Abstract
Staphylococcus aureus is a major cause of a variety of both local and systemic infections. It can invade human host cells, a process that may account for disseminated and recurrent infections. S. aureus postinvasion events in nonprofessional phagocytes are only partially understood. While morphological data suggest a phagosomal escape, there is a lack of corroborating functional data. Using a combination of pH determination and morphological techniques, we have tested the integrity of Staphylococcus-containing phagosomes in 293 (HEK-293), HeLa, and EA.hy926 cells over time. Rapid acidification of S. aureus-containing phagosomes occurred and was sustained for up to 24 h. All S. aureus strains tested displayed equally sustained intraphagosomal pH levels without exhibiting any correlation with pH level and hemolytic activity. The membrane morphology of the phagosomal compartment was heterogeneous, even under conditions where acidic pH was fully maintained, an observation incompatible with phagolysosomal membrane destruction. As an exception, S. aureus strain 6850 showed a reduced phagosomal acidification signal 6 h after invasion. Additionally, only strain 6850 failed to localize to LAMP-1-positive vesicles in HeLa cells, although this was observed only rarely. Several other strongly beta-hemolytic strains did not modulate phagolysosomal pH, suggesting that S. aureus α-toxin and β-toxin are not sufficient for this process. Taken together, our data suggest that S. aureus-containing phagolysosomes generally remain functionally intact in nonprofessional phagocytes, thereby contrasting with transmission electron micrographic results.
Staphylococcus aureus, a Gram-positive bacterium, is a leading cause of important nosocomial and community-acquired infections. It causes both local and systemic diseases, ranging from minor wound infections and abscesses to life-threatening conditions, such as endocarditis, osteomyelitis, sepsis, and toxic shock syndrome (5, 21, 29). Although previously considered an exclusively extracellular pathogen, S. aureus has been shown to invade different types of mammalian host cells (3, 36) via a zipper-like mechanism similar to professional phagocytosis (7, 10, 27; reviewed in references 47 and 47a). In addition, prolonged survival in professional phagocytes, such as macrophages (25) and neutrophil granulocytes (51), has been observed in vitro, and transmission of infection by infected neutrophils has been experimentally shown in vivo (14). Clinical studies also indicate a possible role for an intracellular staphylococcal reservoir in recurring diseases, such as rhinosinusitis (6, 38).
Although a number of studies aimed at characterizing the intracellular life-style of S. aureus, the fate of internalized bacteria remains still unclear, with several reports of contrasting behavior regarding a phagosomal escape. Supporting the existence of the intact phagosome, colocalization studies using lysosomal dyes, such as LysoTracker, have shown the presence of S. aureus within acidic cellular compartments following internalization. To one extreme, live imaging of internalized S. aureus strains, including both normal and small colony variants (SCV) in human umbilical vein endothelial cells (HUVEC), showed that a fraction of bacteria can also survive within the phagosome for up to 5 days without any signs of an escape (43). In contrast, electron micrographs (EMs) of infected bovine mammary epithelial cells (MAC-T) suggested that S. aureus strain Novel is located freely in the cytoplasm 3 h after internalization (3). Furthermore, a colocalization experiment involving LysoTracker as a marker of phagosomal acidification demonstrated a lack of acidification of phagosomes in HeLa cells containing wild-type staphylococci, with intact expression of the accessory global regulator agr (42). Similarly, wild-type staphylococci were also rarely positive for LAMP-2, a characteristic marker of phagolysosomes. From this study, it was concluded that the subsequent transition to an autophagosome provided the grounds for an escape by S. aureus from the membrane-bound confinements that ultimately leads to a caspase-independent death of the host cell (42).
S. aureus α-toxin, a 33.2-kDa polypeptide encoded by hla, has recently been reported to be a key factor in the phagosomal escape of S. aureus in CFT-1 cells, a human cystic fibrosis airway epithelial cell line (20). Secreted as a water-soluble monomer, α-toxin assembles on cell membranes to form a water-filled pore, thus unfolding cytotoxic, hemolytic, and dermonecrotic activities (49).
So far, the studies on potential phagosomal escape by S. aureus were essentially based on fluorescence or electron microscopic methods to investigate the subcellular localization (3, 19, 42, 43). In spite of these morphological data, there is a considerable lack of functional data for the fate of S. aureus-containing phagosomes (SACP). During phagosomal maturation, the phagosomal compartment is acidified and maintained at a low pH by vacuolar ATPases (V-ATPases) (reviewed in reference 15). A V-ATPase has been shown to be active in 293 cells (28). Thus, monitoring of phagosomal pH over time is a valuable tool for measuring the integrity and functionality of the phagosomal compartment.
The main goals of this study were (i) to obtain functional evidence for the destruction of the phagolysosomes in human nonprofessional phagocytes and (ii) to establish whether a phagosomal escape is a general property of S. aureus. Additionally, we tested the role of the global regulator SigB, the alternative S. aureus σ-factor, in phagosomal modulation.
MATERIALS AND METHODS
Cell lines and growth conditions.
HeLa (human cervix carcinoma) and 293 (human embryonic kidney) cells were obtained from the German Collection of Microorganisms and Cell Culture (DSMZ). The human endothelial cell line EA.hy926 (8) was kindly provided by Volker Gerke (Münster, Germany). All cell lines, including the murine macrophage-derived cell line J774, were maintained in Dulbecco's modified Eagle's medium (DMEM)-Nut Mix F-12 medium nutrient mix (Gibco) supplemented with 10% fetal calf serum (FCS; Biochrom, Berlin, Germany), 50 IU ml−1 penicillin, and 50 mg ml−1 streptomycin (PAA Laboratories, Pasching, Austria) under standard tissue culture conditions at 37°C and with 5% CO2. The cells were split by trypsinization (trypsin-EDTA) twice a week. The cell lines were routinely tested for Mycoplasma infection every 6 to 8 weeks using a PCR Mycoplasma test kit from AppliChem (Darmstadt, Germany).
Bacterial strains and fluorescein isothiocyanate (FITC) labeling.
The staphylococcal strains used in this study are listed in Table 1. Strain 6850 is a clinical isolate that caused bacteremia and metastatic abscesses, produces β-lactamase (39), and had been used earlier to study small colony variants (2). It is a strong α-toxin producer, as shown by transcriptomics (11) and phenotypically by β-hemolysis on blood agar. It also produces β-toxin and δ-toxin. We have shown previously that it is cytotoxic for HUVEC (16) and human mesothelial cells (17). We have reported that 6850 is strongly invasive (∼120% to 150% of Cowan I for 293 cells and Ea.hy926 cells) (18). Invasiveness for HeLa cells was similar (unpublished data). The surface displays of fibronectin-binding proteins (FnBPs) were comparable for 6850 and Cowan I (18). Phenotypical control of the bacterial colonies grown on sheep blood agar was performed. To compare hemolytic activities, S. aureus strains were plated on horse blood and sheep blood agar for detection of α-toxin and β-toxin, respectively. Agar plates were incubated overnight at 37°C, cooled at 4°C for 2 h, and left at room temperature for another 2 h prior to transillumination analysis of hemolysis.
TABLE 1.
Bacterial strains used in this study
| Organism and strain(s) | Property(ies)a | Reference(s) or source |
|---|---|---|
| S. aureus | ||
| Cowan I | Phenotypically without α- and β-toxin; t076 | ATCC 12598 |
| 6850 | MSSA, wild-type isolate; t185 | 2, 39 |
| Novel | Bovine mastitis isolate; t2414 | 48 |
| Wood 46 | Strong α-toxin producer; protein A-deficient; t211 | ATCC 10832 |
| BS 507, BS 513 | MRSA; clinical isolates (nose); t2814 (BS 507); t063 (BS 513) | This study |
| BS 800, BS 890 | MSSA; clinical isolates (nose); t1014 (BS 800); t3010 (BS 890) | This study |
| BS 891 | Reverted SCV; clinical isolate (nose); related to BS 890; t3010 | This study |
| RN4220 | Phenotypically without α- and δ-toxin; t211 | 24 |
| S. carnosus | ||
| TM300 | No S. aureus virulence factors | 41 |
| TM300 (pFNBA4) | Expression of FnBPA (Cmr) | 45 |
Designations beginning with “t” followed by a number denote spa type (e.g., “t076” denotes spa type t076).
Mueller-Hinton (MH) broth (without agitation) and MH agar were used for the routine bacterial culture (overnight) at 37°C. The medium was supplemented with chloramphenicol (10 μg ml−1) wherever appropriate.
Staphylococci were labeled with FITC (dissolved in dimethyl sulfoxide [DMSO]) for 30 min, as described previously (22, 46), and resuspended in phosphate-buffered saline (PBS) supplemented with 1% human serum albumin (HSA). Prior to infection, the optical densities at 540 nm (OD540) of different FITC-labeled bacterial suspensions were adjusted to 1. Control experiments performed with suspensions of live bacteria yielded the equivalent of 1 × 108 CFU ml−1 per 1 OD540 unit for strain Cowan I (not shown).
Host cell infection.
Adherent host cells were seeded and grown overnight in a 24-well culture dish to subconfluency (3 × 105 cells/well for 293 cells and 2 × 105 cells/well for HeLa, EA.hy926, and J774 cells). Prior to infections, cells were washed with DMEM-Nut Mix F-12 medium nutrient mix containing 10% FCS, 50 IU ml−1 penicillin, and 50 mg ml−1 streptomycin (complete medium). The cell culture medium was subsequently replaced with 500 μl of invasion medium (10 mM HEPES [pH 7] in DMEM-Nut Mix F-12 medium nutrient mix with 1% HSA). The cells were cooled on ice for 10 min, and 50 μl of FITC-labeled staphylococcal suspensions was added to each well. Infection was synchronized by sedimentation of the bacteria at 4°C for 1 h, and invasion was subsequently started by shifting to 37°C. Following incubation for 55 min, extracellular bacteria were eliminated by removal of the invasion medium and incubation with medium supplemented with lysostaphin at 37°C for 10 min (final concentration, 20 μg ml−1; AMBI, Lawrence, NY). Cells were further incubated in complete medium for various intervals, as indicated in the figures.
Flow cytometry-based pH monitoring.
Cells were trypsinized, transferred to 5-ml round-bottom 12- by 75-mm polystyrene (fluorescence-activated cell sorting [FACS]) tubes (Falcon; BD, Heidelberg, Germany), pelleted, and resuspended in PBS with 1% HSA. To obtain dose-response curves, neutralizing agents (NH4Cl, chloroquine, and bafilomycin A1 [LC Laboratories, Woburn, MA]) were added to the cells at designated concentrations and incubated at 37°C for 15 min (bafilomycin A1 and NH4Cl) or 20 min (chloroquine) prior to flow cytometric measurement.
Each sample was aliquoted and incubated with and without 50 μM monensin for 15 min at 37°C prior to analysis. Quenching of FITC fluorescence by low pH was used as a marker for acidification. The difference in Fl-1 fluorescence signals (expressed as the difference between numbers of arbitrary fluorescence units [ΔAFU]) in samples without monensin and the monensin-treated counterpart was used as a readout for pH (see below).
Samples were analyzed with a FACSCalibur (Becton Dickinson). For each cell type, a preset fixed forward scatter and side scatter (FSC-SSC) gating and a fixed amplification were used, and fluorescence in the Fl-1 channel was analyzed with propidium iodide (PI; Fluka Biochemika, Buchs, Switzerland) exclusion in Fl-3 after addition of 5 mg ml−1 PI. Histograms of fluorescence (Fl-1 height) of FSC-SSC-gated cells were obtained after acquisition; fluorescence intensity markers (M) were preset generally to include <2% of the cells of the respective negative controls (cells only).
The quenching of FITC fluorescence (excitation at 488 nm/emission at 520 nm) is proportional to acidic pH. The invasion rate with monensin (total internalized bacteria), expressed as numbers of arbitrary fluorescence units (AFU), was determined according to the formula AFU = (mean Fl-1 height of cells in M) × (percentage of cells in M)/1,000 and normalized to the mean fluorescence intensity of each corresponding bacterial preparation, as previously described (22, 46).
The difference in AFU (ΔAFU) in the presence or absence of monensin, normalized to the invasion rate [ΔAFU = (AFU−monensin − AFU+monensin)/AFU+monensin × 10,000], was used as readout for pH. Larger negative values correspond to more-acidic (i.e., lower) pH values.
Bacterial viability and lysostaphin protection assay.
Bacterial viability was determined by serial dilution and plating following incubation with 100 μg ml−1 FITC in 0.1 M NaHCO3 buffer (pH 9.0) or components of this solution for 1 h. As a control, bacteria were incubated in PBS containing 1% HSA. Bacteria were washed in 1% HSA-PBS, appropriate dilutions were plated on MH agar and incubated at 37°C overnight, and the CFU were counted.
To further exclude any negative effect of FITC labeling on the intracellular viability of the bacteria, a lysostaphin protection assay was performed. Cells were infected with FITC-labeled staphylococci as described above, including addition of 20 μg ml−1 lysostaphin (see above, “Host cell infection”). Infection with unlabeled bacteria served as a control. After 2 or 6 h of overall infection time, cells were lysed by adding 1 ml of sterile distilled water, and serial dilutions of the lysates were made in 1% HSA-PBS. Appropriate dilutions were plated in triplicate on MH agar and incubated overnight at 37°C, and the number of internalized bacteria was calculated from the mean numbers of CFU counted on the plates.
Electron microscopy.
For transmission electron microscopy (TEM), duplicates of samples analyzed by flow cytometry (293 and HeLa cells) were pooled and transferred to 1.5 ml Eppendorf tubes.
For Karnovsky fixation, 500 μl Karnovsky fixative (2.5% glutaraldehyde, 2% paraformaldehyde, and 0.1 M sodium cacodylate buffer, pH 7.2) was added, cells were centrifuged (10 min at 1,000 rpm), and the pellet was incubated in the fixative for 30 min at 4°C. After three washing steps, the cells were left in the washing solution (0.1 M cacodylate [Hartenstein, Würzburg, Germany]) overnight at 4°C before further processing. For enhanced membrane preservation, samples were washed with PBS once and then fixed in cold 3% KMnO4 (in distilled water) for 20 min at room temperature. All samples were then washed two times in 50 mM Sörensen buffer (87.8 mM Na2HPO4 and 12.2 mM KH2PO4; final pH 7.4), were incubated for 45 min in 2.5% glutaraldehyde with 50 mM KCl, 2.5 mM CaCl2, and 50 mM cacodylate (pH 7.4), were washed three times with 50 mM cacodylate, and were left overnight at 4°C in cacodylate before further processing. Subsequently, the samples were fixed for 2 h at 4°C with 2% osmium tetroxide (Roth, Karlsruhe, Germany) in 50 mM sodium cacodylate (pH 7.4) and stained overnight with 0.5% aqueous uranyl acetate. Specimens were dehydrated and embedded in Epon 812 (Serva, Heidelberg, Germany), and ultrathin sections were cut. At least 5 randomly chosen sections of each specimen were examined over the complete extension of the section (>10 fields per section at ×8,000 magnification) using an EM10 and an EM900 electron microscope (Zeiss, Oberkochen, Germany). Photographic negatives were scanned, and representative fields of interest were selected and adjusted to similar brightness and contrast levels using Adobe Photoshop CS.
Confocal fluorescence microscopic measurements of SACP pH.
For infection, SNARF1-labeled staphylococci were used. For direct labeling with SNARF1-succinimidyl ester (SNARF1-SE; Invitrogen, Karlsruhe, Germany), the bacteria were incubated in PBS containing 125 μg ml−1 SNARF1-SE for 1 h at room temperature, harvested by centrifugation at 3,000 × g for 10 min, resuspended in PBS, and incubated with shaking at 37°C for 10 min. After centrifugation, the bacterial pellet was resuspended in invasion medium and added to the host cells. Images were acquired by a Zeiss LSM510 META confocal laser scanning microscope with 8-bit depth in line-switching multitrack mode following excitation of SNARF1 at 561 nm. Using the 405/488/561-nm dichroic mirror and the META detector, two parts of the pH-dependent ratiometric emission spectrum of SNARF1 were recorded, covering the ranges of 571 to 603 nm (S2) and 635 to 753 nm (S1). Each line was scanned 8 times and averaged. Three independent experiments were performed, and in each, approximately 25 infected phagosomes were analyzed per sample (numbers of fields of view differed).
Image processing was performed using LSM 510 software (Carl Zeiss GmbH, Jena, Germany) and ImageJ (1). The acquired channels were processed with a 7- by 7-pixel low-pass filter, and a threshold of 25 was applied to remove background noise. A combined binary mask for both channels was multiplied with the filtered channel images. The final ratio image was created by image arithmetic (S2/S1 × 10). Only the mean intensity of SNARF1 ratio signals in the direct vicinity of staphylococci was analyzed. A more detailed description of the method can be found in reference 12.
Fluorescence microscopy.
Fluorescence microscopy was carried out with a Zeiss LSM510 META confocal microscope (Carl Zeiss, Jena, Germany). Confocal images, unless noted otherwise, represent ∼1-μm-thick confocal slices of the specimen. Yellow fluorescent protein (YFP) was excited by the 514-nm line of the argon ion laser with a 514-nm dichroic mirror, and emitted fluorescence was detected with the META detector, using a detection range of 518 nm to 550 nm. For LAMP1-YFP-based visualization of phagosomal localization, the beam path for SNARF1-SE fluorescence contained the 561-nm line of a yellow diode laser, a 561-nm dichroic mirror, and a 575-nm long-pass filter. Nonconfocal transmitted light images were generated in parallel to YFP detection by the 514-nm line of the argon ion laser with a transmitted-light detector below the specimen/focal plane. At least five fields were analyzed in three independent experiments; shown are representative fields.
Statistical analysis.
Data from flow cytometric analysis and lysostaphin protection and recovery assays were expressed as means ± standard errors of the means (SEM) of results from at least three independent experiments, performed in duplicate and triplicate, respectively. Microfluorometry data were presented as means ± standard deviations (SD). For statistical analysis, the means of results from replicates were compared. Independent samples with symmetric, nonhomogeneous distribution without equal variances were assumed. Consequently, a one-sided Welch t test was performed, using Microsoft Excel 2003. A significant difference was assumed if the P value for the null hypothesis (H0) (i.e., no reduction in ΔAFU) was ≤0.01.
RESULTS
Monitoring of intraphagosomal pH by reversible fluorescence quenching of FITC-labeled staphylococci.
The reversible quenching of FITC fluorescence can be used to monitor the pH of SACP after invasion, using FITC-labeled staphylococci as a probe. Synchronization and pulsing of invasion assays with FITC-labeled S. aureus were performed by removing extracellular bacteria using the glycyl-glycyl endopeptidase lysostaphin. The efficiency of the lysostaphin killing of extracellular staphylococci was confirmed by plating infection media, which were sterile (data not shown). In subsequent flow cytometric analyses, FITC fluorescence signals were obtained from parallel aliquots of samples, one of which was treated with the neutralizing ionophore monensin (34, 46), whereas the other aliquot was not treated. The quenching of FITC fluorescence in untreated samples is dependent on phagolysosomal pH, while the sample with monensin treatment is expected to have maximum fluorescence intensity due to a neutral environment. Therefore, the signal can be expressed as the difference in the number of arbitrary fluorescence units (ΔAFU) between monensin-treated and untreated samples, which correlates with SACP acidification.
To validate the ability of the assay to detect pH changes in SACP, increasing concentrations of neutralizing agents were added to 293 cells infected with live FITC-labeled S. aureus strain Cowan I. Addition of either NH4Cl or chloroquine showed dose-dependent reduction of ΔAFU, demonstrating a quantitative and sensitive detection of intraphagosomal pH changes (Fig. 1 A and B). Similar dose-response curves were obtained with chloroquine, fixed S. aureus strain Cowan I (data not shown), and prevention of phagosomal acidification with the addition of increasing concentrations of the V-ATPase inhibitor. As expected, brefeldin A, an inhibitor of anterograde vesicular transport, had no effect on pH (Fig. 1D).
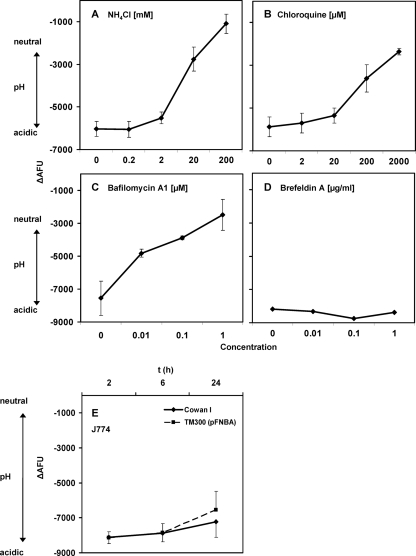
FIG. 1.
Validation of the flow cytometric pH assay. Dose-response curves with neutralizing agents were obtained after infection of 293 cells with live FITC-labeled S. aureus strain Cowan I. After sedimentation and incubation for 2 h at 37°C, increasing concentrations of neutralizing agents were added to the samples. Cell-associated FITC fluorescence was subsequently measured as numbers of arbitrary fluorescence units using flow cytometry. The ΔAFU values of samples with and without addition of the neutralizing agent monensin were calculated, and values were normalized for invasion (see Materials and Methods). Higher negative ΔAFU values correlate with lower pH values. Similar results were found in HeLa cells (data not shown). x axes indicate the concentrations of NH4Cl (A), chloroquine (B), bafilomycin A1 (C), and brefeldin A (D). Data shown are means of results from 3 independent experiments performed in duplicate ± SEM (for panel D, n = 1). The assay also showed acidification in professional phagocytes. In phagocytosis experiments with J774 after 2, 6, and 24 h of infection (E), substantial ΔAFU values were found. Data shown are means of results from 3 independent experiments performed in duplicate ± SEM. t, time point.
Initial experiments with fixed Staphylococcus carnosus TM300 (pFNBA4), a noninvasive staphylococcal strain expressing the S. aureus invasin FnBPA, using confocal microscopy confirmed that FITC-labeled bacteria were localized in an acidic compartment of HeLa cells after 6 h (data not shown). These controls further corroborate the data obtained by fluorescence quenching in flow cytometry, demonstrating that staphylococci were localized in an acidic compartment (Fig. 1; see also Fig. 2 and 3).
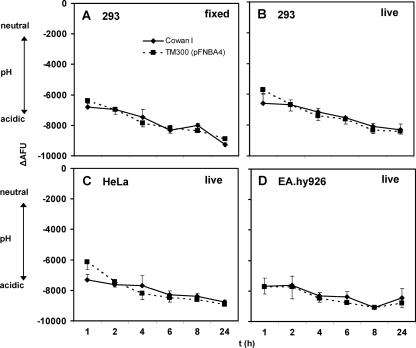
FIG. 2.
Persistent acidification of Staphylococcus-containing phagosomes for up to 24 h after infection in different nonprofessional phagocytes. Acidification was analyzed for 293 (A and B), HeLa (C), and EA.hy926 (D) cells (see the legend to Fig. 1 and Materials and Methods) at the indicated time points following infection with fixed (A) or live (B to D) staphylococci. In all tested cell lines, fluorescence quenching, indicative of low pH, was maintained for 24 h. Data shown are means of results from 3 independent experiments performed in duplicate ± SEM.
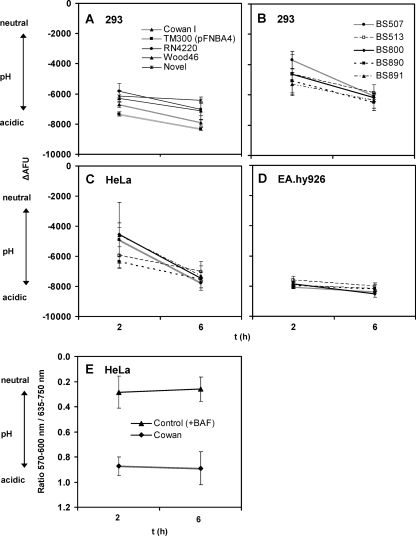
FIG. 3.
Maintenance of phagosomal pH for wild-type S. aureus strains for up to 6 h. The strains tested include those of reverted SCV (BS 891), methicillin-resistant S. aureus (MRSA) (BS 507 and BS 513), and methicillin-susceptible S. aureus (MSSA) (BS 800 and BS 890). Acidification was analyzed at 2 and 6 h after infection of 293 (A, B), HeLa (C), and EA.hy926 (D) cells (see the legend to Fig. 1 and Materials and Methods). For all strains, ΔAFU values were larger at 6 h than at 2 h, indicating a sustained acidic phagosomal pH. All clinical isolates except BS 891 are beta-hemolytic on sheep blood agar, consistent with α-toxin and/or β-toxin production. Microfluorometric data show sustained acidification of Cowan I in HeLa cells in contrast to the level for the control, with the V-ATPase blocked by the inhibitor bafilomycin A1 (E). Flow cytometric data shown are means of results from at least three independent experiments performed in duplicate ± SEM (A to D). Microfluorometric data shown are means of results from 3 independent experiments ± SD (E).
In order to exclude a potential influence of FITC labeling on the viability of intracellular bacteria, we performed quantitative plating experiments with FITC-labeled and unlabeled bacteria. Incubation of different staphylococcal strains in FITC-staining solution or its components without FITC resulted in similar levels of recovery in both groups, as determined by CFU counting. This indicated that the viability of the staphylococci was not reduced by the labeling procedure (Table 2). In addition, lysostaphin protection assays showed comparable levels of intracellular viability with and without FITC labeling (Table 3). Taken together, these results excluded an adverse effect of FITC labeling on both viability and potential intracellular replication of staphylococci.
TABLE 2.
Viability of bacteria before and after FITC labelinga
| Condition | 108 CFU ± SEM (% of control) for: |
|
|---|---|---|
| Cowan I | 6850 | |
| HSA-PBS at 1% (control) | 3.33 ± 0.27 (100) | 3.05 ± 0.18 (100) |
| Carbonate buffer at 0.1 M | 3.54 ± 0.26 (106) | 3.56 ± 0.28 (117) |
| Carbonate buffer at 0.1 M plus 10% DMSO | 3.62 ± 0.78 (109) | 3.20 ± 0.63 (105) |
| FITC staining buffer (complete) | 3.38 ± 0.28 (102) | 3.35 ± 0.52 (110) |
Staphylococci were resuspended in the media indicated. FITC staining buffer consisted of 0.1 M NaHCO3 buffer (pH 9.0) supplemented with 100 μg/ml FITC (isomer I; Molecular Probes), dissolved in DMSO (10% [vol/vol] final concentration). Data shown are means ± SEM of results from 4 independent experiments performed in duplicate. The incubation time was 30 min. Statistical analysis using Welch's t test did not show a statistically significant difference between FITC-labeled bacteria and controls (all P values were >0.5).
TABLE 3.
Antibiotic protection assay for intracellular viability of FITC-labeled and unlabeled bacteriaa
| Strain | Presence of FITC | ‰ recovery of inoculum ± SEM (% of unlabeled control without FITC) at: |
|
|---|---|---|---|
| 2 h | 6 h | ||
| Cowan I | − | 0.32 ± 0.08 (100) | 0.49 ± 0.15 (100) |
| + | 0.32 ± 0.12 (100) | 0.38 ± 0.08 (77) | |
| TM300 (pFNBA4) | − | 0.48 ± 0.20 (100) | 1.00 ± 0.52 (100) |
| + | 0.45 ± 0.28 (94) | 1.67 ± 0.80 (167) | |
| 6850 | − | 0.33 ± 0.11 (100) | 0.53 ± 0.14 (100) |
| + | 0.46 ± 0.20 (140) | 0.72 ± 0.33 (136) | |
In order to account for minor variations in the actual staphylococcal suspensions, the fraction of the inoculum recovered from 293 cells is indicated as a per mille of the inoculum (i.e., of the number of input CFU) at 2 h and 6 h. The calculated MOI was ∼10, with inoculums of ∼5 × 106 CFU/well. Data are shown as means ± SEM of results from 5 (for Cowan I and 6850) and 3 [for TM300 (pFNBA4)] independent experiments performed in duplicate. Values for unlabeled controls were set to 100%, and recovery of labeled bacteria is expressed as a percentage of this control value in order to compare both values. Percent values were calculated from the raw data and subsequently rounded to integers. Statistical analysis using Welch's t test did not show a statistically significant difference between FITC-labeled and unlabeled bacteria (all P values were >0.25).
Having established the positive and negative controls for the determination of intraphagosomal pH based on FITC quenching, we were interested in determining the acidification status of phagosomes in professional phagocytes after infection with staphylococci. To achieve this objective, J774 cells, a cell line derived from murine macrophages, were infected with FITC-labeled S. aureus strain Cowan I and a control strain (S. carnosus) and analyzed by flow cytometry at the time points indicated in Fig. 1. The ΔAFU values obtained after the analysis showed that phagosomes of J774 cells had a sustained low pH at up to 24 h postinfection (Fig. 1E). Furthermore, ΔAFU values in J774 cells were within a range of −9,000 to −7,000 and thus similar to ΔAFU values observed in nonprofessional phagocytes, e.g., 293 cells (Fig. 1). In addition to revealing the acidification status of the phagosomes in all tested cell lines, these results also demonstrated that the flow cytometric assay was able to detect a low intraphagosomal pH in SACP and that this assay was even sensitive enough to detect smaller pH changes.
Maintenance of profound acidification of nonprofessional phagosomes in three different cell lines for up to 24 h.
Phagosomes of nonprofessional phagocytes undergo maturation in a manner similar to that found in professional phagocytes, such as macrophages and neutrophil granulocytes. One functional characteristic in this process is a rapid acidification of the compartment mediated by V-ATPase. As phagosomes of J774 macrophage cells maintained acidic pH even up to 24 h (Fig. 1E), we wanted to ascertain if the nonprofessional phagocytes also behave in a similar fashion after infection with S. aureus. Therefore, we monitored the pH of phagosomes in three different human cell lines, including 293 (human embryonic kidney), HeLa (human cervix carcinoma), and EA.hy926 (HUVEC fusion) cells that were infected with FITC-labeled staphylococci for 1, 2, 4, 6, 8, and 24 h to observe the intraphagosomal pH course over time. Profoundly negative ΔAFU levels, corresponding to an acidic pH, were already observed after 1 h postinfection and remained low for 24 h (Fig. 2). Fixed S. aureus strain Cowan I and S. carnosus strain TM300 (pFNBA4) served as negative controls for phagosomal escape in 293 cells (Fig. 2A). As expected, the pH of phagosomes containing fixed staphylococci was acidic and remained low over the observed time course. The development of pH over time of phagolysosomes harboring live staphylococci, over a time course, exhibited identical patterns in all the cell lines and both the bacterial strains tested (Fig. 2B to D). This finding demonstrated sustained phagosomal acidification without any indication of a possible phagosomal escape by S. aureus, which otherwise would have neutralized the intraphagosomal environment.
General maintenance of functional integrity of phagosomes for a number of S. aureus strains in all three cell lines tested.
The sustained intraphagosomal acidity and phagosome membrane integrity observed for S. aureus strain Cowan I and S. carnosus strain TM300, harboring pFNBA4 (Fig. 2), prompted us to determine whether the phagosomal escape observed by other laboratories is strain specific. Previous studies have reported the disappearance of a membrane surrounding intracellular staphylococci 3 h after internalization (3). Therefore, we expected a change in pH to be detectable upon comparison of the ΔAFU values observed 2 and 6 h after internalization. To account for potential strain differences, we used a set of well-defined laboratory strains as well as a number of phenotypically different clinical isolates. The strains investigated include strain Novel, which has been used in the first report on phagosomal escape by S. aureus (3). In contrast to our expectation, none of the tested strains showed a neutralization of the acidic pH (Fig. 3 A to D), suggesting the absence of phagosomal escape. In contrast, the intraphagosomal pH was found to be low and became even more acidic with time. Statistical analysis of the ΔAFU values confirmed the apparent observation that there was no significant reduction in intraphagosomal acidification between 2 and 6 h. As we observed this in all strains tested, regardless of their beta-hemolytic activity, there was no detectable correlation between beta-hemolytic activity and phagosomal acidification. Furthermore, phagosomal pHs were rather similar in all the three cell lines used, especially at 6-h time points (Fig. 3), with EA.hy926 demonstrating the fastest and most pronounced acidification (Fig. 3D). Control experiments performed with 293 cells for strain Cowan I, using inoculums up to 4-fold higher than usual, yielded no difference for pH data at 2 h and 6 h (data not shown).
To corroborate the obtained flow cytometric data with a different method for pH measurement, we determined the intraphagosomal pH using the ratiometric pH-sensitive dye SNARF1 succinimidyl ester (SNARF1-SE). Bacteria were labeled with SNARF1-SE before infecting HeLa cells. At the time points indicated in Fig. 3E, the images were acquired by a confocal laser scanning microscope. The red (635 to 753 nm; S1) and the yellow fluorescence (571 to 603 nm; S2) spectra of SNARF1-labeled bacteria were collected, and a ratio channel was created. Following the analysis of the recorded fluorescence spectra, we found that the lumina of infected phagolysosomes were acidic (Fig. 3E). As predicted, a control where samples were neutralized by employing the V-type ATPase inhibitor bafilomycin A1 exhibited neutral pH.
Lack of clear correlation between morphological membrane alterations and functional differences.
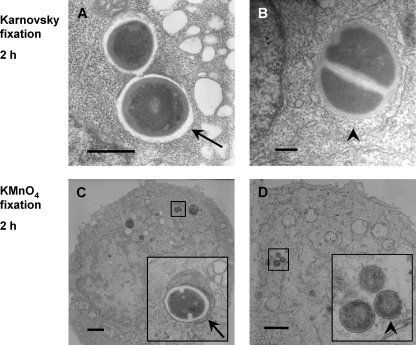
To detect possible morphological membrane changes in the SACP, and as a means to visually complement the functional data obtained for the membrane integrity of the phagosome, we used Karnovsky fixation and subsequent TEM. 293 cells from the same pool were used for TEM after flow cytometric analysis. The host cells contained only few staphylococci, confirming the low multiplicity of infection (MOI) used for infection (calculated MOI, ∼10). Intracellular individual cocci of live S. aureus strain Cowan I were either found in membrane-bound compartments or free, without any visible membrane around the bacteria (Fig. 4 A and B). Both conditions could be detected as early as 2 h after invasion. Similar results were found in electron micrographs of infected HeLa cells (data not shown). As a second approach, sample preparation was performed using KMnO4, which is known to achieve a better visualization of cellular and intracellular membranes. The electron micrographs taken under these conditions equally showed both staphylococci confined in a compartment as well as intracellular bacteria without a visible surrounding membrane (Fig. 4C and D). Thus, no difference could be seen when cells were fixed under conditions preserving all cellular components (Karnovsky fixation) or, preferentially, membranes (KMnO4 fixation) (Fig. 4). However, the morphological findings could not be correlated with the functional data, as the pH assay suggested that the SACP remained intact and acidified over the period of investigation postinfection.
FIG. 4.
Intracellular staphylococci both with and without apparent phagosomal membrane at 2 h. Following infection of 293 cells for 2 h with live Cowan I, samples were prepared for TEM using Karnovsky fixation (A, B). In addition, fixation with KMnO4 was performed for enhanced membrane visualization (C, D). Electron micrographs taken from cells after 2 h of infection showed both intracellular staphylococci with visible phagosomal membrane (arrows) and bacteria around which a compartment cannot be clearly discerned (arrowheads). Thus, no correlation could be found between morphology and flow cytometric pH data. Scale bar = 0.5 μm (A, B) or 1 μm (C, D).
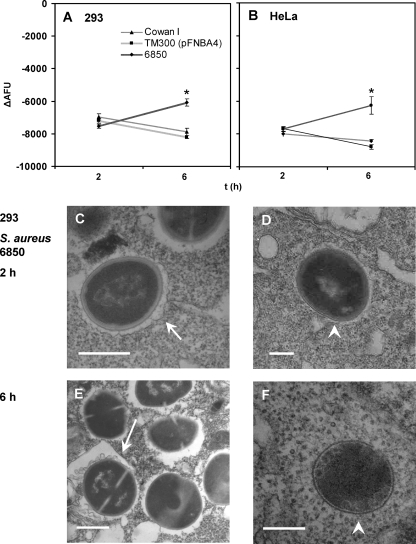
Partial modulation of the phagosomal acidification signal by strain 6850.
Of all tested staphylococcal strains, only S. aureus strain 6850 modulated the recorded phagolysosomal pH signal. In both 293 and HeLa cells, the signal (negative ΔAFU values) at 6 h was consistently smaller than that observed at 2 h (Fig. 5 A and B) and indicated a change in pH from acidic at 2 h postinfection to less acidic at 6 h within the SACP. The measurements of infected cells at 24 h gave results similar to those observed at 6 h. However, due to an increasing rate of cell death, these data were difficult to interpret. Cell death was more prominent in HeLa cells than in 293 cells (data not shown). Strain 6850 also displayed similar behavior in EA.hy926 cells (data not shown). Unlike for the modulation of the phagosome by strain 6850, phagosomes containing other bacterial strains, including Cowan I and TM300 (pFNBA4), consistently maintained an acidic environment, which became even more acidic with time.
FIG. 5.
Reduced phagosomal acidification signal for S. aureus strain 6850 at 6 h. Acidification was analyzed at 2 and 6 h after infection for 293 and HeLa cells (see the legend to Fig. 1 and Materials and Methods). In contrast to what was found for all other wild-type strains tested, the ΔAFU values for 6850 at 6 h are reduced compared to those observed at 2 h. Electron micrographs of 293 cells infected with 6850 for 2 and 6 h showed both intracellular bacteria with (arrows) (C and E) and without (arrowheads) (D and F) surrounding membrane. Scale bar = 0.5 μm. Data shown are means of results from 20 (A) or 6 (B) independent experiments performed in duplicate ± SEM. An asterisk represents statistical significance between ΔAFU values at 2 and 6 h for 6850 (P < 0.01).
The electron micrographs obtained in parallel with the flow cytometric pH measurements, however, did not provide additional evidence for phagosomal escape by strain 6850. As with strain Cowan I, strain 6850 was found in both states, that of being membrane enclosed and that of having an apparent cytoplasmic location, regardless of the investigated time point of the infection (2 and 6 h after infection) (Fig. 5C to F).
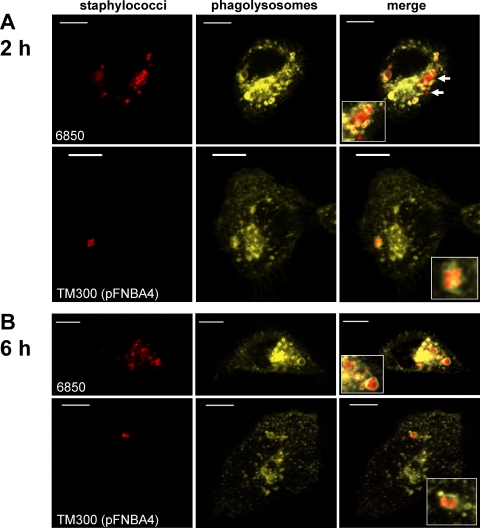
Phagolysosomal LAMP-1 decoration in strain 6850.
As S. aureus strain 6850 was the only investigated strain that was able to modulate the intraphagosomal pH, we were curious to examine phagolysosomal LAMP-1 decoration. Recently, a study reported a crucial role for α-toxin in phagosomal escape (20), using LAMP decoration. Staphylococcal α-toxin had been implicated in phagolysosomal escape in cystic fibrosis airway epithelial cells (20) but not in cells complemented with a functional cystic fibrosis transmembrane conductance regulator (CFTR) (12, 20). Thus, due to the absence of any correlation between the hemolytic activity and the ability to modulate the phagosomal pH, our previous results (12) suggest the existence of an effector other than α-toxin, involved in the modulation of phagosomal pH by strain 6850 (Fig. 6 A). We examined whether the bacteria were surrounded by a phagolysosomal membrane by infecting HeLa cells that expressed the lysosomal membrane marker LAMP-1 fused to YFP (12). Of all investigated strains, only strain 6850 resulted in a few bacteria that were devoid of a LAMP1-YFP-labeled membrane envelope (Fig. 6). Thus, it appears that only a fraction of phagosomes was affected by phagolysosomal modulation by 6850.
FIG. 6.
Phagolysosomal localization of staphylococci throughout a 6-h infection as indicated by the fluorescent marker LAMP-1-YFP. Representative images of live HeLa-cells expressing LAMP-1-YFP are shown at 2 h (A) and 6 h (B) after infection, with different staphylococcal strains stained with SNARF1-SE. In rare cases, S. aureus 6850 is not surrounded by a LAMP-1-YFP-positive membrane (arrows), whereas all other strains show continuous localization within HeLa phagolysosomes. Bacteria were stained with SNARF1-SE. Red, staphylococci; yellow, LAMP-1-YFP. Bar = 10 μm.
DISCUSSION
Phagosomal escape by a pathogen is an important virulence strategy for systemic infections, including spread to neighboring cells and tissues. Like several other pathogenic bacterial species, S. aureus has been reported to escape from the phagosome and to be found freely in the cytoplasm following invasion of nonprofessional phagocytes (3, 19, 20). However, the absence of a clearly discernible structure corresponding to phagolysosomal membranes does not necessarily reflect the destruction of the membrane. Thus, we used a combination of several techniques to obtain both functional and morphological data in order to determine the fate of the phagosomal membrane in three different human cell lines (293, HeLa, and EA.hy926) (Fig. 2, 3, and 5). The flow cytometric measurements of phagosomal acidification of SACP in a large cell population were complemented by pH measurement of individual SACP on a single-cell level (Fig. 3). To avoid potential limitation by strain-specific results, we used a number of well-studied laboratory strains as well as clinical isolates with different phenotypic characteristics (Fig. 2, 3, and 5).
In the current investigation, we demonstrated that FITC-labeled staphylococci can be used as a probe to study the intraphagosomal pH of nonprofessional phagocytes using flow cytometry (Fig. 1 and 2). In all cell lines tested, a rapid acidification of the phagosomal lumen was observed within 2 h postinfection (Fig. 2, 3, 5, and 6), and preliminary data suggest that a pronounced acidification already occurs within 1 h (data not shown). The assay was validated by a number of experimental approaches: (i) exogenous neutralization led to a dose-dependent reduction in the signal (Fig. 1A to D); (ii) in a murine macrophage cell line, acidification was pronounced (Fig. 1E), which is consistent with the recent literature (15, 35); and (iii) microfluorometry similarly showed a persistently low pH in HeLa cells for up to 6 h, as observed by flow cytometry (Fig. 3). Phagolysosomal acidification was generally maintained for up to 24 h, regardless of whether bacteria were fixed or live (Fig. 2 and 3). The only exception to this was found for S. aureus strain 6850, which showed a decreased level of phagolysosomal acidification 6 h after invasion in comparison to the level for the 2-h time point. Combined with imaging on light and electron microscopic levels, our data suggest that S. aureus does not generally modulate phagolysosomal acidification. However, this does not exclude that further strains may be able to modulate phagolysosomal acidification. When modulation occurs, it appears not to be an all-or-nothing event (Fig. 5).
Notably, we did not observe a clear correlation between morphological alterations of phagosomal structures in TEM and functional data (Fig. 4 and 5); although TEM studies employing different fixation techniques suggest the absence of the phagolysosomal membrane, the acidification status of the phagolysosomes does not change. The lack of a clear correlation between findings from the microscopic images and functional data is even more striking, since we used two different approaches to visualize phagosomal membrane structures, conventional Karnovsky fixation and KMnO4 fixation (30), which has been shown to highlight membrane structures in eukaryotic (33) as well as prokaryotic (50) cells.
Our findings are consistent with previous reports in which a phagosomal escape was not observed but a longer intracellular persistence, especially of SCV, was mainly found (43). In contrast, earlier studies have reported phagosomal escape by S. aureus strain Novel in MAC-T cells (3) and indicated that S. aureus escapes more efficiently in cystic fibrosis cells than in their intact, i.e., CFTR-repaired counterparts (19). Therefore, we hypothesize that a phagosomal escape may be cell line specific and may be facilitated by cellular defects or stress conditions.
Among other cells, we have tested EA.hy926 cells, an endothelial cell line (Fig. 2 and 3). These cells proved highly susceptible for staphylococcal invasion, and staphylococci survived within these cells for the complete observation time. The ability to invade and survive within endothelial cells may play an important role in the pathogenesis of intravascular infections, as has been suggested for experimental endocarditis (40). It was reported previously that HeLa cells cannot acidify phagosomes containing agr+ S. aureus strains (42). However, we were not able to detect major differences between the nonprofessional phagocytes tested, whereas a pronounced difference was detectable between two types of professional phagocytes, J774 macrophages (Fig. 1E) and neutrophil granulocytes (data not shown), as reported previously (15, 35). Whether the apparent discrepancy is due to a different technical approach remains to be determined, as the former result has been obtained using lysosomotropic fluorescent probes.
The exceptional phenotype of strain 6850 was most remarkable and stable. Due to the fact that the phagosomal pH determination by flow cytometry was based on a population measurement (i.e., the population of phagosomes in a population of host cells), the reduction in the signal (ΔAFU) over time with respect to strain 6850 could be explained by three scenarios: (i) a partial but rather equal neutralization of all phagosomes; (ii) the destruction of a proportion of phagosomes, leaving the rest of the phagosomes unaffected; and (iii) a combination of these events.
Whereas the TEM data yielded heterogeneous data with staphylococci partly enveloped by host cell membranes and partly without apparent phagolysosomal structures, acidification of phagolysosomes was uncompromised (Fig. 4 and 5). S. aureus 6850, however, was observed to fractionate in a LAMP1-YFP-positive and -negative population within one host cell, which renders the first scenario unlikely (Fig. 6). The rarity of bacteria without such bounding membranes can explain the relatively small decrease in ΔAFU in the pH analysis. The destruction of only a small proportion of phagosomes could thus be responsible for minor changes in ΔAFU, which was readily detected with the flow cytometric assay.
Interestingly, there was no difference between strongly beta-hemolytic strains (e.g., Wood 46, BS513, and 6850 ΔsigB) and nonhemolytic strains (e.g., Cowan I and BS507) (Fig. 3), suggesting no major role for α-hemolysin. This is in accordance with our most recent data obtained by anhydrotetracyclin-dependent heterologous expression of α-toxin within phagolysosomes of airway epithelial cells (S9 cells). In this system, there was no detectable evidence for a phagosomal destruction by α-toxin (12), suggesting that α-toxin alone may not be sufficient for a phagosomal escape. Furthermore, strain Wood 46 is a naturally occurring sigB mutant due to a stop codon mutation (23). Despite the fact that Wood 46 is a strong producer of α-toxin, owing to negative regulation of α-toxin by SigB (53), this strain does not exhibit any phagosomal modulation (Fig. 3A). Similarly, the sigB mutant of strain 6850 showed a more pronounced β-hemolysis due to having higher levels of α-toxin and β-toxin production than the wild type but was unable to alter phagolysosomal acidification (data not shown). In line with this, the α-toxin-deficient mutant of strain 8325-4 should show an even stronger acidification than the wild type, which was not the case (data not shown). Whereas the involvement of the agr system in phagosomal escape has been reported (20), our data suggest that α-toxin may not be generally sufficient to mediate a phagosomal escape. This view is supported by a comparative genomic analysis of noninternalized and internalized S. aureus strain 6850, which showed that expression of hla was downregulated intracellularly (11). However, presently we cannot formally rule out that α-toxin may act together with other factors.
Since strain Wood 46 is a naturally occurring sigB mutant, we sought to investigate a potential role for SigB with the established flow cytometric analysis. For that purpose, we engineered a sigB-deficient isogenic mutant of strain 6850 and compared it to the wild type. 6850 ΔsigB showed a more pronounced β-hemolysis due to having roughly 2-fold-higher levels of α-toxin and β-toxin production than the wild type but was unable to alter phagolysosomal acidification in 293 cells (data not shown), suggesting the requirement for sigB in phagosomal modulation by 6850. However, two complementation approaches using two different plasmids were not successful. We have used (i) p3086, containing sigB under the control of a tet-inducible promoter (37), and (ii) pIK64, containing an 800-bp fragment (including sigB) under the very strong xyl-inducible promoter (26). The former yielded a heterogeneous phenotype (i.e., variable hemolytic activity and pigmentation) on blood agar, indicating variable sigB activity, and reduced growth upon selection and induction. This precluded its use in the pH assay in this study, since this assay is based on a population measurement. The second plasmid yielded a homogeneous phenotype on blood agar. However, intracellular induction was not functionally successful, conceivably due to the presence of repressing glucose concentrations in cell culture media. Thus, we currently cannot exclude a potential polar effect of the mutation.
In summary, the presented data strongly suggest that only few of the isolates and laboratory strains of S. aureus are capable of phagolysosomal modulation. Whereas α-toxin is produced in an agr-dependent fashion, expression is reduced in SigB-positive staphylococci (4, 13, 26), and overproducers of the pore-forming toxin do not show enhanced escape rates. Since phagosomal escape has been implicated in the induction of cell death (3, 9, 31, 32, 52) and intracellular persistence (11), e.g., of SCV (44), a more detailed understanding of the molecular processes is necessary. The nature of the effector(s) responsible for the phagolysosomal modulation (and potential escape) and the exact underlying mechanism remain to be determined.
Acknowledgments
We thank Nadine Leitschuh, Karina Lamprecht, Silvia Dittmann, and Kerstin Paprotka for expert technical assistance, Daniela Bunsen and Claudia Gehrig for preparation of EM samples, and Christoph Schoen for statistical advice. We thank Georg Krohne for discussing the electron microscopic data, Markus Bischoff for helpful discussions, and Matthias Frosch for general support.
This work has been funded by Deutsche Forschungsgemeinschaft grant SFB-TR34 C6 (B.S.) and in part by the German Ministry for Science and Research (BMBF) within the program “Entrepreneurial Regions: Competence Centers” under code ZIK011 (B.G. and M.F.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 7 June 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Balwit, J. M., P. van Langevelde, J. M. Vann, and R. A. Proctor. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033-1037. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W., C. A. Wesson, L. E. Liou, L. K. Fox, G. A. Bohach, and W. R. Trumble. 1998. Intracellular Staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infect. Immun. 66:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesney, P. J. 1989. Clinical aspects and spectrum of illness of toxic shock syndrome: overview. Rev. Infect. Dis. 11(Suppl 1):S1-S7. [PubMed] [Google Scholar]
- 6.Clement, S., P. Vaudaux, P. Francois, J. Schrenzel, E. Huggler, S. Kampf, C. Chaponnier, D. Lew, and J. S. Lacroix. 2005. Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent Staphylococcus aureus rhinosinusitis. J. Infect. Dis. 192:1023-1028. [DOI] [PubMed] [Google Scholar]
- 7.Dziewanowska, K., J. M. Patti, C. F. Deobald, K. W. Bayles, W. R. Trumble, and G. A. Bohach. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgell, C. J., C. C. McDonald, and J. B. Graham. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. U. S. A. 80:3734-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esen, M., B. Schreiner, V. Jendrossek, F. Lang, K. Fassbender, H. Grassme, and E. Gulbins. 2001. Mechanisms of Staphylococcus aureus induced apoptosis of human endothelial cells. Apoptosis 6:431-439. [DOI] [PubMed] [Google Scholar]
- 10.Fowler, T., E. R. Wann, D. Joh, S. Johansson, T. J. Foster, and M. Höök. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79:672-679. [DOI] [PubMed] [Google Scholar]
- 11.Garzoni, C., P. Francois, A. Huyghe, S. Couzinet, C. Tapparel, Y. Charbonnier, A. Renzoni, S. Lucchini, D. P. Lew, P. Vaudaux, W. L. Kelley, and J. Schrenzel. 2007. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giese, B., S. Dittmann, K. Paprotka, K. Levin, A. Weltrowski, D. Biehler, T. T. Lâm, B. Sinha, and M. J. Fraunholz. 2009. Staphylococcal α-toxin is not sufficient to mediate escape from phagolysosomes in upper airway epithelial cells. Infect. Immun. 77:3611-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gresham, H. D., J. H. Lowrance, T. E. Caver, B. S. Wilson, A. L. Cheung, and F. P. Lindberg. 2000. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J. Immunol. 164:3713-3722. [DOI] [PubMed] [Google Scholar]
- 15.Haas, A. 2007. The phagosome: compartment with a license to kill. Traffic 8:311-330. [DOI] [PubMed] [Google Scholar]
- 16.Haslinger-Löffler, B., B. C. Kahl, M. Grundmeier, K. Strangfeld, B. Wagner, U. Fischer, A. L. Cheung, G. Peters, K. Schulze-Osthoff, and B. Sinha. 2005. Multiple virulence factors are required for Staphylococcus aureus-induced apoptosis in endothelial cells. Cell. Microbiol. 7:1087-1097. [DOI] [PubMed] [Google Scholar]
- 17.Haslinger-Löffler, B., B. Wagner, M. Bruck, K. Strangfeld, M. Grundmeier, U. Fischer, W. Volker, G. Peters, K. Schulze-Osthoff, and B. Sinha. 2006. Staphylococcus aureus induces caspase-independent cell death in human peritoneal mesothelial cells. Kidney Int. 70:1089-1098. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, M., D. Schäfer, K. M. Juuti, G. Peters, B. Haslinger-Löffler, P. I. Kuusela, and B. Sinha. 2009. Expression of Pls (plasmin sensitive) in Staphylococcus aureus negative for pls reduces adherence and cellular invasion and acts by steric hindrance. J. Infect. Dis. 200:107-117. [DOI] [PubMed] [Google Scholar]
- 19.Jarry, T. M., and A. L. Cheung. 2006. Staphylococcus aureus escapes more efficiently from the phagosome of a cystic fibrosis bronchial epithelial cell line than from its normal counterpart. Infect. Immun. 74:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarry, T. M., G. Memmi, and A. L. Cheung. 2008. The expression of alpha-haemolysin is required for Staphylococcus aureus phagosomal escape after internalization in CFT-1 cells. Cell. Microbiol. 10:1801-1814. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, C. M. 1993. Staphylococcus aureus binding to cardiac endothelial cells is partly mediated by a 130 kilodalton glycoprotein. J. Lab. Clin. Med. 121:675-682. [PubMed] [Google Scholar]
- 22.Juuti, K. M., B. Sinha, C. Werbick, G. Peters, and P. I. Kuusela. 2004. Reduced adherence and host cell invasion by methicillin-resistant Staphylococcus aureus expressing the surface protein Pls. J. Infect. Dis. 189:1574-1584. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson-Kanth, A., K. Tegmark-Wisell, S. Arvidson, and J. Oscarsson. 2006. Natural human isolates of Staphylococcus aureus selected for high production of proteases and alpha-hemolysin are σB deficient. Int. J. Med. Microbiol. 296:229-236. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 25.Kubica, M., K. Guzik, J. Koziel, M. Zarebski, W. Richter, B. Gajkowska, A. Golda, A. Maciag-Gudowska, K. Brix, L. Shaw, T. Foster, and J. Potempa. 2008. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 3:e1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor SigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammers, A., P. J. Nuijten, and H. E. Smith. 1999. The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol. Lett. 180:103-109. [DOI] [PubMed] [Google Scholar]
- 28.Lang, K., C. Wagner, G. Haddad, O. Burnekova, and J. Geibel. 2003. Intracellular pH activates membrane-bound Na+/H+ exchanger and vacuolar H+-ATPase in human embryonic kidney (HEK) cells. Cell. Physiol. Biochem. 13:257-262. [DOI] [PubMed] [Google Scholar]
- 29.Lowy, F. D., J. Fant, L. L. Higgins, S. K. Ogawa, and V. B. Hatcher. 1988. Staphylococcus aureus—human endothelial cell interactions. J. Ultrastruct. Mol. Struct. Res. 98:137-146. [DOI] [PubMed] [Google Scholar]
- 30.Luft, J. H. 1956. Permanganate—a new fixative for electron microscopy. J. Biophys. Biochem. Cytol. 2:799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menzies, B. E., and I. Kourteva. 1998. Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infect. Immun. 66:5994-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menzies, B. E., and I. Kourteva. 2000. Staphylococcus aureus α-toxin induces apoptosis in endothelial cells. FEMS Immunol. Med. Microbiol. 29:39-45. [DOI] [PubMed] [Google Scholar]
- 33.Mollenhauer, H. H. 1959. Permanganate fixation of plant cells. J. Biophys. Biochem. Cytol. 6:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mollenhauer, H. H., D. J. Morre, and L. D. Rowe. 1990. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim. Biophys. Acta 1031:225-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nauseef, W. M. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219:88-102. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa, S. K., E. R. Yurberg, V. B. Hatcher, M. A. Levitt, and F. D. Lowy. 1985. Bacterial adherence to human endothelial cells in vitro. Infect. Immun. 50:218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pané-Farré, J., B. Jonas, S. W. Hardwick, K. Gronau, R. J. Lewis, M. Hecker, and S. Engelmann. 2009. Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J. Bacteriol. 191:2561-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plouin-Gaudon, I., S. Clement, E. Huggler, C. Chaponnier, P. Francois, D. Lew, J. Schrenzel, P. Vaudaux, and J. S. Lacroix. 2006. Intracellular residency is frequently associated with recurrent Staphylococcus aureus rhinosinusitis. Rhinology 44:249-254. [PubMed] [Google Scholar]
- 39.Proctor, R. A., S. C. Dalal, B. Kahl, D. Brar, G. Peters, and W. W. Nichols. 2002. Two diarylurea electron transport inhibitors reduce Staphylococcus aureus hemolytic activity and protect cultured endothelial cells from lysis. Antimicrob. Agents Chemother. 46:2333-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Que, Y. A., J. A. Häfliger, L. Piroth, P. Francois, E. Widmer, J. M. Entenza, B. Sinha, M. Herrmann, P. Francioli, P. Vaudaux, and P. Moreillon. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201:1627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleifer, K. H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus—Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 42.Schnaith, A., H. Kashkar, S. A. Leggio, K. Addicks, M. Krönke, and O. Krut. 2007. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 282:2695-2706. [DOI] [PubMed] [Google Scholar]
- 43.Schröder, A., R. Kland, A. Peschel, C. von Eiff, and M. Aepfelbacher. 2006. Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: small colony variants are able to survive in lysosomes. Med. Microbiol. Immunol. 195:185-194. [DOI] [PubMed] [Google Scholar]
- 44.Sendi, P., and R. A. Proctor. 2009. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 17:54-58. [DOI] [PubMed] [Google Scholar]
- 45.Sinha, B., P. Francois, Y. A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinha, B., P. P. Francois, O. Nüsse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 47.Sinha, B., and M. Herrmann. 2005. Mechanism and consequences of invasion of endothelial cells by Staphylococcus aureus. Thromb. Haemost. 94:266-277. [DOI] [PubMed] [Google Scholar]
- 47a.Sinha, B., and M. Fraunholz. 2010. Staphylococcus aureus host cell invasion and post-invasion events. Int. J. Med. Microbiol. 300:170-175. [DOI] [PubMed] [Google Scholar]
- 48.Smith, T. H., L. K. Fox, and J. R. Middleton. 1998. Outbreak of mastitis caused by one strain of Staphylococcus aureus in a closed dairy herd. J. Am. Vet. Med. Assoc. 212:553-556. [PubMed] [Google Scholar]
- 49.Song, L., M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J. E. Gouaux. 1996. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859-1866. [DOI] [PubMed] [Google Scholar]
- 50.Tokuyasu, K., and E. Yamada. 1959. Fine structure of Bacillus subtilis. I. Fixation. J. Biophys. Biochem. Cytol. 5:123-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Said-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907-3919. [DOI] [PubMed] [Google Scholar]
- 52.Wesson, C. A., L. E. Liou, K. M. Todd, G. A. Bohach, W. R. Trumble, and K. W. Bayles. 1998. Staphylococcus aureus agr and sar global regulators influence internalization and induction of apoptosis. Infect. Immun. 66:5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Höper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and σB. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]