Abstract
Rhodococcus equi, a facultative intracellular pathogen of macrophages, causes severe, life-threatening pneumonia in young foals and in people with underlying immune deficiencies. R. equi virulence is dependent on the presence of a large virulence plasmid that houses a pathogenicity island (PAI) encoding a novel family of surface-localized and secreted proteins of largely unknown function termed the virulence-associated proteins (VapACDEFGHI). To date, vapA and its positive regulators virR and orf8 are the only experimentally established virulence genes residing on the virulence plasmid. In this study, a PAI deletion mutant was constructed and, as anticipated, was attenuated for growth both in macrophages and in mice due to the absence of vapA expression. Expression of vapA in the PAI mutant from a constitutive promoter, thereby eliminating the requirement for the PAI-encoded vapA regulators, resulted in delayed bacterial clearance in vivo, yet full virulence was not restored, indicating that additional virulence genes are indeed located within the deleted pathogenicity island region. Based on previous reports demonstrating that the PAI-carried gene vapG is highly upregulated in macrophages and in the lungs of R. equi-infected foals, we hypothesized that vapG could be an important virulence factor. However, analysis of a marked vapG deletion mutant determined the gene to be dispensable for growth in macrophages and in vivo in mice.
Rhodococcus equi is a soilborne, facultative intracellular Gram-positive pathogen belonging to the order Actinomycetales. While R. equi has been reported to infect various numbers of different animal hosts (e.g., pigs, sheep, and cattle) (24), the greatest burden of R. equi disease is in its primary host, young foals between the ages of 1 and 6 months, in which mortality rates, if untreated, can approach 80% (7, 9, 22, 39). Even with treatment, ∼30% fatality rates have been reported in foals infected with this organism (1, 12). Currently, there are no commercially available vaccines licensed to prevent disease caused by R. equi. In addition to being an important cause of serious disease in foals, this bacterium has also emerged as an opportunistic pathogen of immunocompromised humans, with conditions such as HIV infection, organ transplantation, chemotherapy, steroid therapy, diabetes mellitus, and alcoholism seen as risk factors for disease (2, 6, 19, 21, 30). In immunocompromised patients, the mortality rate is 50 to 55% among HIV-infected individuals, compared to 11% in immunocompetent patients (8, 13, 18). In both susceptible foals and humans, R. equi disease commonly presents as a pyogranulomatous pneumonia (14), although numerous other clinical manifestations may occur concomitantly. Extrapulmonary disorders (EPDs) in foals infected with R. equi, such as abdominal lymphadenitis, ulcerative enterotyphlocolitis, and pyogranulomatous hepatitis, are associated with a significant reduction in survival rates (28).
The ability of R. equi to resist macrophage killing and replicate intracellularly is linked to possession of large plasmid which, in all equine isolates and 30% of human isolates, is approximately 80 kb in size. R. equi strains cured of the plasmid are avirulent in foals and murine models of infection and are unable to replicate in macrophages cultured in vitro (14, 35). Sequencing and annotation of the virulence plasmids from two distinct foal isolates, R. equi strains 103+ and 33701, have revealed that the plasmid has 73 coding sequences (CDSs), including eight pseudogenes (20, 33). Based on homology searches, the virulence plasmid can be divided into four regions, specifically the areas of replication/partitioning, conjugation, and unknown function and an ∼21-kb region with features typical of a pathogenicity island (PAI) (20, 33). The PAI is believed to have been acquired through horizontal gene transfer from a source of unknown origin. Computational analysis of the PAI has revealed that it contains 26 putative CDSs, including the unique and R. equi-specific family of proteins called the Vap (virulence associated protein) family (20). There are six full-length vap genes, (vapA, -C, -D, -E, -G, and -H) and three vap pseudogenes (vapF, -I, and -X) (20, 23, 33). These pseudogenes are thought not to encode functional proteins, as they exhibit either substantial truncations and/or frameshift mutations in their coding sequences.
All strains of R. equi isolated from infected foals are positive for VapA, an immunodominant lipoprotein located on the bacterial surface (34, 35). VapA is required for intracellular growth in macrophages (17), in which it may aid in preventing fusion of R. equi-containing vacuoles with lysosomes (10, 36, 38). An R. equi vapA deletion mutant is unable to establish a persistent infection and is rapidly cleared by severe combined immunodeficient (SCID) mice (17). Two genes, virR and orf8, also carried in the pathogenicity island region are needed for optimal vapA transcription. (25, 29). VirR is a LysR-type transcriptional regulator which binds to the vapA promoter (29), while homology analyses suggest that Orf8 is an orphan two-component response-type regulator (33). Loss of either regulator results in attenuation of the organism (25).
Although vapA is necessary, it is not sufficient for virulence, since expression of wild-type levels of VapA in a plasmid-cured derivative of a virulent strain does not restore virulence in mice or foals (11), indicating that virulence determinants in addition to vapA reside on the virulence plasmid. To date, with the exception of the key regulators of vapA expression, virR and orf8 (26, 29), the location and identity of these other virulence determinants remain unknown, and determining such has been the focus of this work. Here we describe the construction of a PAI deletion mutant which was used as a molecular tool to determine the location of plasmid-encoded virulence factors, aside from vapA. Our data show that these determinants do indeed reside within the PAI region. Based on sequence and expression data, we and others have hypothesized that vapG could be an important virulence determinant of R. equi (4, 5, 16, 25). To assess this hypothesis experimentally, a marked deletion mutant of vapG was constructed using a two-step allelic exchange strategy, and the effect of the deletion on the ability of the mutant strain to survive and replicate in macrophages and in vivo in mice was evaluated.
MATERIALS AND METHODS
Bacterial strains.
The strains used are listed in Table 1. Virulent R. equi strain 103+ was originally isolated from a foal with R. equi pneumonia and was kindly provided by J. Prescott (Guelph, Ontario, Canada). Avirulent R. equi strain 103− was derived by serial subculture of strain 103+ at 37°C to facilitate loss of the virulence plasmid (17). The vapA deletion mutant on the 103+ strain background was derived as described earlier (17). The standard culture medium used was brain heart infusion (BHI) (Difco Laboratories, Detroit, MI) broth or agar, and unless otherwise noted, cultures were incubated at 30°C to help maintain the presence of the R. equi virulence plasmid. Antibiotics, when necessary, were used at the following concentrations: apramycin, 80 μg/ml; hygromycin, 180 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Escherichia coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Zymo Research |
| Rhodococcus equi strains | ||
| 103+ | Wild-type strain with 81-kb virulence plasmid | 11 |
| 103− | Plasmid-cured variant of strain 103+ | 17 |
| 103−/vapA | Strain 103− transformed with pMV261-vapA | This study |
| 103+ ΔvapA | Strain 103+ with deletion of the vapA gene | 17 |
| 103+ ΔvapA/vapA | vapA deletion mutant transformed with pMV261-vapA | This study |
| ΔPAI | Strain 103+ with deletion of pathogenicity island | This study |
| ΔPAI/vapA | PAI mutant transformed with pMV261-vapA | This study |
| 103+ ΔvapG | Strain 103+ with deletion of the vapG gene | This study |
| 103+ ΔvapG/vapG | vapG deletion mutant transformed with pSET152.vapG | This study |
| Plasmids | ||
| pBluescript II | Ampr αlacZ′ | Stratagene |
| pSJ42 | pBluescript II digested with SspI and DraIII | This study |
| pSJ47 | pSJ42 with fragment downstream of orf22 (coordinates 22802-23501) | This study |
| pSJ48 | pSJ47 with fragment upstream of orf62 (coordinates 77178-77742) | This study |
| pSJ49 | pSJ48 with 1.1-kb Aprr-encoding fragment from pSC146 | This study |
| pSJ53 | pSJ49 with 3.5-kb lacZ-containing fragment from pMV261-lacZ2 | This study |
| pSC146 | Mycobacterium-E. coli shuttle plasmid, ts oriM AprrHimar1 | 3 |
| pMV261 | Mycobacterium-E. coli shuttle plasmid with HygroriM hsp60 | 32 |
| pMV261-lacZ2 | pMV261 with lacZ | 31 |
| pMV261-vapA | pMV261 with vapA cloned in frame downstream of hsp60 promoter | 15 |
| pSelAct | AprrlacZ codA:upp | 37 |
| pJM10 | pSelAct with fragment downstream of vapG (coordinates 693-1545) | This study |
| pJM11.1 | pJM10 with fragment upstream of vapG (coordinates 2042-2711) | This study |
| pJM11.1hyg | pJM11.1 with 1.4-kb Hygr-encoding fragment from pMV261 | This study |
| pMV261-lacZ2ΔoriM | pMV261-lacZ2 lacking oriM | This study |
| pGBC14.1lacZ | pMV261-lacZ2ΔoriM with flanks of vapG, Hygr | This study |
| pSET152 | phiC31 integrase, attP Aprr | 15 |
| pETDuet-1 | AmprlacI MCS1 MCS2 | Novagen |
| pSET152-AMH | pSET152 with hsp60 from pMV261 and MCS2 from pETDuet-1 MCS2 | This study |
| pSET152.vapG | pSET152-AMH with vapG-containing fragment (coordinates 1516-2061) | This study |
Antisera.
Rabbit polyclonal serum to R. equi was raised as previously described (14). Rabbit polyclonal serum to VapA was generated as reported earlier (17).
Plasmid construction.
The plasmids used are listed in Table 1. The first cloning step to construct the suicide plasmid used in making the pathogenicity island mutant was restriction digestion of pBlueScript (Stratagene, La Jolla, CA) with SspI and DraIII to remove all restriction sites between them. The remaining vector fragment was recircularized to obtain pSJ42. Next, a 700-bp DNA fragment located immediately downstream of orf22 was amplified with primers ORF22(f) and ORF22(r) (Table 2) and cloned into the EcoRV site of pSJ42 to yield pSJ47. A 583-bp amplicon covering the region upstream of orf62 was amplified with primers ORF62u(f) and ORF62u(r) (Table 2) and cloned into pSJ47 digested with XbaI to give pSJ48. Subsequently, a 1.1-kb DNA fragment carrying the apramycin resistance gene aacC4 was obtained by digesting pSC146 (3) with PstI and PmlI and was end repaired with T4 DNA polymerase. It was then cloned into pSJ48 which had been digested with AgeI and SgrAI. This resulted in pSJ49, which carried aac4 flanked by DNA upstream of orf62 on its 5′ end and downstream of orf22 on its 3′ end. Finally, a 3.5-kb DNA fragment containing the Escherichia coli lacZ gene was obtained by digesting pMV261-lacZ2 (31) with PstI and DraI, followed by end repair. It was cloned in the ScaI site of pSJ49 after its sticky ends were filled with Klenow fragment to obtain pSJ53.
TABLE 2.
Primers used in this study
| Primer namea | Sequenceb | Reference |
|---|---|---|
| VapA-hsp60-FI | 5′-CGC TGG CCA CTC TTC ACA AGA CG-3′ | This study |
| VapA-hsp60-RII | 5′-CTA TGG CCA CTA GGC GTT GTG CCA-3′ | This study |
| VapC1III | 5′-TAT CAA TCT AGA TTG ATG ACG ACG GTC GAG-3′ | 17 |
| VapC1cIV | 5′-ATA TAT GTT AAC TCA CAC CAA ATG CCA TCG-3′ | 17 |
| VapD1V | 5′-ATA TAT TCT AGA ATG GGT CGG AAG GTA AAC-3′ | 17 |
| VapD1cVI | 5′-ATA TAT GTT AAC ACT TGT TCC TCA CGC AGC-3′ | 17 |
| VapE1 | 5′-ATA TAT TCT AGA GTC GCG CTT GAA GTG CGG-3′ | 17 |
| VapE1c | 5′-ATA TAT GTT AAC CAG CTA TCG CCA GGC G-3′ | 17 |
| VapF1VII | 5′-ATA TAT TCT AGA CTG ACG ATA GCT GGG CCT-3′ | 17 |
| VapF1cVIII | 5′-ATA TAT GTT AAC CAA TCA TTG CGC TAA CAC-3′ | 17 |
| REVP2 | 5′-GAC CTG TTC ATA GCC GAG-3′ | 17 |
| REVP2c | 5′-TCG TCC TCG ATC CGC TGC-3′ | 17 |
| REVP4 | 5′-ATC GTC GCG ATC TGC TGA-3′ | 17 |
| REVP4c | 5′-TCA TTC TCG CGG TTG TGC-3′ | 17 |
| REVP5 | 5′-CTT TGC AGT CGG CCT GAG-3′ | 17 |
| REVP5c | 5′-GAA GAT GAG TAG CAC TGT C-3′ | 17 |
| REVP6 | 5′-GAG AGT TCA GTT TCG CGG-3′ | 17 |
| REVP6c | 5′-CCT TTC CAT TGG TGT CTT C-3′ | 17 |
| REtrbA1 | 5′-GCG TCA GTG CGA CAG TGA TG-3′ | 17 |
| REtrbA1c | 5′-TCG GAG TCA GGT CGG AGG-3′ | 17 |
| ORF61(f) | 5′-TGC GTC CTC CCG CCC GTC-3′ | This study |
| ORF61(r) | 5′-ATG CGC CCA TCC TAT TGC-3′ | This study |
| ORF62-1(f) | 5′-GTT CGA TCT TCC GTT CTG-3′ | This study |
| ORF22-1(r) | 5′-CAT CGC TAC GGC ACA ACC-3′ | This study |
| ORF22(f) | 5′-AGT GCA GAG AAG CAG AGC-3′ | This study |
| ORF22(r) | 5′-ACC CAC ATC AGC CAA CT-3′ | This study |
| ORF62u(f) | 5′-ACG CGC TCT ACC TGC TCG-3′ | This study |
| ORF62u(r) | 5′-CTT TTG CCC TTC TGG CAT GG-3′ | This study |
| vapG-up-F | 5′-AAT TCG GCC GTC TCT GGC TTC ATA ACA C-3′ | This study |
| vapG-up-R | 5′-AAT AGG ATC CAA TAG GAC GGC GCA CTC-3′ | This study |
| vapG-down-F | 5′-AAT AGG ATC CAA GGG TCC GAA CAC TCA C-3′ | This study |
| vapG-down-R | 5′-AAG AGA ATT CAG TGC CGC TCC TAC GTT G-3′ | This study |
| vapG-comp-NdeI | 5′-GCG CCA TAT GTC GTG AGT GTT CGG ACC C-3′ | This study |
| vapG-comp-EcoRV | 5′-ATA AGA TAT CAC GCG CCG ACT GTG AG-3′ | This study |
| vapG-F | 5′-AAG AAC ATG TGC GCC GTC CTA TTG-3′ | This study |
| vapG-int-FIX | 5′-CCG CCA GAA TCA CCA GTA AAC-3′ | This study |
| vapG-int-RX | 5′-GCG AAC GCG GAA ACT TCA ATG-3′ | This study |
| vapG/podG-R | 5′-AAT AGG TAC CCG TTC GGA GGT ATT CG-3′ | This study |
| HSP60_R | 5′-CCC ATA TGC CAT TGC GAA GTC-3′ | This study |
| F_HSP60 | 5′-ATG GAT CCC TCC GTT GTA GTG CTT GTG-3′ | This study |
| gyrB-qPCR-F | 5′-GTC GAG CAG GGT CAC GTG TA-3′ | This study |
| gyrB-qPCR-R | 5′-AGC TCC TTT GCG TTC ATC T-3′ | This study |
| MCS-F | 5′-CGG GAT CCG GGA TAT ACA TAT GG-3′ | This study |
| MCS-R | 5′-CGG CCG TTT AAA TTA ACT CGA GGG TAC CG-3′ | This study |
| ORF6F-RTPCRXI | 5′-AGC TGT GCC TGC AAC ATT CG-3′ | This study |
| ORF6R-RTPCRXII | 5′-CTA CGC TAC ATC GCC TAT CC-3′ | This study |
Superscripts are the abbreviations used in Fig. 1A.
Restriction enzyme sites are in italic.
The vapA expression plasmid pMV261-vapA (15) was constructed by in-frame cloning of a vapA-containing fragment immediately downstream of the constitutive mycobacterial hsp60 promoter in the vector pMV261 (32) to allow for vapA expression independently of its two regulators, virR and orf8, which would be absent in the PAI deletion mutant strain.
To generate the suicide plasmid to construct the vapG mutant, an 858-bp region downstream of vapG was amplified by PCR using primer pairs vapG-up-F and vapG-up-R (Table 2). The PCR product was digested with restriction enzymes EagI and BamHI and cloned into pSelAct (37) (a gift generously provided by R. van der Geize) which had been digested with the same enzymes, giving rise to pJM10. A 669-bp region of DNA upstream of vapG was amplified using the primers vapG-down-F and vapG-down-R (Table 2) and digested with BamHI and EcoRI. This fragment was then ligated to the downstream fragment by digesting pJM10 with BamHI and EcoRI, giving rise to the plasmid pJM11. The mycobacterial shuttle plasmid pMV261.hyg (11) was digested with DraI and HpaI to yield a blunt-ended 1.4-kb fragment containing a hygromycin resistance cassette (hyg). The hygromycin-containing fragment was then cloned in between the two vapG flanks in pJM11 by digesting pJM11 with BamHI and blunt ending the overhangs by Klenow treatment, giving rise to pJM11.hyg. The plasmid pJM11.hyg was then digested with PvuII to yield a blunt-ended 3.3-kb fragment containing both vapG flanks ligated to the hygromycin resistance cassette. This fragment was then cloned into pMV261.lacZ2 which had been previously digested with SgrAI and KpnI and Klenow treated to create compatible blunt ends. Digestion with SgrAI and KpnI removed the oriM from pMV261.lacZ2, thus allowing the resulting plasmid, pGBC14.1lacZ, to be used as a suicide plasmid to generate the vapG mutant. To confirm the deletion of vapG, we performed PCR using several primer pairs, including vapG-up-F and vapG-down-R, vapG-F and vapG/podG-R, and vapG-int-F and vapG-int-R. Further confirmation of the deletion of vapG was achieved by reverse transcriptase PCR (RT-PCR) and DNA sequencing.
To create the complementing plasmid for the vapG mutation, a 36-bp fragment from pETDuet-1 (Novagen, Madison, WI) containing the multiple-cloning site (MCS-2) was amplified using primers MCS-F and MCS-R. The PCR product was digested with BamHI and DraI and cloned into pSET152 (15) which had been previously digested with BamHI and EcoRV, giving rise to pSET152-AM. The plasmid pSET152-AM was subsequently digested with BamHI and NdeI to allow for the insertion of a constitutive hsp60 promoter as a 442-bp fragment amplified from pMV261 using primers F_HSP60 and HSP60_R, giving rise to the integrating plasmid, pSET152-AMH. Next, a 553-bp vapG-containing fragment was amplified by PCR using the primers vapG-comp-NdeI and vapG-comp-EcoRV. The resulting PCR fragment was digested with NdeI and EcoRV and cloned in frame immediately downstream of the hsp60 promoter in the integrating plasmid, pSET152-AMH. PCR and RT-PCR were used to confirm the complementation of the vapG mutant with a wild-type copy of vapG.
Construction of R. equi mutants.
The suicide vector pSJ53, containing flanking regions of the PAI ligated to either side of the gene encoding apramycin resistance and the lacZ gene encoding β-galactosidase on its backbone, was used to derive the pathogenicity island mutant of R. equi 103+ in which nearly 25.6 kb spanning ORF62 through ORF22 (coordinates 77178 to 23501) was deleted, using the method described by Jain et al. (17). In brief, pSJ53 was electroporated into R. equi, and transformants were selected on BHI plates supplemented with apramycin (80 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml), the chromogenic substrate of β-galactosidase. Blue transformants were screened by PCR (data not shown) to confirm them as single-crossover intermediates in which the entire pSJ53 plasmid had integrated into the virulence plasmid at either of the flanks of the PAI. Two blue colonies that were PCR positive for the single crossover were serially subcultured in BHI containing apramycin at 30°C to facilitate a second recombination event between the wild-type PAI and its mutated version resulting in a looping out of the intervening vector sequence, including the lacZ gene. Following plating on BHI agar supplemented with apramycin and X-Gal, the resulting white colonies were analyzed by PCR to confirm the loss of the PAI region and retention of the virulence plasmid backbone (17). A similar two-step allelic exchange procedure was performed to construct the vapG deletion mutant using the suicide vector pGBC14.1lacZ, except that hygromycin at 180 μg/ml was used for antibiotic selection.
Electroporation of R. equi.
R. equi strains were grown in 200 ml BHI broth to an optical density at 600 nm (OD600) of ∼0.8. Bacteria were pelleted and washed twice with an equal volume of cold distilled water (dH2O) before finally being resuspended in 10 ml of cold 10% glycerol made in dH2O. Aliquots (400 μl) of the cells were made and stored at −80°C. For electroporation, an aliquot of cells was mixed with 0.5 μg of plasmid DNA and placed in a prechilled 0.2-cm electroporation cuvette. Electroporation was performed using a Gene Pulser (Bio-Rad) set at 2.5 kV, 25 μF, 1,000 Ω, and single pulse. Immediately after electroporation, 1 ml of BHI broth supplemented with 0.5 M sucrose was added to the cuvette. Bacteria were then incubated for 1 h at 30°C and subsequently plated on BHI agar supplemented with the appropriate antibiotics.
RNA isolation.
Five-milliliter bacterial cultures were grown to mid-logarithmic phase (OD600 of 0.8), after which 1 ml of culture (∼1 × 108 bacteria) was removed and incubated with 2 volumes RNAprotect Bacteria reagent (Qiagen, Valencia, CA) and bacteria subsequently harvested by centrifugation at 4,000 × g for 10 min. The cells were resuspended in 700 μl RLT buffer (RNeasy mini kit; Qiagen) and added to 0.1-mm-diameter acid-washed zirconium/silica beads (BioSpec, Bartlesville, OK). The bacterial samples were lysed for 5 min at maximum speed with a Mini BeadBeater-1 (BioSpec) cell disruptor. Total RNA was subsequently isolated using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions.
RT-PCR.
Total RNA in each sample was quantified by UV spectrophotometry, and cDNA was synthesized using the ThermoScript RT-PCR system (Invitrogen, Carlsbad, CA) from equal amounts (1 μg) of RNA template using 50 ng of the supplied random hexamer primers and 15 U of ThermoScript RT enzyme according to the manufacturer's instructions. One-tenth of the cDNA reaction mixture (2 μl) was then used as template for standard PCR amplification with Pfu DNA polymerase (Stratagene, La Jolla, CA) using primer pairs specific for vapG (vapG-int-F and vapG-int-R) or the internal housekeeping gene control, gyrB (gyrB-qPCR-F and gyrB-qPCR-R), as listed in Table 2.
Western blot analysis.
Bacterial strains were grown overnight at 37°C in 5 ml BHI, and the optical density at 600 nm was adjusted to 1.0. A 1-ml aliquot from each suspension was pelleted in a microcentrifuge set at 13,000 rpm for 5 min. The supernatant was discarded and the bacterial pellet resuspended in 100 μl SDS gel loading buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM dithiothreitol [DTT]) and boiled for 10 min at 100°C. After the sample was allowed to cool, 10 μl of each sample was loaded onto a precast 8 to 16% Tris-glycine 1.0-mm polyacrylamide gel (Invitrogen, Carlsbad, CA) and run at 80 V for 30 min, followed by 1 h at 120 V. Following electrophoresis, proteins were transferred to a 0.45-μm nitrocellulose membrane using a Bio-Rad Transblot apparatus set at 150 mA for 3 h. Once transfer was complete, the membrane was immersed in blocking buffer (Pierce Scientific, Rockford, IL) overnight at 4°C. Labeling of VapA protein was done with a 1:1,000 dilution of rabbit anti-VapA antiserum for 60 min at room temperature, followed by repeated washing with Tris wash buffer (Pierce Scientific) to remove unbound antibody. The membrane was then incubated in a 1:20,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Pierce Scientific) for 60 min at room temperature. Following repeated washes, detection of protein was performed using Pierce SuperSignal Pico ECL kit (Pierce Scientific) as per the manufacturer's instructions.
Flow cytometry.
After overnight culture at 37°C in BHI, bacteria were washed twice and resuspended in cation-free phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). Approximately 108 CFU of bacteria were incubated at room temperature with a 1:500 dilution of rabbit immune serum containing anti-VapA polyclonal antiserum. After being washed three times with PBS containing 1% BSA, bacteria were stained by incubation at room temperature with a 1:500 dilution of Alexa 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Invitrogen) for 45 min. Bacteria were then washed three times with PBS supplemented with 1% BSA, fixed in 2% paraformaldehyde, and analyzed on a Becton Dickinson LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Negative controls included unstained bacteria, the addition of secondary antibody without prior incubation in primary antibody, and the use of preimmune serum as an IgG control.
Macrophages and macrophage cell lines.
Murine RAW264.7 macrophages were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine and incubated at 37°C in 5% CO2. For intracellular growth assays, macrophages were seeded in 24-well tissue culture plates at 1 × 105 cells per well. Equine alveolar macrophages were obtained by bronchoalveolar lavage (BAL) of healthy adult horses. Horses were sedated intravenously with xylazine (0.4 mg/kg) (Butler Co., Dublin, OH) and butorphanol tartrate (Torbugesic, 0.02 mg/kg) (Fort Dodge Animal Health, Fort Dodge, IA). A sterile 1.8-m BAL catheter (Jorvet, Loveland, CO) was passed via nasal approach until wedged into a bronchus. The lavage solution consisted of four aliquots of 60-ml physiologic saline (0.9% NaCl) solution infused and aspirated immediately. The resulting BAL fluid exudate cells were washed twice in PBS supplemented with 9 mM EDTA, penicillin G (100 U/ml), and streptomycin (100 μg/ml). Cells (5 × 105) were incubated for 4 h at 37°C and 6% CO2 in minimal essential media alpha (αMEM) (HyClone, Waltham, MA) supplemented with 12% donor horse serum (DHS) (HyClone), 2 mM glutamine, and penicillin G-streptomycin in DHS-coated 24-well plates. Following incubation, adherent macrophages were washed twice with warm αMEM to remove nonadherent cells and incubated for a further 3 h in antibiotic-free medium. Macrophages were then washed again prior to infection with R. equi.
Intracellular growth of R. equi in macrophages.
Overnight broth cultures of bacteria at an optical density at 600 nm of 1.0 (∼2 × 108 CFU/ml) were pelleted, washed once with PBS, and resuspended in PBS. Murine macrophage monolayers were washed once with warm DMEM, and the medium was replaced with fresh DMEM supplemented with 10% FCS and 2 mM glutamine. Bacteria were added at a multiplicity of infection (MOI) of ∼10 bacteria per macrophage. Bacterial incubation with macrophages proceeded for 45 min at 37°C, and then the monolayers were washed repeatedly with prewarmed DMEM to remove unbound bacteria. After an additional 15-min incubation period to allow bound bacteria to be internalized, the monolayers were washed again, and the medium was replaced with complete DMEM supplemented with 20 μg/ml of amikacin sulfate. At various times postinfection, macrophage monolayers were washed repeatedly, and 500 μl sterile water was then added to lyse the macrophages upon further incubation at 37°C for 20 min. Bacterial growth was determined by dilution plating of lysates onto BHI agar plates. CFU were enumerated after 48 h of incubation at 37°C.
Bacterial survival in equine macrophages was determined by immunofluorescent staining, as previously described (17). Basically, R. equi strains were grown in BHI broth overnight at 37°C. The bacteria were pelleted and diluted in αMEM supplemented with non-heat-inactivated DHS to a concentration of 1 × 107 CFU/ml. Equine alveolar macrophage monolayers were cultured in 24-well plates (∼1 × 105/well) and infected at an MOI of 1:1 as described above. At 1, 24, and 48 h after infection, monolayers were washed once with PBS, fixed with 100% methanol for 45 min at 4°C, washed twice, and stored in 1× PBS with 5% FCS. Bacteria associated with macrophages were stained by incubating monolayers with rabbit anti-R. equi antibodies for 1 h at room temperature. After being washed with 1× PBS-5% FCS three times, monolayers were incubated with Alexa 488-conjugated goat anti-rabbit IgG (Invitrogen, Carlsbad, CA) for 1 h in the dark at room temperature. Finally, the macrophage monolayers were washed four times as before and examined under a Zeiss Axiovert 200 M fluorescence microscope. Two hundred macrophages per well were counted, and the number of bacteria associated with these cells was determined for each strain at each time point. Any cell containing 10 or more bacteria was simply scored as having 10 bacteria due to the difficulty in reliably quantifying large bacterial numbers within an individual macrophage.
Infection of mice.
Female severe combined immunodeficient (SCID) mice were obtained from either Charles River (Wilmington, MA) or Jackson Laboratories (Bar Harbor, ME). Mice were received at 6 weeks of age and were used when they were between the ages of 8 and 10 weeks. For the infection of mice, frozen aliquots of the bacterial strains were thawed and grown for 1 h at 37°C in BHI broth. Bacteria were pelleted and resuspended in PBS. Depending on the experiment, groups of mice were infected intravenously through the tail vein with approximately 5 × 105 or 5 × 106 bacteria. The total number of bacteria injected was confirmed retrospectively by dilution plating of the injection stock. At various times postinfection, four mice from each group were euthanized, and their spleens, livers, and lungs were removed. Each organ was placed in sterile PBS and homogenized with a tissue homogenizer (Seward, Bohemia, NY). Serial 10-fold dilutions of the homogenate were plated onto BHI agar, and CFU counts were determined after 48 h of incubation at 37°C.
Statistical analysis.
Statistical analyses were performed using the SPSS version 17.0.1 statistical package (SPSS Inc., Chicago, IL). Normal distribution and equal variance of the data were assessed using the Shapiro-Wilk and Levene tests, respectively. Comparison of the means in fold replication of intracellular bacterial numbers between bacterial strains was done using a one-way analysis of variance (ANOVA). When appropriate, multiple pairwise comparisons were done using Tukey's honestly significant difference (HSD) test. Significance was set at a P value of <0.05.
RESULTS
Creation of a pathogenicity island deletion mutant of R. equi.
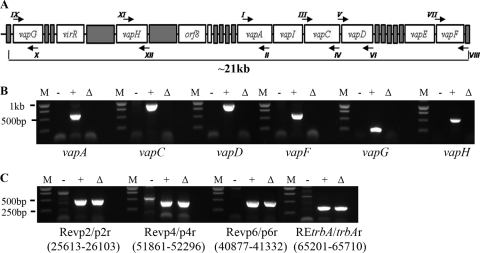
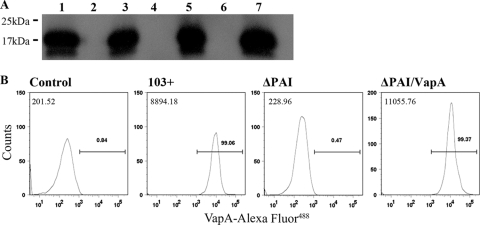
With the exception of the virulence factor vapA and its two regulators virR and orf8, no other virulence determinants on the virulence plasmid have been conclusively identified to date. It is known that vapA is not sufficient for virulence, since expression of vapA alone in a plasmidless strain does not restore the virulence phenotype. We sought to narrow the search for other plasmid-encoded virulence factors by addressing whether additional virulence determinants reside within the pathogenicity island (PAI) or, alternatively, are positioned outside this area within the plasmid backbone. Therefore, an R. equi strain in which we deleted an ∼26-kb stretch of genetic sequence that encompassed the entire pathogenicity island extending from orf62 to orf22 was constructed. A two-step knockout strategy employing a suicide vector with a single-crossover intermediate step was used as previously described (17). To confirm the absence of PAI-carried genes (i.e., vapA, vapC, vapD, vapF, vapG, and vapH) in the PAI mutant, PCR analysis using primer pairs (Fig. 1 A) that would specifically and individually amplify these genes was performed. As shown in Fig. 1B, no amplicons were obtained for any of these primer pairs in the PAI mutant, just as seen with the plasmid-cured strain 103−, used as a negative control, thus confirming their absence in the ΔPAI strain. In contrast, all primer pairs yielded an amplicon of the expected size when wild-type 103+ DNA was used as the template. To ensure that no gross alterations in the virulence plasmid backbone had taken place during the recombination events leading to the recovery of the PAI mutant, we performed PCR analysis using four primer pair sets annealing to different regions outside the PAI. As shown in Fig. 1C, both wild-type 103+ and the PAI mutant gave PCR products of the correct length for each of the primer pairs used, whereas no specific products were obtained with 103− DNA as template. Having deleted the entire pathogenicity island, the PAI deletion mutant was then used as a vehicle for constitutive expression of vapA to assist in pinpointing the likely location of plasmid regions required for virulence. To do so, the PAI mutant was transformed with an episomal recombinant plasmid, pMV261-vapA, in which the virulence determinant vapA was expressed from the mycobacterial heat shock promoter hsp60, thus allowing for constitutive expression of vapA independently of its two regulators, virR and orf8. The resulting strain, ΔPAI vapA, was analyzed by Western blotting using specific polyclonal antiserum to VapA to confirm the presence of VapA protein. As shown in Fig. 2 A, the PAI mutant (lane 6) lacks VapA, whereas the PAI mutant transformed with pMV261-vapA (lane 7) produces VapA protein at levels comparable to those of R. equi wild-type strain 103+ exposed to inducing conditions (lane 1). To ensure that the VapA protein produced in the PAI mutant was located on the bacterial cell surface and was not inappropriately confined to the cytosol due to possible disruption of normal protein targeting in the mutant, surface-localized VapA was quantified by flow cytometry. Using rabbit polyclonal antiserum as primary antibody and fluorescently conjugated goat anti-rabbit IgG as secondary antibody, the ΔPAI vapA strain was confirmed to have wild-type amounts of VapA on its surface (Fig. 2B).
FIG. 1.
Confirmation of the PAI deletion. (A) Schematic of the pathogenicity island and locations of the PCR primers (arrows) used in analysis of the PAI mutant. (B) Results of PCR analysis of the PAI mutant using primers annealing within the deleted region. Primers specific for either vapA, -C, -D, -F, -G, or -H were used to confirm the deletion in the mutant. PCR amplicons for these primer pairs are shown. Total DNA from R. equi 103− (lanes −), R. equi 103+ (lane +), and the PAI mutant (lanes Δ) was used as the template in PCRs. DNA standards (lanes M) are shown to the left of each group of lanes. (C) Results of PCR analysis of the virulence plasmid backbone in the PAI mutant. Primer pairs annealing to different regions spanning the virulence plasmid outside the deleted pathogenicity island region were used in PCRs using the same template DNAs as for panel A. Annealing site coordinates are shown in parentheses under the respective primer pair.
FIG. 2.
Expression of VapA in the PAI mutant. (A) Western blot analysis. Whole-cell extracts were prepared from R. equi 103+ (lane 1), virulence plasmid-cured 103− (lane 2), 103−/vapA (lane 3), 103+ ΔvapA (lane 4), 103+ ΔvapA/vapA (lane 5), ΔPAI (lane 6), and ΔPAI/vapA (lane 7), all cultured at 37°C as described in Materials and Methods. The presence of VapA protein was detected with rabbit polyclonal antiserum to VapA. Sizes of protein molecular mass standards are indicated on the left. (B) Flow cytometry profile showing the expression of VapA on the surface of R. equi. Bacteria were grown in liquid broth at 37°C, washed, and stained with a polyclonal antiserum to VapA. The relative median fluorescence values of wild-type isolate 103+, the PAI mutant strain (ΔPAI), and the recombinant pMV261-vapA-transformed PAI mutant strain (ΔPAI/vapA) were compared. Analyses included a comparison to bacteria stained with an irrelevant antibody (control; preimmune serum). The number in the upper left corner of each histogram represents the median fluorescence intensity for each strain. The values above the gate reflect the percentage of VapA-positive cells within the population analyzed.
VapA does not restore the virulence phenotype to the PAI mutant.
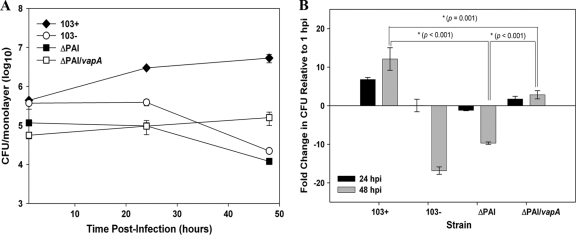
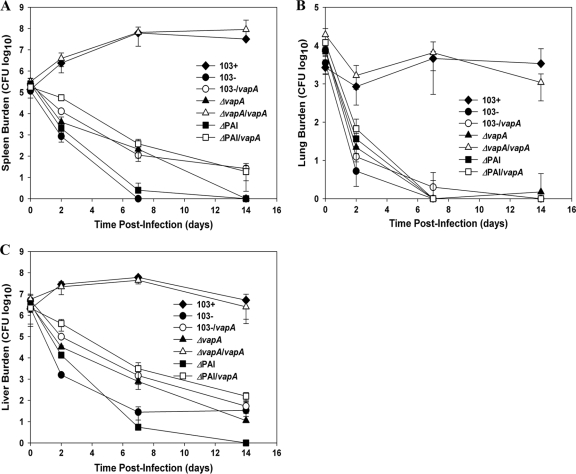
Given that the PAI mutant does not possess the crucial virulence determinant vapA, the expression of which is essential for replication of R. equi in in vitro-cultured macrophages and in vivo in mice and foals (17), it was anticipated that the strain would be compromised in both settings. As expected, the PAI mutant was fully attenuated, displaying intramacrophage growth kinetics that mirrored those of the avirulent plasmid-cured strain 103− (Fig. 3 A), both of which showed ∼10-fold or greater reduction in CFU at 48 h postinfection (hpi) (Fig. 3B). The latter is in contrast to the growth of wild-type bacteria, where an ∼10-fold increase in macrophage-associated CFU was apparent at 48 h. We next assessed whether or not the intracellular replication potential of the PAI mutant could be restored by expression of vapA in the mutant (Fig. 2A and B). It was reasoned that if production of wild-type levels of VapA protein in the PAI mutant did not restore intracellular growth, it would indicate that additional virulence determinants reside within the pathogenicity island of R. equi. The results show that the production of VapA protein allowed the PAI mutant to persist in macrophages, with the vapA-expressing strain showing a significant positive difference in fold growth compared to the PAI mutant (3-fold increase versus 10-fold decrease, respectively; P < 0.001) at 48 hpi. Although expression of vapA in the PAI mutant allowed the strain to survive better in macrophages than the PAI mutant without vapA, the strain was still not able to efficiently replicate, with a significant difference (P = 0.001) in CFU at 48 hpi compared to wild-type 103+ (Fig. 3A and B). Similarly, production of VapA protein by the PAI mutant also allowed it to survive slightly longer in vivo in SCID mice, which are incapable of clearing an infection of wild-type R. equi (Fig. 4). While the PAI mutant showed rapid clearance at a rate almost identical to that of the plasmid-cured strain 103−, the vapA-expressing PAI mutant displayed delayed clearance in the spleens and livers (Fig. 4A and C) relative to the PAI mutant. At 14 days postinfection, the vapA-expressing PAI mutant had a 158-fold-higher CFU count in the liver (Fig. 4C) and an ∼20-fold higher CFU in the spleen than the PAI mutant. Nonetheless, production of VapA was not enough; bacterial numbers of the vapA-expressing PAI mutant still showed an ∼3-log-unit decline in both the spleen (Fig. 4A) and liver (Fig. 4C) and complete clearance in the lung (Fig. 4B) by 7 days postinfection. In contrast, wild-type bacteria had increased 454-fold in the spleen (Fig. 4A) and 30-fold in the liver (Fig. 4C) and held steady in the lung (Fig. 4B) during this same time frame. The inability of vapA expression to restore the virulence phenotype to the PAI mutant either in macrophages or in vivo in mice was not due to some defect in the pMV261-vapA expression plasmid, as complete restoration of bacterial growth in macrophages (data not shown) and replication and persistence in mice (Fig. 4) was observed in a vapA deletion mutant transformed with this complementing plasmid.
FIG. 3.
Expression of vapA fails to restore the intracellular growth defect of the PAI mutant. (A) RAW264.7 macrophage monolayers were infected with R. equi strains 103+, 103−, ΔPAI, and ΔPAI/vapA at an MOI of 10:1. Following uptake and repeated washing to remove unbound bacteria and the addition of antibiotic to kill any remaining extracellular bacteria, triplicate macrophage monolayers were lysed at 1 h, 24 h, and 48 h postinfection. Lysates were plated onto BHI agar plates to determine the associated CFU. The data are expressed as means ± standard deviations. The graphs shown here are representative of three independent experiments. (B) Fold change in the CFU of intracellular bacteria at 24 h and 48 h postinfection relative to 1 h postinfection (hpi). A positive ratio reflects an increase in bacterial CFU over time resulting from bacterial replication in macrophages, whereas a negative ratio reflects a decrease in bacterial number over time. Values shown are the means ± standard deviations for triplicate monolayers from an individual experiment. The data shown are representative of three independent experiments.
FIG. 4.
Expression of vapA fails to rescue the PAI mutant of R. equi from clearance in SCID mice. SCID mice were injected intravenously with 5 × 106 CFU of R. equi 103+, 103−, 103−/vapA, 103+ ΔvapA, 103+ ΔvapA/vapA, ΔPAI, and ΔPAI/vapA. At 0, 2, 7, and 14 days postinfection, mice were humanely sacrificed and organs removed aseptically. The total numbers of bacteria in the spleens (A), lungs (B), and livers (C) were determined by dilution plating of organ homogenates. Each point on the graph represents the mean ± standard deviation of bacterial counts for four mice.
Creation of a vapG mutant and vapG-complementing strain of R. equi.
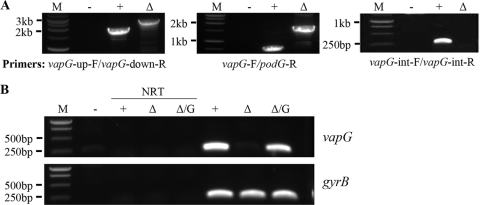
The apparent failure of vapA expression to restore wild-type virulence to the PAI mutant suggested that vapA is not the only virulence determinant within the pathogenicity island region and that additional virulence determinants reside in the island. Because it possessed characteristics typical of virulence factors, it was hypothesized that another vap gene, vapG, was a logical candidate for an R. equi virulence determinate. To test this hypothesis, an R. equi strain with the vapG gene deleted by means of a two-step knockout strategy similar to that as described above for the PAI mutant was constructed. PCR analysis with specific diagnostic primer pair sets confirmed that the gene for vapG had been replaced by the 1.4-kb DNA fragment encoding hygromycin resistance (Fig. 5 A). For example, PCR analysis using primer pair vapG-up-F and vapG-down-R yielded a 2.1-kb amplicon in wild-type 103+ and the anticipated 2.9-kb product in the mutant (Fig. 5A). Likewise, primer pair vapG-F-vapG/podG-R gave ∼700-bp and ∼1.6-kb amplicons in the wild type and the ΔvapG mutant, as expected. Finally, using primer pair vapG-int-F and vapG-int-R, which anneal at sites internal to vapG, the expected ∼250-bp amplicon was observed using wild-type DNA as the template, while no product was obtained with ΔvapG mutant DNA. To further confirm deletion of the vapG gene, we performed RT-PCR on total RNA extracts using an internal vapG primer pair (vapG-int-F and -R) as a probe for the presence or absence vapG transcripts. The results of the RT-PCR (Fig. 5B) confirmed the absence of vapG transcripts in the vapG mutant. Furthermore, it was verified that transformation of the vapG mutant with a wild-type copy of vapG resulted in restoration of vapG gene expression (Fig. 5B).
FIG. 5.
Confirmation of deletion of vapG. (A) PCR analysis of the vapG mutant. Primer pairs that specifically amplify regions across the deletion site were used in PCRs in which total DNA from R. equi 103+ (+) and the vapG mutant (Δ) served as the template. Standard DNA markers are indicated on the left (M). A no-template control (−) is also shown. (B) RT-PCR analysis of vapG expression. Total RNA was extracted from R. equi strains 103+ (+), the vapG mutant (Δ), and the complemented vapG mutant (Δ/G). cDNA was synthesized using equivalent concentrations of total RNA as the template. The presence of vapG and gyrB (internal control) was assessed by PCR using primer pairs specific for internal regions of each of the genes. Standard DNA markers are indicated on the left (M). No-template controls (−) and no-RT controls (NRT) are shown.
vapG is dispensable for growth in macrophages.
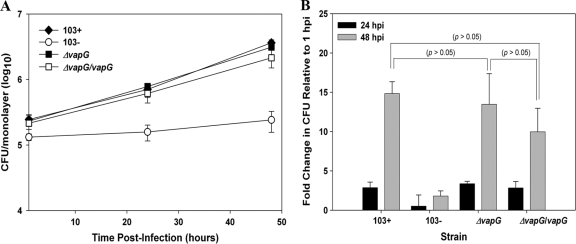
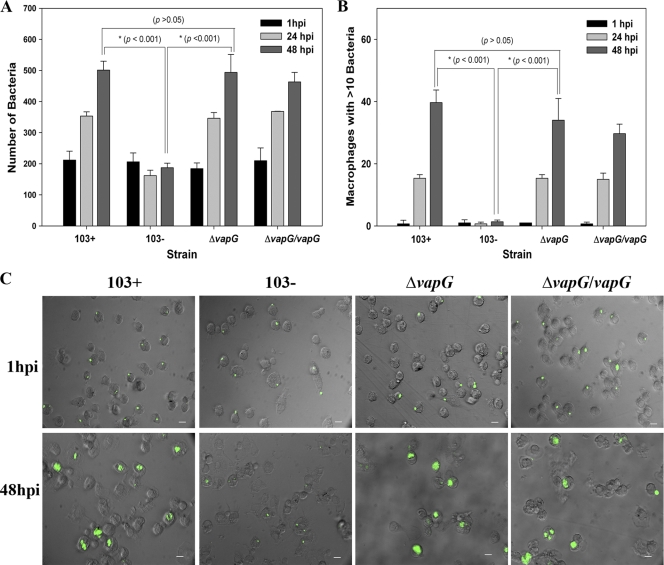
To evaluate whether deletion of vapG affected the ability of R. equi to replicate intracellularly in macrophages, murine RAW264.7 macrophages were infected with the vapG mutant, and its intracellular growth capacity relative to those of wild-type R. equi strain 103+ and the virulence plasmid-cured strain, 103−, was assessed (Fig. 6 A and B). As expected, the plasmid-cured strain failed to replicate in the macrophages (Fig. 6A), whereas the virulent strain 103+ multiplied ∼15-fold over a 48-h period. The growth of the vapG mutant closely mimicked that of the wild type, as did the growth of the vapG-complemented strain, whose numbers increased ∼13-fold and 10-fold, respectively, over the same time period (Fig. 6B). No significant difference was observed between 103+ and the vapG mutant at any time point postinfection, demonstrating that the deletion of vapG does not adversely affect the growth of R. equi in murine macrophages. Identical results were obtained using murine primary bone marrow-derived macrophages (data not shown). To exclude the possibility that VapG is required solely for growth in macrophages of its native host, we assessed the growth of the vapG mutant in equine alveolar macrophages. Similar to findings with the murine macrophages, the vapG mutant grew as well as wild-type bacteria in equine macrophages. In these cells, the two bacterial strains exhibited indistinguishable growth curves, reflected by identical increases in the total number of bacteria associated with the macrophage monolayer (P > 0.05) (Fig. 7 A) and in the number of macrophages with >10 bacteria per macrophage over time (P > 0.05) (Fig. 7B) when quantified by immunofluorescent counting as previously described (14). In contrast, the plasmid-cured strain failed to replicate in the macrophages (Fig. 7A and B). Representative photomicrographs of equine macrophage monolayers infected with wild-type 103+, the vapG mutant, the complemented vapG mutant strain, or the plasmid-cured strain are provided in Fig. 7C
FIG. 6.
vapG is dispensable for growth in murine macrophages. (A) Intracellular replication of R. equi strains in RAW264.7 macrophages. Macrophage monolayers were infected with R. equi 103+, 103−, 103+ ΔvapG, and 103+ ΔvapG/vapG, and bacterial intracellular replication was followed as described in the legend to Fig. 3. (B) Fold change in intracellular bacterial CFU among the various strains at 24 h and 48 h postinfection (hpi). Values shown are the means ± standard deviations for monolayers assessed in triplicate. The data shown here are representative of three independent experiments.
FIG. 7.
vapG is dispensable for growth in equine alveolar macrophages. Intracellular replication of R. equi strains in equine alveolar macrophages is shown. (A) The number of bacteria associated with 200 macrophages was determined at 1 h, 24 h, and 48 h postinfection by fluorescence microscopy. (B) Number of macrophages with 10 or more intracellular bacteria at the indicated time points postinfection. Values shown are the means ± standard deviation for monolayers assessed in triplicate. The data shown here are representative of three independent experiments. (C) Representative epifluorescence microscopy images of equine macrophages infected with R. equi strains 103+, 103−, 103+ ΔvapG, and 103+ ΔvapG/vapG taken at 1 h, 24 h, and 48 h postinfection. Scale bar, 10 μm; magnification, ×400.
vapG is dispensable for survival and growth in vivo.
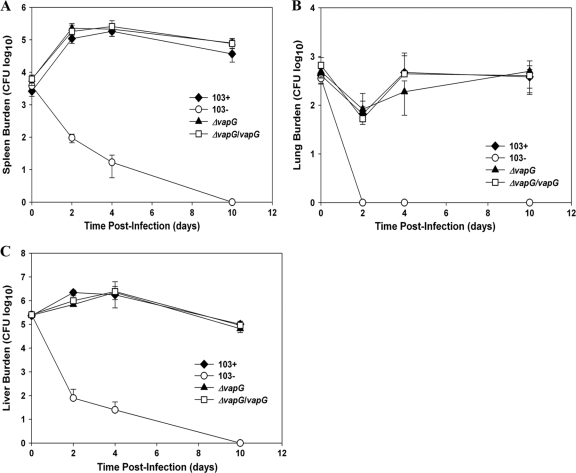
Although loss of vapG did not affect the macrophage growth capabilities of the bacteria, we reasoned that vapG might be necessary specifically for survival in vivo, as has been observed to be the case with some virulence proteins of Mycobacterium tuberculosis (27). To determine this, SCID mice were challenged with the vapG mutant and the clearance of the mutant relative to those of the wild-type and plasmid-cured controls was monitored. Analysis of bacterial organ burden showed that the vapG mutant replicated as efficiently as its wild-type parent, displaying an ∼40-fold increase in bacterial number in the spleen (Fig. 8 A) and a 9-fold increase in numbers in the liver (Fig. 8C) at 48 hpi, a level of burden which persisted for the duration of the infection. Furthermore, the vapG mutant was able to persist in the lungs (Fig. 8B) of infected mice to the same degree as wild-type R. equi. Clearance of the vapG mutant complemented with a wild-type copy of vapG integrated on the chromosome was indistinguishable from those of the vapG mutant and wild-type R. equi 103+ at all time points postinfection, whereas the plasmid-cured strain was rapidly cleared. The data demonstrate that vapG is dispensable for bacterial survival and replication in vivo in mice.
FIG. 8.
vapG is dispensable for survival in vivo. SCID mice were infected intravenously with 5 × 105 CFU of R. equi 103+, 103−, 103+ ΔvapG, and 103+ ΔvapG/vapG. At 0, 2, 7, and 10 days postinfection, mice were humanely sacrificed and organs removed and homogenized. The total numbers of bacteria in the spleens (A), lungs (B), and livers (C) were determined by dilution plating of the organ homogenates. Each point on the graph represents the mean ± standard deviation of bacterial CFU of four mice.
DISCUSSION
Although necessary, VapA is not sufficient for R. equi virulence. Microarray analysis by Ren and Prescott demonstrated that only genes within the pathogenicity island were differentially regulated during intramacrophage growth, while virulence plasmid genes outside this region showed no significant changes in transcription (25). This led to the hypothesis that the additional plasmid-derived virulence factors were most likely to reside within the pathogenicity island, rather than in the plasmid backbone. To address the hypothesis, a pathogenicity island deletion strain of R. equi, removing ∼26 kb of plasmid sequence encompassing orf62 to orf22, was constructed. The mutant strain was then used as the background for constitutive expression of vapA, reasoning that the phenotype of the VapA-producing PAI strain would yield insight into the locations of these additional virulence factors. Attenuated growth of the pathogenicity island mutant in macrophages and accelerated clearance of the mutant in vivo in SCID mice was observed, as expected, since the strain lacked vapA. While constitutive expression of vapA by the PAI deletion mutant did help to delay its clearance in both macrophages and mice, the virulence phenotype was not restored. The findings, therefore, were supportive of the hypothesis that still further virulence determinants were to be found within the deleted PAI region.
We had earlier shown that a mutant strain deleted for the pathogenicity island genes vapA, -I, -C, -D, -E, and -F and five genes of unknown function (orf16 to -18, 0680, and 0740) is complemented by in trans expression of vapA alone, suggesting that these 10 genes are not essential for virulence (17). Likewise, orf10, another gene of unknown function, is similarly dispensable, since its deletion had no effect on R. equi clearance in mice (25, 26). Of the remaining unanalyzed PAI genes, it was noted that vapG possessed a number of characteristics similar to those of established virulence determinants. Specifically, it is an extracellular protein sharing 55% similarity with VapA. It possesses a clear signal sequence indicating that it is secreted, as are many proteins involved in host-pathogen interactions, although it remains to be established whether VapG is surface localized. Expression of vapG is induced by many environmental signals, including oxidative stress (4), increased temperature (25), low iron and magnesium concentrations, and reduced pH (25), conditions found within the local environment of host phagosomes. Induction of vapG in response to any of these signals could perhaps indicate a role for this protein in the bacterial response to such stresses within the macrophage. Further support for this idea is provided by the findings that vapG expression is highly upregulated in foal macrophages (25) and that it is the most highly expressed vap gene (with ∼5-fold-higher expression than vapA) in the lungs of foals infected with R. equi (16). Therefore, vapG was considered by us and others to be a highly probable virulence determinant candidate. To test this, a vapG deletion stain was constructed using an allelic exchange methodology (17) established by this laboratory. Analysis of the vapG mutant in both murine and equine macrophages showed that deletion of the gene had no significant effect on the ability of the strain to replicate intracellularly, a somewhat surprising finding given that expression of the gene is significantly increased in that environment (25). When assessed in the SCID mouse infection model, it was observed that the vapG mutant displayed in vivo survival and replication kinetics indistinguishable from those of wild-type R. equi. Thus, vapG is dispensable for growth in in vitro-cultured macrophages and in vivo in mice, data which upon first look suggest that it plays little if any role in virulence of the organism.
There are certain caveats which preclude the complete exclusion of a role for VapG in disease pathogenesis. It is possible that VapG is important during the earliest phase of infection, for example, during initial interactions with the host respiratory tract (e.g., epithelial cells), perhaps facilitating initial attachment and adhesion events. Alternatively, VapG may function in aiding dissemination from the lung following respiratory infection. Neither of these two possibilities can be assessed by direct infection of in vitro-grown macrophages or intravenously challenged animals. Furthermore, while VapG appears to be dispensable in the murine infection model, it may play a role in survival and propagation of bacteria in the lungs of infected foals, the natural hosts of R. equi. The significantly high level of vapG expression in the lungs of infected foals is supportive of the hypothesis that VapG is needed during infection of susceptible horses. An in vivo equine infection model could definitely evaluate the requirement for this protein in R. equi disease; however, such analysis is beyond the scope of this work.
The data suggest that virulence determinants, aside from vapA and its two regulators (virR and orf8), reside within the pathogenicity island. While it is formally a possibility that VirR and Orf8 influence genes on the plasmid backbone, the probability of this is viewed to be low given that expression of these backbone genes remains unchanged during intracellular growth, while the expression of the regulators is increased (25). Full-length PAI genes that to our knowledge have yet to be assessed for a role in virulence include orf1 (encoding a putative Lsr2 protein), orf3 (putative S-adenosylmethionine [SAM]-dependent methyltransferase) (20), orf5 (putative major facilitator superfamily transporter) (20), orf6 (vapH; unknown function), orf7 (unknown function), orf9 (hypothetical protein), orf11 (hypothetical protein), and orf21 (chorismate mutase, scm2) (20). Work to establish which of those genes are needed for virulence is currently under way.
Acknowledgments
We thank James Barber, Jessica Miller, David Quiceno, Artem Rogovskyy, and Meredith Tatum for technical assistance. We are grateful to Vibhay Tripathi for thoughtful discussions regarding this work and to Kari Fine for assistance with the statistical analysis. We extend appreciation to Miriam Braunstein for critiquing the manuscript and to Steeve Giguère and Tracy Sturgill, who generously dedicated their time and resources to perform the bronchoalveolar lavages and who also reviewed the manuscript.
This study was funded by the National Institutes of Health (R01 AI060469).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 3 May 2010.
REFERENCES
- 1.Ainsworth, D. M., S. W. Eicker, A. E. Yeagar, C. R. Sweeney, L. Viel, D. Tesarowski, J. P. Lavoie, A. Hoffman, M. R. Paradis, S. M. Reed, H. N. Erb, E. Davidow, and M. Nalevanko. 1998. Associations between physical examination, laboratory, and radiographic findings and outcome and subsequent racing performance of foals with Rhodococcus equi infection: 115 cases (1984-1992). J. Am. Vet. Med. Assoc. 213:510-515. [PubMed] [Google Scholar]
- 2.Arlotti, M., G. Zoboli, G. L. Moscatelli, G. Magnani, R. Maserati, V. Borghi, M. Andreoni, M. Libanore, L. Bonazzi, A. Piscina, and R. Ciammarughi. 1996. Rhodococcus equi infection in HIV-positive subjects: a retrospective analysis of 24 cases. Scand. J. Infect. Dis. 28:463-467. [DOI] [PubMed] [Google Scholar]
- 3.Ashour, J., and M. K. Hondalus. 2003. Phenotypic mutants of the intracellular actinomycete Rhodococcus equi created by in vivo Himar1 transposon mutagenesis. J. Bacteriol. 185:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2002. H2O2, which causes macrophage-related stress, triggers induction of expression of virulence-associated plasmid determinants in Rhodococcus equi. Infect. Immun. 70:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2001. Induction of vap genes encoded by the virulence plasmid of Rhodococcus equi during acid tolerance response. Res. Microbiol. 152:439-449. [DOI] [PubMed] [Google Scholar]
- 6.Borghi, E., M. La Francesca, L. Gazzola, G. Marchetti, S. Zonato, P. Foa, A. d'Arminio Monforte, and G. Morace. 2008. Rhodococcus equi infection in a patient with spinocellular carcinoma of unknown origin. J. Med. Microbiol. 57:1431-1433. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, N. D., M. S. O'Conor, M. K. Chaffin, and R. J. Martens. 2005. Farm characteristics and management practices associated with development of Rhodococcus equi pneumonia in foals. J. Am. Vet. Med. Assoc. 226:404-413. [DOI] [PubMed] [Google Scholar]
- 8.Cornish, N., and J. A. Washington. 1999. Rhodococcus equi infections: clinical features and laboratory diagnosis. Curr. Clin. Top. Infect. Dis. 19:198-215. [PubMed] [Google Scholar]
- 9.Elissalde, G. S., H. W. Renshaw, and J. A. Walberg. 1980. Corynebacterium equi: an interhost review with emphasis on the foal. Comp. Immunol. Microbiol. Infect. Dis. 3:433-445. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Mora, E., M. Polidori, A. Luhrmann, U. E. Schaible, and A. Haas. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635-653. [DOI] [PubMed] [Google Scholar]
- 11.Giguere, S., M. K. Hondalus, J. A. Yager, P. Darrah, D. M. Mosser, and J. F. Prescott. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect. Immun. 67:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giguere, S., S. Jacks, G. D. Roberts, J. Hernandez, M. T. Long, and C. Ellis. 2004. Retrospective comparison of azithromycin, clarithromycin, and erythromycin for the treatment of foals with Rhodococcus equi pneumonia. J. Vet. Intern. Med. 18:568-573. [DOI] [PubMed] [Google Scholar]
- 13.Harvey, R. L., and J. C. Sunstrum. 1991. Rhodococcus equi infection in patients with and without human immunodeficiency virus infection. Rev. Infect. Dis. 13:139-145. [DOI] [PubMed] [Google Scholar]
- 14.Hondalus, M. K., and D. M. Mosser. 1994. Survival and replication of Rhodococcus equi in macrophages. Infect. Immun. 62:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Y., and M. K. Hondalus. 2008. Site-specific integration of Streptomyces PhiC31 integrase-based vectors in the chromosome of Rhodococcus equi. FEMS Microbiol. Lett. 287:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacks, S., S. Giguere, and J. F. Prescott. 2007. In vivo expression of and cell-mediated immune responses to the plasmid-encoded virulence-associated proteins of Rhodococcus equi in foals. Clin. Vaccine Immunol. 14:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain, S., B. R. Bloom, and M. K. Hondalus. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. Mol. Microbiol. 50:115-128. [DOI] [PubMed] [Google Scholar]
- 18.Kedlaya, I., and M. Ing. 2008. Rhodococcus equi. http://emedicine.medscape.com/article/235466-overview. [DOI] [PubMed]
- 19.Lasky, J. A., N. Pulkingham, M. A. Powers, and D. T. Durack. 1991. Rhodococcus equi causing human pulmonary infection: review of 29 cases. South. Med. J. 84:1217-1220. [DOI] [PubMed] [Google Scholar]
- 20.Letek, M., A. A. Ocampo-Sosa, M. Sanders, U. Fogarty, T. Buckley, D. P. Leadon, P. Gonzalez, M. Scortti, W. G. Meijer, J. Parkhill, S. Bentley, and J. A. Vazquez-Boland. 2008. Evolution of the Rhodococcus equi vap pathogenicity island seen through comparison of host-associated vapA and vapB virulence plasmids. J. Bacteriol. 190:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz, P., A. Burillo, J. Palomo, M. Rodriguez-Creixems, and E. Bouza. 1998. Rhodococcus equi infection in transplant recipients: case report and review of the literature. Transplantation 65:449-453. [DOI] [PubMed] [Google Scholar]
- 22.Muscatello, G., D. P. Leadon, M. Klayt, A. Ocampo-Sosa, D. A. Lewis, U. Fogarty, T. Buckley, J. R. Gilkerson, W. G. Meijer, and J. A. Vazquez-Boland. 2007. Rhodococcus equi infection in foals: the science of ‘rattles’. Equine Vet. J. 39:470-478. [DOI] [PubMed] [Google Scholar]
- 23.Polidori, M., and A. Haas. 2006. VapI, a new member of the Rhodococcus equi Vap family. Antonie Van Leeuwenhoek 90:299-304. [DOI] [PubMed] [Google Scholar]
- 24.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren, J., and J. F. Prescott. 2003. Analysis of virulence plasmid gene expression of intra-macrophage and in vitro grown Rhodococcus equi ATCC 33701. Vet. Microbiol. 94:167-182. [DOI] [PubMed] [Google Scholar]
- 26.Ren, J., and J. F. Prescott. 2004. The effect of mutation on Rhodococcus equi virulence plasmid gene expression and mouse virulence. Vet. Microbiol. 103:219-230. [DOI] [PubMed] [Google Scholar]
- 27.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuss, S. M., M. K. Chaffin, and N. D. Cohen. 2009. Extrapulmonary disorders associated with Rhodococcus equi infection in foals: 150 cases (1987-2007). J. Am. Vet. Med. Assoc. 235:855-863. [DOI] [PubMed] [Google Scholar]
- 29.Russell, D. A., G. A. Byrne, E. P. O'Connell, C. A. Boland, and W. G. Meijer. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J. Bacteriol. 186:5576-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speck, D., I. Koneth, M. Diethelm, and I. Binet. 2008. A pulmonary mass caused by Rhodococcus equi infection in a renal transplant recipient. Nat. Clin. Pract. Nephrol. 4:398-403. [DOI] [PubMed] [Google Scholar]
- 31.Steyn, A. J., J. Joseph, and B. R. Bloom. 2003. Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 47:1075-1089. [DOI] [PubMed] [Google Scholar]
- 32.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 33.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai, S., K. Koike, S. Ohbushi, C. Izumi, and S. Tsubaki. 1991. Identification of 15- to 17-kilodalton antigens associated with virulent Rhodococcus equi. J. Clin. Microbiol. 29:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai, S., T. Sekizaki, T. Ozawa, T. Sugawara, Y. Watanabe, and S. Tsubaki. 1991. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect. Immun. 59:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyooka, K., S. Takai, and T. Kirikae. 2005. Rhodococcus equi can survive a phagolysosomal environment in macrophages by suppressing acidification of the phagolysosome. J. Med. Microbiol. 54:1007-1015. [DOI] [PubMed] [Google Scholar]
- 37.van der Geize, R., W. de Jong, G. I. Hessels, A. W. Grommen, A. A. Jacobs, and L. Dijkhuizen. 2008. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 36:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Bargen, K., M. Polidori, U. Becken, G. Huth, J. F. Prescott, and A. Haas. 2009. Rhodococcus equi virulence-associated protein A is required for diversion of phagosome biogenesis but not for cytotoxicity. Infect. Immun. 77:5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zink, M. C., J. A. Yager, and N. L. Smart. 1986. Corynebacterium equi infections in horses, 1958-1984: a review of 131 cases. Can. Vet. J. 28:213-217. [PMC free article] [PubMed] [Google Scholar]