Abstract
Escherichia coli strains causing avian colibacillosis and human neonatal meningitis, urinary tract infections, and septicemia are collectively known as extraintestinal pathogenic E. coli (ExPEC). Characterization of ExPEC strains using various typing techniques has shown that they harbor many similarities, despite their isolation from different host species, leading to the hypothesis that ExPEC may have zoonotic potential. The present study examined a subset of ExPEC strains: neonatal meningitis E. coli (NMEC) strains and avian-pathogenic E. coli (APEC) strains belonging to the O18 serogroup. The study found that they were not easily differentiated on the basis of multilocus sequence typing, phylogenetic typing, or carriage of large virulence plasmids. Among the APEC strains examined, one strain was found to be an outlier, based on the results of these typing methods, and demonstrated reduced virulence in murine and avian pathogenicity models. Some of the APEC strains tested in a rat model of human neonatal meningitis were able to cause meningitis, demonstrating APEC's ability to cause disease in mammals, lending support to the hypothesis that APEC strains have zoonotic potential. In addition, some NMEC strains were able to cause avian colisepticemia, providing further support for this hypothesis. However, not all of the NMEC and APEC strains tested were able to cause disease in avian and murine hosts, despite the apparent similarities in their known virulence attributes. Thus, it appears that a subset of NMEC and APEC strains harbors zoonotic potential, while other strains do not, suggesting that unknown mechanisms underlie host specificity in some ExPEC strains.
Escherichia coli strains causing extraintestinal disease are known as extraintestinal pathogenic E. coli (ExPEC) and include the uropathogenic E. coli (UPEC), neonatal meningitis E. coli (NMEC), and avian-pathogenic E. coli (APEC) subpathotypes. Recent studies have shown that members of various ExPEC subpathotypes harbor similar virulence-associated genes, despite their isolation from varied hosts and tissues (3, 8, 10, 20, 25, 27, 30, 32), and genomic sequencing of APEC O1 revealed that only 4.5% of the genome was not found in the other ExPEC strains sequenced (17). More recently, a cluster of isolates from human and avian hosts thought to represent potential zoonotic pathogens has been identified (20).
Common among the isolates of this mixed cluster are genes associated with the conserved region of large virulence plasmids, which are a defining trait of the APEC subpathotype (15, 19, 24, 36, 37) and which are essential for APEC virulence (5, 23). Interestingly, a closely related plasmid that was associated with high-level bacteremia in a neonatal rat meningitis model has also been described in an NMEC isolate (30).
Other virulence traits are also shared among ExPEC subpathotypes. Indeed, few traits, if any, appear to be exclusive to a particular ExPEC subpathotype, and in fact, some traits that were thought to be exclusive have been shown to contribute to the pathogenesis of more than one condition (8).
Such similarities in the virulence traits found among APEC and other ExPEC subpathotypes have led to speculation that APEC has zoonotic potential (20, 25, 27) and may be a food-borne source of ExPEC causing disease in humans (10, 14, 18, 22). Indeed, ExPEC strains have been identified in retail foods and poultry products (7, 11, 12, 18), and at least one study has found avian isolates to be indistinguishable from human isolates (10). However, other studies showed that human ExPEC strains were clearly distinct from avian strains (6) and that the consumption of poultry or contact with poultry did not correlate with the colonization of antimicrobial-resistant E. coli (34).
Here, we seek to further test the hypothesis that APEC strains have zoonotic potential. Of particular interest are O18 strains, which are common among human NMEC strains but which are also found among APEC strains (20, 26). In fact, it has been suggested that APEC O18:K1:H7 strains are potential human pathogens (27). Though it has been shown that human ExPEC strains can cause avian colibacillosis similar to that caused by APEC, suggesting that these ExPEC strains are not host specific (26), it has also been reported that E. coli strains from avian septicemia are more virulent to chicks than NMEC strains (33). However, the ability of APEC to cause disease in mammals has not yet been established.
The aim of the present study was to explore the zoonotic potential of NMEC and APEC O18 strains by comparing their plasmid contents, genotypes, phylogenetic group assignments, pulsed-field gel electrophoresis (PFGE) patterns, and sequence types (ST), determined by multilocus sequence typing (MLST), and their abilities to cause disease in the rat model of human neonatal meningitis and chicken models of avian colisepticemia.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study were those of serogroup O18 obtained from a previously described collection of APEC and NMEC isolates (20) and are shown along with the control strains in Table 1.
TABLE 1.
Characteristics of the E. coli strains used in this study
| Strain | Source (reference) | No. of plasmids (size [kb]) | Serogroup | Phylogenetic group | Type by MLST | Presence of genes associated with the conserved virulence region of APEC plasmids |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iutA | sitA | RepFIB | hlyF | ompT | etsAB | iss | iroN | ||||||
| DH5αa | 0 | NTe | A | ST1060 | − | − | − | − | − | − | − | − | |
| APEC 79 | California | 2 (133, 90) | O18 | A | ST23 | − | + | + | + | + | + | + | + |
| APEC 353 | Nebraska | 2 (133, 116) | O18 | B2 | ST95 | + | + | + | + | + | + | + | + |
| APEC 358 | Nebraska | 1 (133) | O18 | B2 | ST95 | + | + | + | + | + | + | + | + |
| APEC 370 | Nebraska | 1 (133) | O18 | B2 | ST95 | + | + | + | + | + | + | + | + |
| APEC 380 | Nebraska | 3 (133, 116, <50) | O18 | B2 | ST95 | + | + | + | + | + | − | + | + |
| NMEC 4 | Netherlands (13) | 0 | O18 | B2 | ST390 | + | + | − | − | − | − | − | + |
| NMEC 15 (SP15) (26) | Netherlands (13) | 2 (143, 116) | O18 | B2 | ST95 | + | + | + | + | + | + | + | − |
| NMEC 18 | Netherlands (13) | 2 (135, <50) | O18 | B2 | ST390 | + | + | + | + | + | + | + | + |
| NMEC 38 (SP38) (26) | Netherlands (13) | 1 (149) | O18 | B2 | ST95 | + | + | + | + | + | + | − | + |
| NMEC 58 | Netherlands (13) | 1 (149) | O18 | B2 | ST416 | + | + | + | + | + | + | + | + |
| NMEC RS218b | (40) | 1 (123) | O18 | B2 | ST95 | − | + | + | − | − | − | − | + |
| NMEC C5b | (40) | 1 (130) | O18 | B2 | ST95 | − | + | + | − | − | − | − | + |
| APEC χ7122c | (24) | 3 (103, 90, 60) | O78 | A | ST23 | + | + | + | + | + | + | + | + |
| APEC O2d | (19) | 2 (180, 101) | O2 | B2 | ST135 | + | + | + | + | + | + | + | + |
Negative control for rat neonatal meningitis model and ELA.
Positive control for rat neonatal meningitis model.
Positive control for chick colisepticemia model.
Positive control for ELA.
NT, nontypeable.
Amplification of virulence plasmid-associated genes.
PCR amplification was performed to determine the presence or absence of a selection of APEC virulence plasmid-associated genes, as described previously (32).
Phylogenetic typing.
Strains were assigned to phylogenetic groups according to the PCR amplification method described by Clermont et al. (2).
MLST.
MLST was performed by following the protocol for E. coli described by Wirth et al. (39) on the MLST website (http://mlst.ucc.ie), with the exception of the primers for mdh, which were replaced with those described elsewhere (28). In addition, the annealing conditions for the fumC and purA reactions were altered to 60°C and 58°C for 30 s, respectively.
PFGE.
PFGE was performed as described previously (31). Salmonella enterica serotype Braenderup H9812 (ATCC BAA-664) was used as the molecular weight size standard. Restriction endonuclease digestion was carried out using 25 U XbaI (Invitrogen) in a final volume of 100 μl at 37°C for 3 h. DNA macrorestriction fragments were resolved over 18 h on 1% SeaKem Gold agarose using a Chef Mapper XA system (Bio-Rad) autoalgorithm function for a small molecular size of 30 kb and a large molecular size of 600 kb. Gels were stained in 1 μg ethidium bromide ml−1 in reagent-grade water for 30 min, and the DNA was visualized by UV transillumination.
Macrorestriction patterns were compared using the BioNumerics Fingerprinting II Informatix software (version 3.0; Bio-Rad). The similarity index of the isolates was calculated using the Dice correlation coefficient option of the software with a position tolerance of 1% and an optimization of 0.5%. The unweighted-pair group method using average linkages (UPGMA) was used to construct the dendrogram.
Preparation of plasmid DNA.
Plasmid DNA was prepared using a Qiagen plasmid minikit, as recommended by the manufacturer. Plasmids were separated by PFGE.
PFGE for large plasmid separation.
PFGE was carried out using a CHEF Mapper XA system (Bio-Rad). Plasmids were separated in 1.0% (wt/vol) DNA-grade agarose (SeaKem), as described previously (36). Plasmid DNA of known sizes from E. coli strains APEC O1 (17), APEC O2 (19), and χ7122 (23, 24) were used as molecular weight standards. Gels were stained in 1.2 μg ethidium bromide ml−1 in distilled water for 1 h, and the DNA was visualized by UV transillumination.
Pathogenicity testing.
All animal experiments were approved by the Iowa State University Institutional Animal Care and Use Committee and were performed in accordance with institutional guidelines.
(i) Rat neonatal meningitis model.
Groups of approximately 12 specific-pathogen-free (SPF), 5-day-old Sprague-Dawley rat pups were given approximately 200 CFU (range, 48 to 312 CFU) of E. coli by the intraperitoneal route (8, 21). The pups were euthanized after 24 h using sodium pentobarbital (Sleepaway; Fort Dodge Laboratories). For bacterial enumeration, blood was collected from the jugular vein and plated on MacConkey agar to indicate septicemia; and cerebrospinal fluid (CSF), collected by cisternal puncture, was plated on MacConkey agar to indicate meningitis. The cerebrums were removed from the rats, fixed in 10% neutral buffered formalin, routinely processed for histopathology, stained with hematoxylin-eosin, and examined for lesions consistent with bacterial meningitis.
(ii) Chicken embryo lethality assay (ELA).
Strains were assessed for lethality in chicken embryos by inoculation of overnight phosphate-buffered saline (PBS)-washed cultures (∼500 CFU) into the allantoic cavities of 12-day-old, embryonated, SPF eggs (29). PBS-inoculated and uninoculated embryos were used as controls. The eggs were candled daily, and embryo deaths were recorded for a period of 4 days.
(iii) Chick colisepticemia model.
Groups of 12 1-day-old Leghorn chicks were inoculated with approximately 107 CFU by the intratracheal (IT) route. The birds were observed for 7 days, and mortalities were recorded. The 8-day-old birds were euthanized by CO2 inhalation and examined for airsacculitis, pericarditis, perihepatitis, and peritonitis. The heart, brain, and air sacs were swabbed and the swabs were cultured on MacConkey agar at 37°C overnight.
RESULTS
Isolation of plasmids from strains.
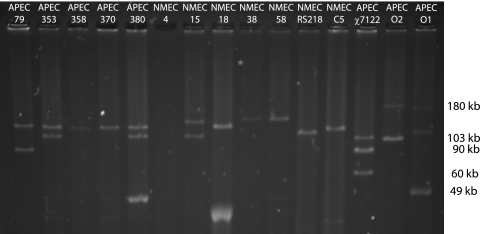
The method used here detects and predicts the sizes of plasmids greater than 50 kb with confidence, and large plasmids ranging in size from 90 kb to 149 kb were identified in all strains except NMEC 4 (Fig. 1). Multiple large plasmids were isolated from some strains, and the number of large plasmids isolated from each strain and their predicted sizes are indicated in Table 1. The relative sizes of the largest plasmid in all APEC strains appeared to be identical (∼133 kb), while the sizes of the plasmids in the NMEC strains varied.
FIG. 1.
Plasmids purified from the test and control strains. Previously sequenced plasmids present in APEC O1, APEC O2, and APEC χ7122 were used as size standards. All strains except NMEC 4 contained at least one large plasmid which correlated 100% with the presence of the RepFIB amplicon. Note that a similarly sized plasmid (∼133 kb) is carried by all APEC O18 isolates.
Amplification of virulence plasmid-associated genes.
The presence of APEC virulence plasmid-associated genes is shown in Table 1. APEC control strains carried all of the genes, while only the RepFIB replicon, sitA, and iroN were amplified from the NMEC control strains, RS218 and C5. The majority of the genes were amplified from the O18 strains of interest in this study with the exception of NMEC 4, which carried only iutA, sitA, and iroN.
Phylogenetic typing.
All of the O18 strains examined except APEC 79 belonged to the B2 phylogenetic group. APEC 79 was assigned to phylogenetic group A (Table 1).
MLST.
MLST revealed that all of the O18 strains investigated except APEC 79 belonged to the ST95 clonal complex. APEC 79 belonged to the ST23 clonal complex. All APEC O18 strains except APEC 79 (ST23) were found to have identical allelic profiles (ST95), and while some variation among the NMEC strains occurred (NMEC 4 and NMEC 18, ST390; NMEC 58, ST416), most were ST95 (Table 1).
PFGE.
Two major clusters with approximately 52% identity were observed after PFGE of XbaI-digested genomic DNA (Fig. 2). The seven NMEC strains, including strains RS218 and C5, clustered together, while most of the APEC strains were found in a separate cluster along with laboratory-adapted E. coli DH5α. However, the NMEC cluster also included ELA-positive control strain APEC O2. Additionally, two outliers were observed in this analysis: APEC 79 and χ7122.
FIG. 2.
PFGE of XbaI-digested DNA from the O18 strains. Strain designations, phylogenetic groups, MLST assignments, and virulence genes associated with the conserved virulence region of APEC plasmids are shown at right. This dendrogram prepared by the unweighted-pair group method using average linkages was generated in BioNumerics software by using the Dice coefficient with a 1.0% band position tolerance. The scale above the dendrogram indicates percent similarity.
Pathogenicity testing. (i) Rat neonatal meningitis model.
Pathogenicity was based on the mortality rates, reisolation rates, and bacterial enumeration observed for each strain (Table 2). No mortalities were observed among the rat pups inoculated with APEC 79, χ7122, or the negative controls, PBS and E. coli DH5α. The low numbers of mortalities observed in groups inoculated with APEC 353, APEC 370, NMEC 4, and the archetypal NMEC strain (strain RS218) were not significantly different from the numbers in the groups where no mortalities were observed. Inoculation with another well-studied NMEC strain, strain C5, resulted in 92% mortality, and the rates of mortality observed in groups inoculated with APEC 380, NMEC 18, NMEC 38, and NMEC 58 were not significantly different from the rate for the group inoculated with NMEC C5.
TABLE 2.
Pathogenicities of E. coli O18 strains in the neonatal rat meningitis model
| Strain | Mortality ratea | Rate of reisolation fromc: |
Inoculum (log10 CFU per animal) | Mean log10 CFU ml−1 |
||
|---|---|---|---|---|---|---|
| Blood of survivors | CSF of survivors | Blood | CSF | |||
| PBS | 0/12a | 0/12 | 0/12 | 0 | 0 | 0 |
| DH5α | 0/12a | 0/12 | 0/12 | 2.68 | 0 | 0 |
| APEC 79 | 0/12a | 7/12 | 2/12 | 3.36 | 2.25 | 1.62 |
| APEC 353 | 2/12ab | 10/10 | 10/10 | 3.33 | >4.13 | >4.62 |
| APEC 358 | 5/12bcf | 7/7 | 7/7 | 3.37 | >4.51 | >4.78 |
| APEC 370 | 1/12ab | 11/11 | 11/11 | 3.41 | >3.60 | >4.69 |
| APEC 380 | 9/12cd | 3/3 | 3/3 | 3.39 | >3.60 | >4.77 |
| NMEC 4 | 3/12af | 9/9 | 9/9 | 3.35 | >3.90 | >4.58 |
| NMEC 15 | 5/12bcf | 7/7 | 7/7 | 3.42 | >3.96 | >4.74 |
| NMEC 18 | 8/12cef | 4/4 | 4/4 | 3.40 | >3.78 | >4.53 |
| NMEC 38 | 9/10de | 1/1 | 1/1 | 3.32 | >3.90 | >5.00 |
| NMEC 58 | 10/11de | 1/1 | 1/1 | 3.49 | >3.90 | >5.00 |
| NMEC C5b | 12/13de | 1/1 | 1/1 | 3.43 | >4.30 | >4.40 |
| NMEC RS218b | 2/13ab | 9/11 | 8/11 | 3.18 | >3.55 | >4.09 |
| APEC χ7122 | 0/13a | 12/13 | 4/13 | 3.14 | 3.02 | 3.73 |
Values with the same superscript lowercase letter are not significantly different (P ≥ 0.05 by Fisher's exact test).
Positive-control strains for the neonatal rat meningitis model.
Data represent the number of animals from which the strain was reisolated/total number of surviving animals.
E. coli was not reisolated from the blood or CSF of rat pups inoculated with PBS or DH5α; however, it was reisolated from both the blood and the CSF of all remaining rat pups in the groups inoculated with E. coli except those in the groups inoculated with APEC 79, χ7122, and RS218.
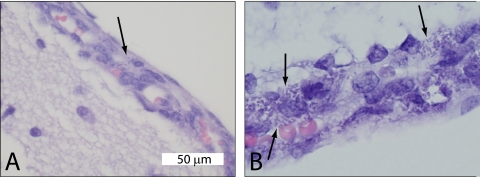
The number of E. coli isolates recovered from the blood and CSF of the rat pups was beyond the countable limits (4 × 103 CFU ml−1 for an entire 50-μl blood sample, 1 × 104 CFU ml−1 for an entire 20-μl CSF sample) for all groups except APEC 79 and χ7122. Histopathology of selected cerebrums showed bacteria in the meninges of rats infected with APEC 353 and NMEC 58, as well as low numbers of neutrophils in the meninges and outer cellular layer in the cerebrum (Fig. 3). No lesions were observed in the cerebrums of rats infected with negative-control strain DH5α.
FIG. 3.
(A) Histopathology of rat brain tissue 24 h postinoculation with APEC 353 showed rod-shaped bacteria (arrow) in the meninges. (B) Histopathology of rat brain tissue 24 h postinoculation with NMEC 58 with rod-shaped bacteria in the meninges (arrows).
(ii) Chicken embryo lethality assay.
The number of mortalities observed among the uninoculated, PBS-inoculated and APEC 79-inoculated eggs did not differ significantly from the number observed among eggs inoculated with the negative control, strain DH5α, while the number of mortalities observed among all the O18 strains except APEC 79 and NMEC 18 were not significantly different from the number observed among APEC O2 positive-control strains (Table 3).
TABLE 3.
Mortality rates among chick embryos inoculated with APEC and NMEC isolates
| Strain | Mortality ratec |
P value vsa: |
|
|---|---|---|---|
| DH5α | APEC O2 | ||
| Uninoculated | 0/6 | 1.000 | <0.001 |
| PBS | 3/10 | 0.300 | 0.001 |
| E. coli DH5α | 2/20 | <0.001 | |
| APEC 79 | 7/20 | 0.127 | <0.001 |
| APEC 353 | 12/20 | 0.002 | 0.064 |
| APEC 358 | 16/20 | <0.001 | 0.661 |
| APEC 380 | 16/20 | <0.001 | 0.661 |
| NMEC 15 | 14/20 | <0.001 | 0.235 |
| NMEC 18 | 11/20 | 0.005 | 0.031 |
| NMEC 38 | 19/20 | <0.001 | 1.000 |
| NMEC 58 | 14/20 | <0.001 | 0.235 |
| APEC O2b | 32/40 | <0.001 | |
P value determined by Fisher's exact test.
Positive control for the ELA.
Data represent the number of embryos that died/total number of embryos tested.
(iii) Chick colisepticemia model.
The pathogenicity of the strains following IT inoculation was determined on the basis of the rates of mortality observed due to E. coli colonization, reisolation of the inoculated E. coli strain from the birds, and the lesion scores attributed to the birds on postmortem examination (Table 4).
TABLE 4.
Pathogenicities of APEC and NMEC isolates in 1-day-old chicks inoculated by the intratracheal route
| Strain | Mortality ratea,b | Reisolation ratea,c | Median (range) lesion scorea |
|---|---|---|---|
| PBS | 0/11a | 2/11a | 0 (0-1)a |
| APEC 353 | 6/12b | 9/12b | 5.5 (0-8)bc |
| APEC 380 | 6/12b | 9/12b | 7 (0-8)bc |
| NMEC 15 | 7/12b | 9/12b | 8 (1-8)c |
| NMEC 58 | 0/12a | 3/12ac | 0 (0-6)ab |
| APEC χ7122 | 6/12b | 8/12bc | 5.5 (0-8)bc |
Values with the same superscript lowercase letter(s) are not significantly different (P ≥ 0.05 by the Kruskal-Wallis or Fisher's exact test).
Data represent the number of chicks that died/total number of chicks tested.
Data represent the number of chicks from which the strain was reisolated/total number of chicks used to test the strain.
NMEC 58 was the only strain tested that did not cause mortality, while inoculation with the other three strains (APEC 353, APEC 380, and NMEC 15) resulted in numbers of mortalities similar to those seen with the positive-control strain APEC χ7122. The rate of reisolation for the group inoculated with NMEC 58 was comparable to that seen for negative-control birds that were inoculated with sterile PBS, while the rates of reisolation for the groups inoculated with χ7122, APEC 353, APEC 380, and NMEC 15 were significantly higher (P ≤ 0.05) than the rates for the group inoculated with PBS. The E. coli strains reisolated from the pericardial sac swabs of two PBS-inoculated birds lacked most of the virulence plasmid-associated genes, suggesting that they were commensal E. coli contaminants.
NMEC 58 was the only strain tested that did not result in a median lesion score significantly higher than that obtained with PBS, and NMEC 15 was the only strain that resulted in a median lesion score significantly higher than that obtained with NMEC 58. The median lesion scores for all the O18 strains were not significantly different from those for positive-control strain APEC χ7122.
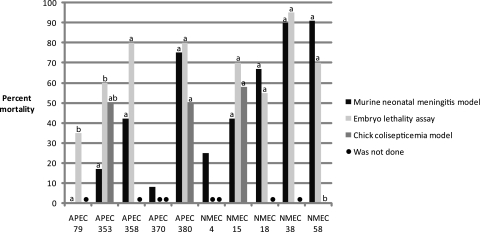
While the mortality rates produced by each test strain appeared to be relatively consistent between different pathogenicity models (Fig. 4), two APEC strains, APEC 79 (P = 0.032) and APEC 353 (P = 0.028), produced significantly lower rates of mortality among neonatal rats than among chick embryos. Conversely, NMEC 58 produced significantly more mortalities (P < 0.001) in both neonatal rats and chick embryos than in 1-day-old chicks.
FIG. 4.
Percent mortality observed for each strain when it was examined using the murine and avian pathogenicity models. For each strain, the same lowercase letter on top of the column indicates that the mortality was not significantly different (P ≥ 0.05 by Fisher's exact test) between pathogenicity models. Note that not every strain was examined in all of the three pathogenicity models (marked by •).
DISCUSSION
Although it has previously been shown that strains isolated from cases of neonatal meningitis are similar to APEC (3, 20, 25, 27, 30) and were able to cause disease in models of avian colisepticemia (26), this is the first study to demonstrate that strains isolated from cases of avian colibacillosis are able to cause disease in a rat model of human neonatal meningitis. These findings are supportive of the hypothesis that APEC may have zoonotic potential and, along with previous reports of APEC being isolated from retail poultry meat (12, 18), reflect the potential for APEC to act as a food-borne source of ExPEC strains causing disease in humans.
The presence of large plasmids, as seen in Fig. 1, in conjunction with PCR amplification of the RepFIB replicon and plasmid-associated virulence genes, suggests that all test strains with the exception of NMEC 4 carry a variant of an APEC virulence plasmid. In NMEC 4, the iutA, sitA, and iroN genes may be located on the chromosome, which has previously been reported in both APEC and NMEC (15, 30, 36). However, while the other NMEC strains studied here, as well as NMEC strain S88 and many others (30), carry most of the genes present in the conserved region of the APEC virulence plasmid, NMEC control strains RS218 and C5 do not, suggesting that they may be atypical NMEC.
The absence of plasmids carrying the conserved virulence region in archetypal NMEC strains RS218 and C5 might suggest that this region is not as important in the manifestation of neonatal meningitis as it is in that of avian colisepticemia. However, NMEC plasmid pS88, carrying the conserved virulence region, has been shown to be associated with high-level bacteremia in a neonatal rat model (30), and inoculation of neonatal rats with NMEC 4 and RS218, both of which lack the typical APEC virulence plasmid, resulted in rates of mortality not significantly different from those for the avirulent controls. It has been shown that APEC plasmids contribute to the ability of E. coli to grow in human urine, cause urinary tract infections in mice (35), and meningitis in rats (16), suggesting that these plasmids contribute to survival within extraintestinal compartments. Certainly, further examination of the contribution of these plasmids to the pathogenesis of meningitis is warranted. However, it is clear that ExPEC's ability to cause meningitis cannot be completely attributed to virulence plasmids, as APEC 79, despite its virulence plasmid, was not lethal to rats, and C5, which does not have a virulence plasmid, was lethal for rats. Indeed, it has previously been suggested that E. coli causes meningitis using different mechanisms (40) and that the pathogenic mechanisms in the so-called archetypal meningitis strains appear to be only partially representative of the pathogenic mechanisms harbored by NMEC isolates (1).
Of the ExPEC O18 strains examined, APEC 79 was the only one not belonging to the B2 phylogenetic group or the ST95 clonal complex. In addition, it was an outlier in the dendrogram constructed from the PFGE profiles and appears to be unrelated to the remaining O18 strains and more closely related to APEC O78 strain χ7122 (phylogenetic group A and ST23). The atypical nature of APEC 79 was reflected in its lack of virulence in chick embryos and in rats, supporting a previous claim that B2 strains have enhanced virulence (9). On the basis of its unusual attributes, it is not surprising that APEC 79 was not found among the cluster of isolates that were thought to represent potential zoonotic pathogens (20), while the other APEC O18 strains examined were.
APEC 79 was frequently depicted as an outlier, suggesting that the molecular-based typing methods undertaken in the present study discriminate similarly. While phylogenetic typing and MLST were unable to discriminate the human ExPEC strains from avian strains, they essentially clustered the strains by host origin after cluster analysis of the PFGE data, suggesting that in the case of ExPEC that PFGE has more discriminatory power than the other typing methods, as seen previously for Salmonella (4). Although the group of APEC strains evaluated in the present study appears to be more related to each other than to NMEC and vice versa, there was also a strong correlation between PFGE relatedness and their geographic site of isolation, making it difficult to determine which factor has more influence on PFGE relatedness.
Both APEC 79 and χ7122 appeared to be of low virulence in the neonatal meningitis model, because inoculation of these strains caused no mortalities and resulted in a limited number of bacteria in the blood and CSF. Interestingly, both strains differed from the other ExPEC strains in this study in the phylogenetic group and MLST to which they were assigned (A and ST23, respectively), indicating that they may be APEC-specific pathogens.
A one-way analysis of variance using the Kruskal-Wallis test showed that there were some significant differences in the blood and CSF counts between groups (<0.0001); however, in some groups, where few pups survived, the sample size was too small for Dunn's multiple-comparison posttest to describe genuine differences. Still, the number of bacteria reisolated from the blood and CSF of rats inoculated with APEC 353, APEC 358, and APEC 370 was significantly higher than the number of DH5α bacteria reisolated, suggesting that all these APEC strains, despite the lower rates of mortality that they cause, were able to cause septicemia and meningitis in neonatal rats. Indeed, histopathology performed on the cerebrums of selected rats postinoculation with NMEC and APEC showed lesions typical of meningitis and the presence of bacteria in the meninges. There appeared to be a high number of bacteria in the cerebrums that did not elicit a strong immune response, which may be due to the short time period that had elapsed since inoculation. This, in addition to the high mortality rates observed with these isolates, suggests that they are highly virulent.
While the number of bacteria present in the blood and CSF were too numerous to count following inoculation of the rats with APEC 353, APEC 370, NMEC 4, and RS218, the associated rates of mortality were low, suggesting that these isolates were of intermediate virulence. It has previously been suggested that the virulence of strain RS218 has decreased over time (26), which could account for its lower level of virulence here. Intriguingly, RS218 appears to lack the virulence plasmid that other O18 strains contain, and the loss of that plasmid may account for RS218's atypical nature and relatively low level of virulence.
Strains APEC 358 and NMEC 15 were significantly more virulent (P ≤ 0.05) for neonatal rats than the groups with no mortalities, yet they were significantly less virulent than strain C5. APEC 380 was the only APEC strain that appeared to be as virulent as NMEC reference strain C5, while three other NMEC strains, NMEC 18, NMEC 38, and NMEC 58, were also highly virulent in the rat model.
While the number of mortalities caused by APEC 79 could not be differentiated from the number that occurred among the negative controls by ELA, the numbers of mortalities caused by all the rest of the strains except NMEC 18 were not significantly different from the numbers that occurred among the embryos inoculated with the positive control, APEC O2. NMEC 18 was significantly different from both the negative and positive controls, and thus, it could be considered of intermediate virulence, according to the results of this assay.
NMEC 58 was shown to be virulent in both the ELA and the rat neonatal meningitis model, but it appeared to be of low virulence in the chick colisepticemia pathogenicity assay. IT inoculation with NMEC 58 resulted in no mortalities and the colonization of only three birds. These results showed that NMEC 58 was less capable of establishing an infection in 1-day-old chicks, while the other strains (APEC 353, APEC 380, and NMEC 15) established infections as or more severe than those seen after inoculation with χ7122. Consistent with a previous study describing a host-specific pathotype (25), the present study appears to have identified a host-specific ExPEC strain of human origin. In contrast, another study by Moulin-Schouleur et al. was unable to find evidence of host specificity within the highly pathogenic subcluster B2-1 (26). Since NMEC 58 appears to possess the full complement of known APEC virulence plasmid-associated genes (Table 1), it does not appear that the virulence plasmid alone is responsible for a broadening of an ExPEC strain's host range from mammals to avian species or vice versa. Identifying the factors that are responsible for host specificity or the lack thereof will enhance our understanding of ExPEC pathogenesis and lead to improved efforts to control ExPEC diseases.
The results of this study suggest that ExPEC may have a core genome which is essential for ExPEC strain survival in extraintestinal environments and that individual strains possess additional genetic information that has been adapted to establish an infection after inoculation by a specific route in a particular host and in a particular tissue type. Similarly, another study found that strains within the same clonal group did not necessarily have comparable virulence attributes and suggested that the acquisition of accessory genes may impart distinct niche-specific growth (38). This issue warrants further attention, and with an increase in the number of ExPEC genomes available, comparative genomics may help to identify the core ExPEC genome and distinguish host- or disease-specific regions.
The ExPEC strains described here were not easily differentiated, despite being isolated from different hosts and varied disease manifestations, reinforcing previous claims of the zoonotic potential of ExPEC. This zoonotic potential was confirmed with the first report of APEC strains causing meningitis in a rat neonatal meningitis model. Many of the O18 strains examined in this study were shown to be highly virulent in avian and murine models, suggesting that they were not host specific and that they were capable of acting as zoonotic pathogens. In contrast, others caused disease in one host species but not the other, indicating that they were host specific. An understanding of the factors contributing to ExPEC's host specificity would better clarify this issue and could help in designing future ExPEC disease control strategies.
Acknowledgments
We are grateful to Roy Curtiss III for providing the APEC χ7122 strain and to Lodewijk Spanjaard, James Johnson, and Kwang Sik Kim for providing the NMEC isolates. We thank Luke Baldwin, Paul Mangiamele, and Kathy Mou for their technical assistance.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Bonacorsi, S., O. Clermont, V. Houdouin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange, and E. Bingen. 2003. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J. Infect. Dis. 187:1895-1906. [DOI] [PubMed] [Google Scholar]
- 2.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E. M. Antao, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Bohnke, H. Steinruck, H. C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163-176. [DOI] [PubMed] [Google Scholar]
- 4.Fakhr, M. K., L. K. Nolan, and C. M. Logue. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginns, C. A., M. L. Benham, L. M. Adams, K. G. Whithear, K. A. Bettelheim, B. S. Crabb, and G. F. Browning. 2000. Colonization of the respiratory tract by a virulent strain of avian Escherichia coli requires carriage of a conjugative plasmid. Infect. Immun. 68:1535-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graziani, C., I. Luzzi, M. Corro, F. Tomei, G. Parisi, M. Giufre, S. Morabito, A. Caprioli, and M. Cerquetti. 2009. Phylogenetic background and virulence genotype of ciprofloxacin-susceptible and ciprofloxacin-resistant Escherichia coli strains of human and avian origin. J. Infect. Dis. 199:1209-1217. [DOI] [PubMed] [Google Scholar]
- 7.Hannah, E. L., J. R. Johnson, F. Angulo, B. Haddadin, J. Williamson, and M. H. Samore. 2009. Molecular analysis of antimicrobial-susceptible and -resistant Escherichia coli from retail meats and human stool and clinical specimens in a rural community setting. Foodborne Pathog. Dis. 6:285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houdouin, V., S. Bonacorsi, N. Brahimi, O. Clermont, X. Nassif, and E. Bingen. 2002. A uropathogenicity island contributes to the pathogenicity of Escherichia coli strains that cause neonatal meningitis. Infect. Immun. 70:5865-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. R., O. Clermont, M. Menard, M. A. Kuskowski, B. Picard, and E. Denamur. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141-1150. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. R., M. A. Kuskowski, M. Menard, A. Gajewski, M. Xercavins, and J. Garau. 2006. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J. Infect. Dis. 194:71-78. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. R., M. A. Kuskowski, K. Smith, T. T. O'Bryan, and S. Tatini. 2005. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 191:1040-1049. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, J. R., A. C. Murray, A. Gajewski, M. Sullivan, P. Snippes, M. A. Kuskowski, and K. E. Smith. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J. R., M. R. Sannes, C. Croy, B. Johnston, C. Clabots, M. A. Kuskowski, J. Bender, K. E. Smith, P. L. Winokur, and E. A. Belongia. 2007. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002-2004. Emerg. Infect. Dis. 13:838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, T. J., D. Jordan, S. Kariyawasam, A. L. Stell, N. P. Bell, Y. M. Wannemuehler, C. F. Alcaron, G. Li, K. A. Tivendale, C. M. Logue, and L. K. Nolan. 2010. Sequence analysis and characterization of a transferrable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect. Immun. 78:1931-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. J., C. M. Logue, Y. Wannemuehler, S. Kariyawasam, C. Doetkott, C. DebRoy, D. G. White, and L. K. Nolan. 2009. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog. Dis. 6:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, T. J., Y. Wannemuehler, S. J. Johnson, A. L. Stell, C. Doetkott, J. R. Johnson, K. S. Kim, L. Spanjaard, and L. K. Nolan. 2008. Comparison of extraintestinal pathogenic Escherichia coli from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Invest. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manges, A. R., S. P. Smith, B. J. Lau, C. J. Nuval, J. N. Eisenberg, P. S. Dietrich, and L. W. Riley. 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog. Dis. 4:419-431. [DOI] [PubMed] [Google Scholar]
- 23.Mellata, M., K. Ameiss, H. Mo, and R. Curtiss III. 2010. Characterization of the contribution to virulence of three large plasmids of avian pathogenic E. coli \'637122 (O78:K80:H9). Infect. Immun. 78:1528-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellata, M., J. W. Touchman, and R. Curtiss. 2009. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli chi7122 (O78:K80:H9). PLoS One 4:e4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mora, A., C. Lopez, G. Dabhi, M. Blanco, J. E. Blanco, M. P. Alonso, A. Herrera, R. Mamani, S. Bonacorsi, M. Moulin-Schouleur, and J. Blanco. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulin-Schouleur, M., M. Reperant, S. Laurent, A. Bree, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 45:3366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M. R. Kao, A. Bree, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 29.Nolan, L. K., R. E. Wooley, J. Brown, K. R. Spears, H. W. Dickerson, and M. Dekich. 1992. Comparison of a complement resistance test, a chicken embryo lethality test, and the chicken lethality test for determining virulence of avian Escherichia coli. Avian Dis. 36:395-397. [PubMed] [Google Scholar]
- 30.Peigne, C., P. Bidet, F. Mahjoub-Messai, C. Plainvert, V. Barbe, C. Medigue, E. Frapy, X. Nassif, E. Denamur, E. Bingen, and S. Bonacorsi. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 77:2272-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 33.Ron, E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28-32. [DOI] [PubMed] [Google Scholar]
- 34.Sannes, M. R., E. A. Belongia, B. Kieke, K. Smith, A. Kieke, M. Vandermause, J. Bender, C. Clabots, P. Winokur, and J. R. Johnson. 2008. Predictors of antimicrobial-resistant Escherichia coli in the feces of vegetarians and newly hospitalized adults in Minnesota and Wisconsin. J. Infect. Dis. 197:430-434. [DOI] [PubMed] [Google Scholar]
- 35.Skyberg, J. A., T. J. Johnson, J. R. Johnson, C. Clabots, C. M. Logue, and L. K. Nolan. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74:6287-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tivendale, K. A., J. L. Allen, and G. F. Browning. 2009. Plasmid-borne virulence-associated genes have a conserved organization in virulent strains of avian pathogenic Escherichia coli. J. Clin. Microbiol. 47:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tivendale, K. A., A. H. Noormohammadi, J. L. Allen, and G. F. Browning. 2009. The conserved portion of the putative virulence region contributes to virulence of avian pathogenic Escherichia coli. Microbiology 155:450-460. [DOI] [PubMed] [Google Scholar]
- 38.Wiles, T. J., J. M. Bower, M. J. Redd, and M. A. Mulvey. 2009. Use of zebrafish to probe the divergent virulence potentials and toxin requirements of extraintestinal pathogenic Escherichia coli. PLoS Pathog. 5:e1000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao, Y., Y. Xie, and K. S. Kim. 2006. Genomic comparison of Escherichia coli K1 strains isolated from the cerebrospinal fluid of patients with meningitis. Infect. Immun. 74:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]