRa). The PCR products were purified and labeled using a digoxigenin (DIG) gel-shift kit (second generation; Roche), according to the manufacturer's instructions. The labeled fragment was incubated at 25°C for 15 min with various amounts of purified KdpE in 10 μl of incubation buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 3 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM dithiothreitol). After incubation, the mixtures were added into 2.5 μl of gel loading buffer and were then electrophoresed in a 4% native polyacrylamide gel in 0.5× TBE (Tris-borate-EDTA) buffer. The band shifts were detected and analyzed according to the manufacturer's instructions.
S. aureus survival in human blood and in U937 monocytic cells.
Heparinized venous blood samples were collected from healthy donors who provided written consent to participate in the study. A previous study showed that S. aureus cells grown on solid medium generated more CP5 than S. aureus cells grown in broth (56). The strains were harvested from the TSB plates after they were cultured at 37°C for 16 h, washed twice in phosphate-buffered saline (PBS), and suspended to an optical density at 600 nm of 0.8. Heparinized human blood (1 ml) was inoculated with 1 × 106 CFU of S. aureus and was incubated at 37°C with shaking (250 rpm). A total of 5 × 106 U937 monocytic cells were mixed with 2 × 106 CFU of S. aureus opsonized with 10% normal human serum and incubated at 37°C under an atmosphere of 5% CO2 with intermittent shaking. Bacterial cells were diluted to the appropriate concentration for testing at the required intervals, and the number of CFU was calculated from the plate counts determined in duplicate on TSB agar. The percentage of S. aureus CFU that survived was determined by comparing the bacterial burden in each sample after the indicated time with the bacterial burden at the start of the assay (0 h).
Microarray data accession number.
The microarray data were submitted to the CIBEX database (http://cibex.nig.ac.jp) with accession number CBX128.
RESULTS
The luxS gene is functional and is required for AI-2 production in S. aureus.
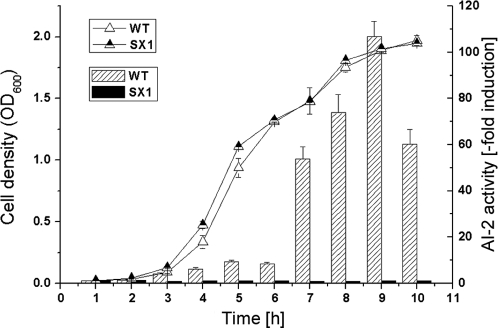
S. aureus NCTC8325 genome sequence analysis indicated that the strain contained a putative luxS gene. To determine its ability to produce the signal molecule AI-2, supernatants from the S. aureus wild type and the luxS mutant were tested for AI-2 activity using V. harveyi reporter strain BB170. The AI-2 activity in the culture supernatant of the wild type was detectable from the mid-exponential phase onward, reaching maximum levels during the beginning of the stationary phase and then decreasing, whereas no AI-2 activity was detectable in the supernatant of the luxS mutant during any phase (Fig. 1). Meanwhile, the abolished AI-2 production pathway could be restored by a complementary plasmid containing luxS (data not shown). This suggests that LuxS is a key determinant in the AI-2 production process in S. aureus, and these results are consistent with those of a previous study of AI-2 production in S. aureus (19).
FIG. 1.
Growth and extracellular AI-2 production. Cultures of S. aureus NCTC8325 wild type (WT) and the luxS mutant (SX1) were inoculated into LB medium at time zero and at the indicated intervals. Growth was monitored, and aliquots were taken and filtered to remove cells. AI-2 activity in the supernatant was measured using the V. harveyi BB170 bioassay. The data shown represent the mean values of four parallel AI-2 bioassays. The fold induction of AI-2 in the wild type exceeded 100 when the OD600 was 1.8.
Identification of genes under LuxS/AI-2 regulation.
To characterize the gene transcriptional profiles influenced by LuxS/AI-2, DNA microarray assays were performed using parental strain NCTC8325, the luxS mutant, and the luxS mutant with exogenous AI-2. Cells were grown in LB medium to an OD600 of 2.1. A 1.6-fold induction ratio as a cutoff limit was used to compare the transcriptional profiles between the wild type and the luxS mutant. Microarray data indicated that 34 genes were induced and 24 genes were repressed. Among these regulated genes, several major classes were associated with the metabolism, signal transduction, and virulence of S. aureus. Table 3 shows the main genes influenced by LuxS. Genes regulated by luxS are mainly associated with metabolism, which is in accord with the findings of a previous study, suggesting that LuxS plays a role in sulfur metabolism in S. aureus (19). Interestingly, genes related to the synthesis of the virulence determinant CP, composed of a cap gene cluster, were regulated by LuxS and AI-2. Meanwhile, the two-component system gene kdpDE was upregulated by luxS deletion and was restored by the addition of exogenous AI-2. We further analyzed the transcription of a set of genes by real-time RT-PCR, and the results corresponded to the microarray data (see Fig. S1 in the supplemental material).
TABLE 3.
Main genes affected by LuxS and AI-2
| Gene function and identifier | Gene product | Log2 ratio |
|
|---|---|---|---|
| Mutant/WT | Mutant + AI-2/WT | ||
| Metabolism genes | |||
| SAOUHSC_00898 | Argininosuccinate lyase | −0.7 | −0.6 |
| SAOUHSC_01540 | Prophage L54a, HNH endonuclease family protein | −1 | −0.1 |
| SAOUHSC_00311 | Ascorbate-specific PTSa enzyme IIB | 1 | 2.5 |
| SAOUHSC_01532 | SLT open reading frame 110-like protein | 0.8 | −0.1 |
| SAOUHSC_01536 | Scaffolding protease | 1 | 0.4 |
| SAOUHSC_02311 | Potassium-translocating P-type ATPase, B subunit, putative | 1.2 | 0.8 |
| SAOUHSC_02374 | HmrA, aminobenzoyl-glutamate utilization protein B, putative | 2 | 1.9 |
| SAOUHSC_02468 | Acetolactate synthase | 0.8 | 0 |
| SAOUHSC_00310 | Ascorbate-specific PTS enzyme IIC | 2.5 | 2.7 |
| SAOUHSC_00312 | PTS IIA component | 2.6 | 2.8 |
| SAOUHSC_02933 | Betaine aldehyde dehydrogenase | 0.9 | 0.5 |
| SAOUHSC_02173 | Amidase, lytic enzyme | 1.5 | 1.3 |
| Membrane component genes | |||
| SAOUHSC_00062 | Integral membrane domain-containing protein | −0.9 | −0.3 |
| SAOUHSC_02821 | Membrane-spanning protein, putative | −0.7 | −0.6 |
| SAOUHSC_01386 | Phosphate ABC transporter, permease protein, putative | −0.9 | −0.1 |
| SAOUHSC_01084 | Cell wall surface anchor protein | −0.7 | −0.2 |
| SAOUHSC_02310 | KdpC, potassium-transporting ATPase, C subunit | 0.9 | 0.6 |
| SAOUHSC_02312 | KdpA, potassium-transporting ATPase, A subunit | 1.4 | 0.8 |
| SAOUHSC_02864 | Ferrous iron transport protein B | 0.8 | 0.6 |
| Regulator genes | |||
| SAOUHSC_00915 | Sua5/YciO/YrdC/YwlC family protein | −0.9 | 0.2 |
| SAOUHSC_03046 | Helix-turn-helix domain-containing protein | −0.7 | −0.4 |
| SAOUHSC_03046 | Helix-turn-helix domain-containing protein | −0.7 | −0.4 |
| SAOUHSC_03046 | Helix-turn-helix domain-containing protein | −0.7 | −0.4 |
| Virulence genes | |||
| SAOUHSC_00399 | Superantigen-like protein | −1 | 0.1 |
| SAOUHSC_00433 | Pathogenicity island protein | −0.8 | −1.3 |
| SAOUHSC_00384 | Superantigen-like protein | −0.9 | −0.7 |
| SAOUHSC_00389 | Superantigen-like protein | −0.9 | −1.1 |
| SAOUHSC_00121 | CapH | 0.9 | 0.7 |
| SAOUHSC_00122 | CapI | 1.0 | 0.8 |
| SAOUHSC_00123 | CapJ | 0.8 | 0.8 |
| SAOUHSC_00124 | CapK | 1 | 0.9 |
| SAOUHSC_00119 | CapF | 1 | 0.7 |
| SAOUHSC_00120 | CapG | 1 | 0.6 |
| SAOUHSC_00125 | CapL | 0.8 | 0.8 |
| SAOUHSC_00126 | CapM | 0.9 | 0.9 |
| SAOUHSC_00127 | CapN | 1 | 0.6 |
| SAOUHSC_00117 | CapD | 0.7 | 0.3 |
| SAOUHSC_00118 | CapE | 0.7 | 0.6 |
| SAOUHSC_01705 | Enterotoxin family protein | 0.7 | 0.9 |
| Hypothetical protein genes | |||
| SAOUHSC_00817 | Hypothetical protein | −0.8 | −0.2 |
| SAOUHSC_01295 | Hypothetical protein | −0.7 | −0.2 |
| SAOUHSC_01296 | Hypothetical protein | −0.7 | −0.3 |
| SAOUHSC_01297 | Hypothetical protein | −0.9 | −0.3 |
| SAOUHSC_01533 | Hypothetical protein | −1.2 | 0.8 |
| SAOUHSC_02141 | Hypothetical protein | −0.7 | −0.7 |
| SAOUHSC_00966 | Hypothetical protein | −0.8 | −0.2 |
| SAOUHSC_02688 | Hypothetical protein | −0.8 | −0.5 |
| SAOUHSC_00323 | Hypothetical protein | −0.7 | −0.1 |
| SAOUHSC_00830 | Hypothetical protein | −0.8 | −0.3 |
| SAOUHSC_02596 | Hypothetical protein | −0.9 | −0.6 |
| SAOUHSC_00043 | Hypothetical protein | −0.7 | −0.5 |
| SAOUHSC_01548 | Hypothetical protein | 0.9 | 1.3 |
| SAOUHSC_02933 | Hypothetical protein | 0.8 | 1.1 |
| SAOUHSC_02761 | Hypothetical protein | 0.8 | 1 |
| SAOUHSC_01921 | Hypothetical protein | 0.7 | 0.5 |
| SAOUHSC_00971 | Hypothetical protein | 0.7 | 0.8 |
| SAOUHSC_02787 | Hypothetical protein | 0.7 | 0.3 |
| SAOUHSC_02865 | Hypothetical protein | 0.7 | 0.3 |
PTS, phosphotransferase system.
S. aureus AI-2 signaling associated with the KdpDE system to regulate CP expression.
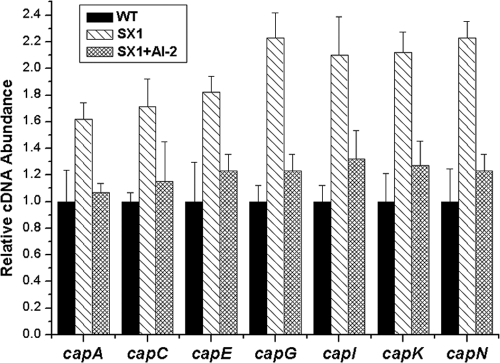
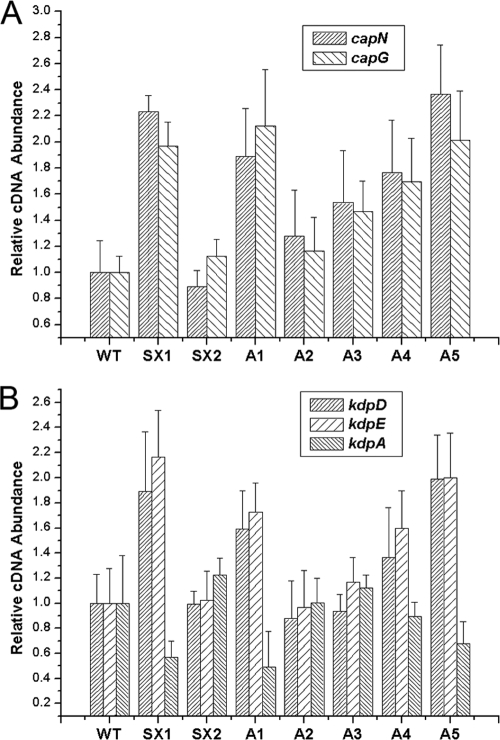
Both microarray and real-time RT-PCR data indicated that the transcript levels of the CP synthesis-related genes from capA to capP were increased in the luxS mutant and were restored by exogenous AI-2 (Fig. 2). However, the transcript levels of various regulatory elements known to modulate CP synthesis in S. aureus, such as agrA, sarA, sbcDC, rnaIII, arlRS, ccpA, mgrA, saeRS, and spoVG, displayed no apparent changes (see Fig. S2 in the supplemental material), suggesting that LuxS/AI-2 modulated cap gene transcription through another mechanism. Interestingly, the transcript levels of the two-component system genes kdpD and kdpE displayed the same tendency as cap gene transcription in these strains (see Fig. S2 in the supplemental material). To further determine whether this alteration was achieved by AI-2 signaling, the pre-AI-2 molecule DPD was used to complement the luxS mutant over a range of concentrations from 3.9 nM to 39 μM. The DPD threshold concentrations were 39 nM to 3.9 μM, and the transcript levels of both the cap gene and the kdpDE genes were restored in the luxS mutant under the threshold concentrations (Fig. 3A and B). These results strongly indicated that the lower transcription level of the cap and kdpDE genes in the wild type was a consequence of AI-2-mediated signaling. Meanwhile, these results indicated that the optimal DPD concentration has a direct link to the expression of KdpDE and CP in S. aureus. The correct concentrations of autoinducer may be critical for cells to make a proper response, and reaching an appropriate AI-2 threshold level is important for the luxS mutant to restore the wild-type phenotype in S. aureus, which is consistent with the finding of other studies of AI-2 (1, 22, 48). On the other hand, kdpA transcription was inversely regulated by kdpDE by AI-2 quorum sensing (Fig. 3B), suggesting that KdpE may suppress KdpABC expression in S. aureus through the same pathway found in E. coli.
FIG. 2.
Comparative measurement of transcription of CP synthesis-related genes. The relative transcript levels of the CP synthesis-related genes were determined in the S. aureus NCTC 8325 wild type (WT), the luxS mutant (SX1), and the luxS mutant with 39 nM AI-2 complementation (SX1 + AI-2). Strains were grown in LB medium with shaking at 37°C to an OD600 of 2.1. AI-2 was added to the luxS mutant at the inoculation time to a final concentration of 39 nM. The RNA was extracted and transcription was quantified by real-time RT-PCR for the CP synthesis-related genes (represented by capA, capC, capE, capG, capI, capK, and capN). The quantity of cap cDNA measured by real-time PCR was normalized to the abundance of 16S cDNA within each reaction mixture. Error bars indicate the variance between triplicate samples within the real-time PCR. The relative cDNA abundance in the wild-type samples was arbitrarily assigned a value of 1.
FIG. 3.
Transcriptional regulation of the cap gene and kdpDE gene expression by LuxS/AI-2. (A) Relative transcript levels of the cap gene (represented by capN and capG). (B) Relative transcript levels of the kdpD, kdpE, and kdpA genes. The levels of transcription of these genes were measured by real-time RT-PCR in the S. aureus NCTC8325 wild type (WT), the luxS mutant (SX1), the luxS mutant with a plasmid containing luxS for genetic complementation (SX2), and the luxS mutant with 3.9 nM to 39 μM AI-2 for complementation (A1, 3.9 nM; A2, 39 nM; A3, 390 nM; A4, 3.9 μM; A5, 39 μM).
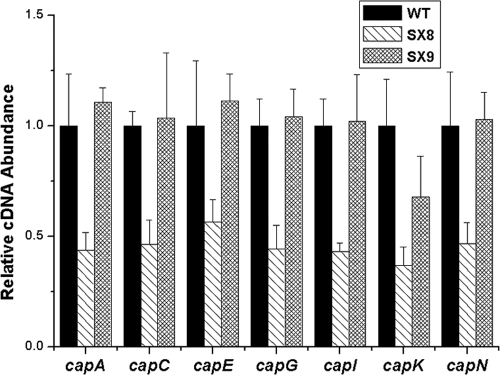
Inactivation of the two-component regulatory system gene kdpDE resulted in decreased transcript level of cap.
The results presented above suggested that the KdpDE two-component system may mediate the AI-2 QS regulation of cap gene transcription. We then performed kdpDE mutational analysis to determine whether deletion of kdpDE would affect cap gene transcription. An allelic replacement mutant of kdpDE was constructed and found to have no obvious growth defects (data not shown). This strain was then tested for cap gene transcription by real-time RT-PCR. The transcript levels of the cap genes were decreased in the kdpDE mutant compared to those in the wild type, and this decrease could be restored by genetic complementation (Fig. 4). These results suggest that the KdpDE system could affect cap gene transcription.
FIG. 4.
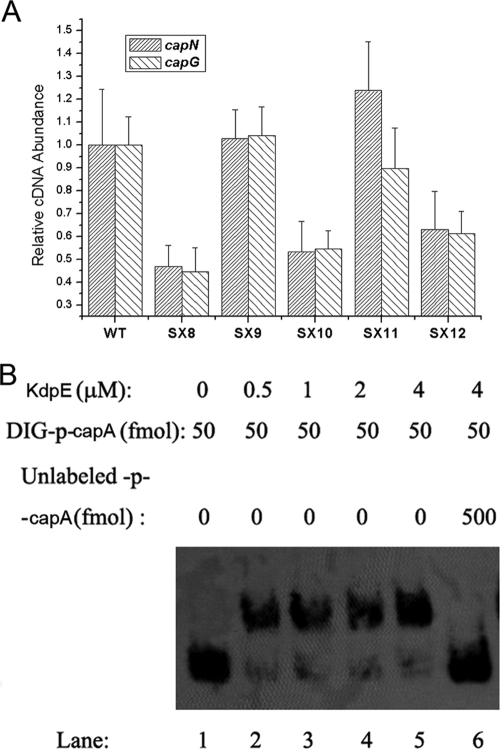
Transcriptional regulation of CP synthesis by KdpDE. The relative transcript levels of CP synthesis-related genes (represented by capA, capC, capE, capG, capI, capK, and capN) were measured by real-time RT-PCR in the S. aureus NCTC8325 wild type (WT), the kdpDE mutant (SX8), and the kdpDE mutant with a plasmid containing kdpDE (SX9).
KdpDE-regulated cap transcription through the KdpE phosphorylation pathway.
KdpD and KdpE are cotranscribed and belong to a family of sensor kinase and response regulator systems with no characterized function in S. aureus. In E. coli, they mediate the transcriptional activation of the kdp operon to control KdpFABC expression through phosphorylation of the signal transmission pathway. To further explore the mechanism of the involvement of KdpDE in cap gene transcription, we constructed the kdpE mutant strain and assessed the role of the phosphorylation site of KdpE in cap gene transcription. The results of real-time RT-PCR showed that deletion of kdpE resulted in the decreased transcription of cap, which could be restored by complementation with a plasmid with kdpE, whereas it could not be restored by the plasmid with kdpE whose phosphorylation site was defective (Fig. 5A), indicating that KdpE regulates the transcription of cap through a phosphorylation pathway. These observations showed for the first time that KdpDE functions as a two-component system in S. aureus and that KdpE relies on phosphorylation to function, just like other response regulators.
FIG. 5.
Transcriptional regulation of cap gene expression by KdpE. (A) Relative transcript levels of the cap gene (represented by capN and capG) in the S. aureus NCTC8325 wild type (WT), the kdpDE mutant (SX8), the kdpDE mutant with a plasmid containing kdpDE (SX9), the kdpE mutant (SX10), the kdpE mutant with a plasmid containing kdpE (SX11), and the kdpE mutant with a plasmid encoding KdpE with a mutated Asp-phosphorylation site (SX12). (B) Binding of KdpE protein to the promoter of cap. The ability of KdpE to bind to the cap promoter was determined by gel-shift assay. Increasing amounts of KdpE were incubated with excess DIG-labeled probes. Lanes 1 to 6, KdpE concentrations of 0, 0.5, 1, 2, 4, and 4 μM, respectively; the amount of DIG-labeled probe in each case was 50 fmol (the concentration was 5 nM). In lane 6, besides the labeled probes, 500 fmol of unlabeled probes was incubated with the KdpE protein.
To further investigate whether KdpE regulates the transcription of cap directly, we performed a gel-shift assay. The purified His6-tagged KdpE protein was used to bind the DIG-labeled fragment of the cap promoter. The purified KdpE protein bound to the cap promoter region in a dose-dependent fashion (Fig. 5B). The retarded protein-DNA complex was detected at KdpE concentrations of 0.5 to 4 μM. Unlabeled cap promoter DNA, added in excess (10-fold in a molar ratio) as a specific competitor, was found to reduce the formation of the protein-DNA complex. Meanwhile, retarded protein-DNA complex was not detected when the DIG-labeled random DNA sequence was used as a probe (data not shown). These results indicate that KdpE can specifically bind to the cap promoter region in vitro.
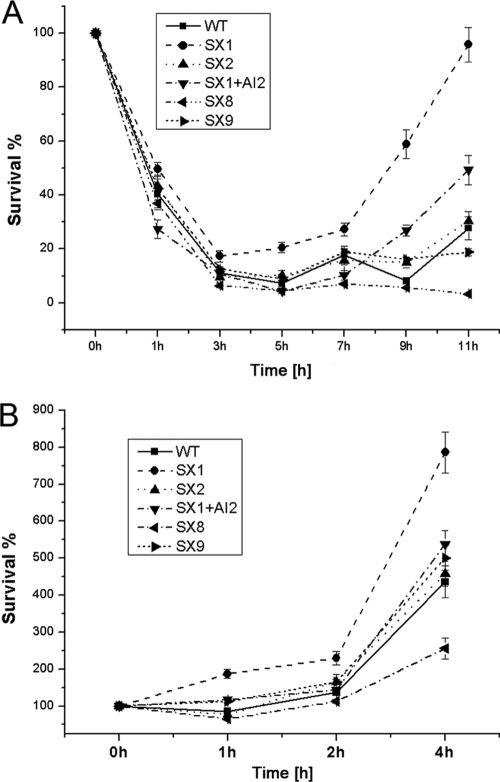
Absence of luxS increased S. aureus survival in human blood and human U937 monocytic cells, and absence of kdpDE attenuated S. aureus survival.
Previous studies have shown that the expression of CP enhances bacterial virulence in a mouse bacteremia model or in macrophages owing to its antiphagocytic nature (42, 56) and that several two-component systems are upregulated by S. aureus during human neutrophil phagocytosis, including the KdpDE system (58). Therefore, we examined the effects of inactivation of luxS, the addition of AI-2 to the luxS mutant, and the absence of kdpDE on the survival of S. aureus in human whole blood and human U937 monocytic cells. The survival and growth of the luxS mutant were higher when it was cultured in whole human blood (Fig. 6A), although there was no statistically significant difference in survival between the wild type, the kdpDE mutant, and several complemented strains during 5 h of incubation. After 7 h incubation, the rate of survival of the luxS mutant was significantly higher, whereas the rate of survival of the kdpDE mutant was reduced accordingly. After 11 h of incubation, the mean rate of survival ± standard deviation (n = 4) of the wild type was 27.6% ± 4.3%, that of the luxS mutant was 95.8% ± 6.4%, and that of the kdpDE mutant was 5.4% ± 3.3%. When these strains were cultured with human U937 monocytic cells, they demonstrated a tendency for survival similar to that in blood (Fig. 6B). Of importance, complementation of the luxS mutant with exogenous AI-2, the plasmid containing luxS, and the kdpDE strain restored the wild-type phenotype. These results indicated that the AI-2 signaling and the KdpDE system were involved in bacterial virulence in invasive S. aureus infection. The altered survival of S. aureus in human blood and monocytic cells correlated with the changes in the transcript levels of CP. The data further validated our hypothesis that S. aureus AI-2 quorum sensing regulates CP synthesis and virulence through the KdpDE two-component regulatory system.
FIG. 6.
Alteration of survival of S. aureus in human blood and in U937 monocytic cells due to absence of luxS or kdpDE. (A) Rates of survival for the S. aureus NCTC8325 wild type (WT), the luxS mutant (SX1), the luxS mutant with a plasmid containing luxS (SX2), the luxS mutant with 39 nM AI-2 (SX1 + AI-2), the kdpDE mutant (SX8), and the kdpDE mutant with a plasmid containing kdpDE (SX9). The strains were incubated in heparinized human blood, and the results are from five separate blood donors. (B) S. aureus survival when it was cultured with U937 monocytic cells. The percentage of S. aureus CFU that survived was determined as described in Materials and Methods.
DISCUSSION
AI-2 has been recognized to be a universal language for bacterial communication in recent years. However, knowledge of whether AI-2 serves as a signal molecule in Gram-positive bacteria, especially in S. aureus, remains elusive, mostly because no homologue of the known AI-2 receptor has been identified in the bacterium. A previous study has shown that the luxS gene in S. aureus was transcribed throughout growth and that AI-2 was produced in rich medium under aerobic and anaerobic conditions, peaking during the transition to the stationary phase (19). In addition, the luxS mutants exhibited a growth defect when they were grown in a limited medium, and this defect could be reduced only by transformation with the complementation vector rather than by using the AI-2 produced by this organism. No cross talk was displayed between the Agr and LuxS/AI-2 systems, and AI-2 did not exert a feedback effect on its own production in S. aureus. Furthermore, virulent phenotypic differences existed between the wild type and the luxS mutant. However, the parental phenotype could not be restored by the complementation vector or in vitro-synthesized AI-2, so they attributed this phenotype to second-site mutations. On the basis of these reasons, they drew the conclusion that LuxS played a role in metabolism but not QS. Undoubtedly, considering the metabolic role of LuxS in the activated methyl cycle, it was the complement vector but not AI-2 that restored the growth of luxS mutants in a sulfur-limited medium. However, it seems inaccurate to deny the QS function of AI-2 just because it cannot complement the virulent phenotypic alteration that is caused by second-site mutations. Our study demonstrated the involvement of LuxS in the regulation of CP production in S. aureus via a signaling process. Our data show that inactivation of luxS resulted in enhancement of the transcriptional level of the CP synthesis gene and resistance of this organism to uptake and killing by phagocytes. Of importance, exogenous AI-2 can complement these alterations, which indicated that the luxS mutation in S. aureus affected these phenotypes in an AI-2-dependent manner. These data lead us to conclude that the change in cap gene expression between the wild type and the luxS mutant could mainly be due to the role of AI-2 as a signal rather than due to the defect of methionine metabolism. Moreover, it is interesting to note that the complementation effect changed with the exogenous AI-2 concentrations, indicating that a concentration-sensing and -responding mechanism is involved, which is consistent with most QS processes.
Although it has been reported that the LuxS/AI-2 system plays important roles in cellular functions in a variety of bacteria, the detailed mechanism through which AI-2 functions has been investigated in only a few species. In V. harveyi, AI-2 detection and transduction rely on a two-component system. Borate-AI-2 is recognized by a soluble periplasmic protein that interacts with cytoplasmic response regulator proteins, thus triggering a sensor-kinase couple to finally control the expression of the luciferase structural operon (luxCDABE) (9, 31, 52). In E. coli and S. Typhimurium, AI-2 is first imported into the cell by the LsrR transporter, and then gene expression control is initiated (40, 55). In another Gram-positive bacterium, Streptococcus mutans, AI-2-regulated genes were identified, even though no homologue of the known AI-2 receptor has been recognized in that organism (37, 54). Our study demonstrated that LuxS/AI-2 can regulate cellular functions through a two-component system, KdpDE, whose role in S. aureus has not been well characterized. Inactivation of luxS results in a clear increase in the levels of transcription of kdpD and kdpE, which encode the sensor histidine kinase KdpD and the response regulator KdpE, respectively. Furthermore, gel-shift assays show that the regulatory protein KdpE can bind to the promoter region of the cap operon. These observations not only reveal that AI-2 regulates CP production through KdpDE but also provide new clues to the function of the KdpDE system in S. aureus. It is reported that a furanone could enhance staphylococcal biofilm formation by luxS repression (25), and we suppose that it may potentially involve the KdpDE two component system, although in that case the experimental evidence is missing. The exact mechanism by which LuxS/AI-2 interacts with KdpDE requires further study. Besides, knowledge of which forms of DPD are active in S. aureus remains elusive, since conversion between DPD derivatives is spontaneous and fast, making isolation or synthesis of each compound for biological activity testing almost impossible. Actually, explicit determination of the active form of DPD came only from biochemical and structural studies of various AI-2 receptors (16, 22, 40).
It is known that S. aureus normally produces surface proteins and polysaccharides during the exponential growth phase and secreted proteins during the stationary phase. In the invasive process, the surface proteins and polysaccharides are virulence determinants that are required to colonize host tissues and initiate the infection, and the secreted proteins are virulence factors that spread to adjacent tissues (46). Changes in the protein expression profile are controlled by a complicated regulatory network. In this study, we found that AI-2 has a negative effect on CP production in S. aureus, which leads us to propose that the LuxS/AI-2 QS system might participate in the downregulation of surface components after the colonization of host tissues.
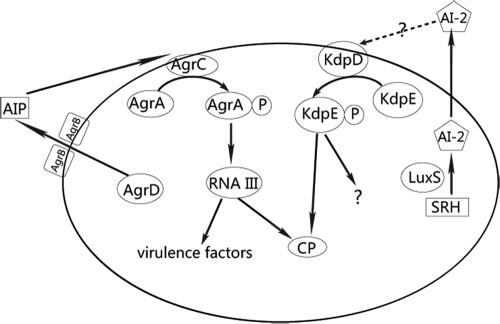
As a major determinant of the virulence of S. aureus, it has been suggested that CP is regulated in a complicated way by a variety of factors in S. aureus, such as spoVG, arlRS, agr, ccpA, mgr, sae, and sarA. However, our results showed no significant differences in the transcriptional levels of these regulators between the luxS mutant and the wild type, indicating that the LuxS/AI-2 QS system does not merge with the characterized regulatory network of CP but might function in a novel and distinct pathway. It is known that the agr QS system regulates many virulence-associated traits in S. aureus. In contrast, according to our experimental data, the LuxS/AI-2 QS system exhibits clear involvement only in CP production (Fig. 7). We thus propose that the LuxS/AI-2 QS system might play a supplementary role in the overall QS process in S. aureus, although the details await further investigation.
FIG. 7.
The QS systems in S. aureus. The Agr QS system regulates many virulence-associated traits in S. aureus, and the LuxS/AI-2 QS system exhibits clear involvement in CP production.
Supplementary Material
Acknowledgments
We thank our colleagues D. Hong, X. Zhang, R. Ma, and Y. You for their technical assistance. We thank the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing the bacterial strains.
This work was supported by the National Natural Science Foundation of China (grants 30970118 and 30721002).
Editor: A. Camilli
Footnotes
Published ahead of print on 24 May 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ahmed, N. A., F. C. Petersen, and A. A. Scheie. 2007. AI-2 quorum sensing affects antibiotic susceptibility in Streptococcus anginosus. J. Antimicrob. Chemother. 60:49-53. [DOI] [PubMed] [Google Scholar]
- 2.Altendorf, K., A. Siebers, and W. Epstein. 1992. The KDP ATPase of Escherichia coli. Ann. N. Y. Acad. Sci. 671:228-243. [DOI] [PubMed] [Google Scholar]
- 3.Archer, G. L., and M. W. Climo. 2001. Staphylococcus aureus bacteremia—consider the source. N. Engl. J. Med. 344:55-56. [DOI] [PubMed] [Google Scholar]
- 4.Bacon Schneider, K., T. M. Palmer, and A. D. Grossman. 2002. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballal, A., B. Basu, and S. K. Apte. 2007. The Kdp-ATPase system and its regulation. J. Biosci. 32:559-568. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, J. F., and J. A. Hoch. 1998. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob. Agents Chemother. 42:1529-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 8.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 9.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 10.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 11.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhasin, N., A. Albus, F. Michon, P. J. Livolsi, J. S. Park, and J. C. Lee. 1998. Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol. Microbiol. 27:9-21. [DOI] [PubMed] [Google Scholar]
- 14.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 15.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 17.Chen, Z., T. T. Luong, and C. Y. Lee. 2007. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J. Bacteriol. 189:7343-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doherty, N., M. T. Holden, S. N. Qazi, P. Williams, and K. Winzer. 2006. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 188:2885-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fattom, A. I., J. Sarwar, A. Ortiz, and R. Naso. 1996. A Staphylococcus aureus capsular polysaccharide (CP) vaccine and CP-specific antibodies protect mice against bacterial challenge. Infect. Immun. 64:1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federle, M. J. 2009. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib. Microbiol. 16:18-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 25.Kuehl, R., S. Al-Bataineh, O. Gordon, R. Luginbuehl, M. Otto, M. Textor, and R. Landmann. 2009. Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob. Agents Chemother. 53:4159-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. Y., and J. C. Lee. 2000. Staphylococcal capsule, p. 361-366. In V. A. Fischetti, R. P. Novick, J. J. Ferriti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 28.Lee, J. C., J. S. Park, S. E. Shepherd, V. Carey, and A. Fattom. 1997. Protective efficacy of antibodies to the Staphylococcus aureus type 5 capsular polysaccharide in a modified model of endocarditis in rats. Infect. Immun. 65:4146-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., C. Attila, L. Wang, T. K. Wood, J. J. Valdes, and W. E. Bentley. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189:6011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, M., A. E. Villaruz, V. Vadyvaloo, D. E. Sturdevant, and M. Otto. 2008. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 32.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 33.Luong, T., S. Sau, M. Gomez, J. C. Lee, and C. Y. Lee. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luong, T. T., and C. Y. Lee. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 35.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meier, S., C. Goerke, C. Wolz, K. Seidl, D. Homerova, B. Schulthess, J. Kormanec, B. Berger-Bachi, and M. Bischoff. 2007. sigmaB and the sigmaB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect. Immun. 75:4562-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt, J., J. Kreth, W. Shi, and F. Qi. 2005. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57:960-969. [DOI] [PubMed] [Google Scholar]
- 38.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 39.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 40.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 41.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson, I. M., J. C. Lee, T. Bremell, C. Ryden, and A. Tarkowski. 1997. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect. Immun. 65:4216-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palazzolo-Ballance, A. M., M. L. Reniere, K. R. Braughton, D. E. Sturdevant, M. Otto, B. N. Kreiswirth, E. P. Skaar, and F. R. DeLeo. 2008. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180:500-509. [DOI] [PubMed] [Google Scholar]
- 44.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 46.Pragman, A. A., and P. M. Schlievert. 2004. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 42:147-154. [DOI] [PubMed] [Google Scholar]
- 47.Rezzonico, F., and B. Duffy. 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 49.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143(Pt 7):2395-2405. [DOI] [PubMed] [Google Scholar]
- 50.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 51.Seidl, K., M. Stucki, M. Ruegg, C. Goerke, C. Wolz, L. Harris, B. Berger-Bachi, and M. Bischoff. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semmelhack, M. F., S. R. Campagna, M. J. Federle, and B. L. Bassler. 2005. An expeditious synthesis of DPD and boron binding studies. Org. Lett. 7:569-572. [DOI] [PubMed] [Google Scholar]
- 53.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Dobler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 56.Thakker, M., J. S. Park, V. Carey, and J. C. Lee. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 58.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Said-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907-3919. [DOI] [PubMed] [Google Scholar]
- 59.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 60.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 62.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 63.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 64.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 65.Xu, L., H. Li, C. Vuong, V. Vadyvaloo, J. Wang, Y. Yao, M. Otto, and Q. Gao. 2006. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.