Abstract
Salmonella enterica subsp. enterica serovar Enteritidis is a leading causative agent of gastroenteritis in humans. This pathogen also colonizes the intestinal tracts of poultry and can spread systemically in chickens. Transfer to humans usually occurs through undercooked or improperly handled poultry meat or eggs. The bacterial twin-arginine transport (Tat) pathway is responsible for the translocation of folded proteins across the cytoplasmic membrane. In order to study the role of the Tat system in the infection and colonization of chickens by Salmonella Enteritidis, we constructed chromosomal deletion mutants of the tatB and tatC genes, which are essential components of the Tat translocon. We observed that the tat mutations affected bacterial cell morphology, motility, and sensitivity to albomycin, sodium dodecyl sulfate (SDS), and EDTA. In addition, the mutant strains showed reduced invasion of polarized Caco-2 cells. The wild-type phenotype was restored in all our Salmonella Enteritidis tat mutants by introducing episomal copies of the tatABC genes. When tested in chickens by use of a Salmonella Enteritidis ΔtatB strain, the Tat system inactivation did not substantially affect cecal colonization, but it delayed systemic infection. Taken together, our data demonstrated that the Tat system plays a role in Salmonella Enteritidis pathogenesis.
Salmonella is a major concern in human health, because it is one of the leading causative agents of gastroenteritis. Humans usually become infected by ingestion of contaminated eggs and undercooked chicken meat (14).
In poultry, Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium are considered most important. Salmonella usually causes asymptomatic infection in birds, but outbreaks with high levels of mortality and symptomatic infection have occurred in birds less than 2 weeks old (20, 46). Infection of poultry is generally characterized by ingestion of the bacteria and colonization of intestinal mucosa. Those bacteria penetrate the intestinal mucosa and spread systemically (15, 21, 22, 37).
Adhesion and invasion of epithelial cells are complex multifactorial processes. Often, a number of different virulence factors contribute to the survivability and successful infection of a microorganism in a given host. This is particularly true for many enterobacteria, including Salmonella. Therefore, it is important to investigate new potential virulence determinants and to study their roles in vitro and in vivo.
In most bacteria, including the Gram-negative salmonellae, enzymatically active respiratory enzymes have to be transported across the impermeable cytoplasmic membrane. Since these respiratory enzymes often contain cofactors, they need to be translocated in a prefolded, sometimes oligomeric state. This can be accomplished by the twin-arginine transport (Tat) system, which recognizes and translocates into the periplasm only polypeptides with a specific N-terminal signal sequence containing a “twin-arginine” motif. The Tat system is well characterized in Escherichia coli. There an operon containing tatABCD genes is found with an additional gene, tatE, located elsewhere in the chromosome. Functionally, TatA, TatB, and TatC are found to form two separate complexes in the inner membrane (8, 65). For the export of a target protein, the twin-arginine motif in its signal sequence is recognized by TatC (1), followed by association of TatA with the complex and transport through a pore formed by TatA protomers (24, 48). Of the other two proteins, TatE is very similar to and functionally interchangeable with TatA (34), while TatD plays an unknown role, demonstrating some DNase activity and involvement in the degradation of misfolded FeS proteins (49, 53, 64, 74).
A number of studies regarding the function of the Tat system at the molecular level have demonstrated its importance in a wide variety of cellular functions (44, 63). However, there is limited information regarding the involvement of the Tat system in virulence. In Pseudomonas aeruginosa, the Tat system may affect virulence by the secretion of stress response- or pathogenesis-related factors (51). Another report finds the Tat system to be a virulence determinant in Agrobacterium tumefaciens (19). In Escherichia coli O157:H7, tatABC deletion resulted in a loss of motility on soft agar plates, which was considered to be due to impaired secretion of Shiga toxin 1 and H7 flagellin, both known as major virulence factors (56).
The present study aims to clarify the impact and possible role of the Tat system in Salmonella Enteritidis virulence. In order to accomplish this, Tat system mutants were subjected to phenotypic assays and invasion assays using polarized human epithelial colorectal adenocarcinoma (Caco-2) cells and were also tested for their abilities to colonize chickens successfully. The results indicated that the Tat system plays an important role not only in cell invasion but also in the systemic spread of Salmonella Enteritidis in chickens.
MATERIALS AND METHODS
Search for putative Tat substrates by DNA sequence analysis.
The full genome sequence of S. Enteritidis strain Sal18, used in this study, is not available. However, we have evidence that the Sal18 genome is highly similar to the S. enterica serovar Enteritidis PT4 NCTC 13349 full-length sequence, provided by the Wellcome Trust Sanger Institute, United Kingdom. Previously, we have designed primers based on the published S. Enteritidis PT4 NCTC 13349 sequence in order to amplify >50 gene loci of interest from various chromosomal regions of Sal18 by PCR. So far, all tested primers resulted in the expected amplification products. A comparison of numerous sequenced fragments from Sal18 with the corresponding regions from PT4 resulted in 95 to 100% identity at the DNA level (data not shown). Therefore, the complete S. Enteritidis PT4 NCTC 13349 sequence was searched for candidate proteins to be translocated by the Tat system. Programs were run as provided by the TatP 1.0 server (Technical University of Denmark; http://www.cbs.dtu.dk/services/TatP) (6) and the TatFind server (http://signalfind.org/tatfind.html) (59).
Construction of mutants.
The mutants generated in this study were derivatives of a Salmonella Enteritidis strain (Sal18) and were constructed using the lambda (λ) red system as described previously (17). Briefly, a PCR product was generated by using primers that complemented flanking regions of an antibiotic resistance cassette with an overhang of at least 50 nucleotides homologous to the region of interest in the Sal18 wild-type (WT) strain. The S. Enteritidis PT4 NCTC 13349 sequence was used as a reference. Plasmids pKD3 (17) and pBR322 (9), containing a chloramphenicol resistance and a tetracycline resistance cassette, respectively, were used as templates for the antibiotic resistance cassettes. The PCR conditions used were 94°C for 3 min; 35 cycles of 94°C (30 s), 53°C (45 s), and 72°C (1.5 min); and finally 72°C for 7 min. The primers used are listed in Table 1. The PCR product was transformed by electroporation into competent wild-type Sal18 cells containing plasmid pKD46, which expressed the λ red recombinase. Bacterial cells were resuspended in SOC medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 20 mM MgCl2, 20 mM glucose) and were allowed to grow for 3 h at 37°C with shaking. The bacteria were then plated onto Luria-Bertani (LB) broth agar plates containing 9 μg ml−1 of chloramphenicol (or 7 μg ml−1of tetracycline) and were incubated overnight at 37°C. The clones obtained were streaked onto LB agar plates containing 30 μg ml−1of chloramphenicol (or 15 μg ml−1of tetracycline) and were subjected to the same incubation conditions as in the previous step. The constructs were confirmed by PCR using a 2720 Thermal Cycler (Applied Biosystems) and by sequence analysis (3730 XL DNA Analyzer; Applied Biosystems). The primers used in this step are listed in Table 1. For removal of the antibiotic cassette, plasmid pCP20 was electroporated into a ΔtatB or ΔtatC strain with the chloramphenicol resistance cassette (LS37 and LS55, respectively). The strain was grown in SOC medium for 1 h at 30°C. The bacterial cells were then plated onto LB plates containing ampicillin at a concentration of 100 μg/ml and were incubated overnight at 30°C. One colony was picked and diluted in 1 ml of phosphate-buffered saline (PBS). Serial dilutions were then made, plated into LB agar plates, and incubated overnight at 42°C. Subsequently, 20 colonies were randomly picked among all dilutions plated and were restreaked onto LB agar plates, LB agar plates containing 100 μg/ml of ampicillin, or LB agar plates containing chloramphenicol at a concentration of 30 μg/ml. The positive clones (chloramphenicol and ampicillin sensitive) were then confirmed by PCR using the respective confirmatory primers listed in Table 1. The strains used in this study are listed in Table 2.
TABLE 1.
Primers used in this study
| Primer | Length (bp) | Nucleotide sequence |
|---|---|---|
| TatB FWa | 70 | 5′-GAAGCGAAAAAGGAAGACGCTAAAAGCCAAGATAAAGAGCAGGTATAATCCATATGAATATCCACCTTAG-3′ |
| TatB RVa | 70 | 5′-CAACTCGATCAGATGCGTGATAAGCGGTTGAGTATCTTCTACAGCCATGTGTGTAGGCTGGAGCTGCTTC-3′ |
| TatC FWa | 70 | 5′-GCTGCGCCTGTTGTCGAATCTTCCCCCTCGTCGAGTGATAAACCGTAAACCATATGAATATCCACCTTAG-3′ |
| TatC RVa | 70 | 5′-ATCAAACATGCTTGCCCCCATATGACAACCGCCCTGGCGGGCGGTTGTGTGTGTAGGCTGAGCTGTTC-3′ |
| TatB FWb | 20 | 5′-TAATGTGTATAATGCGGCC-3′ |
| TatB RVb | 20 | 5′-ACGACCAGACGACGCTCATG-3′ |
| TatC FWb | 20 | 5′-CCCTGCCGCCGCTGAAACAC-3′ |
| TatC RVb | 19 | 5′-TTTGCAAACTGGCTACTGG-3′ |
| EcoRITatA-C FWc | 23 | 5′-GAATTCTGGCTGGTTGGCTGGCG-3′ |
| HindIIITatA-C RVc | 22 | 5′-AAGCTTAACGCCAATATCAAAC-3′ |
| EcoRITatA-D FWc | 22 | 5′-GAATTCTTACTCGTCAACCGCC-3′ |
| HindIIITatA-D RVc | 24 | 5′-AAGCTTCGCGTTCGATGTTACTGC-3′ |
λ red primer.
Confirmatory primer.
Primer used for amplification and cloning of the tatABC and tatABCD regions.
TABLE 2.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype and/or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| MC4100 | F−lacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 | 13 |
| B1LK0 | MC4100 tatC | 65 |
| S. Enteritidis | ||
| Sal18 | S. enterica serovar Enteritidis 301 PT13 | 55 |
| LS21 | Like Sal18, but ΔSPI-1::cat | 18 |
| LS25 | Like Sal18, but attTn7::cat | This study |
| LS29 | Like Sal18, but attTn7::tet | This study |
| LS37 | Like Sal18, but ΔtatB::cat | This study |
| LS39 | Like Sal18, but ΔtatB, cat removed | This study |
| LS55 | Like Sal18, but ΔtatC::cat | This study |
| LS106 | Like Sal18, but ΔtatC, cat removed | This study |
| LS107 | Like LS39, but with pLP196 | This study |
| LS108 | Like LS39, but with pLP198 | This study |
| LS114 | Like LS39, but with pLP199 | This study |
| LS117 | Like LS106, but with pLP198 | This study |
| LS118 | Like LS106, but with pLP199 | This study |
| Plasmids | ||
| pBR322 | Apr Tcr ColElOri; 4,361 bp | 9 |
| pKD3 | repR6Kγ AprFRT CmrFRT | 17 |
| pKD46 | reppSC101ts Apr ParaBADγβexo | 17 |
| pCP20 | Apr CmrcI857 PRflp pSC101 oriTS | 17 |
| pLP196 | pBR322 + tatA+B+C′ (truncated tatC) | This study |
| pLP198 | pBR322 + tatA+B+C+ | This study |
| pLP199 | pBR322 + tatA+B+C+D+ | This study |
Congo red binding assay.
Salmonella strains were tested for their abilities to bind Congo red dye. Briefly, 3 μl of a bacterial culture grown overnight in LB medium was diluted in an equal volume of distilled water. The mixture was spotted onto a Congo red agar plate (1% Bacto tryptone, 0.5% yeast extract, 40 μg ml−1 Congo red dye, 1.5% agar) and incubated at 28°C for 24 h. In addition, the overnight culture of bacteria was also streaked onto Congo red plates and incubated under the same conditions (47, 58).
Albomycin sensitivity assay.
Bacterial strains were tested for albomycin sensitivity as previously described (40). Briefly, 100 μl of an overnight bacterial culture was inoculated into 5 ml of 0.5% molten agar (45°C). The mixture was then immediately poured onto an LB agar plate. After solidification of the soft agar, a paper disc containing 10 μl albomycin solution at 1 mg/ml or serial dilutions thereof was placed on the plate, and the inhibition of growth was observed after 8 to 12 h of incubation at 37°C. Alternatively, the albomycin solution was spotted directly onto the bacterium-containing top agar.
Motility assay.
To test for motility, 10 μl of an LB broth culture with an absorbance at 600 nm of 0.8 was placed in a stab in LB plates containing 0.3% agar. The plates were incubated at 37°C without being inverted, and the diameter of the hazy zone formed by the migrating bacteria was measured at 3 and 6 h of incubation.
Cell morphology.
Cell morphology was determined by microscopic examination of a Gram stain (28) of bacteria that had been grown at 37°C in LB medium or on LB plates.
Sensitivity assays.
Sensitivities to EDTA and sodium dodecyl sulfate (SDS) were determined for the ΔtatB and ΔtatC mutant strains and were compared to those of the wild-type strain Sal18. Briefly, 100 μl of an overnight culture was added to 5 ml of soft agar (0.5%). This mixture was then poured onto an LB plate. Filter paper discs containing 10 μl of SDS (5 or 10%) or EDTA (0.1 M, 0.02 M, or 0.004 M) were placed on the solidified medium and incubated overnight at 37°C without inversion. Alternatively, the solutions were spotted directly onto the bacterium-containing top agar. The inhibition of growth was assessed by measuring the clear zones around the discs. In addition, the SDS sensitivities of the wild-type strain Sal18 and strain LS37 were assessed in a broth assay. An overnight culture of the bacteria was diluted 100 times in LB broth containing 5% SDS. The culture was incubated with shaking at 37°C, and the absorbance (OD600) was determined for 7 h after inoculation.
Caco-2 cell invasion assay. (i) Cell culture preparation.
Caco-2 cells, an immortalized line of human epithelial colorectal adenocarcinoma cells, were grown in 75-cm2 flasks (BD Falcon), in HyClone medium (Dulbecco's modified Eagle medium [DMEM]/high glucose; Fisher Scientific) enriched with 10% inactivated fetal bovine serum (FBS; Seracare) and 1% essential amino acids (Invitrogen). The cells were incubated at 37°C under 5% CO2 until they became confluent. Cells were harvested, and 2 × 105 cells were seeded in a 6.5-mm-diameter (pore size, 0.4 μm) transwell plate (Fisher Scientific) and grown until the transepithelial resistance (TER), determined using the Millicell-ERS (electrical resistance system) (Millipore), reached approximately 700 to 900 Ω, indicating that the cells had become polarized.
(ii) Invasion assay.
The invasion assay has been described previously (18, 45). Bacterial cultures were grown overnight and inoculated into fresh LB broth at a 1/100 ratio. The bacterial cells were allowed to grow to an absorbance (OD600) of 0.7. After a wash with DMEM, the bacteria were inoculated apically onto the polarized Caco-2 cells at a multiplicity of infection (MOI) of 100 and were incubated for 1 h at 37°C under 5% CO2. The bacteria were removed, and the cell culture was washed with 200 μl of PBS (136 mM NaCl, 2.5 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) three times. After the wash, DMEM containing 400 μg/ml of gentamicin was added, and the Caco-2 cells were incubated for 2 h under 5% CO2. Cells were again washed with PBSA; 1% Triton X-100 was added; and the cells were incubated for 15 min at 37°C. Serial dilutions were plated onto LB agar alone or onto LB agar plus the appropriate antibiotic (5 μg/ml of tetracycline or 34 μg/ml of chloramphenicol) and were incubated overnight at 37°C. For competition experiments using bacterial strains expressing tetracycline or chloramphenicol resistance determinants, the lysed Caco-2 cells were plated on both tetracycline- and chloramphenicol-containing LB plates. Bacterial colonies were counted and CFU per milliliter calculated. The number of wild-type bacteria that invaded the polarized cells was considered to be 100%, and the invasiveness of the mutant strain was calculated as a percentage of that. In competition experiments, a competitive index of invasion (CII) was calculated. The CII is defined as the ratio (in colony-forming units) of the mutant wild type strains divided by their ratio in the input (initial mix added to the cell monolayer) (69). Three replicates of each bacterial strain were done per assay, and each assay was repeated at least three times.
Cloning of the tatABCD genes.
To complement the chromosomal gene knockouts in strains LS37 (ΔtatB) and LS55 (ΔtatC) with the corresponding plasmid-encoded functional wild-type genes, a 1.9-kb EcoRI and HindIII PCR fragment containing tatA+B+C′ (tatC truncated due to frameshift), a 1.9-kb EcoRI and HindIII PCR fragment encoding tatA+B+C+, or a 2.4-kb EcoRI and HindIII PCR fragment carrying tatA+B+C+D+ was cloned into vector pBR322 (9), resulting in plasmid pLP196, pLP198, or pLP199, respectively. Primers complementing the flanking regions of the tatABCD operon (according to the S. enterica serovar Enteritidis PT4 NCTC 13349 sequence provided by the Wellcome Trust Sanger Institute, United Kingdom) and also containing the appropriate restriction enzyme recognition sites were designed. The PCR product and pBR322 were digested with EcoRI and HindIII, purified, and ligated. The resulting plasmids were then electroporated into the mutant strains, yielding the complemented tatB and tatC mutants (Table 2).
In vivo analysis of bacterial colonization and systemic spread. (i) Salmonella Enteritidis inocula.
A wild-type Sal18 attTn7::cat strain (LS25) and a ΔtatB strain (LS37) were used. The bacteria were grown in LB broth overnight at 37°C with shaking and were used to seed the same medium at a 1/100 dilution. The strains were incubated using the previous condition until an absorbance (OD600) of 0.7 was reached. The cells were harvested by centrifugation at 6,371 × g for 15 min at 4°C. The pellet was resuspended in PBS at a concentration of 5 ×109 CFU/ml. The viable cell counts were confirmed by serial dilution and plating on LB plates. An aliquot of the culture was used to perform PCR with specific primers to confirm that the bacteria had not reverted to wild type. At the conclusion of the experiment, PCR was performed on the bacteria recovered from chicken tissues and cecal contents to confirm the identities of the strains.
(ii) Chickens.
Specific-pathogen-free (SPF) eggs were obtained from Charles River Laboratories and were incubated at the Poultry Science Building at the University of Saskatchewan, Saskatoon, Canada. Once hatched, the Leghorn chickens were kept in isolation rooms at the Vaccine and Infectious Disease Organization (VIDO) animal care facility. When the chickens were 1 day old, cloacal swabs were obtained and plated on Brilliant Green Agar (BGA) to confirm that the birds were Salmonella free. The birds had access to water and food ad libitum. All procedures with animals were done according to the protocol approved by the University of Saskatchewan Committee on Animal Care.
(iii) Experimental challenge of chickens with Salmonella Enteritidis.
Two animal trials were carried out. In each trial, three groups of 90 birds were used. Groups 1 and 2 were inoculated via oral gavage with 5 × 109 CFU/0.5 ml of strains LS25 and LS37, respectively, and group 3 received 0.5 ml of PBS. In trial 1, the birds were challenged at the age of 7 days, and in trial 2, they were challenged at the age of 4 days. In both trials, on days 1, 2, and 4 postchallenge, 10 birds from each group were euthanized per day. Cecum contents, livers, and spleens were collected in sterilized preweighed tubes containing saline (0.85% sodium chloride). The samples were then weighed, and the livers and spleens were homogenized. The resulting homogenates were then plated on BGA in serial dilutions and were incubated at 37°C for approximately 24 h. The CFU per gram of sample was calculated. In addition, 1 ml of each sample was inoculated into 5 ml of selenite broth and was incubated at 37°C for 24 h. After this, the enriched samples were streaked onto BGA plates and incubated at 37°C for 24 h.
Statistical analysis.
For the Caco-2 coinfection and cell invasion assay, the calculated CII was the mean from three independent experiments ± standard error. Each CII was analyzed using Student's t test, and the null hypothesis was that each CII was not different from 1. Bacterial counts in the cecum, liver, and spleen following challenge with wild-type versus mutant strains were compared using the Mann-Whitney U test. The enrichment data were analyzed using the chi-square test, and comparison between groups was performed using Fisher's exact test. A P value less than 0.05 was considered significant.
RESULTS
Putative substrates of the Salmonella Enteritidis Tat system.
In order to identify putative substrates of the Tat system, particularly in a search for candidate virulence factors translocated by this pathway, we applied the Tat prediction programs that are available online, TatP (6) and TatFind (59), to the S. Enteritidis PT4 NCTC 13349 sequence. The TatP program predicted 25 proteins with putative twin-arginine signal sequences, whereas the TatFind program predicted 27 “true” Tat substrates. Most candidates were detected by both programs. A substantial number of the Tat-transported candidates from S. Enteritidis represent proteins with high similarity to Tat substrates in Escherichia coli K-12. In E. coli 28 polypeptides are known or predicted to harbor N-terminal twin-arginine signal peptides (http://www.lifesci.dundee.ac.uk/groups/tracy_palmer/docs/signals.htm). The TatFind search in S. Enteritidis revealed 20 candidates with significant similarity to 10 proteins of the group from E. coli, namely, HyaA (2 copies), HybO, HybA, NapA, NapG, TorA, DmsA (3 copies), FdnG, FdoG, and YedY. Many of these proteins function in oxidation (hydrogen, formate) or reduction (nitrite, nitrate, dimethyl sulfoxide [DMSO], trimethylamine oxide [TMAO]) and are able to bind redox cofactors (e.g., molybdopterin, molybdopterin guanine dinucleotide, iron-sulfur clusters). In addition, polypeptides were predicted as Tat substrates that showed striking similarity to SufI, MdoD, AmiA, AmiC, FhuD, and YcbK of E. coli. A search using the TatP program gave the same picture with only two exceptions: first, an additional DmsA equivalent was predicted, and second, the periplasmic hydroxamate binding protein FhuD was not recognized as a putative Tat substrate.
The following S. Enteritidis proteins that had not been identified as Tat substrates in E. coli were predicted by TatP and/or TatFind: two citrate lyase beta chain proteins (corresponding accession numbers, YP_002242226.1 and YP_002242739.1), two putative sulfatases (YP_002242248.1 and YP_002242921.1), tetrathionate reductase subunit A (YP_002243762.1), a thiosulfate reductase precursor (YP_002244151.1), a putative colanic acid biosynthesis protein (YP_002244186.1), a putative inner membrane protein (YP_002245645.1), and a cytochrome c-type biogenesis protein (YP_002246083.1). Since none of these nine polypeptides suggested an obvious link to cell motility or virulence mechanisms, they were not investigated further by us.
Sal18 tat mutants display altered cell morphology.
As previously discussed, TatA, TatB, and TatC are essential proteins for a fully functional Tat system. In order to study the possible role of the Tat system in invasion and infection, we concentrated on tatB and tatC, two loci that are known to be essential for a functional twin-arginine secretion system in E. coli (50, 52, 66, 73).
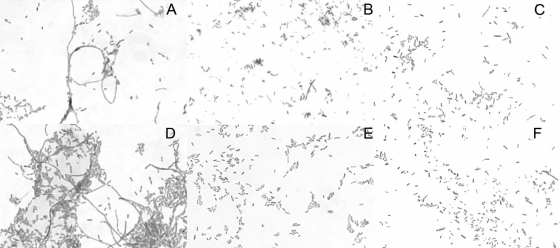
Cells of the wild-type Sal18 strain (LS25) and the ΔtatB::cat (LS37), ΔtatB (LS39), ΔtatC::cat (LS55), and ΔtatC (LS106) mutant strains were examined. In addition, strains LS39 and LS106 were also complemented with plasmid pLP198 (tatA+B+C+) or pLP199 (tatA+B+C+D+). While the Sal18 wild-type cells presented the classic rod-like shape characteristic of most Salmonella strains, the ΔtatB (LS37, LS39) and ΔtatC (LS55, LS106) mutant cells were elongated in shape, often occurring in a chain-like formation. The complemented strains had restored the wild-type morphology, independently of the plasmid used for complementation (Fig. 1). The phenotypic analysis of bacterial cell morphology is summarized in Table 3.
FIG. 1.
The Salmonella Enteritidis wild-type strain Sal18 and its ΔtatB and ΔtatC derivatives were Gram stained, and the cell morphology was observed. The wild-type cells, as well as the plasmid-complemented cells, displayed the classic rod-like shape of Gram-negative bacteria, while the mutant cells showed an elongated shape and chain formation. (A) ΔtatB, cat removed (LS39); (B) ΔtatB pLP198 (tatA+B+C+); (C) ΔtatB pLP199 (tatA+B+C+D+); (D) ΔtatC, cat removed (LS106); (E) ΔtatC pLP198 (tatA+B+C+); (F) wild-type Sal18.
TABLE 3.
Phenotypic traits of S. Enteriditis wild-type, mutant, and complemented mutant strains
| Strain | Relevant genotype | Plasmid-encoded tat genes | Cell morphology | Inhibitiona by EDTA at: |
Albomycin sensitivityb | ||
|---|---|---|---|---|---|---|---|
| 0.5 M | 0.1 M | 0.02 M | |||||
| Sal18 | WT | No | Normal | T (2.7) | T (1.3) | S (3.5) | |
| LS25 | WT attTn7::cat | No | Normal | T (2.7) | ND | ND | S (3.5) |
| LS37 | ΔtatB::cat | No | Elongated | C (2.0) | C (1.5) | C (1.0) | R |
| LS39 | ΔtatB | No | Elongated | C (2.7) | C (2.1) | T (1) | R |
| LS107 | ΔtatB | tatA+B+C′ | NDc | ND | ND | ND | (R) |
| LS108 | ΔtatB | tatA+B+C+ | Normal | T (2.7) | T (2.1) | T (1.2) | S (3.2) |
| LS114 | ΔtatB | tatA+B+C+D+ | Normal | T (2.7) | T (2.1) | T (1.2) | S (3.2) |
| LS106 | ΔtatC | No | Elongated | C (3.1) | C (2.3) | T (1) | R |
| LS117 | ΔtatC | tatA+B+C+ | Normal | T (2.7) | T (2.1) | T (1.2) | S (3.2) |
| LS118 | ΔtatC | tatA+B+C+D+ | Normal | T (2.7) | T (2.1) | T (1.2) | S (3.2) |
C, clear inhibition zone; T, turbid, weak inhibition zone. Numbers in parentheses are diameters of the inhibition zone (in centimeters).
R, resistant; (R), partially resistant; S, sensitive. Numbers in parentheses are diameters of the clear inhibition zone (in centimeters).
ND, not determined.
Mutations in tatB and tatC affect bacterial cell motility.
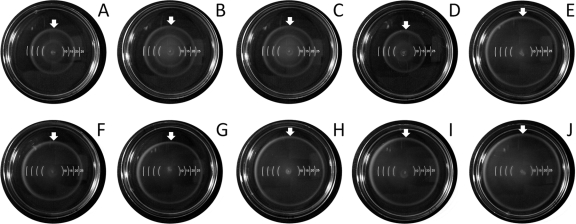
Under certain conditions, flagella are reported to contribute to the virulence of enteropathogenic bacteria, as they promote chemotaxis, host colonization, and host cell invasion (38, 61, 71). In order to analyze the impact of the Tat system on flagellar function in Salmonella Enteritidis, the motilities of strains LS37 (ΔtatB::cat), LS39 (ΔtatB, cat removed), LS55 (ΔtatC::cat), and LS106 (ΔtatC, cat removed) were assessed in comparison to those of the wild-type Sal18 and Sal18 attTn7::cat (LS25) strains on soft LB agar plates. We compared the results for the strains with or without antibiotic cassettes in order to rule out any polar effect caused by the antibiotic cassette insertion. After 3 h of incubation, it was observed that the two wild-type strains had similar swimming zones with diameters of 12 mm and 11 mm, while the LS37 (ΔtatB::cat) and LS55 (ΔtatC::cat) mutants were less motile (swimming zones measuring 7 mm in diameter). Strains LS39 (ΔtatB, cat removed) and LS106 (ΔtatC, cat removed) displayed the same swimming zone diameters as the strains containing the antibiotic cassette (7 mm). The complemented strains LS39/pLP198 (tatA+B+C+), LS39/pLP199 (tatA+B+C+D+), LS106/pLP198 (tatA+B+C+), and LS106/pLP199 (tatA+B+C+D+) recovered the wild-type phenotype; their swimming zone diameters ranged from 11 mm to 13 mm. When the same plates were observed after 6 h of incubation, the wild-type strains Sal18 and LS25 produced swimming zones of 32 mm and 31 mm, respectively, while the motilities of LS37 (ΔtatB::cat) and LS55 (ΔtatC::cat) were restricted to swimming zones of 18 mm in diameter. Strains LS39 (ΔtatB, cat removed) and LS106 (ΔtatC, cat removed) presented the same motility as strains that contained an antibiotic cassette (18 mm). The complemented strains LS39/pLP198 (tatA+B+C+), LS39/pLP199 (tatA+B+C+D+), LS106/pLP198 (tatA+B+C+), and LS106/pLP199 (tatA+B+C+D+) recovered the wild-type phenotype; their swimming zone diameters were 28 mm, 28 mm, 27 mm, and 27 mm, respectively. The results are presented in Fig. 2. These data indicate that inactivation of tatB and tatC has an impact on cell motility. They also demonstrate that the insertion of an antibiotic cassette did not interfere with the expected phenotype, and therefore, it is unlikely that a polar effect on tatD or genes located downstream caused the altered motility in the mutated strain. Moreover, our data also show that tatD does not play an essential role for the functionality of the Tat system, since complementation of ΔtatB or ΔtatC with an episomal copy of the tatA, tatB, and tatC genes was enough to restore the wild-type phenotype.
FIG. 2.
Swimming motility results for wild-type Salmonella Enteritidis Sal18 and the different mutant derivatives and plasmid-complemented mutants in 0.3% agar after 6 h of incubation. (A) ΔtatB::cat (LS37); (B) ΔtatB, Cm removed (LS39); (C) ΔtatC::cat (LS55); (D) ΔtatC, cat removed; (E) Sal18 attTn7::cat (LS25); (F) ΔtatB pLP198 (tatA+B+C+); (G) ΔtatB pLP199 (tatA+B+C+D+); (H) wild-type Sal18; (I) ΔtatC pLP198 (tatA+B+C+); (J) ΔtatC pLP199 (tatA+B+C+D+). Arrows point to the maximum radius of the swimming zone.
The Tat system does not influence the expression of curli.
Curli are proteinaceous, highly aggregative extracellular fibers present on the surfaces of enteric bacteria and are involved in biofilm formation and host colonization (4, 26, 27, 62). In theory, curli-mediated adhesion or motility may affect the invasiveness of Salmonella Enteriditis. In order to determine if the Tat system has an effect on the biogenesis of curli, the wild-type Sal18 strain (LS25) and the mutants LS37 (ΔtatB) and LS55 (ΔtatC) were tested for their abilities to bind Congo red dye. We were not able to observe any differences in the expression of curli on the samples tested.
The tatB and tatC mutants show increased sensitivity to EDTA.
Salmonella Enteriditis strains Sal18, LS25 (WT attTn7::cat), LS37 (ΔtatB), LS39 (ΔtatB), and LS106 (ΔtatC) and E. coli strain B1LK0 (tatC) were tested for their sensitivities to EDTA. The ΔtatB strain with the presence or absence of the antibiotic cassette was tested as a control to rule out any polar effect caused by the antibiotic cassette insertion in the bacterial chromosome. Filter paper discs containing different concentrations of EDTA (0.5 M, 0.1 M, 0.02 M, and 0.004 M) were placed on LB agar seeded with the different strains. The results are summarized in Table 3. We observed that E. coli strain B1LK0 (tatC) was more sensitive to EDTA than the ΔtatB (LS37, LS39) and ΔtatC (LS106) derivatives of Salmonella Enteritidis. However, we did not observe substantial differences in growth inhibition among strains LS37, LS39, and LS106.
Sensitivity to SDS.
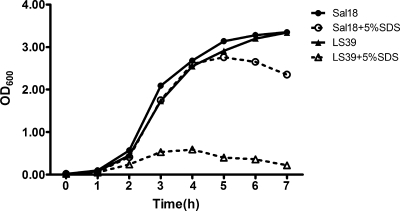
Strains LS25 (wild type) and LS37 (ΔtatB) were tested for their sensitivities to SDS by two different approaches. In the first approach, the wild-type and ΔtatB strains were seeded in LB agar plates, and growth inhibition around the paper filter discs containing either 5 or 10% SDS was assessed. With both concentrations, we observed a more pronounced inhibition zone around the SDS-soaked discs on the plates seeded with the ΔtatB strain than on those seeded with the wild type. However, with both strains, some isolated colonies were observed within the halo of inhibition. In the second approach, the strains were grown in LB broth containing 5% SDS, and the absorbance (OD600) was measured at every hour for 7 h after inoculation. Both strains grew exponentially until 4 to 5 h. After that, the absorbance (OD600) decreased (Fig. 3). To further understand if there was a drop in bacterial viability, we plated the cells onto LB agar plates and examined them by microscopy. The ΔtatB bacterial cells formed aggregates, which is most likely the reason for the lower absorbance found with the ΔtatB mutant culture, but we did not find drastically reduced viability for the ΔtatB or wild-type strains when plated (after vigorous vortexing of the culture). Both the ΔtatB strain and the wild-type strain grew normally in the absence of SDS (Fig. 3) without forming aggregates.
FIG. 3.
Wild-type Salmonella Enteritidis Sal18 and LS37 (ΔtatB::cat) grown in LB broth without (solid lines) and with (dashed lines) 5% SDS. The absorbance was measured every hour after bacterial inoculation.
Mutants lacking a functional Tat system are resistant to the antibiotic albomycin.
In the course of this study, we have developed a fast plate assay in our laboratory to determine if the Tat system is functional. Several years ago, the group of T. Palmer demonstrated that the E. coli FhuD protein is a substrate for the twin-arginine transport system (32). Our test is based on two observations: first, the translocation of the ferric-hydroxamate binding protein FhuD into the periplasm is strictly dependent on a functional Tat system; second, in order to be active, the antibiotic albomycin, which has structural homology to siderophores of the hydroxamate type, needs to be imported into the cytoplasm via an ABC transporter with FhuD as the periplasmic component. As a consequence, mutants with deletions in the tatB and tatC genes, which are unable to export FhuD into the periplasm, should be resistant to this antibiotic. This hypothesis was tested on LB agar plates with E. coli B1LK0 (tatC), which was resistant to albomycin, whereas the control strain, MC4100, displayed a zone of growth inhibition around a filter paper disc impregnated with albomycin, as outlined in Materials and Methods.
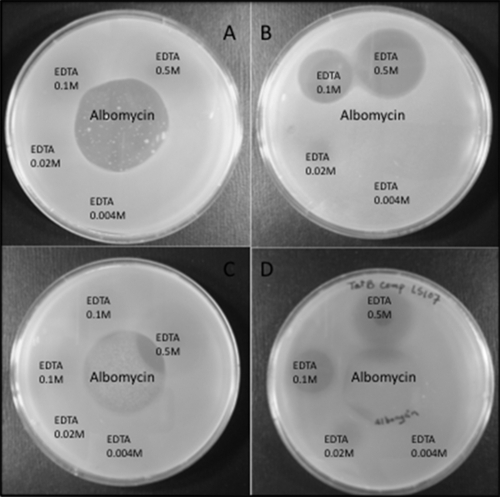
Salmonella Enteritidis strains Sal18 (wild type), LS37 (ΔtatB::cat), LS39 (ΔtatB, cat removed), and LS106 (ΔtatC, cat removed) were subjected to the albomycin sensitivity assay. The wild-type strain presented a clear halo of growth inhibition with a diameter of 7 mm around the impregnated filter paper disc. In contrast, the ΔtatB and ΔtatC mutants showed cell growth right up to the antibiotic-containing disc, demonstrating resistance to albomycin. Plasmid pLP196, expressing wild-type TatA and TatB and a C-terminally truncated TatC protein, mediated only a partial phenotypic complementation of the ΔtatB strain. Plasmids pLP198 and pLP199, which harbor the functional tatABC genes and the complete tatABCD operon, respectively, restored the wild-type phenotype in all tatB and tatC mutants (except for the E. coli tatC mutant, which remained albomycin resistant) (Table 3; Fig. 4). All phenotypic analysis is summarized in Table 3.
FIG. 4.
Albomycin and EDTA sensitivity assay results. The center of the plate was loaded with albomycin surrounded by EDTA with concentrations of 0.5 M, 0.1 M, 0.02 M, and 0.004 M in the peripheries of the plates. The plates were seeded with either Sal18 (A), LS39 (ΔtatB) (B), LS39 (ΔtatB) plus pLP199 (tatA+B+C+D+) (C), or LS39 (ΔtatB) plus pLP196 (tatA+B+C′ [truncated tatC]) (D).
A functional Tat system is important for invasion of polarized Caco-2 cells in vitro.
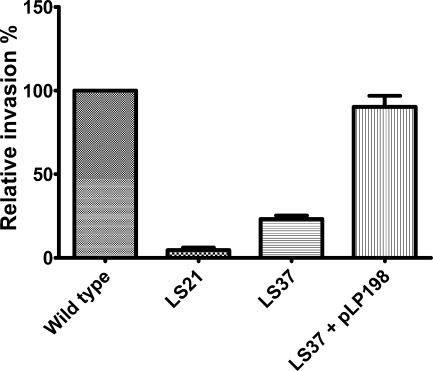
In order to assess their cell invasion capabilities, strains LS29 (WT attTn7::tet) and LS37 (ΔtatB::cat) were used to infect polarized Caco-2 cells. In addition, the reference strain LS21 (ΔSPI-1::cat), previously used by our group and known to be defective in Caco-2 cell invasion (18), was used as an internal control to demonstrate that the assay was working as anticipated. E. coli DH5α, which is not able to invade cells, was used as a negative control in each experiment. In the single-infection experiments, the invasiveness of each strain was calculated as a percentage of the wild-type strain invasion. The ΔtatB mutant had an invasiveness of 23.23% of the wild-type strain invasion in polarized Caco-2 cells, while the ΔSPI-1 strain had an invasiveness of 4.82% of the wild-type strain invasion (Fig. 5). As another control of the experiment, the ΔtatB strain complemented with either plasmid pLP198 (tatA+B+C+) or plasmid pLP199 (tatA+B+C+D+) was also tested and presented an invasiveness of 89.71% (Fig. 5) or 136% of the wild-type strain invasion.
FIG. 5.
Single infection of the Caco-2 polarized cell line with wild-type S. Enteritidis Sal18, LS37 (ΔtatB::cat), or LS37 (ΔtatB::cat) complemented with plasmid pLP198 harboring the tatABC genes. The ΔSPI::cat strain was used as a control. The invasiveness of the mutant strains is measured as a percentage of that of the wild-type strain. Each bar corresponds to an average of three experiments performed in triplicate.
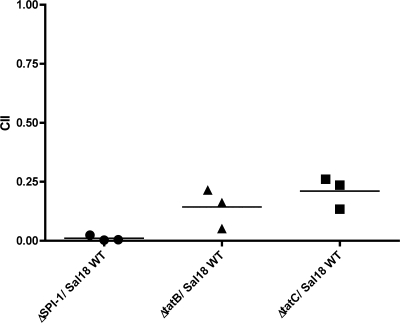
A competitive assay was performed in which cells were coinfected with the ΔtatB, ΔtatC, or ΔSPI-1 mutant strain and the wild-type strain at a 1:1 ratio. In this type of assay, the wild-type Sal18 strain functions as an internal control, thus minimizing variations between assays and providing more-reproducible results (69). A CII (competitive index in invasion) was calculated as previously described (69). We observed CIIs of 0.010, 0.143, and 0.210 for the groups coinfected with the wild type and the ΔSPI-1, ΔtatB, or ΔtatC strain, respectively (Fig. 6). The CII of each of the three different groups was statistically significantly different from 1 (P, ≤0.003).
FIG. 6.
Coinfection of polarized Caco-2 cells with the wild-type strain LS29 (Sal18 attTn7::tet) and derivatives LS37 (ΔtatB::cat) and LS55 (ΔtatC::cat) using antibiotic markers to distinguish the strains. Values significantly different from 1 indicate impaired cell invasion. Strain LS21 (ΔSPI-1::cat) was used as a control. The values for each strain correspond to averages of three experiments performed in triplicate.
A functional Tat system is important for effective systemic spread of Salmonella Enteritidis in chickens.
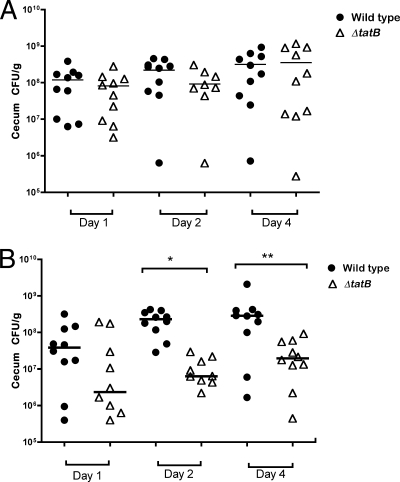
In order to study the impact of the Tat system on the ability of Salmonella Enteritidis to colonize the intestines of chickens and invade their internal organs, 4-day old and 7-day old Leghorn chickens were challenged with 5 × 109 CFU of either LS25 (wild-type Sal18 attTn7::cat) or LS37 (ΔtatB::cat). When cecal colonization was studied in birds challenged at the age of 4 days, no statistically significant difference was observed regardless of the time (postchallenge) of sampling (P, >0.05) (Fig. 7 A). However, when the experiment was repeated with 7-day-old birds, there was a statistically significant difference between colonization of the cecum by the wild type and that by the ΔtatB mutant on day 2 (P, <0.0001) and day 4 (P, 0.01) postchallenge (Fig. 7B), as determined by direct counts.
FIG. 7.
Direct bacterial counts (as determined after plating on Brilliant Green Agar plates and incubation for 24 h at 37°C) in cecal contents on days 1, 2, and 4 postchallenge from birds challenged at the age of 4 days (A) or 7 days (B). Horizontal bars represent median values. Each symbol represents the CFU obtained from samples of an individual bird. The single asterisk indicates a significant statistical difference between the group challenged with wild-type LS29 (Sal18 attTn7::tet) and that challenged with the ΔtatB::cat strain on day 2 (P < 0.0001); the double asterisk indicates a statistical difference between the two groups on day 4 (P = 0.01).
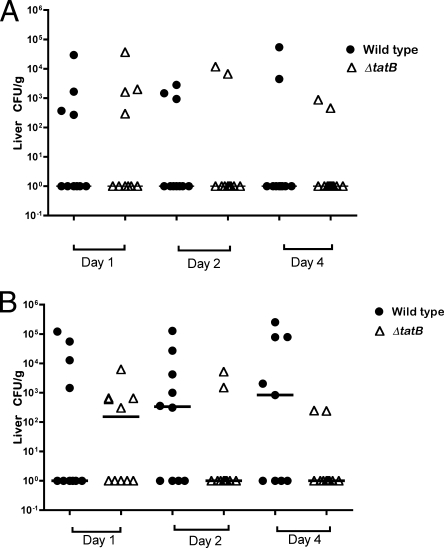
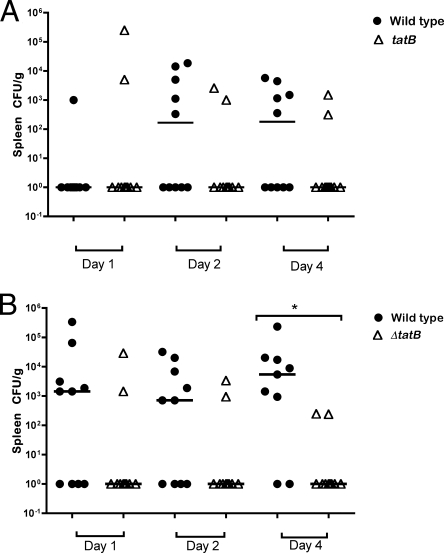
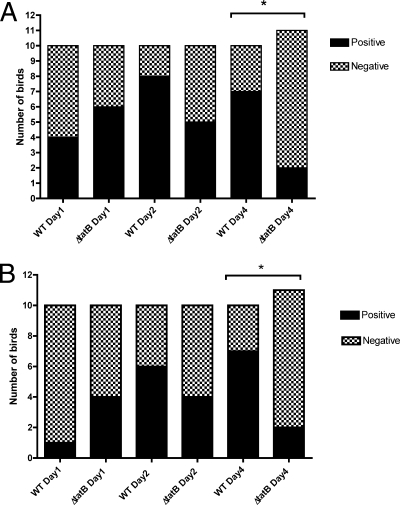
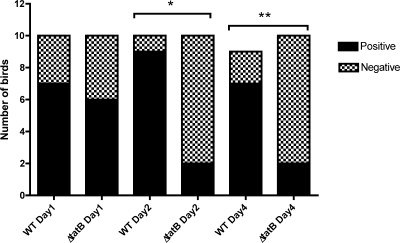
Initially, systemic infection was assessed by diluting the homogenized organ samples and plating for viable cell counts without enrichment. No statistically significant difference between the abilities of the wild-type Sal18 and ΔtatB mutant strains to colonize the livers of birds challenged at the age of 4 or 7 days was observed when the birds were sampled on day 2 or day 4 postchallenge (Fig. 8 A and B). However, there appeared to be a trend for the ΔtatB-challenged group to clear the systemic infection faster, as demonstrated by the lower bacterial counts and the lower number of infected birds sampled on day 2 or day 4 postchallenge in both trials (Fig. 8A and B). Regarding the analysis of bacterial counts from the spleen, there was a statistically significant difference between the group challenged at the age of 7 days with the wild-type strain and the group challenged with the ΔtatB mutant when the birds were sampled on day 4 postchallenge (P, 0.004) (Fig. 9 B). In addition, in birds challenged at the age of 7 days and sampled 2 days postchallenge. and in birds challenged at the age of 4 days and sampled 4 days after challenge, there was a trend for the ΔtatB mutant to be present in lower numbers than the wild-type strain in the spleens of the birds challenged on the same days (P, 0.08 and 0.10, respectively) (Fig. 9A and B). Systemic infection was also assessed after enrichment of liver and spleen samples in selenite broth. Throughout the course of the trial, in birds challenged at the age of 4 days, the number of spleens positive for the wild-type bacteria increased considerably, while the number of positive spleens in birds challenged with the ΔtatB mutant tended to decrease slightly (Fig. 10 B). With respect to systemic spread, after enrichment of samples, the birds challenged at the age of 7 days showed a statistically significant difference between the group that received the wild-type bacteria and the group that received the ΔtatB mutant when sampled at 2 or 4 days postchallenge (P, 0.003 and 0.02) (Fig. 11). This was also observed for birds challenged at the age of 4 days and sampled 4 days postchallenge (P, 0.03) (Fig. 10). Taken together, these data indicated that the absence of a functional Tat system plays a role in Salmonella Enteritidis cecum colonization and is also important for effective and prolonged systemic infection of chickens.
FIG. 8.
Direct bacterial counts (as determined after plating on Brilliant Green agar plates and incubation for 24 h at 37°C) in the liver on days 1, 2, and 4 postchallenge from birds challenged at the age of 4 days (A) or 7 days (B). Horizontal bars represent median values. Each symbol represents the CFU obtained from samples of an individual bird. There is a trend for the group challenged with LS37 (ΔtatB::cat) to clear the infection faster on days 2 and 4 (P, 0.11 and 0.05, respectively).
FIG. 9.
Direct bacterial counts (as determined after plating on Brilliant Green Agar plates and incubation for 24 h at 37°C) in the spleen on days 1, 2, and 4 postchallenge from birds challenged at the age of 4 days (A) or 7 days (B). Horizontal bars represent the median values for different birds. There was a trend for the group challenged with LS37 (ΔtatB::cat) at the age of 4 days to clear the infection faster (P = 0.10). Each symbol represents the CFU obtained from samples of an individual bird. The asterisk indicates a significant statistical difference between the groups challenged with wild-type LS29 (Sal18 attTn7::tet) and LS37 (ΔtatB::cat) on day 4 (P = 0.004). The group challenged with LS37 (ΔtatB::cat) at the age of 7 days presented a trend to clear systemic infection faster on day 2 (P = 0.08).
FIG. 10.
(A) Enrichment results for livers and spleens from chickens challenged at the age of 4 days. The asterisk represents a significant statistical difference between the groups challenged with wild-type LS29 (Sal18 attTn7::tet) versus LS37 (ΔtatB::cat) on day 4 postchallenge (P = 0.03). (B) Enrichment results for spleens from chickens challenged at the age of 4 days. The asterisk represents a significant statistical difference between the groups challenged with wild-type LS29 (Sal18 attTn7::tet) versus LS37 (ΔtatB::cat) on day 4 postchallenge (P = 0.03).
FIG. 11.
Enrichment results for livers and spleens from chickens challenged at the age of 7 days. Asterisks represent significant statistical differences between the groups challenged with wild-type LS29 (Sal18 attTn7::tet) versus LS37 (ΔtatB::cat) on days 2 (*) (P = 0.003) and 4 (**) (P = 0.02) postchallenge.
DISCUSSION
There has been a sustained interest in the identification of potential virulence factors in Salmonella Enteritidis in order to develop more-effective strategies to control this pathogen. The Tat system is a specialized secretion pathway in prokaryotes that is responsible for the transport of certain proteins in a folded conformation across the cytoplasmic membrane (7, 53). There are several reports demonstrating a link between the Tat system and virulence in various pathogens, such as E. coli O157, Pseudomonas aeruginosa, Yersinia pseudotuberculosis, Pseudomonas syringae, Ralstonia solanacearum, and Agrobacterium tumefaciens (12, 19, 25, 43, 51, 56). However, to our knowledge, there are no reports concerning the role of the Salmonella Enteritidis Tat system in infection and virulence. In this study we investigated that relationship.
The morphological changes in the S. Enteritidis ΔtatB and ΔtatC strains, including an elongated shape and chain formation, were similar to those observed in a previously characterized E. coli tatC mutant strain. The change in morphology could not be attributed to the antibiotic cassette present in both the ΔtatB (LS37) and the wild-type (LS25) strain, since the wild-type strain had the normal cell morphology. Further support for this conclusion was found in the observation that both ΔtatB mutants showed the same phenotype, independently of the presence or absence of the antibiotic cassette (LS37 and LS39). It has been shown previously that a disturbed Tat pathway can affect cell morphology, growth and biofilm formation (29), and cell wall integrity (19, 60). Cell wall amidases are responsible for cleaving the murein septum during cell division (30, 31), and a previous study of E. coli tatABC mutants demonstrated that the mislocalization of the cell wall amidases AmiA and AmiC led to the same morphotype as that observed in our study (33, 67). We did not specifically investigate the targeting of the cell wall amidases, but we found that AmiA, AmiC, and SufI, which is also assigned to play a role in cell division (66), were predicted as Tat substrates when we applied the TatP and TatFind programs to the S. Enteritidis genome.
The ΔtatB (LS37) strain was more sensitive to SDS than the wild-type strain when tested on LB plates and showed reduced growth compared to that of the wild-type strain in LB medium supplemented with 5% SDS (Fig. 3). Our results are in agreement with a previous report on E. coli that demonstrated the hypersensitivity of ΔtatB and ΔtatC mutant strains to SDS, although Salmonella Enteriditis was clearly less sensitive to SDS than E. coli. We also tested the different strains for EDTA sensitivity. While the wild-type Salmonella Enteritidis was resistant to EDTA, LS37 (ΔtatB::cat), LS39 (ΔtatB Δcat), and LS106 (ΔtatC Δcat) were sensitive proportionally to the EDTA concentration. The divalent cations Ca2+ and Mg2+ are important for the stability of the outer membrane; EDTA acts as a chelator of these cations, which in turn may affect the integrity of the outer membrane. The ΔtatB and ΔtatC mutants may be sensitive due to a compromised outer membrane structure. These results also demonstrated that the insertion of the chloramphenicol cassette did not influence the final phenotype, since the ΔtatB mutants with or without the chloramphenicol cassette were equally sensitive to EDTA. In addition, we also observed that E. coli strain B1LK0 (tatC) was more sensitive to EDTA than the S. Enteriditis tat mutants. This result is in agreement with a previous study that reported that this strain was hypersensitive to SDS (70).
In order to display its inhibitory activity, albomycin needs to be imported into the cytoplasm of bacteria and subsequently to be cleaved by peptidase N. One moiety of the molecule is the active part (thioribosyl pyrimidine derivative) of the antibiotic, whereas the remaining domain shows structural similarity to ferrichrome, a siderophore of the hydroxamate type. In E. coli and Salmonella, albomycin is actively imported by the ferrihydroxamate uptake (fhu) system (10, 23). Transport across the outer membrane is mediated by the TonB-dependent receptor FhuA, and the subsequent translocation into the cytoplasm involves FhuD in the periplasm, as well as a complex in the cytoplasmic membrane composed of FhuB and FhuC (11). The periplasmic location of the binding protein FhuD is essential for the uptake of ferric hydroxamates and albomycin (39, 41, 57). Two secretory pathways could be responsible for the export of FhuD into the periplasm: the Sec pathway, also called the general secretory pathway (GSP), which translocates unfolded polypeptide chains, and the Tat pathway, which transports folded proteins. Based on a previous study of E. coli, which demonstrated a Tat-dependent periplasmic localization of the ferrihydroxamate binding protein (32), we assumed that FhuD in Salmonella Enteritidis would be translocated into the periplasm via the Tat system. However, it was not known if FhuD was also able to enter the Sec pathway as an alternative route. In order to analyze this, we developed an albomycin sensitivity plate assay. Initially, E. coli strains MC4100 (wild type) and B1LK0 (MC4100 tatC) were tested. As anticipated, the wild type was sensitive and the tatC mutant was resistant to albomycin. We also tested the S. Enteriditis ΔtatB strain (LS37) and a ΔtatB strain with the chloramphenicol cassette removed (LS39), as well as the wild-type strain (LS25). The wild type was sensitive, while both tat mutants proved resistant to the antibiotic. These results confirm that the phenotype conferred on the LS37 strain was indeed the result of the tatB deletion and was not due to polar effects caused by the gene replacement or to the expression of the cat gene. More importantly, these data indicate that FhuD uses the Tat system exclusively to reach the periplasm of Salmonella Enteritidis. Amino acid sequences resembling the twin-arginine consensus motif were detected in the FhuD proteins of a number of Gram-negative and Gram-positive bacteria (W. Köster, unpublished data). It remains unclear why FhuD is the only known periplasmic siderophore binding protein to be exported through the Tat system. Other members of the siderophore binding protein family do not have an obvious Tat consensus region in their signal sequences, suggesting that they are exported by the general secretion pathway.
In addition, we tested the strains for their abilities to invade polarized Caco-2 cell monolayers. In the single-strain invasion assay, we compared the ΔtatB mutant, the ΔSPI-1 mutant, and E. coli DH5α with our wild-type Salmonella strain. Impaired invasion by the ΔSPI-1 mutant has been described by our group previously and was observed once again (18). The ΔtatB mutant also had decreased invasion capability (23.23%) in relation to the Salmonella Enteritidis wild-type strain (100%) (Fig. 5), suggesting that this mutation plays a role in Caco-2 cell invasion. To eliminate the variability of the cells within each experiment, polarized Caco-2 cells were coinfected with a ΔtatB or ΔtatC mutant and the wild-type Sal18 strain. In these competition assays, we observed a sharp decline in invasion by both Tat mutants (LS37 and LS39) (Fig. 6). These results confirm the decrease in invasiveness observed in the original assays. To our knowledge, this is the first time this assay has been used to analyze the invasiveness of Tat system mutants. Cell invasion has been associated with several bacterial factors, including the type 3 secretion system (T3SS), flagella, and fimbriae (18, 42, 54). We speculated that the decreased invasion presented by the Tat mutants could be an effect of the impairment of a phenotypic trait, such as curli-mediated adhesion or motility. Several Salmonella enterica serovars express, on their surfaces, thin adhesive fimbrial structures called curli (16). Curli have been shown to be involved in attachment to and invasion of host cells, as well as in biofilm formation, and are therefore considered a virulence determinant (5). We did not observe any differences in curli expression among the ΔtatB and ΔtatC mutants and the wild-type Salmonella strain, suggesting that the Tat system does not interfere with curli assembly. In addition, the Caco-2 invasion assays were performed under conditions (e.g., 37°C) that were not favorable for optimal expression of curli. These facts eliminate an altered curli phenotype as a possible reason for the impaired invasiveness of Δtat mutants in Caco-2 cells. In many Gram-negative bacteria, including Salmonella Typhimurium, motility is important for full virulence, since it enables the bacterium to cross the thick and viscous mucus layer in the intestinal epithelium and promotes the contact of the bacterium with the cell surface (35, 68). The Salmonella Enteritidis flagellum is considered a potential virulence factor in poultry (2, 3, 54). In our study, we observed that the ΔtatB::cat (LS37) and ΔtatC::cat (LS55) mutants had impaired motility in comparison to the wild-type Sal18 attTn7::cat strain (LS25). This is in agreement with previous studies showing that an E. coli O157:H7 tatABC mutant was nonmotile (56) and that a ΔtatC mutant of Pseudomonas aeruginosa was less motile than the wild-type strain (51). No proteins involved in flagellar biogenesis that carry Tat signal sequences have been identified (56). Regarding our Caco-2 invasion experiments, Tat system impairment may play a direct or indirect role in the several systems responsible for bacterial invasion, such as SPI-1 and flagella. However, further studies are necessary to confirm this hypothesis.
For all Salmonella tat mutants tested in this study, the tatABC region expressed on a plasmid was both necessary and sufficient for the restoration of the phenotype to wild type with respect to morphology, motility, sensitivity to EDTA, SDS, and albomycin, and invasive properties in polarized Caco-2 cells. Expression of the complete plasmid-encoded tatABCD operon revealed the same effect, indicating that tatD does not play a major role in the assembly of the functional Tat system. The partial complementation of the ΔtatB mutant by pLP196 could be the result of low-level expression of chromosomally encoded TatC or could be due to a residual function of the plasmid-encoded truncated TatC. Moreover, it is noteworthy that inhibitory effects of over-stoichiometric TatB concentrations have been described previously (66).
Based on these findings, we conclude that the replacements of the chromosomal tatB and tatC genes by the chloramphenicol resistance cassette (and its subsequent removal) caused polar effects on tat genes located downstream, as one would anticipate. However, those effects seem to be restricted to the tat operon, and there is no evidence that the observed phenotypes of the tat mutant strains (LS37, LS39, LS55, and LS106) are caused by secondary mutations elsewhere in the chromosome.
In order to evaluate the effect of a nonfunctional Tat system on Salmonella Enteritidis infection in a poultry model, we challenged 4-day-old and 7-day-old SPF Leghorn chickens with the wild-type Sal18 and ΔtatB mutant strains administered via oral gavage. Younger chickens were used because they are more susceptible to Salmonella (72). We used a relatively high dose (5 × 109 CFU/g) of Salmonella Enteritidis to ensure adequate levels of systemic spread by the wild-type bacteria. It is noteworthy that the plating efficiency obtained with Brilliant Green Agar (BGA) is only 40%, in contrast to that for other standard rich media such as LB medium (data not shown), which decreases the levels of Salmonella Enteritidis detection in the samples and therefore has an impact on our results. However, if BGA were not used as a selective medium, in many cases—notably cecal content counts—there would be a complete overgrowth of flora. We observed that the level of colonization of the ceca in chickens challenged at the age of 4 days did not differ between the wild-type Sal18- and ΔtatB mutant-treated groups on any of the days of sampling. In order to determine if the age of birds at the time of challenge had an effect on colonization, we challenged birds at the age of 7 days. In the older birds, we observed a difference between the group challenged with the wild type and the group challenged with ΔtatB bacteria. These differences were statistically significant 2 and 4 days postchallenge, indicating that the Tat system of Salmonella Enteritidis may play a role in cecum colonization in chickens (Fig. 7A and B). The ability of Salmonella Enteritidis to colonize the cecum and subsequently invade intestinal cells seems to be a synergistic process, since inactivation of single genes of important and recognized virulence systems in bacteria or even entire pathogenicity islands had little or no effect on intestinal colonization by Salmonella Enteritidis or other Salmonella species (18, 36).
For 4-day-old birds, we analyzed the systemic infection and observed that the spleens of birds challenged with the wild-type Sal18 strain had higher bacterial loads than those of birds challenged with the ΔtatB mutant when sampled 4 days postchallenge (Fig. 9A and B). Additionally, there was a tendency of the bacterial counts from the wild-type group to remain somewhat leveled, while the bacterial numbers in the ΔtatB group tended to decline throughout the period. Also, the wild-type Sal18 strain infected a higher number of birds than the ΔtatB mutant. This trend was confirmed by the systemic colonization enrichment data. Higher numbers of birds challenged at the age of 4 days were positive for the wild-type Sal18 strain on day 4 postchallenge (Fig. 9). In addition, among chicks challenged at the age of 7 days, higher numbers of birds given the wild-type Sal18 strain than of birds given the ΔtatB strain were positive on days 2 and 4 postchallenge (Fig. 10). These data indicate that the Tat system is involved in the processing of a protein that takes part in Salmonella interaction with the host during systemic spread of the bacterium, since we could clearly observe a delay in systemic infection presented by the ΔtatB mutants.
There have been several reports relating to the Tat system and virulence. In a rat model, a Pseudomonas aeruginosa tatC mutant did not cause chronic lung infection (51). A tatC mutant of Yersinia paratuberculosis showed impaired colonization of lymphoid organs in mice (43). Our data indicate that an impaired Tat system reduces cecal colonization, delays systemic infection, and decreases the efficiency of sustained systemic colonization. To our knowledge, this is the first report of the role of the Salmonella Enteritidis Tat system in both cecal colonization and systemic spread and persistence in chickens. These data provide evidence that the Tat pathway is important in virulence in yet another bacterial species and animal model. Since no obvious Tat-translocated virulence candidates were found with the help of the TatP and TatFind analyses, the exact mechanism by which a nonfunctional Tat system might interfere with cell adhesion, invasion, and survival in a host organism remains to be elucidated.
In conclusion we have demonstrated that a functional Tat pathway is essential for optimal cell division, motility, FhuD-mediated iron acquisition, and cell wall integrity in Salmonella Enteriditis. The Tat system plays an important role in the invasiveness of Salmonella Enteriditis for polarized Caco-2 cells, in the colonization of the cecum, and in sustaining systemic colonization in chickens.
Acknowledgments
We thank D. Wilson and the Clinical Research Group at VIDO for assistance with animal experiments and T. Palmer (University of Dundee, United Kingdom) for kindly providing E. coli strains. We thank H. Townsend for statistical analysis and manuscript review.
This research was supported by the National Science and Engineering Research Council of Canada (NSERC), Bioniche Life Sciences, and the Poultry Industry Council (PIC). A.A.P. is the holder of an NSERC Senior Industrial Research Chair, and W.K. is the holder of an NSERC Associate Industrial Research Chair. C.S.M. holds a Post-Doctoral Fellowship from the Saskatchewan Health Research Foundation (SHRF).
Editor: F. C. Fang
Footnotes
Published ahead of print on 24 May 2010.
Published with permission of the Director of VIDO as journal series number 549.
REFERENCES
- 1.Alami, M., I. Luke, S. Deitermann, G. Eisner, H. G. Koch, J. Brunner, and M. Müller. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12:937-946. [DOI] [PubMed] [Google Scholar]
- 2.Allen-Vercoe, E., A. R. Sayers, and M. J. Woodward. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen-Vercoe, E., and M. J. Woodward. 1999. The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J. Med. Microbiol. 48:771-780. [DOI] [PubMed] [Google Scholar]
- 4.Austin, J. W., G. Sanders, W. W. Kay, and S. K. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295-301. [DOI] [PubMed] [Google Scholar]
- 5.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 8.Bolhuis, A., J. E. Mathers, J. D. Thomas, C. M. Barrett, and C. Robinson. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276:20213-20219. [DOI] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 10.Braun, V., R. Gross, W. Köster, and L. Zimmermann. 1983. Plasmid and chromosomal mutants in the iron(III)-aerobactin transport system of Escherichia coli. Use of streptonigrin for selection. Mol. Gen. Genet. 192:131-139. [DOI] [PubMed] [Google Scholar]
- 11.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Met. Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 12.Caldelari, I., S. Mann, C. Crooks, and T. Palmer. 2006. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol. Plant Microbe Interact. 19:200-212. [DOI] [PubMed] [Google Scholar]
- 13.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalker, R. B., and M. J. Blaser. 1988. A review of human salmonellosis: III. Magnitude of Salmonella infection in the United States. Rev. Infect. Dis. 10:111-124. [DOI] [PubMed] [Google Scholar]
- 15.Collins, J. E. 1997. Impact of changing consumer lifestyles on the emergence/reemergence of foodborne pathogens. Emerg. Infect. Dis. 3:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desin, T. S., P. K. Lam, B. Koch, C. Mickael, E. Berberov, A. L. Wisner, H. G. Townsend, A. A. Potter, and W. Köster. 2009. Salmonella enterica serovar Enteritidis pathogenicity island 1 is not essential for but facilitates rapid systemic spread in chickens. Infect. Immun. 77:2866-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, Z., and P. J. Christie. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duchet-Suchaux, M., P. Lechopier, J. Marly, P. Bernardet, R. Delaunay, and P. Pardon. 1995. Quantification of experimental Salmonella enteritidis carrier state in B13 Leghorn chicks. Avian Dis. 39:796-803. [PubMed] [Google Scholar]
- 21.Egli, T., W. Köster, and L. Meile. 2002. Pathogenic microbes in water and food: changes and challenges. FEMS Microbiol. Rev. 26:111-112. [DOI] [PubMed] [Google Scholar]
- 22.Fadl, A. A., K. S. Venkitanarayanan, and M. I. Khan. 2002. Identification of Salmonella enteritidis outer membrane proteins expressed during attachment to human intestinal epithelial cells. J. Appl. Microbiol. 92:180-186. [DOI] [PubMed] [Google Scholar]
- 23.Fecker, L., and V. Braun. 1983. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J. Bacteriol. 156:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gohlke, U., L. Pullan, C. A. McDevitt, I. Porcelli, E. de Leeuw, T. Palmer, H. R. Saibil, and B. C. Berks. 2005. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U. S. A. 102:10482-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, E. T., D. G. Brown, J. K. Swanson, and C. Allen. 2007. Using the Ralstonia solanacearum Tat secretome to identify bacterial wilt virulence factors. Appl. Environ. Microbiol. 73:3779-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gophna, U., T. A. Oelschlaeger, J. Hacker, and E. Z. Ron. 2002. Role of fibronectin in curli-mediated internalization. FEMS Microbiol. Lett. 212:55-58. [DOI] [PubMed] [Google Scholar]
- 28.Gram, H. C. 1884. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschritte der Medizin 2:185-189. [Google Scholar]
- 29.Harrison, J. J., H. Ceri, E. A. Badry, N. J. Roper, K. L. Tomlin, and R. J. Turner. 2005. Effects of the twin-arginine translocase on the structure and antimicrobial susceptibility of Escherichia coli biofilms. Can. J. Microbiol. 51:671-683. [DOI] [PubMed] [Google Scholar]
- 30.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Höltje. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 31.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ize, B., I. Porcelli, S. Lucchini, J. C. Hinton, B. C. Berks, and T. Palmer. 2004. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J. Biol. Chem. 279:47543-47554. [DOI] [PubMed] [Google Scholar]
- 33.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 34.Jack, R. L., F. Sargent, B. C. Berks, G. Sawers, and T. Palmer. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, M. A., S. D. Hulme, P. A. Barrow, and P. Wigley. 2007. The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol. 36:199-203. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen, F., R. Bailey, S. Williams, P. Henderson, D. R. Wareing, F. J. Bolton, J. A. Frost, L. Ward, and T. J. Humphrey. 2002. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int. J. Food Microbiol. 76:151-164. [DOI] [PubMed] [Google Scholar]
- 38.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 39.Köster, W., and V. Braun. 1990. Iron(III) hydroxamate transport into Escherichia coli. Substrate binding to the periplasmic FhuD protein. J. Biol. Chem. 265:21407-21410. [PubMed] [Google Scholar]
- 40.Köster, W., and V. Braun. 1986. Iron hydroxamate transport of Escherichia coli: nucleotide sequence of the fhuB gene and identification of the protein. Mol. Gen. Genet. 204:435-442. [DOI] [PubMed] [Google Scholar]
- 41.Köster, W., and V. Braun. 1989. Iron-hydroxamate transport into Escherichia coli K12: localization of FhuD in the periplasm and of FhuB in the cytoplasmic membrane. Mol. Gen. Genet. 217:233-239. [DOI] [PubMed] [Google Scholar]
- 42.Krogfelt, K. A. 1991. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesins of Escherichia coli. Rev. Infect. Dis. 13:721-735. [DOI] [PubMed] [Google Scholar]
- 43.Lavander, M., S. K. Ericsson, J. E. Broms, and A. Forsberg. 2006. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect. Immun. 74:1768-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leake, M. C., N. P. Greene, R. M. Godun, T. Granjon, G. Buchanan, S. Chen, R. M. Berry, T. Palmer, and B. C. Berks. 2008. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U. S. A. 105:15376-15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. U. S. A. 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lister, S. A. 1988. Salmonella enteritidis infection in broilers and broiler breeders. Vet. Rec. 123:350. [DOI] [PubMed] [Google Scholar]
- 47.Malcova, M., H. Hradecka, R. Karpiskova, and I. Rychlik. 2008. Biofilm formation in field strains of Salmonella enterica serovar Typhimurium: identification of a new colony morphology type and the role of SGI1 in biofilm formation. Vet. Microbiol. 129:360-366. [DOI] [PubMed] [Google Scholar]
- 48.Mangels, D., J. Mathers, A. Bolhuis, and C. Robinson. 2005. The core TatABC complex of the twin-arginine translocase in Escherichia coli: TatC drives assembly whereas TatA is essential for stability. J. Mol. Biol. 345:415-423. [DOI] [PubMed] [Google Scholar]
- 49.Matos, C. F., A. Di Cola, and C. Robinson. 2009. TatD is a central component of a Tat translocon-initiated quality control system for exported FeS proteins in Escherichia coli. EMBO Rep. 10:474-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller, M. 2005. Twin-arginine-specific protein export in Escherichia coli. Res. Microbiol. 156:131-136. [DOI] [PubMed] [Google Scholar]
- 51.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oresnik, I. J., C. L. Ladner, and R. J. Turner. 2001. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40:323-331. [DOI] [PubMed] [Google Scholar]
- 53.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 54.Parker, C. T., and J. Guard-Petter. 2001. Contribution of flagella and invasion proteins to pathogenesis of Salmonella enterica serovar Enteritidis in chicks. FEMS Microbiol. Lett. 204:287-291. [DOI] [PubMed] [Google Scholar]
- 55.Poppe, C. 2000. Salmonella infections in the domestic fowl, p. 107-132. In C. Wray and A. Wray (ed.), Salmonella in domestic animals. CABI Publishing, Wallingford, Oxon, United Kingdom.
- 56.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L. F. Wu. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohrbach, M. R., S. Paul, and W. Köster. 1995. In vivo reconstitution of an active siderophore transport system by a binding protein derivative lacking a signal sequence. Mol. Gen. Genet. 248:33-42. [DOI] [PubMed] [Google Scholar]
- 58.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose, R. W., T. Brüser, J. C. Kissinger, and M. Pohlschröder. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45:943-950. [DOI] [PubMed] [Google Scholar]
- 60.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy, K., G. M. Hilliard, D. J. Hamilton, J. Luo, M. M. Ostmann, and J. M. Fleckenstein. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saldana, Z., J. Xicohtencatl-Cortes, F. Avelino, A. D. Phillips, J. B. Kaper, J. L. Puente, and J. A. Giron. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11:992-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sargent, F., B. C. Berks, and T. Palmer. 2006. Pathfinders and trailblazers: a prokaryotic targeting system for transport of folded proteins. FEMS Microbiol. Lett. 254:198-207. [DOI] [PubMed] [Google Scholar]
- 64.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sargent, F., U. Gohlke, E. De Leeuw, N. R. Stanley, T. Palmer, H. R. Saibil, and B. C. Berks. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 268:3361-3367. [DOI] [PubMed] [Google Scholar]
- 66.Sargent, F., N. R. Stanley, B. C. Berks, and T. Palmer. 1999. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073-36082. [DOI] [PubMed] [Google Scholar]
- 67.Schmiel, D. H., and V. L. Miller. 1999. Bacterial phospholipases and pathogenesis. Microbes Infect. 1:1103-1112. [DOI] [PubMed] [Google Scholar]
- 68.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segura, I., J. Casadesus, and F. Ramos-Morales. 2004. Use of mixed infections to study cell invasion and intracellular proliferation of Salmonella enterica in eukaryotic cell cultures. J. Microbiol. Methods 56:83-91. [DOI] [PubMed] [Google Scholar]
- 70.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stecher, B., M. Barthel, M. C. Schlumberger, L. Haberli, W. Rabsch, M. Kremer, and W. D. Hardt. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell.Microbiol. 10:1166-1180. [DOI] [PubMed] [Google Scholar]
- 72.Van Immerseel, F., U. Methner, I. Rychlik, B. Nagy, P. Velge, G. Martin, N. Foster, R. Ducatelle, and P. A. Barrow. 2005. Vaccination and early protection against non-host-specific Salmonella serotypes in poultry: exploitation of innate immunity and microbial activity. Epidemiol. Infect. 133:959-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. A. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 74.Wexler, M., F. Sargent, R. L. Jack, N. R. Stanley, E. G. Bogsch, C. Robinson, B. C. Berks, and T. Palmer. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275:16717-16722. [DOI] [PubMed] [Google Scholar]